Abstract
Selenoprotein H (SelH) is one of the 25 so far identified selenoproteins. Selenoproteins may function as antioxidants, heavy metal antidotes, and neural survival factors. Previous studies have shown that overexpression of SelH in HT22 cells protected the cells from UVB irradiation-induced death by reducing superoxide formation. The objective of this study was to determine the effects of SelH on cell signaling pathways after UVB irradiation. We exposed both human SelH- and vector-transfected HT22 cells to UVB irradiation and collected samples at 5- and 17-hrs of recovery. Cell viability was assessed, as well as protein levels of caspase-3,-8,-9, apoptosis-inducing factor (AIF), P53, nuclear respiratory factor-1 (NRF-1) and heat shock protein 40 (HSP40). Mitochondrial membrane potential was determined by flow cytometry. Overexpression of SelH protected cells against UVB-induced injury by blockade of the mitochondria-initiated cell death pathway, prevention of mitochondrial membrane depolarization, and suppression of the increase of p53. Furthermore, overexpression of SelH increased levels of NRF-1, an antioxidant, and HSP40, a protein chaperone that repairs denatured protein. We conclude that SelH protects neurons against UVB-induced damage by inhibiting apoptotic cell death pathways, by preventing mitochondrial depolarization, and by promoting cell survival pathways.
Keywords: Neuronal death, Apoptosis, Mitochondrion, Ultraviolet, Selenoprotein, Signal transduction
Introduction
Selenoprotein H (SelH) is one of the 25 so far identified selenoproteins (Burk and Hill, 2005; Chen and Berry, 2003). Many selenoproteins, including SelH, have been shown to contain a CXXU motif, indicative of a redox active protein and a significant subset of the mammalian selenoproetome has been associated with cellular detoxification functions (Gromer et al., 2005) Some well-known selenoproteins include perioxidases and reductases such as glutathione peroxidase (GPx), phospholipid hydroperoxide glutathione peroxidases (PHGPXs) and thioredoxin reductases (TRs). Although the functions of selenoproteins are not fully defined, studies have shown that selenoproteins may function as antioxidants, heavy metal antidotes, and neural survival factors. For example, selenoprotein P catalyzes the reduction of phospholipid hydroperoxide by glutathione (GSH) (Saito et al., 1999) or thioredoxin (Takebe et al., 2002), functions as a peroxynitrite scavenger (Arteel et al., 1998) and promotes cell survival (Hirashima et al., 2003; Yan & Barrett, 1998). Disruption of the selenoprotein P gene in mice resulted in increased susceptibility to oxidative stress and impaired neurological function (Hill et al., 2003; Schomburg et al., 2003).
Although SelH has not been studied as extensively as selenoprotein P, emerging evidence has indicated that silencing drosophila SelH (dSelH) reduces, and overexpression of dSelH increases, cell viability and antioxidant concentrations (Morozova et al;, 2003). Furthermore, down-regulating SelH by RNA interference was shown to increase the sensitivity of mouse lung cancer LCC1 cells to hydrogen peroxide challenge (Novoselov et al., 2007). On the contrary, overexpression of SelH in the murine hippocampal neuronal HT22 cell line resulted in higher levels of glutathione, total antioxidant capacities, and glutathione peroxidase enzyme activity than control cells after treatment with l-buthionine(S,R)-sulfoximine to deplete glutathione (Panee et al., 2007). We have previously shown that overexpression of human SelH (hSelH) in HT22 cells protected cells from UVB irradiation induced death by reducing superoxide formation (Ben Jilani et al., 2007). The objective of this study was to determine the effects of hSelH on cell signaling pathways and mitochondrial membrane potentials relative to UVB irradiation. We exposed both SelH-transfected HT22 (SelH-HT22) cells and vector-transfected HT22 (Vector-HT22) cells to UVB irradiation, and then measured cell viability, protein levels of cleaved caspases, AIF, p53, and mitochondrial membrane potential. We also identified changes in two pro-survival proteins, NRF-1 and HSP40. Our data showed that overexpression of SelH protected cells against UVB-induced injury by inhibiting cell death pathways, preventing mitochondrial membrane depolarization, and promoting cell survival pathways.
Materials and methods
Cell Maintenance and Treatment
Stably transfected murine hippocampal HT22 neuronal cells which carried either the MSCV expression vector alone (vector-HT22) or encoded hSelH (SelH-HT22) were obtained from Dr. Panee at the University of Hawaii. The transfection procedures and efficacy of transfection have been previously reported (Ben Jilani et al., 2007, Panee et al., 2007). The hSelH mRNA levels are about 34-fold higher than the gene levels of endogenous mSelH (Panee et al., 2007). Cells were propagated in Dulbecco’s Modified Eagle Medium (DMEM) containing 10% fetal bovine serum (FBS), 2 mM glutamine, and 200 mM streptomycin/penicillin (Invitrogen) and then maintained at 90%–95% relative humidity in 5% CO2 at 37°C. The culture medium was renewed every 3 days. For cell viability assays, cells were seeded in 6-well cell culture plates (Corning, Aton, MA, USA) and were allowed to reach 80 % optical confluency prior to UVB treatments. All experiments were performed in triplicate or repeated on at least three occasions.
UVB Irradiation
Cells were seeded in 96 or 24 well plates and cultured to 80% cell confluence. Prior to UVB irradiation, the cultures were washed twice with cold PBS to remove residual serum and non-attached cells. Cells were incubated in serum-free medium and exposed to 7J/cm2 dose of UVB radiation from a Fisher UV Transilluminator FB-TI-88A over a period of 5 min. After UVB radiation, cells were returned to the culture incubator for various periods of recovery at 37°C.
Cell Viability Assay
The percentage of viable cells was determined using propidium iodide exclusion and flow cytometry (Dolbeare et al., 1990) on a FACSAria™ flow cytometer (Becton Dickinson, San Jose, CA) at 17 hrs following UBV challenge.
