Abstract
The clinical, pathologic, and molecular features of pleomorphic lobular carcinoma in situ (PLCIS) and the relationship of PLCIS to classic LCIS (CLCIS) are poorly defined. In this study, we analyzed 31 cases of PLCIS (13 apocrine and 18 non-apocrine subtypes) and compared the clinical, pathologic, immunophenotypic and genetic characteristics of these cases with those of 24 cases of CLCIS. Biomarker expression was examined using immunostaining form E-cadherin, GCDFP-15, ER, PR, AR, HER2, CK5/6, and Ki67. Array-based comparative genomic hybridization (aCGH) to assess genomic alterations was performed usingm microdissected formalin-fixed paraffin-embedded samples.
Patients with PLCIS presented with mammographic abnormalities. Histologically, the tumor cells were dyshesive and showed pleomorphic nuclei, and there was often associated necrosis and microcalcifications. All lesions were E-cadherin negative. Compared to CLCIS, PLCIS showed significantly higher Ki67 index, lower ER and PR expression, and higher incidence of HER2 gene amplification. The majority of PLCIS and CLCIS demonstrated loss of 16q and gain of 1q. Apocrine PLCIS had significantly more genomic alterations than CLCIS and non-apocrine PLCIS.
Although lack of E-cadherin expression and the 16q loss and 1q gain-aCGH pattern support a relationship to CLCIS, PLCIS has clinical, mammographic, histologic, immunophenotypic and genetic features that distinguish it from CLCIS. The histologic features, biomarker profile, and genomic instability observed in PLCIS suggest a more aggressive phenotype than CLCIS. However, clinical follow-up studies will be required to define the natural history and most appropriate management of these lesions.
Keywords: Pleomorphic lobular carcinoma in situ, lobular carcinoma in situ, lobular neoplasia, array-based comparative genomic hybridization, biomarkers
INTRODUCTION
Mammary carcinoma in situ is classified as ductal carcinoma in situ (DCIS) or lobular carcinoma in situ (LCIS) based on a combination of architectural and cytologic features. LCIS is typically an incidental microscopic finding and is rarely identified clinically, by mammographic screening, or by gross pathologic examination. It is often multicentric in the breast and frequently bilateral. Traditionally, this lesion has been regarded as a marker of increased risk for invasive breast cancer development (15, 21). However, a recent study comparing chromosomal alterations in LCIS and synchronous invasive lobular carcinoma has demonstrated a clonal relationship between most of the paired lesions, suggesting a precursor-product relation between LCIS and invasive lobular carcinoma (13). Histologically, LCIS is most often composed of a solid proliferation of monotonous, small, dyshesive cells that have a low proliferative rate, are ER- and PR-positive, and rarely, if ever, show amplification of the HER2 gene or HER2 protein overexpression (18). LCIS lesions are further characterized by loss of expression of the adhesion molecule E-cadherin, most commonly due to mutation or deletion of CDH1 locus on chromosome 16q (26).
The widespread use of screening mammography has resulted in a dramatic increase of newly diagnosed mammary carcinomas in situ (3) including carcinoma in situ lesions that exhibit histologic features that deviate from those of classical DCIS and LCIS, thereby resulting in problems in their categorization. One such variant has been designated pleomorphic lobular carcinoma in situ (PLCIS), a lesion in which the histologic features overlap between classic LCIS and DCIS. PLCIS was originally described and is commonly seen in association with invasive pleomorphic lobular carcinoma (7, 17). However, it may also present as an isolated lesion without concurrent invasive disease (i.e. pure PLCIS) (4, 24). Prior to its recognition, this lesion was probably most often classified as high grade DCIS due to the presence of nuclear pleomorphism and the frequent presence of comedo-type necrosis. With the availability of E-cadherin immunohistochemistry, carcinomas in situ showing features of both classic LCIS (loss of cell cohesion and loss of E-cadherin expression) and DCIS (high nuclear grade and/or the presence of necrosis and microcalcifications) have been diagnosed more frequently.
PLCIS lesions pose challenges to breast pathologists, surgeons and oncologists. The most appropriate classification, risk of subsequent invasive carcinoma, natural history and clinical management of this lesion are unknown. In particular, it is unclear whether treatment recommendations for patients with PLCIS should follow those appropriate for patients with DCIS or those for patients with classical forms of LCIS. Studies addressing the clinicopathologic features, biomarkers and genetic alterations of PLCIS have to date been limited.
In this study, we characterized the clinical, mammographic, pathologic, immunophenotypic and genetic characteristics in pure PLCIS and examined whether a molecular relationship exists between PLCIS and CLCIS. The results of this study may provide insights about the biologic potential of PLCIS and therefore may have implications for the clinical management of patients with these lesions.
MATERIALS AND METHODS
Case selection
Thirty-one cases of PLCIS without concurrent or known prior ipsilateral invasive breast carcinoma were identified during review of carcinoma in situ lesions from the Department of Pathology at St. Jude Medical Center, CA; Virginia Mason Medical Center, WA; Institut Bergonie, Bordeaux, France; Netherlands Cancer Institute, Amsterdam, Netherlands; Institut Curie, Paris, France; Beth Israel Deaconess Medical Center, MA; and UCSF Medical Center, CA. For comparison, 24 cases of CLCIS without concurrent or prior invasive cancer were identified from the pathology files of the Department of Pathology at UCSF Medical Center. Pathology slides and clinical data were reviewed. Clinical data extracted from the pathology reports included age, mammographic findings and clinical presentation. Formalin-fixed, paraffin-embedded tissue blocks or unstained slides were obtained for immunohistochemistry and aCGH analysis. Twenty-one cases of PLCIS and twenty cases of CLCIS had sufficient material for both aCGH and immunohistochemistry. Of note, three of the PLCIS cases had been fixed in Hollande solution. While this did not have significant adverse effect on immunohistochemistry, it precluded DNA extraction and aCGH analysis. Comparative analysis of aCGH data was performed between PLCIS and CLCIS. This study was approved by the UCSF institutional review board and the institutional review boards of the contributing hospitals.
