Abstract
Cell adhesion molecules have been implicated as key organizers of synaptic structures, but there is still a need to determine how these molecules facilitate neurotransmitter receptor recruitment to developing synapses. Here, we identify erythrocyte protein band 4.1-like 3 (protein 4.1B) as an intracellular effector molecule of Synaptic Cell Adhesion Molecule 1 (SynCAM1) that is sufficient to recruit NMDA-type receptors (NMDARs) to SynCAM1 adhesion sites in COS7 cells. Protein 4.1B in conjunction with SynCAM1 also increased the frequency of NMDAR-mediated mEPSCs and area of presynaptic contact in an HEK293 cell/ neuron co-culture assay. Studies in cultured hippocampal neurons reveal that manipulation of protein 4.1B expression levels specifically affect NMDAR-mediated activity and localization. Finally, further experimentation in COS7 cells show that SynCAM1 may also interact with protein 4.1N to specifically effect AMPA type receptor (AMPAR) recruitment. Thus, SynCAM1 may recruit both AMPARs and NMDARs by independent mechanisms during synapse formation.
INTRODUCTION
Unraveling the mechanisms by which synapses form during development of the central nervous system is essential to understanding the origin of neurodevelopmental disorders and cognitive impairment (Zoghbi, 2003). Synaptogenesis is a multi-step process that is initiated by contact between two neurons. As this contact becomes adhesive prior to becoming a synapse (Chow and Poo, 1985), it has long been hypothesized that cell adhesion molecules (CAMs) are key to the early events of synaptogenesis (Bloch, 1989). One huge stride forward in our understanding of synapse formation was the realization that CAMs not only mediate adhesion at synapses, but also initiate the recruitment of crucial synaptic components such as synaptic vesicles in the axon and neurotransmitter receptors in the dendrite (Barrow et al., 2009; Biederer et al., 2002; Nam and Chen, 2005; Scheiffele et al., 2000; Sytnyk et al., 2002; for review see Washbourne et al., 2004a).
Recently, a family of immunoglobulin-domain containing CAMs, called Synaptic Cell Adhesion Molecule (SynCAMs), were identified as potent inducers of presynaptic terminals, when expressed in non-neuronal cells and cocultured with neurons (Biederer et al., 2002). This synaptogenic potential is shared with a handful of other CAMs, including the neuroligins (Nlgns) and their presynaptic partners the neurexins (Dean et al., 2003; Scheiffele et al., 2000), netrin-G ligands (NGLs; Kim et al., 2006) and synaptic cell adhesion-like molecules (SALMs; Ko et al., 2006; Wang et al., 2006). While it appears that all of these molecules are able to induce the formation of the presynaptic terminal, their ability to recruit postsynaptic components has been less well studied. To date, the most heavily investigated interactions lie within the intracellular domain of Nlgn1. Nlgn1 can interact with the postsynaptic density protein PSD-95 through a type I PDZ binding motif (Irie et al., 1997), and can recruit NMDA-type glutamate receptors through both the PDZ binding motif and the WW binding domain (Barrow et al., 2009; Iida et al., 2004).
Similarly, SynCAMs also possess intracellular interaction domains including a type II PDZ binding motif and a FERM (4.1, ezrin, radixin, moesin) binding motif (Biederer, 2005a; Biederer et al., 2002). Potential interacting molecules, or effectors, have been identified; however, none of these interactions have been explored for their role in postsynaptic differentiation. In vitro and in yeast-two-hybrid studies, SynCAM1 was shown to bind calcium/calmodulin-dependent serine protein kinase (CASK) (Biederer et al., 2002), Syntenin1 (Biederer et al., 2002; Meyer et al., 2004) and glutamate receptor interacting protein 1 (GRIP1; Meyer et al., 2004) via the C-terminal PDZ-binding domain. All three proteins are thought to play a scaffolding role in recruiting or organizing proteins at a variety of cellular junctions (Funke et al., 2005). In addition, SynCAM1 can bind to erythrocyte protein band 4.1-like 3 (protein 4.1B) via the juxtamembranous FERM binding domain (Yageta et al., 2002), an interaction which is thought to promote cell adhesion. All four molecules (CASK, Syntenin1, GRIP1 and 4.1B) are expressed in the CNS, have multiple protein-protein interaction domains and all could potentially play a role in the development of the postsynaptic specialization.