Mitochondrial Membrane Potential Assay
Cells were grown in 6 well plates to 70% confluence, washed with PBS twice and incubated in serum-free medium for 1 hr prior to treatment. The cells were then challenged with 7J/cm2 of UVB and allowed to recover for 5 hrs prior to assessment of mitochondrial membrane potentials. The mitochondrial membrane potential of control and irradiated samples were examined via flow cytometry using the dye JC-1. Test samples were loaded (30 min at 37°C) with 2.5 μg/ml of JC-1 (Molecular Probes) and data were collected on a BD FACSAria™ cytometer using FACSDiva™ software version 6.1. Controls included “untreated” vector-HT22 and SelH-HT22 cells as well as FCCP-treated samples providing “depolarized” populations of the two cell types. FCCP treated cells were used to produce gates separating cells exhibiting mitochondrial depolarization from those with normally polarized mitochondria. To enhance spectral separation, the red “J-aggregate” fluorescence of normally polarized mitochondria was monitored via a 610nm bandpass filter and the green fluorescence of JC-1 monomer was detected using a 530nm bandpass filter. FCCP (Molecular Probes) at 20μM was applied to non-irradiated samples for 1 hr to generate fully depolarized controls. The resulting data files were analyzed using FlowJo™ software (Tree Star, Inc. Ashland, OR). Nested gates of forward versus side scatter and the predominate forward scatter width peak were used to limit the 2-parameter display of JC-1 fluorescence at 610 and 530 nm. Fluorescence compensation (610 minus 530) was employed during data analysis to further enhance resolution of cells with decreased mitochondrial membrane potential. The resulting 2-parameter plots of JC-1 fluorescence show population compression characteristic of electronic compensation at the lower limits of detection.
Western Blot Analysis
At 17 hrs following UVB treatment, cells were collected and lysed on ice in lysis buffer containing 20 mM Tris pH7.4, 10 mM KCL, 3 mM MgCl2, 0.5% NP40 and complete inhibitors (Millipore). Lysates were centrifuged at 2,000 g for 10 min, and the resulting supernatant was designated as the cytoplasmic fractions. The pellets were washed twice with lysis buffer and resuspended in lysis buffer containing 1%SDS. The resulting lysates were sonicated briefly on ice (Misonix, Ultrasonic Cell Disrupter) and then centrifuged at 20,800 g for 30 min. The supernatants were designated as nuclear fractions. Protein lysates were separated in 10% NuPAGE BT gels (Invitrogen), transferred to PVDF membrane (Millipore) and probed with p53 (Santa Cruz, 1:250 dilution); AIF (Santa Cruz, 1;500 dilution), caspase-3 (Cell Signaling: 1:1000 dilution); caspase-9 (Cell Signaling, 1:1000 dilution); caspase-8 (Millipore, 1:1000 dilution); HSP40 (Assay designs, 1:1000 dilution; NRF-1(Santa Cruz, 1:500 dilution).
Statistical analysis
All data were presented as means ± SD. ANOVA followed by Tukey’s Multiple Comparison Test was used to analyze data. A p value <0.05 was considered as significant.
Results
Cell Viability in Vector-HT22 and SelH-HT22 Cells
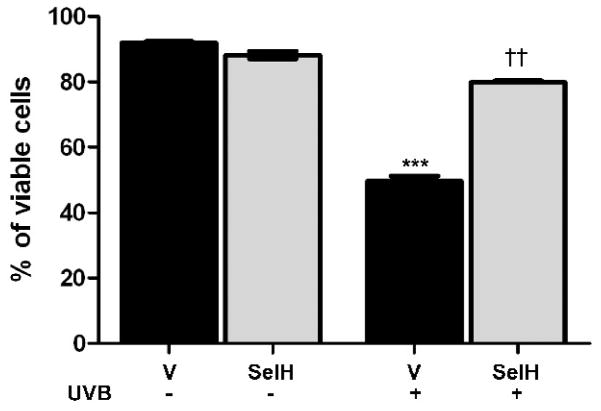
Cell viability was measured using flow cytometric analysis of propidium iodide stained cells (Fig. 1). In cells transfected with vector alone, treatment with UVB irradiation resulted in a close to 50% reduction in cell viability compared with non-UVB treated cells (p<0.001). However, overexpression of SelH significantly protected cells from UVB-induced damage. As a results, 80% of the SelH-HT22 cells remained viable at 17 hrs following irradiation (p<0.01 vs. vector-HT22 cells irradiated by UVB).
Fig. 1.
Cell viability in vector-HT22 and SelH-HT22neuronal cells at 17 hrs following UVB irradiation. Viability reduced after UVB treatment in vector-HT22 cells compared to naïve control. Overexpression of SelH improved cell viability after UVB challenge. ANOVA followed by Tukey’s test, ***p<0.001 vs. sham and ††p<0.01 vs. UVB-treated vector cells.