Histopathology
Hematoxylin and eosin (H&E)-stained sections from PLCIS were reviewed by the two of the study pathologists (YC and SS) to confirm the diagnosis. All of the lesions demonstrated characteristic features of CLCIS including distension of involved spaces by a solid proliferation of dyshesive cells with frequent intracytoplasmic vacuoles and lack of E-cadherin expression. To qualify as PLCIS, it was required that the lesions also showed moderate to marked nuclear pleomorphism and that at least some of the tumor nuclei were ≥ 4 times the size of a lymphocyte, as described by Sneige et al (24). PLCIS lesions were further categorized as either apocrine or non-apocrine types, the former characterized by cells with abundant eosinophilic cytoplasm.
The following histologic features were also evaluated in each case: presence or absence of necrosis, type of necrosis (comedo vs punctate), presence or absence of calcifications, and presence or absence of CLCIS within the same or adjacent terminal duct lobular units (TDLUs). The CLCIS was further categorized as type A or type B according to established criteria (10).
Biomarker Assessment
Immunohistochemistry was performed on tissue sections using the streptavidin-biotin peroxidase method. Antibodies and dilutions used were: ER (clone 1D5, DAKO, Carpinteria, CA/USA) at 1:400 dilution; PR (clone 1A6, Novocastra, Newcastle/UK) at 1:25; HER2 (clone TAB250, Zymed, South San Francisco, CA/USA) at 1:200; Ki-67 (DAKO) at 1:100; CK5/6 (Chemicon, Temecula, CA/USA) at 1:100; androgen receptor (AR) (Biogenex, San Ramon, CA/USA) at 1:80; E-cadherin (Zymed) at 1:1000; and GCDFP-15 (Signet, Dedham, MA/USA) at 1:20.
Antigen retrieval was either by heat-induced epitope retrieval in 10 mM citrate buffer at pH 6.0 (for ER, PR, AR, CK5/6, E-cadherin, GCDFP-15), incubation with Ficin (Zymed) at 37°C (for HER2), or 0.01% trypsin digestion followed by heat treatment in 10 mM citrate buffer (for Ki-67). Slides were blocked in 3% H2O2 and then incubated with the primary antibodies. Binding of primary antibodies was detected by incubation with a biotinylated secondary antibody followed by streptavidin-horseradish peroxidase and 3,3 diaminobenzidine tetra hydrochloride as the chromagen. Finally, the sections were counterstained with hematoxylin.
Scoring of Immunohistochemistry
All immunostains were evaluated by one of the study pathologists (YC).
ER, PR and AR
were scored semi-quantitatively by evaluating the percentage and intensity of stained tumor nuclei using the H-score (23). The staining intensity ranged from 0 to 3+, with 0 representing no staining, 1+ weak staining, 2+ moderate staining, and 3+ strong staining. The percentages of positive tumor cells in each staining intensity category were recorded. The results of ER and PR were expressed as the H-score where: H-score= (1 × %1+) + (2 × %2+) + (3 × %3+). An H-score of > 0 was interpreted as hormone receptor positive.
HER2
was scored by the original HercepTest (DAKO) scoring criteria, using a 0–3+ scale. Tumor cells with staining intensity 0 and 1+ were considered negative for HER2 protein overexpression; those with intensity 2+ were regarded equivocal; and those with staining intensity 3+ were considered positive. Fluorescence in situ hybridization (FISH) analysis (Vysis, Des Plaines, IL/USA) was performed on all cases showing a 2+ or 3+ score by immunohistochemisty. For FISH analysis, cases showing a ratio of HER2:centromere 17 copy number greater than 2.2 were considered positive for HER2 gene amplification. None of the cases with 2+ score on immunohistochemistry in this study showed gene amplification by FISH. Therefore, HER2 positive cases in this study showed both 3+ staining and HER2 gene amplification. Of note, the HER2 status defined above demonstrated complete concordance with profile of chromosome 17 on aCGH.
Ki-67
staining was used to establish a proliferation index. Slides were first scanned at low-power magnification to select CIS foci with highest mitotic activity. 1000 tumor cells were counted in these mitotically active areas. The proliferation index was obtained by the percentage of tumor nuclei that were labeled by Ki-67.
CK5/6
was scored for the intensity (ranging from 0 to 3+) and percentage of the tumor cells with cytoplasmic staining. Lesions showing at least 10% of the tumor cells with moderate or strong staining intensity (2 or 3+) were considered CK5/6 positive.
GCDFP-15
was evaluated for the percentage of tumor cells with cytoplasmic staining. Since positive tumor cells typically demonstrated strong cytoplasmic staining, the intensity was not further graded. Lesions with at least 10% of reactive cells were considered positive for GCDFP-15.
Array CGH
H&E-stained sections from PLCIS and CLCIS were reviewed by one of the study pathologists (YC) and areas of tumor cells were outlined for microdissection.
Microdissection and DNA Extraction
Microdissection and DNA extraction were performed as described previously (13). LCIS lesion areas were estimated in mm2 to calculate the number of unstained slides required for microdissection in order to yield enough DNA. DNA from each extraction was quantitated by TaqMan real-time PCR using a CA repeat probe (9).
Amplification
Prior to labeling, the DNA samples were amplified by random prime extension protocols using components from the BioPrime DNA Labeling System (Invitrogen, Carlsbad, CA). The reaction consisted of 50–100 ng of tumor DNA, or 50 ng of reference genomic DNA (Promega, San Luis Obispo, CA, USA).
Labeling and Hybridization
Human array versions 2.0, 3.1 and 3.2 chromium surface arrays were prepared by the UCSF Cancer Center Array Core. Each array consisted of 2,464 BAC and P1 clones printed in triplicate providing representation of the entire genome at a 1.5 Mb resolution (25).
The amplified DNA was random prime labeled using the BioPrime Kit (Invitrogen, Carlsbad, CA/USA) (protocol at http://cc.ucsf.edu/people/waldman/Protocols). Tumor DNA was labeled with Cy3 conjugated dCTP and normal genomic DNA was labeled with Cy5 conjugated dCTP (GE, Piscataway, NJ/USA), hybridized for 48 hours and washed as previously described (9).