We investigated these potential effectors of SynCAM1 in terms of their ability to recruit glutamate receptors to sites of synaptic adhesion. We focused on NMDARs as they appear to be the first glutamate receptors recruited to synapses during synaptogenesis (Barrow et al., 2009; Liao et al., 1999; McAllister, 2007; Petralia et al., 1999; Washbourne et al., 2002). We identified protein 4.1B as a potent and specific SynCAM1 effector molecule for the recruitment of NMDARs. Surprisingly, we also identified protein 4.1N as a specific SynCAM1 effector for AMPAR recruitment. These results were confirmed by electrophysiological studies in an HEK293 cell/neuronal co-culture assay (Biederer and Scheiffele, 2007; Fu et al., 2003). Imaging and electrophysiological studies of hippocampal neurons in culture demonstrate an important role for protein 4.1B during synapse formation and the recruitment of NMDARs to synapses. Thus, our experiments establish 4.1 proteins as SynCAM1 effector molecules that impact postsynaptic development.
RESULTS
The Cell Adhesion Molecule / Receptor Recruitment Assay (CAMRA)
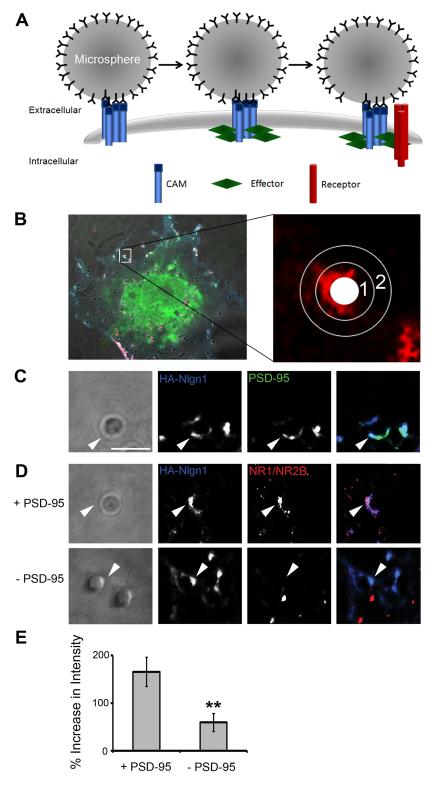
The initial goal of this work was to identify and characterize potential postsynaptic effector molecules for SynCAM1. We therefore characterized an assay that could be used to identify molecules sufficient to recruit glutamate receptors upon clustering of SynCAM1, or any CAM, in a postsynaptic configuration. The approach employed was an adhesion-based recruitment assay that measures microsphere-mediated clustering of recombinant molecules expressed in non-neuronal COS7 cells that we call the Cell Adhesion Molecule / Receptor Recruitment Assay (CAMRA). In the CAMRA, microspheres coated with antibodies against the protein of interest bound and aggregated many copies of a specific CAM that were expressed in COS7 cells (Fig.1A). Subsequently, we visualized co-transfected molecules and determined whether they were also accumulated at the site of microsphere binding (Fig.1B).
Fig. 1.
The Cell Adhesion Molecule / Receptor Recruitment Assay (CAMRA). (A) Model of clustering events in transfected COS7 cells after microsphere application when it is directed against tagged cell adhesion molecules (CAMs) on the surface of cells also expressing an intracellular effector molecule (Effector) and surface neurotransmitter receptors (Receptor). (B) Quantification of surface receptor immunofluorescence intensity around a single microsphere. Enlarged image depicts defined areas in a single channel that are used to determine intensity increases at microsphere. An average of the intensity within the area of annulus1 equals intensity of fluorescence at microsphere, the area of annulus 2 equals intensity of fluorescence in background, while the white circle is area taken by the microsphere. The left panel depicts the microsphere in the context of the whole cell, with all three fluorescence channels, CAM (blue), effector (green) and receptor (red). (C) PSD-95 was recruited to areas with HA-Nlgn1 accumulation in contact with microspheres (arrow head). Scale bar equals 2 μm. (D) NMDARs were recruited to sites of contact with microspheres and accumulations of Nlgn1 only when PSD-95 was co-transfected into COS7 cells. (E) Quantification of intensity increases of surface NMDARs at sites of contact in presence and absence of PSD-95 (165.9 ± 29.6% vs. 59.3 ± 18.1%, p < 0.01, n = 14, error bars represent s.e.m.).