SelH Reduced activated Caspase-3 and -9 after UVB irradiation
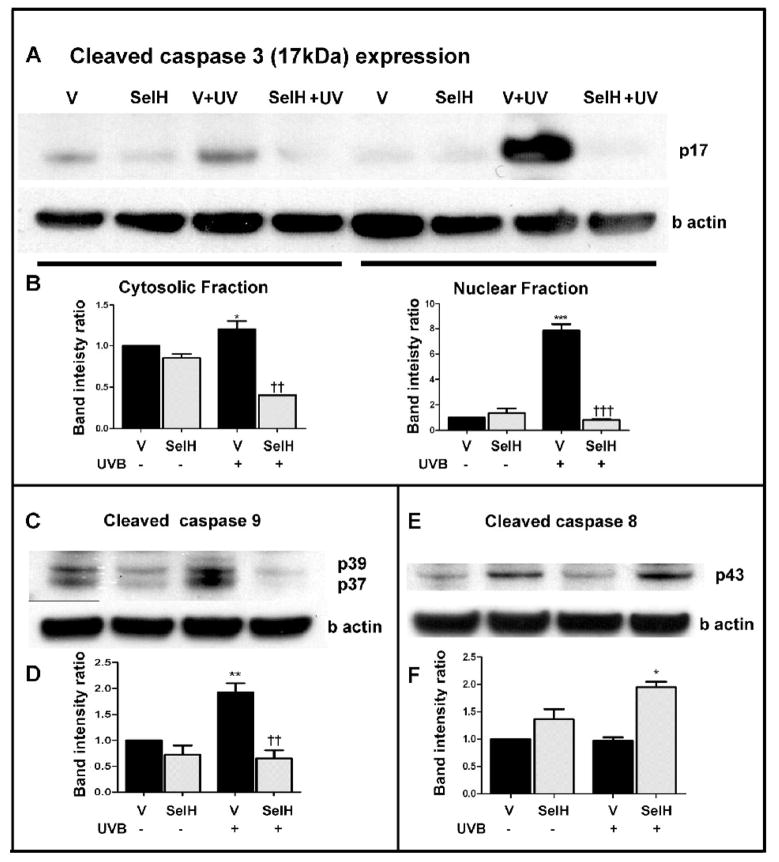
To determine whether overexpression of SelH prevents cell death by inhibiting cell death pathways, we measured active caspase-3 following UVB treatment in both vector-HT22 and SelH-HT22 cells. The results displayed a significant increase in cleaved caspase-3 at 17 hrs in the cytosolic fraction of the vector-HT22 cells (p<0.05 vs. non-irradiated vector-HT22 samples) and such increase was not evident in SelH-HT22 cells (p<0.01 vs. UVB-irradiated vector-HT22 cells). Similarly, in the nuclear fractions from UVB treated cells, an increase in cleaved caspase-3 was identified in vector-HT22 samples (p<0.001) and this increase was ameliorated in SelH-HT22 cells (p<0.001 vs. UVB-irradiated vector-HT22 cells). A representative caspase-3 protein blot and summary bar graphs are given in Figs. 2A & 2B. Levels of AIF were also measured and neither SelH- nor vector-transfected cells displayed increases of AIF in the nuclear fractions after UVB irradiation (data not shown).
Fig. 2.
Western blotting analyses of cleaved caspases at 17 hrs of recovery following 7J/cm2 UVB challenge. Caspase-3 and caspase-9 are significantly increased while caspase-8 is not altered in vector-HT22 cells after UVB challenge. Overexpression of SelH reduced the levels of cleaved caspase-3 and caspase-9. A, Cleaved caspase-3 protein band in both the cytosolic and nuclear fractions; B, semiquantitative changes of cleaved caspase-3 in the cytosolic and nuclear fractions; C, Cleaved caspase-9 protein band in the cytosolic fraction; D, semiquantitative changes of cleaved caspase-9; E, Cleaved caspsase-8 protein band in the cytosolic fraction; F, semiquantitative changes of cleaved caspase-8. Beta-actin was used as an internal protein loading control. Protein band intensity was presented as ratio of cleaved caspases to β actin and value from vector-HT22 cells was converted to 1.0. ANOVA followed by Tukey’s test, *p<0.05, **p<0.01, and ***p<0.001 vs. sham and †† p<0.01, ††† p<0.001 vs. UVB-treated vector-HT22 cells.
To investigate whether the activation of caspase-3 is mediated through the mitochondria-initiated intrinsic cell death pathway or a receptor-ligand triggered cell death pathway, we measured cleaved caspase-9 and caspase-8 levels using Western blotting. The results showed that cleaved caspase-9 levels increased in vector-HT22 cells following UVB irradiation compared with non-irradiated vector-HT22 controls (p<0.01). Overexpression of SelH significantly ameliorated this increase (Figs. 2C & 2D). The baseline levels of capsase-8 were higher in the SelH-HT22 than in the vector-HT22 cells, albeit the difference did not reach a statistical significance. The levels of cleaved caspase-8 were not elevated after UVB irradiation. In SelH-HT22 cells, the active caspase-8 levels were increased when compared to vector-HT22 cells after UVB treatment; however, there were no significant increase when compared to their own baseline levels (Figs. 2E & 2F), suggesting that overexpression of SelH failed to reduce caspase-8 activation.
SelH Stabilized Mitochondrial Membrane Potential
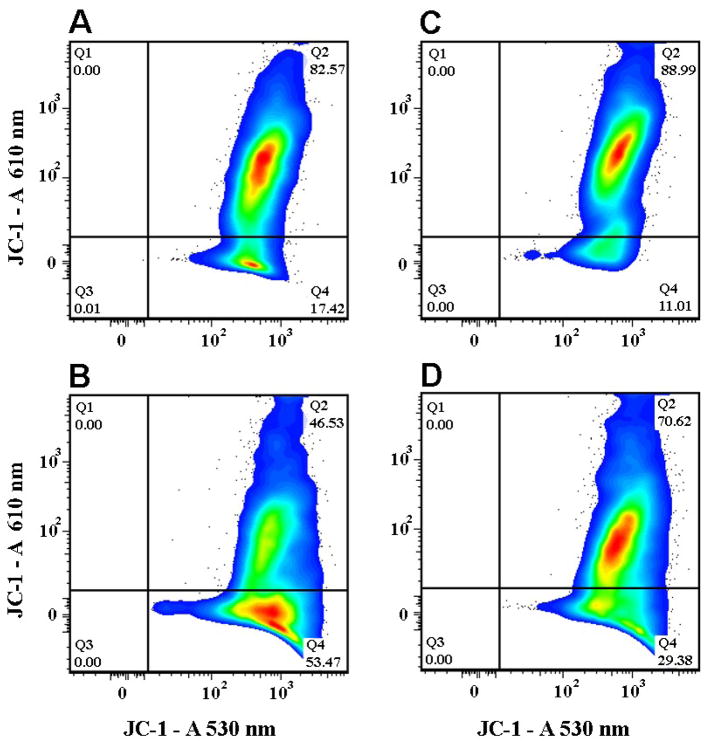
Since the above results suggested that the reduction of UVB-induced cell death was associated with blockade of a mitochondria-initiated cell death pathway, we decided to measure mitochondrial membrane potential using JC-1. The results indicated an early depolarization in vector-HT22 cells following UVB exposure. Thus, as shown in Fig. 3, untreated vector-HT22 cells exhibited depolarized fraction of 17.42% (Fig. 3A). At 5 hrs following UVB treatment, the depolarized population increased to 53.47% (Fig. 3B). In contrast, mitochondrial membrane was maintained in SelH-Ht22 cells at baseline level (11.01%, Fig. 3C) and following UVB treatment (29.38%, Fig. 3D). Compared to vector-HT22 cells, overexpression of SelH prevented mitochondrial membrane depolarization in a large population of cells.