Image Capture and Processing
Arrays were imaged using a CCD camera as described previously (2). Intensity data were acquired through DAPI, cy-3, and cy-5 channels. The SPOT 2.0 software program was used to process the image data (program available at http://cc.ucsf.edu/jain/public). The human DNA sequence draft at http://genome.ucsc.edu (May 2004 freeze) was used to map clones. The log2 ratios for each case were median centered to zero. Thresholds for determining chromosome gain or loss were defined separately for each individual sample using a discrete time hidden Markov model (6) as implemented in the aCGH package of BioConductor open source software (http://www.bioconductor.org), correcting for varying signal-to-noise in each sample. The frequency of gains and losses for a given clone in a group of interest was calculated as the proportion of samples in which a clone was gained or lost in that group. As our CGH arrays do not cover the entire genome, each clone was assigned a genomic distance equal to the sum of half of the distance between its center and that of its two adjacent clones to quantitate the fraction of the genome altered.
Statistical Analysis
The significance of genetic changes among different groups is determined by Student’s t test, ANOVA regression analysis and a MaxT test using permutation analysis to control for family-wise false positive error rates. Briefly, for the MaxT test, a t-statistic is computed to determine the significance of copy number changes between two groups. The group labels are then randomly permutated 1000 times and the maximum t-statistic observed between the groups is recorded. The MaxT adjusted p-value is then calculated by considering the proportion of permutations in which the maximum permutation based t-statistic exceeded the observed statistic for each clone.
For biomarker analysis, Wilcoxon Mann Whitney test and Fisher’s Exact test were used to compare the expression values between the different groups of LCIS. Wilcoxon Mann Whitney test was employed to compare the age distribution among LCIS subtypes. The results were considered statistically significant if the P value was < 0.05.
RESULTS
Clinical and Mammographic Features
All of the PLCIS patients in this study were female with a mean age 55 years (range 40–86 years). Compared to CLCIS (mean age of 50), patients with PLCIS were significantly older (p= 0.03) and this difference was mostly attributed to the apocrine PLCIS. Apocrine PLCIS were seen primarily in postmenopausal women with a mean age of 60 years, whereas the mean age for non-apocrine PLCIS was 51 (p= 0.008).
The majority of PLCIS patients (n=27) presented with abnormal mammographic findings and 3 presented with a palpable mass. In one patient information regarding the mode of presentation was not available. For the 27 cases who presented with abnormal mammograms, the findings included microcalcifications (21 cases), microcalcifications with nodule/architectural distortion/density (4 cases), and architectural distortion/density (2 cases).
Pathologic Features
PLCIS lesions demonstrated overlapping histologic features between CLCIS and DCIS (Figure 1). All 31 cases shared some features of CLCIS such as a solid proliferation of dyshesive cells that filled and distended TDLUs. Pagetoid involvement of ducts was noted in the majority of the cases. In addition, in 27 cases (87%) there were synchronous foci of CLCIS. The CLCIS cells were usually a mixture of type A and type B cytology and they were found both in close proximity to the PLCIS (sometimes in the same TDLU) and/or breast tissue away from the PLCIS. However, PLCIS lesions also demonstrated some features considered to be more typical of DCIS including considerable nuclear enlargement (≥ 4x of lymphocyte) and moderate to marked nuclear pleomorphism (>2–3x variation in nuclear size). Further, necrosis was identified in 22 lesions (71%) (comedo-type in 18 and punctate in 4), and in 29 cases (94%) calcifications were present in association with the lesion.
Figure 1.
Pathologic features of pleomorphic lobular carcinoma in situ (PLCIS). A. Low power photomicrograph showing massive expansion of ducts and lobules by neoplastic cells, with associated comedo necrosis and calcifications (Original magnification 40×); B. High power view of PLCIS with apocrine cytology showing dyshesive cells with nuclear pleomorphism and abundant eosinophilic cytoplasm (400×); C. High power view of PLCIS with non-apocrine cytology (400×).
Of note, these PLCIS cases did not represent a homogeneous group, displaying some variation in cytologic features. In particular, 13 cases stood out as a distinct subtype characterized by large cells with abundant eosinophilic cytoplasm imparting an apocrine appearance (a variant we termed apocrine PLCIS). These lesions typically exhibited foci of comedo necrosis. The combination of large cells with abundant cytoplasm, nuclear pleomorphism, and comedo necrosis produced a histologic appearance that could readily be mistaken for DCIS (Figure 1B). However, the cells in these cases were dyshesive and had frequent intracytoplasmic vacuoles that in some instances were large enough to produce signet ring cell forms.
Biomarker Expression
Biomarker expression was examined in the different groups of LCIS lesions using immunostains for E-cadherin, ER, PR, AR, HER2, Ki-67, CK5/6 and GCDFP-15 and FISH analysis for HER2 gene amplification. The results are summarized in Table 1 and in Supplemental Digital Content 1 (see Table, which shows clinical-pathologic features of lobular carcinoma in situ, http://links.lww.com/PAS/A31) and representative images are illustrated in Figure 2.
TABLE 1.
Summary and Comparison a of Biomarkers among Lobular Carcinoma In Situ (LCIS) Subtypes
| ER b | PR b | AR b | mean Ki-67 | HER2 e | GCDFP-15 e | CK5/6 e | |
|---|---|---|---|---|---|---|---|
| CLCIS (N=21) | 248 (100%) |
228 (100%) |
246 (100%) |
4.2% | 0% | Not done | 19% |
| All PLCIS (N=31) |
109 c (66%) |
94 c (62%) |
199 (100%) |
11.5% c | 13% | 74% | 23% |
| Non apocrine PLCIS (N=18) |
160c (100%) |
129c (94%) |
168 c (100%) |
9.9% c | 0% | 50% | 28% |
| Apocrine PLCIS (N=13) |
47 c,d (23%) |
45 c,d (17%) |
246 d (100%) |
13.9% c | 31% c,d | 100% d | 17% |
CLCIS, classic LCIS; PLCIS, pleomorphic PLCIS.
Wilcoxon Mann Whitney test (t-test) for ER, PR, AR and Ki-67; Fisher’s exact test (chi-square test) for HER2, GCDFP-15 and CK5/6
Expressed as mean H score (% of cases positive)
p <0.05 for comparison with CLCIS
p <0.05 apocrine vs non-apocrine PLCIS
% of cases positive
Figure 2.