First, we tested the CAMRA utilizing a well characterized protein complex known to interact at the synapse: Nlgn1, PSD-95 and the NMDAR composed of NR1 and NR2B subunits. Nlgn1 interacts with PSD-95 (Irie et al., 1997) and this interaction appears to regulate NMDAR recruitment to Nlgn1 clusters in cultured hippocampal neurons (Nam and Chen, 2005). Specifically, we asked whether microsphere-directed aggregation of an HA-tagged Nlgn1 (HA-Nlgn1) would promote GFP-tagged PSD-95 (PSD-95-GFP) co-accumulation. Cells transfected with these molecules were incubated with anti-HA antibody-coated microspheres for 1 hour at 37°C. We chose this time as we have previously determined that aggregation of PSD-95 to Nlgn1 clusters takes on the order of one hour in cortical neurons (Barrow et al., 2009). After fixation and immunolabeling, the fluorescence intensities of extracellular HA and internal GFP were measured at microspheres (Fig.1B, annulus 1) and compared to background intensity levels (Fig.1B, annulus 2). We observed a 123.9 ± 10.9% (n = 15) increase in fluorescence intensity of PSD-95-GFP at microspheres relative to background (Fig.1C, arrowheads). Therefore, we conclude that PSD-95-GFP was recruited to sites of HA-Nlgn1 clustering at microspheres, thus reflecting a relationship previously described (Irie et al., 1997). Further, this effect on the recruitment of PSD-95 is specific to the interaction with Nlgn1 as PSD-95-GFP did not accumulate at HA-SynCAM1 mediated sites of contact with microspheres relative to background (11.9 ± 4.5%, n = 15; Fig.2C and Supplemental Fig.1).
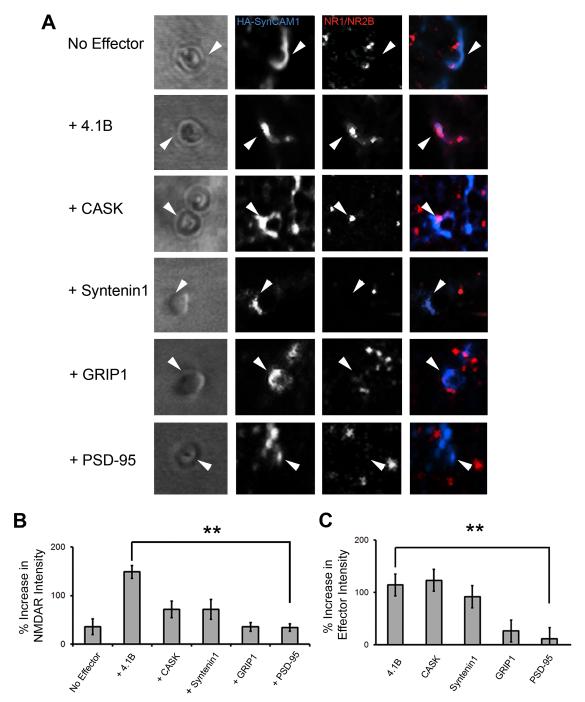
Fig. 2.
Protein 4.1B is a potent SynCAM1 effector molecule, recruiting NMDARs to sites of adhesion with microsphere. (A) Close up images of individual microspheres applied to cells transfected with SynCAM1 (blue), NMDARs (red) and one of the candidate effectors indicated on the left (not fluorescently labeled in these experiments). Arrowheads indicate examples of contact sites at microsphere where HA-SynCAM1 was aggregated. (B) Quantification of NMDAR recruitment via the candidate effector molecules. Protein 4.1B significantly increased intensity of receptor staining at microspheres relative to control (148.7 ± 13.3% vs. 33.5 ± 7.6%, p < 0.005*, n = 15). (C) Quantification of effector recruitment via SynCAM1 to sites of adhesion at microspheres (p < 0.005*, n = 15, error bars represent s.e.m.; * with correction for multiple comparisons).
Next, we analyzed whether surface-expressed NMDARs (NR1 and GFP-NR2B) localized to sites of HA-Nlgn1 aggregation at microspheres. We chose the NR2B subunits as these receptor subunits are most relevant to the early development of the glutamatergic postsynaptic density (Durand and Konnerth, 1996; Isaac et al., 1997; Washbourne et al., 2002; Wu et al., 1996). We compared conditions in which PSD-95 was either present or absent in the transfected cells (Fig.1D). As predicted, the surface level of NMDARs (as determined by labeling of the extracellular GFP tag) was significantly higher at microspheres when PSD-95 was co-transfected (165.9 ± 29.6% vs. 59.3 ± 18.1%, p < 0.01, n = 14; Fig.1E). Thus, we conclude that the CAMRA allows us to reliably measure the accumulation of an effector molecule (PSD-95) at sites of microsphere-mediated CAM (Nlgn1) clustering, and quantify the concomitant recruitment of NMDARs in the presence of the effector.
Potential SynCAM1 effector molecules
To determine if SynCAM1 could fulfill a similar role in glutamate receptor recruitment via one of its potential effector proteins, we employed the CAMRA to identify effector molecules that were sufficient to increase the accumulation of NMDARs to sites of SynCAM1 clustering. We tested four SynCAM1 binding proteins that had been identified in vitro: protein 4.1B, CASK, Syntenin1 and GRIP1.
COS7 cells were transfected with HA-SynCAM1, NR1, GFP-NR2B and one of the candidate effectors and then we applied microspheres directed to HA-SynCAM1 (Fig.2A,B). In the absence of an effector, there was a small increase in the accumulation of surface NMDARs (surface GFP-NR2B; 35.4 ± 16.33%) at microspheres that had clear accumulations of HA-SynCAM1 (110.56 ± 13.53%, n = 15; Fig.2A,B). This result was analogous to the Nlgn1-NMDAR-only co-transfected condition, and there was no significant difference between those two conditions (p = 0.2136; Fig.1D). Thus, we conclude that in the absence of effectors, the CAMs themselves may only promote small increases in NMDAR recruitment. PSD-95 is not predicted to interact with the intracellular domain of SynCAM1 (Biederer, 2005b; Meyer et al., 2004), therefore, we measured surface NMDAR recruitment to sites of HA-SynCAM1 mediated adhesion in the presence of PSD-95-GFP as a negative control (Fig.2A). Surface NMDAR recruitment under these conditions was not significantly different than HA-SynCAM1 alone (33.5 ± 7.6% vs. 35.4 ± 16.3%, p = 0.6, n = 15; Fig.2B).
Of the candidate effectors examined, only the addition of protein 4.1B produced a dramatic increase in NR1/NR2B accumulation at microsphere-mediated sites of HA-SynCAM1 aggregation relative to control (148.7 ± 13.3% vs. 35.4 ± 16.3%, p < 0.005, n = 15; Fig.2A,B). Surface NMDAR intensities at microspheres applied in the presence of CASK (71.6 ± 16.9%, p = 0.0421, n = 15), Syntenin1 (68.9 ± 20.5%, p = 0.2134, n = 15), or GRIP1 (35.2 ± 9.1%, p = 0.9669, n = 15), were not significantly different from the PSD-95 condition nor each other after correction for multiple comparisons (Fig.2B). However, CASK and Syntenin1 trend towards significance suggesting that there may be a basal level of NMDAR recruitment by these effector proteins, but not by GRIP1 and PSD-95. The effect of protein 4.1B was not mediated by increasing the total amount of HA-SynCAM1 at the plasma membrane (64 ± 5.3% of total HA-SynCAM1 was at the surface versus 73 ± 5.7% with addition of 4.1B, p = 0.12). In conclusion, these experiments revealed that protein 4.1B is a potent effector molecule of SynCAM1 in recruiting NMDARs to microsphere-mediated sites of adhesion.
SynCAM1 interacts with protein 4.1B to specifically recruit NMDARs
Given the weak effect of three of the four potential effector proteins in recruiting NMDARs to SynCAM1 adhesion sites, we explored the nature and specificity of their interactions with SynCAM1 (Fig.2C and Supplemental Fig.1). We compared effector recruitment measures to our negative control: PSD-95-GFP accumulation at microsphere-mediated sites of HA-SynCAM1 clustering. As measured by the percent increase of GFP-tagged effector at microspheres relative to background, HA-SynCAM1 drove the accumulation of 4.1B (114.1 ± 11%, p < 0 .005, n = 15), CASK (123 ± 16.9%, p < 0.005, n = 15) and Syntenin1 (91.8 ± 12.2%, p < 0.005, n = 15) compared to PSD-95 (Fig.2C). HA-SynCAM1 was unable to efficiently recruit GRIP1 in this assay despite previous reports of a direct interaction (26.4 ± 13.3%, p = 0.4068, n = 15). Thus, protein 4.1B, CASK and Syntenin1 were all recruited to sites of microsphere-induced SynCAM1 clustering in COS7 cells to a similar extent, yet protein 4.1B was significantly different in its ability to cause NMDAR accumulation at microspheres. Taken together, this suggests that protein 4.1B possesses the most potent recruitment activity on NMDARs compared to all effectors examined.
To further explore the specificity of the interaction between SynCAM1 and protein 4.1B, we examined aggregation of 4.1B-GFP at microspheres directed towards a mutant HA-SynCAM1 lacking the FERM binding domain (HA-SynCAM1ΔFERMb; Fig.3A). Full length 4.1B-GFP did not co-accumulate at microsphere-induced HA-SynCAM1ΔFERMb aggregation (29.4 ± 9.1%, n = 15; Fig.3C,D). Similarly, 4.1B lacking its FERM domain (4.1BΔFERM) did not accumulate at sites of HA-SynCAM1 aggregation (Fig.3B,C). To confirm that protein 4.1B was recruited specifically to clustered HA-SynCAM1 and not non-specifically to densely packed adhesion sites, we measured 4.1B-GFP accumulation using HA-NLG1 as the targeted CAM. In this case, 4.1B-GFP did not significantly co-localize to microspheres where large amounts of HA-NLG1 were localized (16.7 ± 6.4%, n = 15; Fig.3D). As an additional test of specificity of our CAMRA, we determined whether NMDARs would aggregate at microspheres directed against HA-SynCAM1 in the presence of 4.1BΔFERM. As compared to the negative control, we observed no significant difference in NMDAR accumulation at microspheres under this condition (33.9 ± 7.6% vs. 33.5 ± 7.6%, p = 0.4429, n = 15).
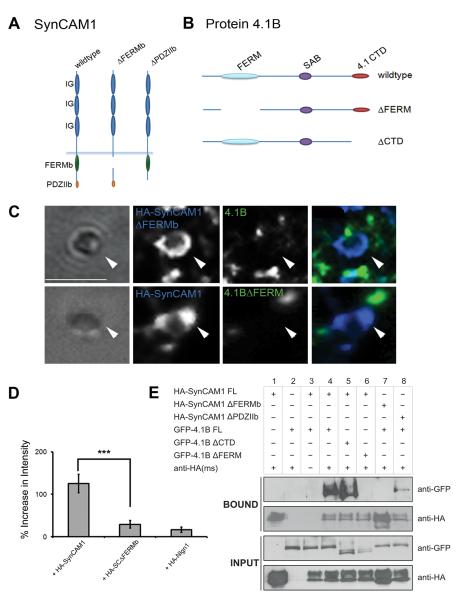
Fig. 3.
Direct interactions between SynCAM1 and protein 4.1B are mediated via key protein-protein interaction domains. (A) Model depicting known protein-protein interaction domains for SynCAM1 and the deletion mutants HA-SynCAM1ΔFERMb and HA-SynCAM1ΔPDZIIb. (B) Model depicting known protein-protein interaction domains for protein 4.1B and the deletion mutants 4.1BΔFERM and 4.1BΔCTD. (C) Images showing the lack of recruitment of protein 4.1B to sites of contact with microspheres when mutant proteins were transfected. (D) Quantification of localization of protein 4.1B to microspheres in the presence of either HA-SynCAM1 full length, HA-SynCAM1 FERMb, or Nlgn1. