Abstract
Psoriasis is a genetically determined inflammatory skin disease. Although the transition from uninvolved into lesional skin is accompanied by changes in the expression of multiple genes, much less is known about the difference between uninvolved skin from psoriatic patients as opposed to skin from normal individuals. Multiple biochemical and morphological changes were reported decades ago in uninvolved psoriatic skin but remain poorly understood. Here we demonstrate dysregulation of 223 transcripts representing 179 unique genes in uninvolved psoriatic skin, 178 of which were not previously known to be altered in their expression. The proteins encoded by these transcripts are involved in lipid metabolism, antimicrobial defenses, epidermal differentiation and control of cutaneous vasculature. Cluster analysis of transcripts with significantly altered expression identified a group of genes involved in lipid metabolism with highly correlated gene expression. Promoter analysis demonstrated enrichment for binding sites of three transcription factors; peroxisome proliferator-activator receptor alpha (PPARA), sterol regulatory element-binding protein (SREBF) and estrogen receptor 2 (ESR2), suggesting that coordinate regulation of lipid metabolic genes may be related to the action of these factors. Taken together, our results identify a “pre-psoriatic” gene expression signature, suggesting decreased lipid biosynthesis and increased innate immunity in uninvolved psoriatic skin.
Keywords: psoriasis, gene expression, microarray, skin, immunology
INTRODUCTION
Psoriasis is a chronic inflammatory and hyperproliferative skin disease, affecting over 6 million Americans (about 2%) at an estimated cost of $1.6 to 3.2 billion annually (Sander et al., 1993). The disease tends to strike early in life, as the majority of cases are diagnosed in individuals less than 30 years of age, and a significant proportion of these cases are in individuals less than 10 years old (Krueger et al., 1984). Along with unsightly cutaneous manifestations with a negative impact on the quality of life (Gupta et al., 1993), psoriasis is accompanied by an inflammatory arthritis affecting up to 40% of patients (Gladman, 1994).
That psoriasis has a genetic basis is undisputed, but many of the causative genes remain to be identified (Gudjonsson and Elder, 2007). Psoriasis is characterized by complex alterations in epidermal growth and differentiation, along with multiple biochemical, immunological, inflammatory, and vascular abnormalities. It has been firmly established that psoriasis is a T-cell mediated disease (Conrad et al., 2007; Nickoloff and Wrone-Smith, 1999) and available data suggest it may have an autoimmune basis (Gudjonsson et al., 2004). Intra-epidermal T-cells are crucial for the development of psoriatic epidermal hyperplasia (Conrad et al., 2007), and it has been postulated that these cells may be reacting against self-antigens presented by HLA-Cw6 (Johnston et al., 2004), which was recently demonstrated to be the major genetic determinant of psoriasis susceptibility (Nair et al., 2008; Nair et al., 2006).
Lesional psoriatic skin (PP) skin has been shown to have a pattern of gene expression that is dramatically different from that of normal (NN) skin of unaffected individuals. Some of the earliest genes identified as having distinctive over-expression in PP skin include transforming growth factor-α (TGF-α) (Elder et al., 1989), tumor necrosis factor-α (TNF-α) (Nickoloff et al., 1991), vascular endothelial growth factor (VEGF) and its receptors (Detmar et al., 1994), proteinase inhibitors such as peptidase inhibitor 3 (SKALP) (Nonomura et al., 1994), hyperproliferation-associated keratins, K16 and K17 (Leigh et al., 1995) and multiple genes mapping to the epidermal differentiation complex (EDC) on chromosome 1q21, including S100A, loricrin, involucrin, small proline-rich region (SPRR), and late cornified envelope (LCE) genes (Zhao and Elder, 1997). Subsequently, microarray studies have been used to characterize large scale gene expression changes in PP skin compared to uninvolved, normal-appearing skin from psoriatic patients (PN skin) and / or NN skin (Bowcock et al., 2001; Kulski et al., 2005; Romanowska et al., 2008; Zhou et al., 2003), or to the involved skin of atopic dermatitis patients (de Jongh et al., 2005; Romanowska et al., 2008). These microarray studies have identified many of the candidates suggested in the original candidate gene studies as well as novel genes not previously implicated in the pathogenesis of psoriasis. However, until now, detailed comparisons of PN vs. NN skin involving large numbers of subjects have been lacking.
PN skin has been shown to have biochemical differences when compared to NN skin, with many of these studies performed over three decades ago (Braun-Falco, 1971; Wilkinson, 1971). Interestingly these morphologic and metabolic alterations in uninvolved skin include processes of lipid metabolism, predominantly in the horny layer of the skin. These were characterized by changes in phospholipid composition and levels and distribution of several hydrolytic enzymes and dehydrogenases (Braun-Falco, 1971; Wilkinson, 1971). Furthermore, increased biosynthesis of arachidonic acid metabolites (Ziboh et al., 1984) and increased blood flow has been noted in PN skin (Klemp and Staberg, 1983) but the etiology of these changes is unknown.
The purpose of this study was to carefully characterize and compare gene expression in normal (NN) vs. unaffected skin (PN) from psoriatic patients. Taking advantage of the large size of our sample, we initially focused on progressive differences between NN vs. PN vs. PP skin in an effort to gain further insight into the disease process. During our analysis, we identified strong patterns of coordinate expression of genes involved in lipid metabolism as well as innate immunity and keratinocyte differentiation in PN vs. NN skin. Taking a bioinformatic approach, we identified three transcription factors that could be responsible for the coordinate expression of these lipid biosynthetic genes.
RESULTS
Lesional psoriatic skin shows markedly different gene expression profile compared to PN and NN skin
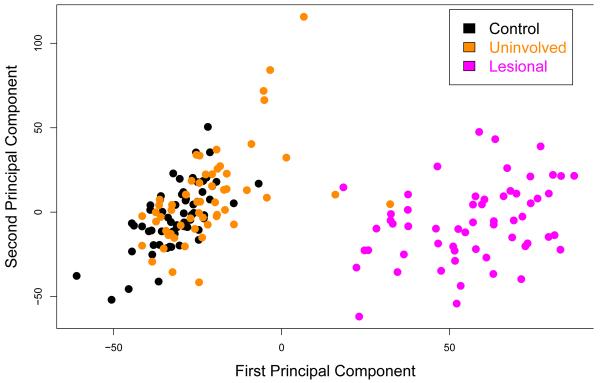
Principal components analysis (PCA) (Figure 1) and unsupervised hierarchical clustering (Figure S1) based on all probes and all samples revealed near-complete separation of the PP samples from both the PN and the NN samples, with only two of 58 PN samples overlapping with the PP samples. However, there was a significant overlap between PN and NN samples, which was confirmed by hierarchical clustering. These analyses indicated a distinct gene expression profile of PP skin that is markedly different from those of PN or NN skin, whereas the difference between PN and NN skin is much more subtle.
Figure 1. Principal component analysis.
The first and second principal components are shown. Two uninvolved samples (2/58) overlapped minimally with the lesional samples whereas large overlap was observed between the control and uninvolved samples.
Uninvolved psoriatic (PN) skin has a large number of differentially regulated genes compared to NN skin
Based on our criteria for differentially regulated transcripts (Materials and Methods), we identified 223 transcripts that were differentially expressed between PN and NN samples (72 up-regulated, 151 down-regulated) (Figure S2). Of these, 201 transcripts represent known genes, 22 of which were redundant, giving a total number of 179 unique differentially regulated genes in uninvolved psoriatic skin (58 genes up-regulated and 121 genes down-regulated). The other 22 were transcripts for hypothetical genes sequences. Based on permutation testing, the mean of the number of differentially expressed transcripts expected under the null hypothesis was 10. Among the most strongly dysregulated genes (Table 1), several of down-regulated transcripts encode proteins involved in fatty acid metabolism, including ALOX15B encoding a 15-lipoxygenase, FADS1 encoding a fatty acid desaturase, and ELOVL3 encoding elongation of very long chain fatty acids-like 3. Other down-regulated transcripts included YWHAE encoding 14-3-3ε, ESR1 encoding estrogen receptor 1, and GAL encoding galanin, which is a vasoactive peptide. Among the most strongly up-regulated genes, several are encoded in the EDC, including SPRR2B, SPRR2G, and SPRR3 and LCED3 encoding proteins involved in terminal differentiation of the epidermis (Marshall et al., 2001) as well as S100A8, S100A9 and S100A7 encoding antimicrobial peptides involved in innate immunity. Other up-regulated genes also encode proteins involved in innate immunity, including DEFB4 encoding human beta defensin-2 (hBD-2), RNASE7 encoding a ribonuclease which, like hBD-2, has broad-spectrum antimicrobial activity (Harder and Schroder, 2002), and IL1F9 encoding the pro-inflammatory cytokine IL-1ε. No matches (0/133) were found between our list of down-regulated transcripts and previously published gene lists (Zhou et al., 2003), and only one (psoriasin, S100A7) of the 58 up-regulated genes identified by our study had been previously reported (Zhou et al., 2003).
Table 1.
Several of the most up- and down-regulated genes in PN vs. NN skin and their progressive changes in NN vs. PN vs. PP skin
| Up-regulated Genes | Fold change NN vs. PN | Fold change PN vs. PP | Fold change NN vs. PP |
|---|---|---|---|
| C10orf99 | 1.811 | 22.862 | 41.393 |
| SPRR2B | 1.779 | 20.231 | 35.982 |
| S100A7 | 1.774 | 10.322 | 18.309 |
| LCE3D | 1.737 | 14.060 | 24.421 |
| SPRR2G | 1.704 | 4.732 | 8.062 |
| WFDC12 | 1.632 | 2.655 | 4.333 |
| S100A9 | 1.565 | 36.174 | 56.601 |
| HAL | 1.526 | 3.095 | 4.723 |
| IL1F9 | 1.498 | 24.258 | 36.338 |
| DEFB4 | 1.469 | 134.078 | 196.997 |
| Down-regulated Genes | Fold change NN vs. PN | Fold change PN vs. PP | Fold change NN vs. PP |
|---|---|---|---|
| ELOVL3 | 1.869 | 3.452 | 6.454 |
| FLJ32569 | 1.854 | 3.703 | 6.867 |
| HSD3B1 | 1.847 | 2.188 | 4.041 |
| MLSTD1 | 1.842 | 2.835 | 5.222 |
| GAL | 1.801 | 3.543 | 6.383 |
| KRT6L | 1.724 | 2.102 | 3.623 |
| THRSP | 1.692 | 4.541 | 7.684 |
| FADS1 | 1.636 | 2.101 | 3.437 |
| MUC7 | 1.566 | 2.294 | 3.737 |
| SCGB2A1 | 1.544 | 2.367 | 3.706 |
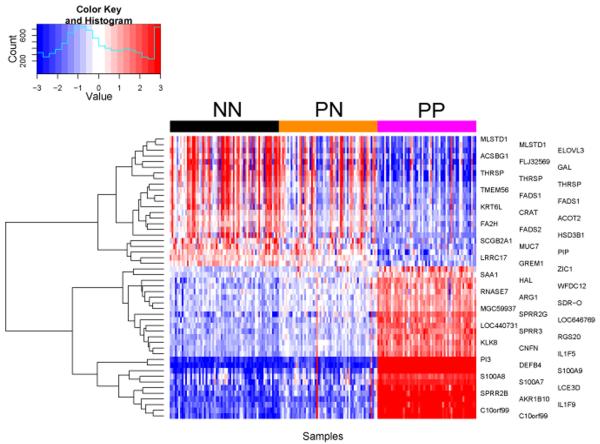
A large group of genes shows progressive changes from NN to PN to PP skin
To identify genes that show progressive change through all three sample groups, we used a threshold ≥ 1.3-fold change and nominal p < 0.05 between PN and NN samples, together with ≥ 2-fold change in the same direction and false discovery rate (FDR) p < 0.05 for comparing PP versus PN skin. Using these criteria, 27 genes manifested a progressive increase in gene expression (PP > PN > NN), whereas 23 genes showed progressive down-regulation (NN > PN > PP, Figure 2, Table 1). As expected, these genes were a subset of those manifesting altered regulation in PN vs. NN skin, including S100A8, S100A9 and S100A7, SPRR2B, SPRR2G, SPRR3, LCE3D, DEFB4, RNASE7, and IL1F9 in the up-regulated group, and GAL and several genes encoding proteins involved fatty acid metabolism in the down-regulated group.
Figure 2. Progressive gene changes in NN vs. PN vs. PP skin.
To analyze genes that show progressive change through all three sample groups we used a threshold of equal or greater than 1.3-fold change and a nominal p-value of less than 0.05 between NN and PN samples, whereas 2-fold change was used comparing lesional versus uninvolved and control skin with an FDR corrected p-value of less than 0.05. The blue color indicates low expression whereas the red color indicates high expression.
Confirmation of differentially expressed genes by QRT-PCR
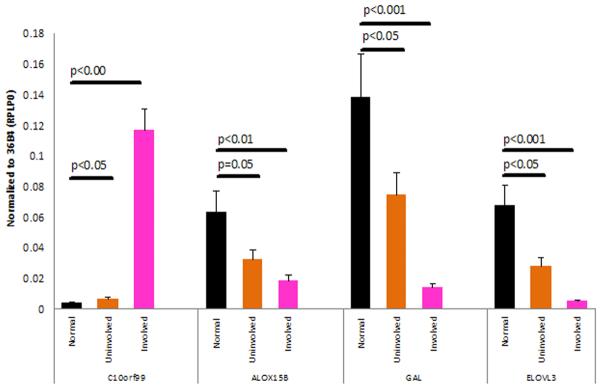
To confirm and validate the microarray results, we performed QRT-PCR for several genes including C10orf99, ALOX15B, GAL and ELOVL3 (n=25 each for NN, PN, and PP skin) (Figure 3). Relative to NN skin, C10orf99 was up-regulated 1.7-fold in PN skin (p<0.05) and 30-fold in PP skin (p<0.0001). In contrast, ALOX15B was down-regulated 2-fold in PN skin (p=0.05), and 3.4-fold in PP skin (p<0.01). GAL was down-regulated 1.9-fold in PN vs. NN skin (p<0.05), and 10-fold down in PP vs. (p<0.01). ELOVL3 was reduced 2.4-fold in PN skin (p<0.05) and 13.4-fold in PP skin (p<0.001). Thus, these results uniformly confirmed the results of microarray analysis.
Figure 3. QRT-PCR analysis of selected transcripts in NN vs. PN vs. PP skin.
QRT-PCR was performed as described in Materials and Methods. Error bars indicate SEM (n=25 in control, uninvolved and lesional groups). P values for various comparisons are indicated above the horizontal bars. On average, C10orf99 was up-regulated by approximately 1.7-fold in uninvolved samples, whereas ALOX15B was down-regulated by 2.0 fold, galanin (GAL) by 1.9-fold and ELOVL3 2.4-fold. C10orf99 was 30-fold up-regulated in lesional skin.
Hierarchical clustering of differentially-expressed genes in PN vs. NN skin
To better assess the gene expression changes we observed in PN skin we performed an unsupervised hierarchical clustering on the 223 differentially-expressed transcripts observed in PN vs. NN skin, either in the PN samples (Figure S3A), or in the NN samples (Figure S3B). The PN samples clustered into two main groups of 27 and 31 samples respectively. By permutation testing, the probabilities of observing by chance a cluster as deeply separated from one upper level cluster as the observed two were calculated to be p = 0.00024 and p = 0.0098, respectively. There was no difference in body mass index (BMI) or age observed between the two clusters. This analysis was repeated for NN skin (Figure S3B). The NN samples clustered into two main groups of 16 and 48 samples respectively, and the probabilities of obtaining the observed clusters by chance were p = 0.00012 and p = 0.24, respectively. Notably, the genes showing the most prominent clustering were the same in both sets of samples, including nine genes involved in lipid metabolism (THRSP, MLSTD1, FADS1, FADS2, SOAT1, ACSBG1, ELOVL3, HSD3B1 and ALOX15B) as well other genes including GAL, KRT6L, TF (encoding transferrin), TMEM56, and FLJ32569. As a group, these genes were expressed at a higher level more frequently in NN skin than in PN skin (compare Figs. 4A and 4B).
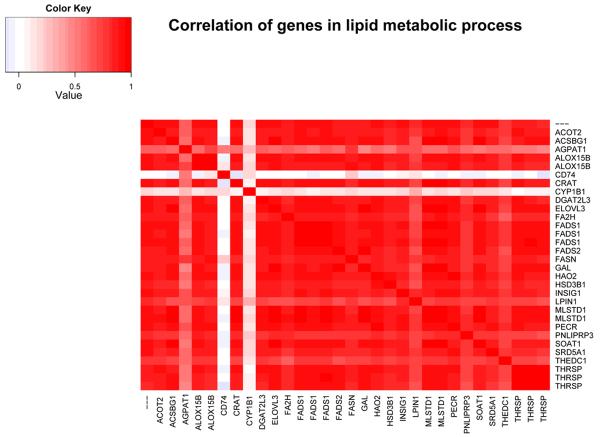
Figure 4. Correlated expression of genes involved lipid metabolism.
Genes shown fall under the GO term “lipid metabolic process”, the term subsuming the largest number of down-regulated transcripts in our sample. We observed a strong correlation between all the genes within this GO category except for CYP1B1 and CD74. CD74 is the invariant polypeptide associated with MHC Class II molecules and has been shown to have a role in enhancing lipid antigen presentation on CD1d. CYP1B1 is a member of the cytochrome P450 family which catalyze many reactions involved in drug metabolism and synthesis of cholesterol, steroids and other lipid. It has been implicated in arachidonic acid metabolism in the cornea (Schwartzman et al., 1987). The most likely reason that these two genes do not show the same correlation as the rest of the group, is that these are only marginally associated with lipid metabolic processes.
Gene Ontology analysis identifies enrichment of several biological processes including defense response and lipid metabolism
To more rigorously identify biological processes encompassing altered patterns of gene regulation in NN vs. PN skin, we utilized Gene Ontology (GO) analysis. Biological processes significantly (p < 0.05) enriched for up-regulated transcripts in PN vs. NN skin include epidermal morphogenesis and development, epidermal cell differentiation, oxygen transport, keratinization and defense response (Table S2), whereas processes significantly enriched for down-regulated transcripts included predominantly lipid and fatty acid metabolism and biosynthetic pathways (Table S2).
Pathway analysis and gene-gene correlation within GO categories reveals strong correlation between individual genes within the lipid metabolic process
We used the Ingenuity Pathway Analysis (IPA) (www.ingenuity.com) tool to perform pathway analysis on the group of genes involved in lipid metabolism. Multiple functions within lipid metabolism were significantly (p<0.01) affected, including quantity, metabolism, conversion denaturation, uptake and synthesis (Figure S4). Because we noticed a tendency for coordinate regulation in the hierarchical clustering analysis, we assessed the correlation of transcript expression between genes falling into significantly up-regulated GO categories. We found high correlation between individual genes within the lipid metabolic process (Figure 4). However, only subgroups of genes within GO categories of defense response (Figure S5) and keratinocyte differentiation were highly correlated (Figure S6).
Transcription factor and promoter analysis shows enrichment for specific transcription factors
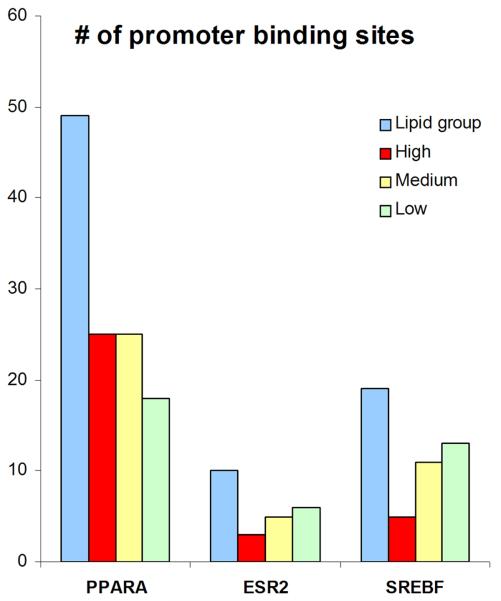
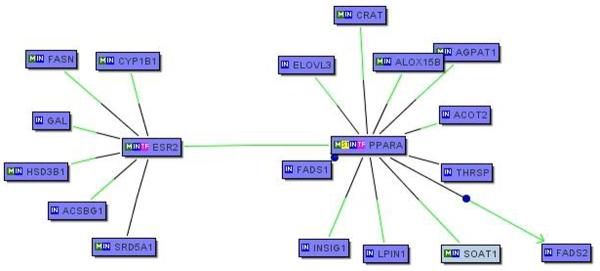
Of the 179 genes that we found to be differentially regulated between PN and NN skin, the largest group belong to the GO category of lipid metabolic process (n = 25) with a very high pairwise correlation (Figure 4). The co-citation feature of the Genomatix software suite (BiblioSphere) was used to identify 21 candidate transcription factors for this group (Table S1). This list of transcription factors did not overlap with any of the 17 transcription factors that we found to be differentially regulated in PN vs. NN skin (data not shown). The most highly represented transcription factors belonged to the family of peroxisome proliferator-activated receptors (PPARs) (Table S1). Using the Genomatix software suite, we analyzed the promoter sequences 500 bp upstream and 100 bp downstream from the transcriptional start sites for each of the lipid metabolic process genes. A total of 98 promoters were found for this group. This was compared to a randomly-selected set of 98 promoter sequences in the three separate groups of control genes obtained from the same microarray analysis. Using the GemsLauncher and MatInspector tools of the Genomatix software, the promoter sequences were analyzed against the 21 candidate transcription factors identified by BiblioSphere. Three of these transcription factors, PPARA, ESR2 and SREBF, showed enriched binding sites in the 25 lipid genes compared to the three control groups (Figure 5). Although there was a slight down-regulation of PPARA on the microarray (1.15-fold, p<0.05) we could not confirm or detect any differences in the expression of these three transcription factors by QRT-PCR (n=30, data not shown). To further interrogate known interactions among these 3 transcription factors and 25 lipid metabolic process genes we uploaded these genes (n=28) into BiblioSphere to examine the gene interaction networks. PPARA and ESR2 connected majority of the genes (17/25) in an extended interaction network (Figure 6).
Figure 5. Bioinformatic analysis of promoter binding sites.
The transcription factors: PPARA, ESR2, and SREBF were found to have increased number of promoter binding sites in the lipid metabolism group compared to three control groups consisting of genes with low, medium and high expression in our dataset.
Figure 6. Pathway analysis of lipid metabolic process genes.
To interrogate known interactions amongst the 25 lipid metabolic process genes that are co-regulated in PN and NN skin and the three transcription factors that show enriched numbers of binding sites in the promoters of these genes, we utilized Genomatix BiblioSphere to build gene interaction networks. Two of the enriched transcription factors, PPARA and ESR, connected most (17/25) of the genes in an extended network.
DISCUSSION
We have previously used this same dataset to evaluate the activation of the Sonic Hedgehog Pathway in psoriasis (Gudjonsson et al., 2009), In this study we compared the gene expression profiles of involved (PP) as well as clinically normal-appearing (PN) skin of psoriatic patients to those observed in normal (NN) skin of non-psoriatic individuals. The large size of our sample allowed us to identify relatively small but significant changes in the transcriptome of PN relative to skin, revealing a coordinated program of gene expression predominantly involving down-regulation of lipid biosynthetic genes. We found 179 genes to be differentially regulated in PN relative to NN skin (Table 1), many of which displayed progressive up- or down-regulation in NN vs. PN vs. PP skin (Figure 2). Only one of these genes has been reported previously. These changes in gene expression confirm biochemical and morphologic observations made on PN skin three decades ago (Braun-Falco, 1971; Wilkinson, 1971) Predominantly found in the stratum corneum, these changes involved the levels and composition of phospholipids, free alpha-amino acids, hydrolytic enzymes and several dehydrogenases. Additional changes in the epidermal lipid composition (Wilkinson, 1971) led to the coining of the term “histochemical parakeratosis” to describe these findings (Braun-Falco, 1971). Taken together, these findings suggest that PN skin might exist in a “pre-psoriatic state”. However, it is important to note that NN skin can also manifest the same coordinated down-regulation of lipid biosynthetic genes observed in PN skin, albeit less frequently (Figures S3A and S3B).
Given the strikingly coordinated expression of these genes (Figure 4), our results prompted us to undertake a systematic search for transcription factors that might underlie this phenomenon. By both GO and pathway analyses, we confirmed significant enrichment for genes involved in lipid metabolism in transcripts that were down-regulated in PN vs. NN skin (Table S2), and showed that many of these genes were further down-regulated in PP skin (Table 1). These results systematically identified a subset of coordinately expressed genes involved in lipid metabolism (Figure 4), which we used to carry out a bioinformatic search for transcription factors that might help to explain our observations (Figures 5 and 6, Figure S7, Table S1). The results of our analysis suggest that these coordinated abnormalities may be related to decreased activity of three transcription factors (PPARA encoding PPAR-α, ESR2 encoding estrogen receptor 2, and SREBF1 encoding sterol regulatory element binding transcription factor 1, (Figure 5). Of these, ESR2 and PPAR-α bioinformatically connected 17 of the 25 lipid biosynthetic genes that we identified as dysregulated in PN vs. NN skin (Figure 6). Although we were unable to detect changes in the mRNA expression of the three candidate transcription factors by QRT-PCR it does not rule out differences in proteins levels or function of these factors. Determination of this would require comprehensive biochemical analyses that were not performed in this study. Interestingly, our search revealed no evidence for activation of inflammatory transcription factors secondary to circulating cytokines or chemokines emanating from active plaques (data not shown).
Dysregulation of transcripts relating to fatty acid signaling and adipocyte differentiation has recently been described in PP vs. PN skin, and related to the activation of peroxisome proliferator-activated receptor δ (PPARδ) (Romanowska et al., 2008). Indeed, we were able to confirm significant dysregulation of 31 of 32 genes reported by Romanowska et al. (Romanowska et al., 2008) to be abnormally expressed in PP vs. PN skin and related to fatty acid signaling (data not shown). Interestingly, PPARδ over-expression has been shown to suppress the activity of PPARγ and PPARα (Shi et al., 2002). Thus, the similarities between our data and that of Romanowska et al (Romanowska et al., 2008) could be related to decreased or suppressed activity of PPARα.
Among the down-regulated lipid biosynthetic gene transcripts that we identified in PN vs. NN skin, ELOVL3 was the most extensively reduced (1.9-fold). It was also progressively down-regulated in NN vs. PN vs. PP skin (Table 1). ELOVL3 is a member of a highly conserved family of microsomal enzymes involved in the formation of very long chain fatty-acids (Jakobsson et al., 2006), which are important constituents of sphingolipids, glycerophospholipids, triacylglycerols, sterol- and wax-esters (Jakobsson et al., 2006). ELOVL3 knockout mice develop thickened skin with excoriations resembling eczematous skin (Westerberg et al., 2004). Measurement of the lipid composition of the epidermis of these mice demonstrated an increase in eicosenoic acid (C20:1) and a significant drop (40%) in C16:0, C18:0 and C18:1 fatty acids (Westerberg et al., 2004), similar to what has been demonstrated for uninvolved psoriatic skin (Wilkinson, 1971). These mice had skin barrier impairment and increased transepidermal water loss (Westerberg et al., 2004). Another progressively down-regulated gene, ACSBG1, encodes the acyl-CoA synthetase bubblegum family member 1. Acyl-CoA synthases carry out a fundamental reaction in fatty acid metabolism; the thioesterification of the acyl group to coenzyme A (CoA) (Watkins, 1997). The fatty acyl-CoA product can have several metabolic fates, including incorporation into phospholipids, mono-, di-, and triacylglycerols, sphingolipids, glycoplids, cholesterol esters or acetylation of proteins (Pei et al., 2006). Fruit flies lacking this gene have elevated tissue levels of saturated very long-chain fatty acids (Pei et al., 2006). Consistent with this observation, skin surface lipids in uninvolved skin of psoriatic patients have been described to have higher amounts of saturated C23 fatty acids (Wilkinson, 1971). THRSP is also progressively down-regulated and encodes SPOT 14, a nuclear protein thought to activate genes encoding the enzymes of fatty acid synthesis (Cunningham et al., 1998; Kinlaw et al., 1992). Its role in skin physiology is presently unexplored. MLSTD1 encodes a fatty acyl-CoA reductase (FAR2) whose expression is limited to tissues that are rich in sebaceous glands, such as skin (Cheng and Russell, 2004). This enzyme converts fatty acids to fatty alcohols, the first step necessary for the formation of ether lipids and waxes. Another down-regulated transcript predominantly expressed in sebaceous glands, HSD3B1, encodes hydroxy-delta-5-steroid dehydrogenase type I (Dumont et al., 1992; Simard et al., 2005). Hydroxy-delta-5-steroid dehydrogenases are required for the biosynthesis of steroid hormones (Simard et al., 2005), and the expression of this enzyme in sebaceous glands allows these tissues to control local concentrations of DHEA and other steroids in the skin (Simard et al., 2005). Interestingly, sebaceous glands are hypoplastic in psoriasis, particularly in scalp lesions (Headington et al., 1989; Wilson et al., 1994). It is possible that this relates to decreased expression of HSD3B1 leading to decreased effect of steroid hormones on sebaceous gland development and maintenance. Two other progressively down-regulated genes participate in the generation of eicosanoids. FADS1 encodes fatty acid desaturase 1, which participates in the conversion of polyunsaturated fatty acids into arachidonic acid, the direct precursor of prostaglandins and leukotrienes (Lewis and Austen, 1984; Schaeffer et al., 2006). ALOX15B is also down-regulated in PN skin and encodes 15-lipoxygenase type 2, another enzyme involved in arachidonic acid metabolism (Jiang et al., 2006). Reduced expression of ALOX15B might explain the longstanding observation that in vitro synthesis of 12-HETE is increased in PN relative to NN skin (Kragballe et al., 1986).
Other transcription factors that have been implicated in formation of the lipid component of the skin barrier during development include KLF4 (Segre et al., 1999) and GATA-3 (de Guzman Strong et al., 2006). Although we did not find evidence for transcriptional involvement of KLF4 or GATA3 in our study (data not shown), psoriasis is characterized by markedly impaired epidermal barrier function (Ghadially et al., 1996). Reminiscent of psoriasis, Elovl3 expression was down-regulated in GATA-3 knockout mice, whereas S100a8, S100a9, Lce, Defb3 (the murine homolog of DEFB4), and protease inhibitor genes were up-regulated (de Guzman Strong et al., 2006). These findings are consistent with the highly integrated functions of the epidermal lipid barrier and the innate immune system in host defense. Antimicrobial peptides such as cathelicidin and beta-defensins are co-packaged along with lipids within epidermal lamellar bodies before their secretion (Aberg et al., 2008; Oren et al., 2003) and coordinate regulation of barrier lipid production and antimicrobial peptides by permeability barrier disruption has been demonstrated (Aberg et al., 2008). The similarity between the genes whose expression is perturbed in psoriasis and in GATA-3 knockout mice are striking, and taken together with the lipid abnormalities we have observed, are strongly suggestive of an incipient defect involving the integrated lipid barrier-innate immune axis in the stratum corneum of PN skin. A somewhat different scenario may take place in atopic dermatitis, in which mutations in the FLG gene encoding filaggrin, a protein critical for corneocyte maturation (Candi et al., 2005), lead to defects in the keratinocyte cornified envelope (Candi et al., 2005; Hudson, 2006). Thus, instead of being caused by a defective protein within keratinocytes (the bricks), as is the case with atopic dermatitis, the stratum corneum defect in psoriasis may involve defective “mortar” between the keratinocytes. Interestingly, psoriasis and atopic dermatitis differ markedly in their expression of innate immune genes, with much lower expression of S100A7, S100A8, S100A9, DEFB4, and PI3 in chronic atopic dermatitis than in psoriasis despite comparable epidermal hyperplasia.(de Jongh et al., 2005) Whether the altered lipid biosynthetic program that we have identified here is also characteristic of atopic dermatitis remains to be determined. It should be noted that the gene expression changes that we observe are relatively small, ranging from 1.3- to 1.86-fold in contrast to up to 200-fold changes we observe in lesional skin (table 1). Such subtle changes in gene expression are not unexpected as the skin in these patients is clinically normal. However, it can make it more difficulty to determine the contribution of individual factors. Other future studies should be directed toward defining psoriasis susceptibility genes and determining whether they are involved in the biological mechanisms underlying coordinated alterations of gene expression programs in PN vs. NN skin.
MATERIALS AND METHODS
Subjects
58 psoriatic subjects and 64 normal healthy controls were enrolled in the study. Informed consent was obtained from all subjects, under protocols approved by the Institutional Review Board of the University of Michigan Medical School. This study was conducted according to the Declaration of Helsinki Principles. The criterion for entry of a case was that the presence of one or more sharply demarcated, erythematous, scaly psoriatic plaques that were not limited to the scalp. In those instances where there was only a single psoriatic plaque, the case was only considered if the plaque occupied more than 1% of total body surface area. Study subjects did not use any systemic anti-psoriatic treatments for 2 weeks prior or topical anti-psoriatic treatments for 1 week prior to biopsy. Gender was balanced in both case and control cohorts. Mean age of controls was 41.1 years (range 18-75) whereas mean age of patients was 48.5 years (range 21-69). The study subjects were recruited from the greater Detroit area and all the patients and controls used in this analysis were Caucasian. Two biopsies were taken under local anesthesia from each psoriatic subject; one 6mm punch biopsy was obtained from PP skin and the other from PN skin, taken at least 10cm away from any active plaque. One or two biopsies were obtained from healthy controls. The PN and NN skin biopsies were always taken from the buttocks or upper thighs.
RNA processing and microarray hybridization
Biopsies were snap-frozen in liquid nitrogen and stored at -80°C until use. Biopsies were crushed with a hammer while still frozen and total RNA isolation was performed using a commercial kit (RNeasy, Qiagen, Chatsworth, CA), employing glass beads (www.biospec.com cat#11079125) for homogenization. RNA quantity and quality was measured on Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA); only samples yielding intact 18S and 28S ribosomal RNA profiles were used. cDNA synthesis and in vitro transcription for probe biotinylation were performed on 5μg of total RNA according to the manufacturer's protocols (Affymetrix, Foster City, CA) . Samples were run on HU133 Plus 2.0 arrays to query expression of ~ 54,000 probes according to the manufacturer's protocol.
QRT-PCR
QRT-PCR was performed on NN samples from 25 normal controls and paired PN and PP samples from 25 psoriatic patients. Primers for the genes ALOX15B, C10orf99, GAL and ELOVL3 were obtained from Superarray Biosciences (Frederick, MD, USA). Results were normalized to the expression of the housekeeping gene; ribosomal protein, large, P0 (RPLP0). The reverse transcription reaction was performed on 0.5ug of RNA template and cDNA was synthesized using anchored-oligo(dT)18 primers as instructed by the manufacturer (Roche Diagnostics, Mannheim, Germany). QRT-PCR was carried out on a LightCyclerTM 2.0 system (Roche Diagnostics, Mannheim, Germany). The reaction profile consisted of an initial denaturation at 95°C for 15 minutes followed by 40 cycles of PCR at 95°C for 10 seconds (denaturation), 58°C for 10 seconds (annealing) and 72°C for 10 seconds (extension). The fluorescence emitted was captured at the end of the extension step of each cycle at 530nm. Primers for ESR2, PPARA and SREBF1 (Applied Biosystems) and QRT-PCR was carried out using an Applied Biosystems 799HT Fast Real Time PCR System (Applied Biosystems Inc, Foster City, CA). QRT-PCR for these 3 primers was performed on 30 NN, 30 PN and 30 PP samples.
Data Analysis and Statistics
The raw data from 180 microarrays were processed using the Robust Multichip Average (RMA) method (Irizarry et al., 2003) and then adjusted to account for gender and batch effects. Hierarchical clustering (using a “complete” agglomeration method) and principal components analysis (PCA) were performed on the adjusted expression data using the publicly available software R (www.r-project.org). Gene expression was contrasted between PN vs. NN based on the following criteria: ≥ 1.3 fold-change in the means of expression in two groups and nominal p-value ≤ 0.05. We used permutation (i.e. permuting subjects' labels) to determine the expected number of differentially expressed genes by chance, and hence estimated the false discovery rate of our differential expression gene list. Those genes that were differentially expressed between PN and NN skin were used to perform hierarchical clustering on both PN and NN skin samples. Significance of the clusters obtained was assessed by permutation testing. Specifically, we permuted sample labels and calculated the probabilities of observing by chance same-sized clusters as deeply separated from the one upper level cluster as the observed clusters for both PN and NN skin.
Gene Ontology
Gene Ontology (GO) category enrichment analysis was performed using the publicly available software DAVID(Database for Annotation, Visualization and Integrated Discovery, http://david.abcc.ncifcrf.gov/). The goal of this analysis was to search for GO terms in molecular function, biological process and cellular component that were significantly enriched in the gene lists obtained above. As this was an exploratory analysis, p-value ≤0.001 was used as the stringent significance criterion and p-value ≤ 0.05 as the loose significance criterion. For those GO categories that were significantly changed we performed gene-gene correlation analysis within each GO term.
Ingenuity Pathway Analysis
Data were analyzed through the use of Ingenuity Pathway Analysis (Ingenuity Systems, www.ingenuity.com). For Network Generation, a data set containing gene identifiers and corresponding expression values was uploaded into the application. Each gene identifier was mapped to its corresponding gene object in the Ingenuity Pathways Knowledge Base. A 1.3-fold cutoff, as described above, was set to identify genes whose expression was significantly differentially regulated. These genes, called focus genes, were overlaid onto a global molecular network developed from information contained in the Ingenuity Pathways Knowledge Base. Networks of these focus genes were then algorithmically generated based on their connectivity.
Promoter analysis
Promoter analysis was performed on genes belonging to the GO category of “lipid metabolic process”, which was the largest group of genes within our list of differentially regulated genes in PN vs. NN skin. As depicted in Figure S7, these genes were uploaded to Genomatix Bibliosphere (www.genomatix.de) and filtered according to co-citation in the literature. This list was subsequently run against all known transcription factors (>600) to identify candidate transcription factors in our dataset. Control genes were arbitrarily selected from our microarray data. The selection criteria was that there was no difference between control and uninvolved skin (<0.3% difference in expression between uninvolved and normal skin and nominal p>0.05). The control group of genes was split into three separate groups based on low expression (40 transcripts, absolute expression values <4), medium expression (40 transcript, absolute expression values between 7 and 8) and high expression (40 transcripts, absolute expression values higher than 11.6). The promoter sequences (500 basepairs (bp) upstream and 100 bp downstream of the transcriptional start site) for the lipid genes and the three control groups were retrieved from the Genomatix promoter database using Gene2Promoter and Eldorado tools of the Genomatix software suite (http://www.genomatix.de). Binding sites in promoter sequences were determined using MatInspector tools of the Genomatix software suite (http://www.genomatix.de). Overrepresented transcription factors were uploaded into BiblioSphere for determination of gene-interaction networks.
Supplementary Material
ACKNOWLEDGEMENTS
The authors express their appreciation for all the research subjects who participated in this study and the assistance of Lynda Hodges and Kathleen McCarthy. The authors thank Philip Stuart for statistical and technical assistance. Dr. Anne Bowcock for providing gene lists referenced in Zhou et al., Physiological Genomics 13:69-78, 2003. This work was supported by grants to J.T.E. (National Institutes of Arthritis, Musculoskeletal and Skin Diseases, R01-AR054966) and J.E.G (American Skin Association, Dermatology Foundation).
Footnotes
CONFLICT OF INTEREST. The authors have no conflicts of interest.
REFERENCES
- Aberg KM, Man MQ, Gallo RL, Ganz T, Crumrine D, Brown BE, et al. Co-regulation and interdependence of the mammalian epidermal permeability and antimicrobial barriers. J Invest Dermatol. 2008;128:917–925. doi: 10.1038/sj.jid.5701099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowcock AM, Shannon W, Du F, Duncan J, Cao K, Aftergut K, et al. Insights into psoriasis and other inflammatory diseases from large-scale gene expression studies. Hum Mol Genet. 2001;10:1793–1805. doi: 10.1093/hmg/10.17.1793. [DOI] [PubMed] [Google Scholar]
- Braun-Falco O. Dynamics of Growth and Regression in Psoriatic Lesions: Alterations in the skin from Normal into a Psoriatic Lesion, and during Regression of Psoriatic Lesions. Stanford University Press; Stanford, California: 1971. pp. 215–237. [Google Scholar]
- Candi E, Schmidt R, Melino G. The cornified envelope: a model of cell death in the skin. Nat Rev Mol Cell Biol. 2005;6:328–340. doi: 10.1038/nrm1619. [DOI] [PubMed] [Google Scholar]
- Cheng JB, Russell DW. Mammalian wax biosynthesis. I. Identification of two fatty acyl- Coenzyme A reductases with different substrate specificities and tissue distributions. J Biol Chem. 2004;279:37789–37797. doi: 10.1074/jbc.M406225200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad C, Boyman O, Tonel G, Tun-Kyi A, Laggner U, de Fougerolles A, et al. Alpha1beta1 integrin is crucial for accumulation of epidermal T cells and the development of psoriasis. Nat Med. 2007;13:836–842. doi: 10.1038/nm1605. [DOI] [PubMed] [Google Scholar]
- Cunningham BA, Moncur JT, Huntington JT, Kinlaw WB. “Spot 14” protein: a metabolic integrator in normal and neoplastic cells. Thyroid. 1998;8:815–825. doi: 10.1089/thy.1998.8.815. [DOI] [PubMed] [Google Scholar]
- de Guzman Strong C, Wertz PW, Wang C, Yang F, Meltzer PS, Andl T, et al. Lipid defect underlies selective skin barrier impairment of an epidermal-specific deletion of Gata-3. J Cell Biol. 2006;175:661–670. doi: 10.1083/jcb.200605057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jongh GJ, Zeeuwen PL, Kucharekova M, Pfundt R, van der Valk PG, Blokx W, et al. High expression levels of keratinocyte antimicrobial proteins in psoriasis compared with atopic dermatitis. J Invest Dermatol. 2005;125:1163–1173. doi: 10.1111/j.0022-202X.2005.23935.x. [DOI] [PubMed] [Google Scholar]
- Detmar M, Brown LF, Claffey KP, Yeo KT, Kocher O, Jackman RW, et al. Overexpression of vascular permeability factor/vascular endothelial growth factor and its receptors in psoriasis. J Exp Med. 1994;180:1141–1146. doi: 10.1084/jem.180.3.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont M, Luu-The V, Dupont E, Pelletier G, Labrie F. Characterization, expression, and immunohistochemical localization of 3 beta-hydroxysteroid dehydrogenase/delta 5-delta 4 isomerase in human skin. J Invest Dermatol. 1992;99:415–421. doi: 10.1111/1523-1747.ep12616131. [DOI] [PubMed] [Google Scholar]
- Elder JT, Fisher GJ, Lindquist PB, Bennett GL, Pittelkow MR, Coffey RJ, Jr., et al. Overexpression of transforming growth factor alpha in psoriatic epidermis. Science. 1989;243:811–814. doi: 10.1126/science.2916128. [DOI] [PubMed] [Google Scholar]
- Ghadially R, Reed JT, Elias PM. Stratum corneum structure and function correlates with phenotype in psoriasis. J Invest Dermatol. 1996;107:558–564. doi: 10.1111/1523-1747.ep12582813. [DOI] [PubMed] [Google Scholar]
- Gladman DD. Natural history of psoriatic arthritis. Baillieres Clin Rheumatol. 1994;8:379–394. doi: 10.1016/s0950-3579(94)80024-3. [DOI] [PubMed] [Google Scholar]
- Gudjonsson JE, Aphale A, Grachtchouk M, Ding J, Nair RP, Wang T, et al. Lack of evidence for activation of the hedgehog pathway in psoriasis. J Invest Dermatol. 2009;129:635–640. doi: 10.1038/jid.2008.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudjonsson JE, Elder JT. Psoriasis. In: Wolff K, Goldsmith LA, Katz SI, Gilchrest BA, Paller AM, Leffell DJ, editors. Fitzpatrick's Dermatology in General Medicine. Vol. 1. McGraw-Hill; New York: 2007. pp. 169–194. [Google Scholar]
- Gudjonsson JE, Johnston A, Sigmundsdottir H, Valdimarsson H. Immunopathogenic mechanisms in psoriasis. Clin Exp Immunol. 2004;135:1–8. doi: 10.1111/j.1365-2249.2004.02310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta MA, Schork NJ, Gupta AK, Kirkby S, Ellis CN. Suicidal ideation in psoriasis. Int J Dermatol. 1993;32:188–190. doi: 10.1111/j.1365-4362.1993.tb02790.x. [DOI] [PubMed] [Google Scholar]
- Harder J, Schroder JM. RNase 7, a novel innate immune defense antimicrobial protein of healthy human skin. J Biol Chem. 2002;277:46779–46784. doi: 10.1074/jbc.M207587200. [DOI] [PubMed] [Google Scholar]
- Headington JT, Gupta AK, Goldfarb MT, Nickoloff BJ, Hamilton TA, Ellis CN, et al. A morphometric and histologic study of the scalp in psoriasis. Paradoxical sebaceous gland atrophy and decreased hair shaft diameters without alopecia. Arch Dermatol. 1989;125:639–642. [PubMed] [Google Scholar]
- Hudson TJ. Skin barrier function and allergic risk. Nat Genet. 2006;38:399–400. doi: 10.1038/ng0406-399. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson A, Westerberg R, Jacobsson A. Fatty acid elongases in mammals: their regulation and roles in metabolism. Prog Lipid Res. 2006;45:237–249. doi: 10.1016/j.plipres.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Jiang WG, Watkins G, Douglas-Jones A, Mansel RE. Reduction of isoforms of 15- lipoxygenase (15-LOX)-1 and 15-LOX-2 in human breast cancer. Prostaglandins Leukot Essent Fatty Acids. 2006;74:235–245. doi: 10.1016/j.plefa.2006.01.009. [DOI] [PubMed] [Google Scholar]
- Johnston A, Gudjonsson JE, Sigmundsdottir H, Love TJ, Valdimarsson H. Peripheral blood T cell responses to keratin peptides that share sequences with streptococcal M proteins are largely restricted to skin-homing CD8(+) T cells. Clin Exp Immunol. 2004;138:83–93. doi: 10.1111/j.1365-2249.2004.00600.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinlaw WB, Tron P, Friedmann AS. Nuclear localization and hepatic zonation of rat “spot 14” protein: immunohistochemical investigation employing anti-fusion protein antibodies. Endocrinology. 1992;131:3120–3122. doi: 10.1210/endo.131.6.1446647. [DOI] [PubMed] [Google Scholar]
- Klemp P, Staberg B. Cutaneous blood flow in psoriasis. J Invest Dermatol. 1983;81:503–506. doi: 10.1111/1523-1747.ep12522836. [DOI] [PubMed] [Google Scholar]
- Kragballe K, Desjarlais L, Duell EA, Voorhees JJ. In vitro synthesis of 12-hydroxy-eicosatetraenoic acid is increased in uninvolved psoriatic epidermis. J Invest Dermatol. 1986;87:47–52. doi: 10.1111/1523-1747.ep12523561. [DOI] [PubMed] [Google Scholar]
- Krueger GG, Bergstresser PR, Lowe NJ, Voorhees JJ, Weinstein GD. Psoriasis. J Am Acad Dermatol. 1984;11:937–947. doi: 10.1016/s0190-9622(84)80018-3. [DOI] [PubMed] [Google Scholar]
- Kulski JK, Kenworthy W, Bellgard M, Taplin R, Okamoto K, Oka A, et al. Gene expression profiling of Japanese psoriatic skin reveals an increased activity in molecular stress and immune response signals. J Mol Med. 2005;83:964–975. doi: 10.1007/s00109-005-0721-x. [DOI] [PubMed] [Google Scholar]
- Leigh IM, Navsaria H, Purkis PE, McKay IA, Bowden PE, Riddle PN. Keratins (K16 and K17) as markers of keratinocyte hyperproliferation in psoriasis in vivo and in vitro. Br J Dermatol. 1995;133:501–511. doi: 10.1111/j.1365-2133.1995.tb02696.x. [DOI] [PubMed] [Google Scholar]
- Lewis RA, Austen KF. The biologically active leukotrienes. Biosynthesis, metabolism, receptors, functions, and pharmacology. J Clin Invest. 1984;73:889–897. doi: 10.1172/JCI111312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall D, Hardman MJ, Nield KM, Byrne C. Differentially expressed late constituents of the epidermal cornified envelope. Proc Natl Acad Sci U S A. 2001;98:13031–13036. doi: 10.1073/pnas.231489198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair RP, Ruether A, Stuart PE, Jenisch S, Tejasvi T, Hiremagalore R, et al. Polymorphisms of the IL12B and IL23R genes are associated with psoriasis. J Invest Dermatol. 2008;128:1653–1661. doi: 10.1038/sj.jid.5701255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair RP, Stuart PE, Nistor I, Hiremagalore R, Chia NV, Jenisch S, et al. Sequence and haplotype analysis supports HLA-C as the psoriasis susceptibility 1 gene. Am J Hum Genet. 2006;78:827–851. doi: 10.1086/503821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickoloff BJ, Karabin GD, Barker JN, Griffiths CE, Sarma V, Mitra RS, et al. Cellular localization of interleukin-8 and its inducer, tumor necrosis factor-alpha in psoriasis. Am J Pathol. 1991;138:129–140. [PMC free article] [PubMed] [Google Scholar]
- Nickoloff BJ, Wrone-Smith T. Injection of pre-psoriatic skin with CD4+ T cells induces psoriasis. Am J Pathol. 1999;155:145–158. doi: 10.1016/S0002-9440(10)65109-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonomura K, Yamanishi K, Yasuno H, Nara K, Hirose S. Up-regulation of elafin/SKALP gene expression in psoriatic epidermis. J Invest Dermatol. 1994;103:88–91. doi: 10.1111/1523-1747.ep12391802. [DOI] [PubMed] [Google Scholar]
- Oren A, Ganz T, Liu L, Meerloo T. In human epidermis, beta-defensin 2 is packaged in lamellar bodies. Exp Mol Pathol. 2003;74:180–182. doi: 10.1016/s0014-4800(02)00023-0. [DOI] [PubMed] [Google Scholar]
- Pei Z, Jia Z, Watkins PA. The second member of the human and murine bubblegum family is a testis- and brainstem-specific acyl-CoA synthetase. J Biol Chem. 2006;281:6632–6641. doi: 10.1074/jbc.M511558200. [DOI] [PubMed] [Google Scholar]
- Romanowska M, al Yacoub N, Seidel H, Donandt S, Gerken H, Phillip S, et al. PPARdelta enhances keratinocyte proliferation in psoriasis and induces heparin-binding EGF-like growth factor. J Invest Dermatol. 2008;128:110–124. doi: 10.1038/sj.jid.5700943. [DOI] [PubMed] [Google Scholar]
- Sander HM, Morris LF, Phillips CM, Harrison PE, Menter A. The annual cost of psoriasis. J Am Acad Dermatol. 1993;28:422–425. doi: 10.1016/0190-9622(93)70062-x. [DOI] [PubMed] [Google Scholar]
- Schaeffer L, Gohlke H, Muller M, Heid IM, Palmer LJ, Kompauer I, et al. Common genetic variants of the FADS1 FADS2 gene cluster and their reconstructed haplotypes are associated with the fatty acid composition in phospholipids. Hum Mol Genet. 2006;15:1745–1756. doi: 10.1093/hmg/ddl117. [DOI] [PubMed] [Google Scholar]
- Schwartzman ML, Balazy M, Masferrer J, Abraham NG, McGiff JC, Murphy RC. 12(R)- hydroxyicosatetraenoic acid: a cytochrome-P450-dependent arachidonate metabolite that inhibits Na+,K+-ATPase in the cornea. Proc Natl Acad Sci U S A. 1987;84:8125–8129. doi: 10.1073/pnas.84.22.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segre JA, Bauer C, Fuchs E. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat Genet. 1999;22:356–360. doi: 10.1038/11926. [DOI] [PubMed] [Google Scholar]
- Shi Y, Hon M, Evans RM. The peroxisome proliferator-activated receptor delta, an integrator of transcriptional repression and nuclear receptor signaling. Proc Natl Acad Sci U S A. 2002;99:2613–2618. doi: 10.1073/pnas.052707099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simard J, Ricketts ML, Gingras S, Soucy P, Feltus FA, Melner MH. Molecular biology of the 3beta-hydroxysteroid dehydrogenase/delta5-delta4 isomerase gene family. Endocr Rev. 2005;26:525–582. doi: 10.1210/er.2002-0050. [DOI] [PubMed] [Google Scholar]
- Watkins PA. Fatty acid activation. Prog Lipid Res. 1997;36:55–83. doi: 10.1016/s0163-7827(97)00004-0. [DOI] [PubMed] [Google Scholar]
- Westerberg R, Tvrdik P, Unden AB, Mansson JE, Norlen L, Jakobsson A, et al. Role for ELOVL3 and fatty acid chain length in development of hair and skin function. J Biol Chem. 2004;279:5621–5629. doi: 10.1074/jbc.M310529200. [DOI] [PubMed] [Google Scholar]
- Wilkinson DI. Lipid Metabolism in Psoriasis. vol. 277-285. Standford University Press; Stanford: 1971. [Google Scholar]
- Wilson CL, Dean D, Lane EB, Dawber RP, Leigh IM. Keratinocyte differentiation in psoriatic scalp: morphology and expression of epithelial keratins. Br J Dermatol. 1994;131:191–200. doi: 10.1111/j.1365-2133.1994.tb08490.x. [DOI] [PubMed] [Google Scholar]
- Zhao XP, Elder JT. Positional cloning of novel skin-specific genes from the human epidermal differentiation complex. Genomics. 1997;45:250–258. doi: 10.1006/geno.1997.4952. [DOI] [PubMed] [Google Scholar]
- Zhou X, Krueger JG, Kao MC, Lee E, Du F, Menter A, et al. Novel mechanisms of T-cell and dendritic cell activation revealed by profiling of psoriasis on the 63,100-element oligonucleotide array. Physiol Genomics. 2003;13:69–78. doi: 10.1152/physiolgenomics.00157.2002. [DOI] [PubMed] [Google Scholar]
- Ziboh VA, Casebolt TL, Marcelo CL, Voorhees JJ. Biosynthesis of lipoxygenase products by enzyme preparations from normal and psoriatic skin. J Invest Dermatol. 1984;83:426–430. doi: 10.1111/1523-1747.ep12273519. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.