Immunophenotype of pleomorphic lobular carcinoma in situ. (Original magnification 400X). A. Absence of staining for E-cadherin (note the positive membranous staining in the adjacent normal ducts); B. Strong cytoplasmic staining demonstrated for GCDFP-15; C. Strong expression of androgen receptor; D. Lack of ER expression (note focal nuclear staining in adjacent normal duct); E. HER2 protein overexpression. F. High Ki-67 labeling index.
All PLCIS and CLCIS cases were E-cadherin negative. Overall 74% of PLCIS were GCDFP-15 positive. GCDFP-15 expression was significantly more frequent in apocrine than non-apocrine types (100% vs 50%, p<0.05). While ER and PR expression was noted in all CLCIS and virtually all non-apocrine PLCIS, only a small fraction of the apocrine PLCIS (~20%) showed ER and PR expression. When compared to CLCIS, PLCIS showed significantly lower levels of ER and PR expression. Even among the non-apocrine PLCIS, the ER and PR expression level as measured by H-score was significantly lower than that in CLCIS. Although AR expression was identified in all LCIS lesions, non-apocrine PLCIS had significantly lower AR levels than both CLCIS and apocrine PLCIS. The average Ki-67 proliferation index was significantly higher in PLCIS than in CLCIS (11.5% vs 4.2%, p<0.05). Furthermore, apocrine PLCIS showed a higher proliferation index than the non apocrine type, but this difference did not reach statistical significance (p=0.06). Overexpression of HER2 was identified in 13% of PLCIS and was not observed in any of the CLCIS cases. Of note, HER2 overexpression was restricted to the apocrine subtype of PLCIS in this cohort. When compared to CLCIS and non- apocrine PLCIS, the differences in the frequency of HER2 overexpression in apocrine PLCIS were statistically significant. There was no difference in the frequency of expression of basal cytokeratin CK5/6 among LCIS subtypes.
aCGH analysis of PLCIS and CLCIS
Array CGH was used to define the genomic copy number profile of 21 PLCIS (8 apocrine and 13 non apocrine subtypes) and 20 CLCIS. A representative aCGH profile from an apocrine PLCIS is shown in Figure 3. The chromosomal alterations of all LCIS in this study are summarized in Table 2 and Supplemental Digital Content 2 (see Table, Supplemental Digital Content 2, http://links.lww.com/PAS/A32) with the clone frequencies depicted in Figure 4. Overall, PLCIS and CLCIS shared similar genomic alterations: both groups were characterized by 1q gain (75% in PLCIS vs 69% in CLCIS) and 16q loss (85% vs 76%, for PLCIS and CLCIS respectively) (Figure 4). Additional changes observed for both groups included loss of 17p (23% vs 10%, for PLCIS and CLCIS respectively). However, some genomic alterations were either only noted in the PLCIS but not the CLCIS such as amplification of HER2 gene at17q11.2–17q12 (10% vs 0%), gain of 16p (14% vs 0%), loss of 8p (5% vs 0%) or more prevalent in the PLCIS such as amplification of cyclin D1 gene at 11q13.3 (14% vs 5%). Of note, amplification of 17q and gain of 16p were only noted in the apocrine but not in the non-apocrine subtype of PLCIS. In addition to 1q gain and 16q loss, recurrent copy number changes for apocrine PLCIS included gains of 16p (36%) and 6p (15%), losses of 3q (22%), 11q (32%), 13q (25%) and 17p (45%), and amplification of cyclin D1 gene (3/8, 38%) and HER2 gene (2/8, 25%); for non apocrine PLCIS, no additional recurrent change was observed.
Figure 3.
The genomic profile of an apocrine subtype of pleomorphic lobular carcinoma in situ by high-resolution array-based comparative genomic hybridization. In this example, the lesion shows gains of 1q and 16p, losses of 11q, 13q, 16q, and 18, and amplification of 11q13.3 (the region containing cyclin D1 gene).
TABLE 2.
Summary of Genomic Changes in Lobular Carcinoma In Situ (LCIS) Subtypes
| CLCIS | PLCIS | Non-Apocrine PLCIS |
Apocrine PLCIS |
|
|---|---|---|---|---|
| Fraction Genome Altered | 0.072 | 0.079 | 0.054♦ | 0.119* |
| Fraction Genome Gained | 0.041 | 0.040 | 0.030♦ | 0.056 |
| Fraction Genome Lost | 0.031 | 0.039 | 0.024♦ | 0.063# |
| Break Points | 6.95 | 7.905 | 5.154♦ | 12.38* |
| Chromosomes with Break Points | 3.8 | 4.19 | 3.154♦ | 5.875* |
| Number of Amplifications | 0.250 | 1.952 | 0.077 | 5.000# |
| Chromosomes with Amplifications | 0.050 | 0.286 | 0.077 | 0.625# |
| Whole Chromosome Changes | 0.150 | 0.191 | 0.000♦ | 0.500# |
CLCIS, classic LCIS ; PLCIS, pleomorphic LCIS.
All the numbers represent “mean value” of the genomic changes.
p< 0.05 apocrine PLCIS vs non apocrine PLCIS
p< 0.06 apocrine PLCIS vs CLCIS
p< 0.05 apocrine PLCIS vs CLCIS
Figure 4.
Frequency plots of genomic copy number gains and losses in pleomorphic lobular carcinoma in situ (PLCIS) and classic LICS (CLCIS). The proportion of LCIS lesions in which each BAC clone is gained (green) or lost (red) is plotted on the y axis according to its genomic location on the×axis. Vertical dotted lines represent chromosome centromeres. A: CLCIS (N=20); B: PLCIS (N=21); C: Non-apocrine PLCIS (N=13); D: Apocrine PLCIS (N=8).