Fig. 3.
Changes in mitochondrial membrane potential in vector- and SelH-HT22 cells at 5 hrs recovery following 7J/cm2 UVB treatment. Cells were loaded with JC-1 and analyzed by flow cytometry to assess affects on relative mitochondrial membrane potential following UVB insult. The data are presented as 2-parameter plots of JC-1 and “J-aggregate” fluorescence, the later exemplified by relatively high fluorescence at 610 nm. Percentages of cells in the non-depolarized “J-aggregate“ fractions (quadrant 2) and depolarized fractions (quadrant 4) are shown. SelH-HT22 cells demonstrated resistance to UVB-mediated mitochondrial depolarization compared to vector-HT22 cell under the same conditions. Panel A, vector-HT22 cells; Panel B, vector-HT22 cells challenged with UVB; Panel C, SelH-HT22 cells; Panel D, SelH-HT22 cells challenged with UVB.
Prevention of p53 translation in SelH-Transfected Cells
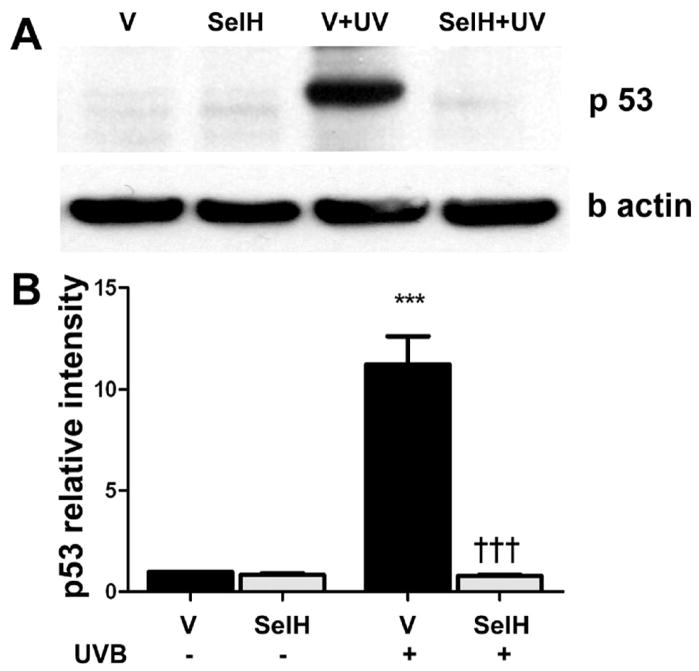
Previous studies have shown that UVB irradiation causes an increase in levels of p53 protein (for review, see Culmsee and Mattsson, 2005). To determine whether overexpression of SelH protects neurons from UVB-induced damage by blocking p53 activation, we measured p53 levels in the nuclear fractions from both vector-HT22 and SelH-HT22 cells. As shown in Fig. 4, a faint 53 kD band was visible in non-UVB treated vector-HT22 cells, while this band was absent in SelH-HT22 cells. After at 17 hrs of recovery following UVB, p53 protein band intensity increased significantly in vector-HT22 cells (p<0.001). In contrast, overexpression of SelH significantly prevented the increase of p53 after UVB-treatment (p< 0.001 vs. UVB-irradiated vector-HT22 samples).
Fig. 4.
Western blot analysis of p53 in the nuclear fraction at 17 hrs of recovery following 7J/cm2 UVB challenge in the vector-HT22 and SelH-HT22 cells. A, representative p53 protein blot; B, semiquantitative changes of p53. Beta-actin was used as an internal protein loading control. P53 protein band intensity was presented as ratio of p53 to β actin and value from vector-HT22 was converted to 1.0. ANOVA followed by Tukey’s test, ***p<0.001 vs. sham and ††† p<0.001 vs. UVB-challenged vector-HT22cells.
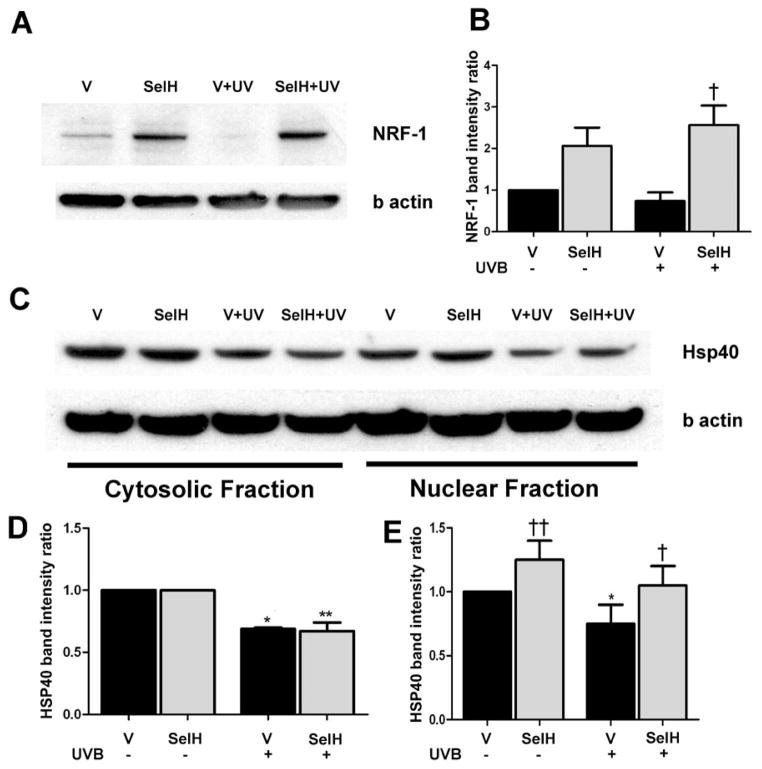
Increases of NRF-1 and HSP40 in SelH-Transfected Cells
The above results suggested that overexpression of SelH blocked the UVB irradiation-driven effects of activation of a mitochondria-initiated cell death pathway by stabilizing mitochondrial membrane potential. These findings prompted us to explore the effects of UVB irradiation and SelH on cell survival signaling pathways. In this part of our study, we focused on 2 proteins, NRF-1 and HSP40, which are responsible for anti-oxidative defense and repair of denatured proteins, respectively. In contrast to the observed expression pattern of caspase-3 and p53, the levels of NRF-1 in the nuclear fractions from vector-HT22 cells were not detectable at 17 hrs following UVB irradiation (Figs. 5A & 5B). Conversely, overexpression of SelH enhanced NRF-1 level following UVB exposure (p<0.05 vs. UVB treated vector-HT22 cells). Notably, elevated levels of NRF-1 were observed in the untreated SelH-HT22 cells compared with the untreated vector-HT22 controls although it did not reach statistical significance.
Fig. 5.
Western blot analysis of NRF-1 and HSP40 at 17 hrs of recovery following 7J/cm2 UVB treatment in the vector-HT22 and SelH-HT22 cells. A, NRF-1 protein bands in the nuclear fraction; B, semiquantitative changes of NRF-1. C, HSP40 protein bands in the cytosolic and nuclear fractions; D, semiquantitative changes of NRF-1. Beta-actin was used as an internal protein loading control. Protein band intensity was presented as ratio of NRF-1 or HSP40 to β-actin and value from vector-HT22 was converted to 1.0. ANOVA followed by Tukey’s test, *p<0.05, **p<0.01 vs. sham and †p<0.05, †† p<0.01 vs. UVB challenged vector-HT22 cells.
In the cytosolic fraction (Figs. 5C& 5D), basal levels of HSP40 in vector-HT22 were the same as in SelH-HT22 cells. After UVB irradiation, levels of HSP40 decreased equally in both vector-HT22 and SelH-HT22 cells as reflected by the band intensity ratio of HSP40 to β-actin (Fig. 5D). In the nuclear fraction (Figs. 5C & 5E), however, HSP40 level was higher in untreated SelH-HT22 cells than in untreated vector-HT22 cells (p<0.01). Following UVB irradiation, HSP40 levels were decreased in vector-HT22 cells (p<0.05) while they were maintained in SelH-HT22 cells (p<0.05 vs. UVB-irradiated vector-HT22 samples).
Discussion
Our results have demonstrated that UVB irradiation triggers death of HT22 neuronal cells by activating a mitochondrial initiated apoptotic cell death pathway which includes activation of caspase-9 and -3 via depolarization of the mitochondrial membrane potential in the early recovery period. In addition, UVB irradiation increases p53 levels and decreases pro-survival factors such as NRF-1 and HSP40. Overexpression of human SelH in HT22 cells reduced UVB-induces neuronal cell death by stabilizing mitochondrial membrane potential, inhibiting the activation of a mitochondrial initiated cell death pathway, and suppressing p53 activation. Furthermore, SelH activates cell survival signaling pathways.
Our previous study has shown that UVB induces cell death in the murine HT22 cells by increasing the production of superoxide (Ben Jilani et al., 2007). In the current study, we have confirmed these previous findings and further demonstrated that UVB irradiation results in cell death through activation of caspase-3, a key player in executing apoptosis. Since both mitochondrial dysfunction and receptor binding activate caspase-3 dependent cell death, we measured cleaved caspase-8 and caspase-9 protein levels to discern the pathways through which caspase-3 was being activated. Since only caspase-9, not caspase-8, was increased after UVB treatment in vector-HT22 cells, this suggests that UVB irradiation activates the mitochondria-initiated cell death pathway, but not the receptor-mediated death pathway. Our results on capase-8 are at variance with published data showing that UVB treatment triggers both intrinsic and extrinsic pathways by activating caspase-9 and -8, and/or interleukin -1 in human keratinocyte cells (Park and Lee, 2007; Simbula-Rosenthal 2006), Jurkat cells derived from human T cell leukemia (Mutou et al., 2008), and the human epidermoid carcinoma cell line A431 (Caricchio et al., 2003). The observed difference may be related to the types of cells and intensity of UVB irradiation employed. It is not known why baseline caspase-8 levels were high in SelH-HT22 cells. However, since caspase-8 levels were not reduced in SelH-HT cells after UVB irradiation, this suggests that SelH does not exert its effect on caspase-8. We also measured AIF levels in the nuclear fraction and observed no significant change, suggesting that AIF does not mediate UVB induced damage in our experimental model.
It is well established that maintenance of membrane potential is a requisite for normal mitochondrial function. Hyperpolarization inhibits the electron transport, resulting in increased ROS production, while depolarization induces the formation of the mitochondrial permeability transition pore (MPTP), allowing release of mitochondrial matrix proteins, subsequently triggering intrinsic cell death pathways. To assess the changes in mitochondrial membrane potential after UVB irradiation, we labeled cells with JC-1 and monitored relative membrane potential using flow cytometry. Using our model, we observed an increased fraction of depolarized mitochondria in vector-HT22 cells at 5 hrs post irradiation. This result, together with our caspases data, suggests that UVB activates caspases by mitochondrial depolarization that favors the formation of MPTP.
Our results further demonstrate that overexpression of SelH blocks the UVB irradiation induced activation of capspase-9 and -3, and stabilizes mitochondrial membrane potential in the early recovery phase, suggesting that a SelH-related function protects the mitochondria and prevents the mitochondria initiated cell death pathway. These results are in line with previous studies providing evidence that overexpression of SelH reduces superoxide formation in UVB irradiated HT22 cells and increases levels of glutathione, glutathione peroxidase activity and antioxidant capacities (Ben Jilani et al., 2007; Panee et al., 2007). It has been established that increases of reactive oxygen species, including superoxide, facilitate the formation of the mitochondrial membrane permeability transition pore (Bernadi, 1996). Taken together, these data suggest that SelH exerts its neuroprotective effects by protecting the mitochondria.
P53 is a tumor suppressor and a transcription factor that modulates the cellular stress response. A range of stress events can activate p53, including DNA damage, hypoxia, withdrawal of trophic factors, hypoglycemia, oxidative stress, viral infection and UV irradiation (Culmsee and Mattson, 2005; Morrison et al., 2003; Timares et al., 2008). Activation of p53 can trigger apoptosis by inducing the expression of p21, BAX, and NH3-only protein PUMA (p53 upregulated modulator of apoptosis), causing mitochondrial membrane permeabilization (Jeffers et al., 2003). Our results showed that p53 protein levels increased significantly after UVB irradiation, in accordance with published results (Burren et al., 1998; She et al., 2000). It has been previously reported that UVB activates p53 through the MAPK-mediated pathways (She et al., 2000). Our results demonstrate that overexpression of SelH prevents the increase of p53 after UVB treatment, thus inhibiting p53-mediated UVB-induced damage.
Although the effect of UVB on NRF-1 has not been reported, one recent study has described a suppressive effect of UVB on NRF-2, NRF-dependent heme oxygenase-1 (HO-1) and phase II detoxifying enzymes in human skin cells (Kokot et al., 2009). Our results indicated that UVB radiation suppressed NRF-1 levels in vector-HT22 cells and overexpression of SelH elevated NRF-1 contents. Our data suggest that SelH increased the resistance of HT22 cells to UVB induced damage by enhancing neuroprotective factor NRF-1. NRF-1 was first identified by Evans and Scarpulla (1989) as the main activating factor for cytochrome c promoter activity (Dhar and Wong-Riley, 2009; Evans and Scarpulla,1989). Recent studies have shown that NRF-1 regulate mitochondrial biogenesis in coordination with nuclear and mitochondrial gene expression, mitochondrial transcriptional factor A (TFAM), and peroxisome proliferator-activated receptor coactivator-1 (PGC-1) (Scarpulla,, 2006, 2008). NRF-1 may also function as a neural growth factor as it promotes neurite outgrowth (Chang et al., 2005). Neuronal stress induced by kainic acid intracerebral injection or hypoxic ischemia resulted in a marked increase in NRF-1 mRNA and protein levels (Hertel et al., 2002; Yin et al., 2008). Mice with homozygous deletion of NRF-1 show reduced mitochondrial DNA content and lethality at day E3.5-E6.5, while heterozygous animals are smaller than normal with a defect in succinate oxidation in the kidney (Huo and Scarpulla, 2001; Scarpulla 2002; for review, see Goffart and Wiesner, 2003).
HSP40, also known as DNAJB1, is a member of the family of heat shock proteins that respond to stress and serve as molecular chaperones preventing irreversible protein aggregation, misfolding, unfolding, translation, and degeneration (Vos et al., 2008). All HSP40 proteins contain the J domain through which they bind to HSP70s and stimulate the ATPase activity of HSP70s (Qiu et al., 2006; Zhao et al., 2008). Therefore, HSP40 determines the activity of HSP70s. Upregulation of HSP70s with heat treatment or stress protein activators protected human keratinocytes from subsequent UVB-induced damage (Merwald et al., 2006; Trautinger et al., 1995, 1996) and neurons from hypoxic/ischemic injuries (Giffard et al., 2004; Yenari et al., 2005; Zhao et al., 2008). UVB is capable of inducing HSP70s; however, there has been no report available on the effects of UVB-treatment and SelH on HSP40. Our results demonstrated a suppression of HSP40 after UVB challenge in vector-Ht22 cells. Overexpression of SelH maintained not only relative high levels of HSP40 in naïve SelH-Ht22 cell but also prevented the fall after UVB irradiation. These data suggest that SelH protected cells from UVB-induced damage by increasing the basal levels of HSP40 and by preventing the reduction of HSP40 after UVB irradiation.
In summary, our data suggest that SelH prevents UVB-induced cell death in two ways. First, SelH helps to maintain the mitochondrial membrane potential and blocks cell death pathways involving caspase-9, -3 and p53. Second, overexpression of SelH promotes cell survival pathways involving NRF-1 and HSP40.
Acknowledgments
The authors greatly appreciate the gift of the vector-HT22 and SelH-HT22 cell lines by Dr. Jun Panee for use in this study. Dr. Li’s laboratory is supported by a grant from National Institute of Health (R01DK075476). The BRITE is partially funded by the Golden Leaf Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arteel GE, Mostert V, Oubrahim H, Briviba K, Abel J, Sies H. Protection by selenoprotein P in human plasma against peroxynitrite-mediated oxidation and nitration. Biol Chem. 1998;379:1201–1205. [PubMed] [Google Scholar]
- Ben Jilani KE, Panee J, He Q, Berry MJ, Li PA. Overexpression of selenoprotein H reduces Ht22 neuronal cell death after UVB irradiation by preventing superoxide formation. Int J Biol Sci. 2007;3:198–204. doi: 10.7150/ijbs.3.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernadi P. The permeability transition pore: control points of a cyclosporin A-sensitive mitochondrial channel involved in cell death. Biochem Biophys Acta. 1996;1275:5–9. doi: 10.1016/0005-2728(96)00041-2. [DOI] [PubMed] [Google Scholar]
- Burk RF, Hill KE. Selenoprotein P: an extracellular protein with unique physical characteristics and a role in selenium homeostasis. Annu Rev Nutr. 2005;25:215–235. doi: 10.1146/annurev.nutr.24.012003.132120. [DOI] [PubMed] [Google Scholar]
- Burren R, Scaletta C, Frenk E, Panizzon RG, Applegate LA. Sunlight and carcinogenesis: expression of p53 and pyrimidine dimers in human skin following UVA I, UVA I + II and solar simulating radiations. Int J Cancer. 1998;76:201–206. doi: 10.1002/(sici)1097-0215(19980413)76:2<201::aid-ijc6>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- Caricchio R, McPhie L, Cohen PL. Ultraviolet B radiation-induced cell death: critical role of ultraviolet dose in inflammation and lupus autoantigen redistribution. J Immunol. 2003;171:5778–5786. doi: 10.4049/jimmunol.171.11.5778. [DOI] [PubMed] [Google Scholar]
- Chang WT, Chen HI, Chiou RJ, Chen CY, Huang AM. A novel function of transcription factor alpha-Pal/NRF-1: increasing neurite outgrowth. Biochem Biophys Res Commun. 2005;334:199–206. doi: 10.1016/j.bbrc.2005.06.079. [DOI] [PubMed] [Google Scholar]
- Chen J, Berry MJ. Selenium and selenoproteins in the brain and brain diseases. J Neurochem. 2003;86:1–12. doi: 10.1046/j.1471-4159.2003.01854.x. [DOI] [PubMed] [Google Scholar]
- Culmsee C, Mattson MP. p53 in neuronal apoptosis. Biochem Biophys Res Commun. 2005;331:761–777. doi: 10.1016/j.bbrc.2005.03.149. [DOI] [PubMed] [Google Scholar]
- Dhar SS, Wong-Riley MT. Coupling of energy metabolism and synaptic transmission at the transcriptional level: role of nuclear respiratory factor 1 in regulating both cytochrome c oxidase and NMDA glutamate receptor subunit genes. J Neurosci. 2009;29:483–492. doi: 10.1523/JNEUROSCI.3704-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolbeare F. Fluorescent staining of enzymes for flow cytometry. Methods Cell Biol. 1990;33:81–88. doi: 10.1016/s0091-679x(08)60513-0. [DOI] [PubMed] [Google Scholar]
- Evans MJ, Scarpulla RC. Interaction of nuclear factors with multiple sites in the somatic cytochrome c promoter. Characterization of upstream NRF-1, ATF, and intron Sp1 recognition sequences. J Biol Chem. 1989;264:14361–14368. [PubMed] [Google Scholar]
- Giffard RG, Xu L, Zhao H, Carrico W, Ouyang Y, Qiao Y, Sapolsky R, Steinberg G, Hu B, Yenari MA. Chaperones, protein aggregation, and brain protection from hypoxic/ischemic injury. J Exp Biol. 2004;207:3213–3220. doi: 10.1242/jeb.01034. [DOI] [PubMed] [Google Scholar]
- Goffart S, Wiesner RJ. Regulation and co-ordination of nuclear gene expression during mitochondrial biogenesis. Exp Physiol. 2003;88:33–40. doi: 10.1113/eph8802500. [DOI] [PubMed] [Google Scholar]
- Gromer S, Eubel JK, Lee BL, Jacob J. Human selenoproteins at a glance. Cell Mol Life Sci. 2005;62:2414–2437. doi: 10.1007/s00018-005-5143-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertel M, Braun S, Durka S, Alzheimer C, Werner S. Upregulation and activation of the Nrf-1 transcription factor in the lesioned hippocampus. Eur J Neurosci. 2002;15:1707–1711. doi: 10.1046/j.1460-9568.2002.01992.x. [DOI] [PubMed] [Google Scholar]
- Hill KE, Zhou J, McMahan WJ, Motley AK, Atkins JF, Gesteland RF, Burk RF. Deletion of selenoprotein P alters distribution of selenium in the mouse. J Biol Chem. 2003;278:13640–13646. doi: 10.1074/jbc.M300755200. [DOI] [PubMed] [Google Scholar]
- Hirashima M, Naruse T, Maeda H, Nozaki C, Saito Y, Takahashi K. Identification of selenoprotein P fragments as a cell-death inhibitory factor. Biol Pharm Bull. 2003;26:794–798. doi: 10.1248/bpb.26.794. [DOI] [PubMed] [Google Scholar]
- Huo L, Scarpulla RC. Mitochondrial DNA instability and peri-implantation lethality associated with targeted disruption of nuclear respiratory factor 1 in mice. Mol Cell Biol. 2001;21:644–654. doi: 10.1128/MCB.21.2.644-654.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffers JR, Parganas E, Lee Y, Yang C, Wang J, Brennan J, MacLean KH, Han J, Chittenden T, Ihle JN, McKinnon PJ, Cleveland JL, Zambetti GP. Puma is an essential mediator of p53-dependent and -independent apoptotic pathways. Cancer Cell. 2003;4:321–328. doi: 10.1016/s1535-6108(03)00244-7. [DOI] [PubMed] [Google Scholar]
- Kokot A, Metze D, Mouchet N, Galibert MD, Schiller M, Luger TA, Bohm M. {alpha}-MSH counteracts the suppressive effect of UVB on Nrf2 and Nrf-dependent gene expression in human skin. Endocrinology. 2009;150:3197–3206. doi: 10.1210/en.2008-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merwald H, Kokesch C, Klosner G, Matsui M, Trautinger F. Induction of the 72-kilodalton heat shock protein and protection from ultraviolet B-induced cell death in human keratinocytes by repetitive exposure to heat shock or 15-deoxy-delta(12,14)-prostaglandin J2. Cell Stress Chaperones. 2006;11:81–88. doi: 10.1379/CSC-89R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozova N, Forry EP, Shahid E, Zavacki AM, Harney JW, Kraytsberg Y, Berry MJ. Antioxidant function of a novel selenoprotein in Drosophila melanogaster. Genes Cells. 2003;8:963–971. doi: 10.1046/j.1365-2443.2003.00687.x. [DOI] [PubMed] [Google Scholar]
- Morrison RS, Kinoshita Y, Johnson MD, Guo W, Garden GA. p53-dependent cell death signaling in neurons. Neurochem Res. 2003;28:15–27. doi: 10.1023/a:1021687810103. [DOI] [PubMed] [Google Scholar]
- Mutou Y, Ibuki Y, Terao Y, Kojima S, Goto R. Induction of apoptosis by UV-irradiated chlorinated bisphenol A in Jurkat cells. Toxicol In Vitro. 2008;22:864–872. doi: 10.1016/j.tiv.2008.01.001. [DOI] [PubMed] [Google Scholar]
- Novoselov SV, Kryukov GV, Xu XM, Carlson BA, Hatfield DL, Gladyshev VN. Selenoprotein H is a nucleolar thioredoxin-like protein with a unique expression pattern. J Biol Chem. 2007;282:11960–11968. doi: 10.1074/jbc.M701605200. [DOI] [PubMed] [Google Scholar]
- Panee J, Stoytcheva ZR, Liu W, Berry MJ. Selenoprotein H is a redox-sensing high mobility group family DNA-binding protein that up-regulates genes involved in glutathione synthesis and phase II detoxification. J Biol Chem. 2007;282:23759–23765. doi: 10.1074/jbc.M702267200. [DOI] [PubMed] [Google Scholar]
- Park K, Lee JH. Photosensitizer effect of curcumin on UVB-irradiated HaCaT cells through activation of caspase pathways. Oncol Rep. 2007;17:537–540. [PubMed] [Google Scholar]
- Qiu XB, Shao YM, Miao S, Wang L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell Mol Life Sci. 2006;63:2560–2570. doi: 10.1007/s00018-006-6192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y, Hayashi T, Tanaka A, Watanabe Y, Suzuki M, Saito E, Takahashi K. Selenoprotein P in human plasma as an extracellular phospholipid hydroperoxide glutathione peroxidase. Isolation and enzymatic characterization of human selenoprotein p. J Biol Chem. 1999;274:2866–2871. doi: 10.1074/jbc.274.5.2866. [DOI] [PubMed] [Google Scholar]
- Scarpulla RC. Nuclear activators and coactivators in mammalian mitochondrial biogenesis. Biochim Biophys Acta. 2002;1576:1–14. doi: 10.1016/s0167-4781(02)00343-3. [DOI] [PubMed] [Google Scholar]
- Scarpulla RC. Nuclear control of respiratory gene expression in mammalian cells. J Cell Biochem. 2006;97:673–683. doi: 10.1002/jcb.20743. [DOI] [PubMed] [Google Scholar]
- Scarpulla RC. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol Rev. 2008;88:611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- Schomburg L, Schweizer U, Holtmann B, Flohe L, Sendtner M, Kohrle J. Gene disruption discloses role of selenoprotein P in selenium delivery to target tissues. Biochem J. 2003;370:397–402. doi: 10.1042/BJ20021853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She QB, Chen N, Dong Z. ERKs and p38 kinase phosphorylate p53 protein at serine 15 in response to UV radiation. J Biol Chem. 2000;275:20444–20449. doi: 10.1074/jbc.M001020200. [DOI] [PubMed] [Google Scholar]
- Simbulan-Rosenthal CM, Daher A, Trabosh V, Chen WC, Gerstel D, Soeda E, Rosenthal DS. Id3 induces a caspase-3- and -9-dependent apoptosis and mediates UVB sensitization of HPV16 E6/7 immortalized human keratinocytes. Oncogene. 2006;25:3649–3660. doi: 10.1038/sj.onc.1209407. [DOI] [PubMed] [Google Scholar]
- Takebe G, Yarimizu J, Saito Y, Hayashi T, Nakamura H, Yodoi J, Nagasawa S, Takahashi K. A comparative study on the hydroperoxide and thiol specificity of the glutathione peroxidase family and selenoprotein P. J Biol Chem. 2002;277:41254–41258. doi: 10.1074/jbc.M202773200. [DOI] [PubMed] [Google Scholar]
- Timares L, Katiyar SK, Elmets CA. DNA damage, apoptosis and langerhans cells--Activators of UV-induced immune tolerance. Photochem Photobiol. 2008;84:422–436. doi: 10.1111/j.1751-1097.2007.00284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautinger F, Kindas-Mugge I, Barlan B, Neuner P, Knobler RM. 72-kD heat shock protein is a mediator of resistance to ultraviolet B light. J Invest Dermatol. 1995;105:160–162. doi: 10.1111/1523-1747.ep12317003. [DOI] [PubMed] [Google Scholar]
- Trautinger F, Kindas-Mugge I, Knobler RM, Honigsmann H. Stress proteins in the cellular response to ultraviolet radiation. J Photochem Photobiol B. 1996;35:141–148. doi: 10.1016/s1011-1344(96)07344-7. [DOI] [PubMed] [Google Scholar]
- Vos MJ, Hageman J, Carra S, Kampinga HH. Structural and functional diversities between members of the human HSPB, HSPH, HSPA, and DNAJ chaperone families. Biochemistry. 2008;47:7001–7011. doi: 10.1021/bi800639z. [DOI] [PubMed] [Google Scholar]
- Yan J, Barrett JN. Purification from bovine serum of a survival-promoting factor for cultured central neurons and its identification as selenoprotein-P. J Neurosci. 1998;18:8682–8691. doi: 10.1523/JNEUROSCI.18-21-08682.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yenari MA, Liu J, Zheng Z, Vexler ZS, Lee JE, Giffard RG. Antiapoptotic and anti-inflammatory mechanisms of heat-shock protein protection. Ann N Y Acad Sci. 2005;1053:74–83. doi: 10.1196/annals.1344.007. [DOI] [PubMed] [Google Scholar]
- Yin W, Signore AP, Iwai M, Cao G, Gao Y, Chen J. Rapidly increased neuronal mitochondrial biogenesis after hypoxic-ischemic brain injury. Stroke. 2008;39:3057–3063. doi: 10.1161/STROKEAHA.108.520114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Braun AP, Braun JE. Biological roles of neural J proteins. Cell Mol Life Sci. 2008;65:2385–2396. doi: 10.1007/s00018-008-8089-z. [DOI] [PMC free article] [PubMed] [Google Scholar]