The extent of genomic alterations was compared between various groups of LCIS (Table 2). As a group, PLCIS showed no significant differences in the fraction genome altered (FGA), fraction genome gained (FGG), fraction genome loss (FGL), number of whole chromosome changes, number of amplifications, or number of break points than the CLCIS group. Likewise, the overall extent of genomic alterations was similar between CLCIS and non-apocrine PLCIS. However, more genomic changes were observed in the apocrine PLCIS. Specifically, apocrine PLCIS had significantly higher FGL (p=0.04), more whole chromosomes changes (p=0.04), more amplifications (p=0.02) and more chromosomes with amplifications (p=0.02) than CLCIS. A trend of increased FGA (p=0.06), increased number of break points (p=0.05), and increased chromosomes with break points (p=0.06) was also noted when apocrine PLCIS was compared to CLCIS. Within PLCIS, apocrine PLCIS also demonstrated more genome alterations than non apocrine type. Apocrine PCLIS had significantly more chromosomal breakpoints (p=0.02), more chromosomes with break points (p=0.03), more whole chromosomes changes (p=0.02), higher FGG (p=0.02), higher FGL (p=0.02) and higher FGA (p=0.01). Of note, amplifications of 17q and 11q and gain of 16p were only noted in the apocrine subtype of PLCIS.
When all cases were clustered in an unsupervised manner, four distinct clusters emerged (Figure 5 and Supplemental Digital Content 3 (see Figure, Supplemental Digital Content 3, which shows the FGA of LCIS Heat Map Cluster, http://links.lww.com/PAS/A33). Clusters 1 and 4 were enriched with CLCIS, cluster 2 with non-apocrine PLCIS, and cluster 3 with apocrine PLCIS. Cases in clusters 1 and 2 were similar and characterized by multiple genomic alterations, with a median fraction genome altered (FGA) 6.2% in cluster 1 and 6.8% in cluster 2. Cluster 4 had overall the least genomic changes with a median FGA 3.1%. Cluster 3 was the amplification cluster and demonstrated genome amplifications involving 17q11.2–17q12 (HER2 gene) and 11q (cyclin D1 gene). This cluster also showed the highest genomic instability with a median FGA at 17.4%. Furthermore, it was enriched in apocrine PLCIS including 50% (4 of the 8 cases) of the analyzed apocrine PLCIS.
Figure 5.
Unsupervised hierarchical clustering of genome copy number profiles measured for 21 pleomorphic lobular carcinoma in situ (PLCIS) and 20 classic LICS (CLCIS). Green indicates increased genome copy number, red indicates decreased copy number, and yellow indicates amplified copy number. The bar to the left indicates chromosome location with chromosome 1pter at the top and 22qter and X at the bottom. The top color bars indicate types of LCIS. In the upper bar, apocrine PLCIS is blue and non apocrine PLCIS is green. In the lower bar, CLCIS is green, and PLCIS is blue. Four clusters emerge: Clusters 1 and 4 are enriched with CLCIS, while cluster 2 with nonapocrine PLCIS and cluster 3 with apocrine PLCIS. Note that cluster 3 is characterized by genome amplification and shows the highest genome instability.
Biomarker expression and aCGH profile in synchronous PLCIS and CLCIS
Although synchronous CLCIS was identified in most of the PLCIS cases, the CLCIS lesions were usually small and scattered and therefore not amenable to aCGH analysis and detailed semiquantitative biomarker measurement. However, evaluation of the biomarker expression in these synchronous CLCIS with comparison to adjacent PLCIS was performed when possible. Of interest, we noted that in 7 of the 10 cases with apocrine PLCIS and negative ER, the synchronous CLCIS were positive for ER, different from the adjacent PLCIS. Furthermore, two of the cases showed distinct HER2 patterns with HER2 overexpression in the apocrine PLCIS and no HER2 overexpression in the adjacent CLCIS. Most of the PLCIS cases had insufficient DNA to perform aCGH analysis on their synchronous CLCIS lesions. However, one case with synchronous CLCIS and apocrine PLCIS was successfully analyzed with the CLCIS showing only 1q gain and 16q loss; the apocrine PLCIS showed additional molecular changes in addition to the 1q gain and 16q loss (results not shown).
DISCUSSION
To our knowledge, this is the largest reported series characterizing pure PLCIS. We found that similar to CLCIS, PLCIS was characterized by 16q loss and 1q gain as well as abrogation of E-cadherin expression, suggesting a relationship between PLCIS and CLCIS. However, PLCIS showed different clinical findings, mammographic presentation and morphologic features than CLCIS. The biomarker expression of PLCIS was also distinct with an overall profile (i.e. decreased ER and PR expression and increased proliferation by Ki-67 labeling index) that is suggestive of a more aggressive phenotype. The distinct apocrine morphologic subtype of PLCIS had especially aggressive phenotypic features.
One of the most striking differences between PLCIS and CLCIS was the clinical presentation. Patients with PLCIS were significantly older than those with CLCIS. If PLCIS and CLCIS were indeed clonally related, it might be speculated that PLCIS requires a longer time for development. Unlike CLCIS which is usually an incidental microscopic finding without a detectable mammographic lesion, PLCIS typically presented as mammographic abnormalities including microcalcifications, architectural distortion and/or density. The clinical presentation and mammographic findings of PLCIS are similar to DCIS and different from CLCIS, as has been described previously (4, 8, 24).
With regard to immunophenotype, we found both similarities and differences between PLCIS and CLCIS. All PLCIS were negative for E-cadherin expression, as is characteristic of CLCIS. However, unlike CLCIS lesions which are consistently positive for ER and PR, negative for HER2, and have low mitotic activity, PLCIS often showed low to negative ER and PR expression, a higher proliferation rate, and an increased incidence of HER2 overexpression. These findings are similar to those reported previously in PLCIS (1, 4, 17, 24). This “unfavorable” biomarker profile suggests that PLCIS represents a biologically more advanced lesion than CLCIS. All PLCIS, especially those of apocrine morphology, were strongly positive for AR. Activation of the AR pathway may be important in the progression of these lesions, and drugs targeting this pathway may constitute a novel therapeutic option for management of PLCIS.
To explore whether the differences in biomarker expression could be attributed to differences in genetic alterations, chromosomal changes were analyzed and compared between PLCIS and CLCIS by high resolution aCGH. The majority of PLCIS in our study demonstrated 1q gain and 16q loss, a molecular signature for lobular carcinomas including CLCIS (13, 16). Based on the more aggressive morphologic features and biomarker profile, a greater degree of genomic instability might be expected in PLCIS. Surprisingly, there was no significant difference in the extent of genomic alterations between the CLCIS and PLCIS groups. One possible explanation for this apparent lack of a difference in the extent of genomic alterations between these two groups may be that in our effort to obtain sufficient DNA for aCGH analysis, the analysis was biased toward cases with relatively extensive CLCIS, which may in turn result in a bias toward more advanced CLCIS lesions (5, 19). Another possibility is that the number of cases studied is insufficient to detect a significant difference. Alternatively, epigenetic alterations such as promoter methylation and/or histone deacetylation (14) rather than genetic changes themselves may be the main mechanisms for alteration in the biomarker expression, and such changes would not be detected by aCGH analysis.
The apocrine subtype of PLCIS did demonstrate more genomic alterations than either CLCIS or non-apocrine PLCIS in our study. Although there was no specific genomic pattern that distinguished apocrine PLCIS from non-apocrine PLCIS or CLCIS, some molecular changes were either only present or more prevalent in apocrine PLCIS, including amplification of 17q11.2–17q12 (the region harboring HER2 gene), amplification of 11q13.3 (the region containing cyclin D1 gene), gain of 16p, and losses of 3q, 11q, 13q and 17p. Of note, many of these recurrent genetic alterations, including amplification of 11q13.3, gain of 16 p and losses of 11q, 13q and 17p, are shared by invasive classic lobular carcinoma and invasive pleomorphic lobular carcinoma as reported previously by our group (13) and Simpson et al. (22). Several important genes are mapped to these chromosomal regions and include p53 gene (17p13.1), MEN1 gene (multiple endocrine neoplasia 1, 11q13), ATM gene (ataxia-telangiectasia mutated, 11q22.3) and CCNF gene (cyclin F, 16p13.3). Furthermore, amplification of HER2 gene and loss of 13q (which harbors RB gene at 13q14.1–14.2) have also been noted in invasive pleomorphic lobular carcinoma (22) but not in invasive classic lobular carcinoma (13). Deregulation of these genes could contribute to the aggressive morphologic features and biomarker profile in apocrine PLCIS.
Although four clusters emerged in the unsupervised clustering of all samples (Figure 5), the various morphologic subtypes of LCIS did not segregate perfectly by aCGH clusters. This finding suggests that there is significant overlap in the pattern of genomic alteration among various LCIS types as defined by morphology and that there does not appear to be a particular genetic signature to define these LCIS subtypes, except the 1q gain and 16q loss for lobular neoplasia as a group. Of note, more apocrine PLCIS appeared in the cluster characterized by amplifications: 4 of 6 cases in this cluster being apocrine PLCIS, or 4 of the 8 apocrine PLCIS analyzed segregated to this cluster. Furthermore, none of the apocrine PLCIS fell into the genetically more stable cluster 4 (with lowest FGA). Data from our aCGH analysis thus suggest that although PLCIS are related to LCIS, they are a genetically heterogeneous group of lesions. The aCGH profiling also suggests that apocrine PLCIS may represent a genetically more distinct group, characterized by more extensive genome alteration, especially amplification.
We previously reported, using a similar aCGH platform and data analysis, the genomic profiles of 24 CLCIS with synchronous invasive lobular carcinoma (13), 20 high-grade DCIS without invasion (12), 35 invasive lobular carcinoma and 164 invasive ductal carcinoma (11). Therefore the degree of genomic instability, as represented by FGA, in different tumor types could be compared. The mean FGA was 0.05 for non-apocrine PLCIS without invasion, 0.07 for CLCIS without invasion, 0.12 for apocrine PLCIS without invasion, 0.13 for CLCIS with invasion, 0.31 for high-grade DCIS without invasion, 0.20 for invasive lobular carcinoma and 0.32 for invasive ductal carcinoma. These comparisons indicate that pure CLCIS and non-apocrine PLCIS have the fewest genetic changes, significantly fewer than CLCIS with invasion. Pure apocrine PLCIS and CLCIS with invasion share a similar degree of genomic complexity. However, both apocrine PLCIS and CLCIS with invasion demonstrate less genomic alteration than invasive lobular carcinoma. Furthermore, apocrine PLCIS displays far less genetic instability than high-grade DCIS. These data support the current notion that CLCIS alone is more of a risk factor than immediate precursor for invasive carcinoma as these lesions harbor fewer genetic changes and thus will require longer time for progression to invasion. However, apocrine PLCIS appears to be a genetically more advanced lesion, similar to CLCIS that has evolved to invasive carcinoma. These data also clearly show that although PLCIS demonstrates some features similar to high-grade DCIS, these lesions have less genomic alteration than high-grade DCIS. Overall, the aCGH profiling changes in PLCIS qualitatively resemble lobular carcinoma more closely than high-grade DCIS.
Apocrine PLCIS appears to be a biologically distinct subtype. The tumor cells show prominent apocrine cytology and strong expression of GCDFP-15, a known marker for apocrine differentiaiton. The lesions are seen predominantly in postmenopausal women with a mean age that is about a decade older than that of patients with both CLCIS and non-apocrine PLCIS. Apocrine PLCIS lesions are mostly (~80%) negative for ER and PR, demonstrate HER2 gene amplification in about one third of cases, and exhibit more genomic alterations. In contrast, the non-apocrine PLCIS are positive for ER and PR and lack HER2 gene amplification. In previous studies on PLCIS including pure PLCIS and those with invasion, all the pure PLCIS were reported to be positive for ER and none showed HER2 gene amplification (4, 24). Those cases appear to be similar to the non-apocrine PLCIS in our study. On the other hand, a recent expression profiling study demonstrated the “molecular apocrine type” in 3 of 4 pleomorphic invasive lobular carcinomas (27), a phenotype that is likely similar to apocrine PLCIS in our study.
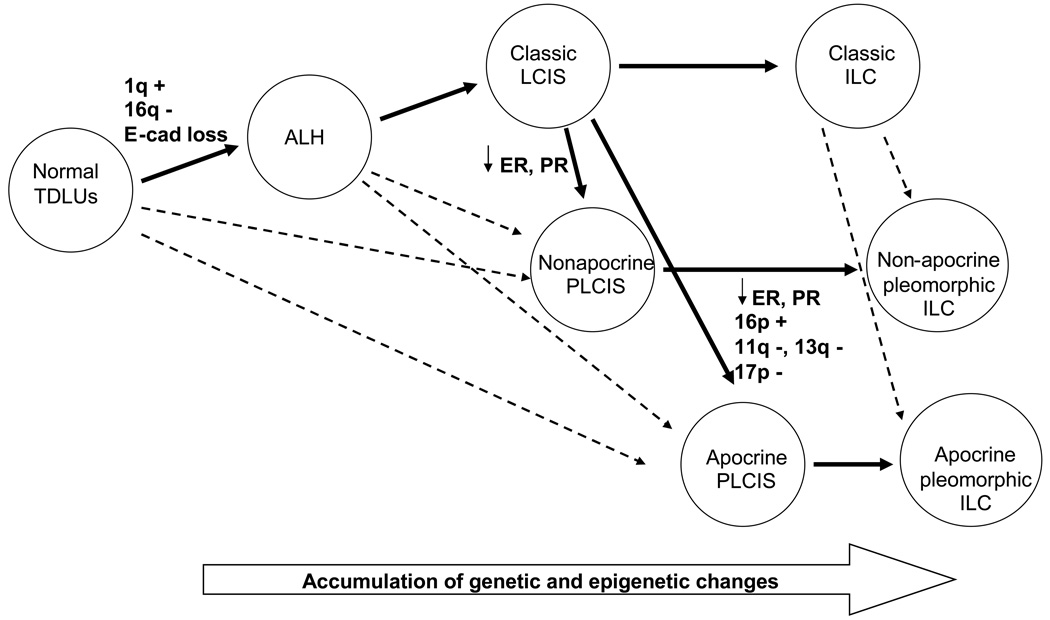
The genetic signature of 1q gain and 16q loss combined with the lack of E-cadherin expression observed in PLCIS supports the lobular lineage for PLCIS along the multi-step cancer progression pathway. It is unclear whether PLCIS arises from CLCIS and represents a genetically more advanced lesion than CLCIS, or if PLCIS originates from a yet unidentified precursor lesion. Cases with synchronous CLCIS and PLCIS will provide a special opportunity to explore the origin for PLCIS. In one informative case with synchronous CLCIS and apocrine PLCIS in our series, both lesions shared 1q gain and 16 q loss with additional chromosomal changes present in the apocrine PLCIS. Our findings are in agreement with a recent report by Reis-Filho et al. (20) and suggest that at least some PLCIS may evolve from the same precursor or through the same genetic pathway as CLCIS. Based on the results of the current study and previous work, we propose a hypothetical pathway for PLCIS along the multi-step model of breast cancer progression (Figure 6). Future studies with more cases to compare the genetic alteration in synchronous CLCIS and PLCIS will help to define more clearly the genetic relationship between PLCIS and CLCIS.
Figure 6.
Hypothesized pathway for lobular neoplasia along a multistep model of breast cancer progression. Solid arrows indicate the predominant pathway, while dashed arrows indicate a potential pathway. TDLUs, terminal duct lobular units; +, gain; −, loss; E-cad, E-cadherin; ALH, atypical lobular hyperplasia; LCIS, lobular carcinoma in situ; ILC, invasive lobular carcinoma; ↓, decreased expression; PLCIS, pleomorphic LCIS.
The natural history of PLCIS is unknown, and thus the optimal management of these lesions is presently unclear. Currently, the bias of most experts is that PLCIS should probably be treated more like DCIS than like CLCIS. However, there are admittedly no clinical follow-up data to support this view which is based on observational data such as those presented here.
In conclusion, PLCIS lacks E-cadherin expression and demonstrates the characteristic 1q gain and 16q loss aCGH pattern supporting a relationship to CLCIS at the molecular level. However, the histologic features, often unfavorable biomarker profile, and extent of genomic alterations are concerning and imply that PLCIS may be a more clinically significant lesion than CLCIS. Furthermore, our study demonstrates clear morphologic and molecular heterogeneity in PLCIS and suggests that the morphologically distinct apocrine PLCIS may be a particularly aggressive lesion. However, clinical follow-up studies will be required to define the natural history and most appropriate management of these lesions.
Supplementary Material
Acknowledgments
Supported by: UCSF Research Evaluation and Allocation Committee (REAC) Grant, Komen Foundation and NIH P50 CA58207.
Abbreviations
- LCIS
lobular carcinoma in situ
- PLCIS
pleomorphic lobular carcinoma in situ
- DCIS
ductal carcinoma in situ
Footnotes
An abstract of this paper was presented at the 94th Annual Meeting of the United States and Canadian Academy of Pathology, Atlanta, San Antonio, TX, USA.
REFERNCES
- 1.Chivukula M, Haynik DM, Brufsky A, et al. Pleomorphic lobular carcinoma in situ (PLCIS) on breast core needle biopsies: clinical significance and immunoprofile. Am J Surg Pathol. 2008;32:1721–1726. doi: 10.1097/PAS.0b013e31817dc3a6. [DOI] [PubMed] [Google Scholar]
- 2.DeVries S, Gray JW, Pinkel D, et al. Comparative genomic hybridization Chapter 4. In: Haines Jonathan L, et al., editors. Current protocols in human genetics/editorial board. 2001. Unit46. [DOI] [PubMed] [Google Scholar]
- 3.Ernster VL, Barclay J. Increases in ductal carcinoma in situ (DCIS) of the breast in relation to mammography: a dilemma. J Natl Cancer Inst Monogr. 1997;151(156) doi: 10.1093/jncimono/1997.22.151. [DOI] [PubMed] [Google Scholar]
- 4.Fadare O, Dadmanesh F, Alvarado-Cabrero I, et al. Lobular intraepithelial neoplasia [lobular carcinoma in situ] with comedo-type necrosis: A clinicopathologic study of 18 cases. Am J Surg Pathol. 2006;30:1445–1453. doi: 10.1097/01.pas.0000213290.58283.82. [DOI] [PubMed] [Google Scholar]
- 5.Fisher ER, Costantino J, Fisher B, et al. Pathologic findings from the National Surgical Adjuvant Breast Project (NSABP) Protocol B-17. Five-year observations concerning lobular carcinoma in situ. Cancer. 1996;78:1403–1416. doi: 10.1002/(SICI)1097-0142(19961001)78:7<1403::AID-CNCR6>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 6.Fridlyand J, Snijders AM, Ylstra B, et al. Breast tumor copy number aberration phenotypes and genomic instability. BMC Cancer. 2006;6:96. doi: 10.1186/1471-2407-6-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frost A, Tsangaris T, Silverberg S. Pleomorphic lobular carcinoma in situ. Pathology Case Review. 1996;1:27–31. [Google Scholar]
- 8.Georgian-Smith D, Lawton TJ. Calcifications of lobular carcinoma in situ of the breast: radiologic-pathologic correlation. AJR Am J Roentgenol. 2001;176:1255–1259. doi: 10.2214/ajr.176.5.1761255. [DOI] [PubMed] [Google Scholar]
- 9.Ginzinger DG, Godfrey TE, Nigro J. Measurement of DNA copy number at microsatellite loci using quantitative PCR analysis. Cancer Res. 2000;60:5405–5409. [PubMed] [Google Scholar]
- 10.Haagensen CD, Lane N, Lattes R, et al. Lobular neoplasia (so-called lobular carcinoma in situ) of the breast. Cancer. 1978;42:737–769. doi: 10.1002/1097-0142(197808)42:2<737::aid-cncr2820420247>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 11.Hwang E, DeVries S, Roydasgupta R, et al. Genomic signature of invasive lobular cancer by DNA array hybridization. Breast Cancer Research and Treatment. 2007;106:S160. [Google Scholar]
- 12.Hwang E, DeVries S, Wa C, et al. Genomic alterations in large, high grade DCIS by array comparative genomic hybridization. Breast Cancer Research and Treatment. 2006;100:S166. [Google Scholar]
- 13.Hwang ES, Nyante SJ, Yi Chen Y, et al. Clonality of lobular carcinoma in situ and synchronous invasive lobular carcinoma. Cancer. 2004;100:2562–2572. doi: 10.1002/cncr.20273. [DOI] [PubMed] [Google Scholar]
- 14.Jones PA. Overview of cancer epigenetics. Semin Hematol. 2005;42:S3–S8. doi: 10.1053/j.seminhematol.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 15.Lakhani SR, Audretsch W, Cleton-Jensen AM, et al. The management of lobular carcinoma in situ (LCIS). Is LCIS the same as ductal carcinoma in situ (DCIS)? Eur J Cancer. 2006;42:2205–2211. doi: 10.1016/j.ejca.2006.03.019. [DOI] [PubMed] [Google Scholar]
- 16.Lu YJ, Osin P, Lakhani SR, et al. Comparative genomic hybridization analysis of lobular carcinoma in situ and atypical lobular hyperplasia and potential roles for gains and losses of genetic material in breast neoplasia. Cancer Res. 1998;58:4721–4727. [PubMed] [Google Scholar]
- 17.Middleton LP, Palacios DM, Bryant BR, et al. Pleomorphic lobular carcinoma: morphology, immunohistochemistry, and molecular analysis. Am J Surg Pathol. 2000;24:1650–1656. doi: 10.1097/00000478-200012000-00009. [DOI] [PubMed] [Google Scholar]
- 18.Mohsin SK, O'Connell P, Allred DC, et al. Biomarker profile and genetic abnormalities in lobular carcinoma in situ. Breast Cancer Res Treat. 2005;90:249–256. doi: 10.1007/s10549-004-4493-8. [DOI] [PubMed] [Google Scholar]
- 19.Page DL, Kidd TE, Jr, Dupont WD, et al. Lobular neoplasia of the breast: higher risk for subsequent invasive cancer predicted by more extensive disease. Hum Pathol. 1991;22:1232–1239. doi: 10.1016/0046-8177(91)90105-x. [DOI] [PubMed] [Google Scholar]
- 20.Reis-Filho JS, Simpson PT, Jones C, et al. Pleomorphic lobular carcinoma of the breast: role of comprehensive molecular pathology in characterization of an entity. J Pathol. 2005;207:1–13. doi: 10.1002/path.1806. [DOI] [PubMed] [Google Scholar]
- 21.Schnitt SJ, Morrow M. Lobular carcinoma in situ: current concepts and controversies. Semin Diagn Pathol. 1999;16:209–223. [PubMed] [Google Scholar]
- 22.Simpson PT, Reis-Filho JS, Lambros MB, et al. Molecular profiling pleomorphic lobular carcinomas of the breast: evidence for a common molecular genetic pathway with classic lobular carcinomas. J Pathol. 2008;215:231–244. doi: 10.1002/path.2358. [DOI] [PubMed] [Google Scholar]
- 23.Snead DR, Bell JA, Dixon AR, et al. Methodology of immunohistological detection of oestrogen receptor in human breast carcinoma in formalin-fixed, paraffin-embedded tissue: a comparison with frozen section methodology. Histopathology. 1993;23:233–238. doi: 10.1111/j.1365-2559.1993.tb01195.x. [DOI] [PubMed] [Google Scholar]
- 24.Sneige N, Wang J, Baker BA, et al. Clinical, histopathologic, and biologic features of pleomorphic lobular (ductal-lobular) carcinoma in situ of the breast: a report of 24 cases. Mod Pathol. 2002;15:1044–1050. doi: 10.1097/01.MP.0000027624.08159.19. [DOI] [PubMed] [Google Scholar]
- 25.Snijders AM, Nowak N, Segraves R, et al. Assembly of microarrays for genome-wide measurement of DNA copy number. Nat Genet. 2001;29:263–264. doi: 10.1038/ng754. [DOI] [PubMed] [Google Scholar]
- 26.Vos CB, Cleton-Jansen AM, Berx G, et al. E-cadherin inactivation in lobular carcinoma in situ of the breast: an early event in tumorigenesis. Br J Cancer. 1997;76:1131–1133. doi: 10.1038/bjc.1997.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weigelt B, Horlings HM, Kreike B, et al. Refinement of breast cancer classification by molecular characterization of histological special types. J Pathol. 2008;216:141–150. doi: 10.1002/path.2407. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.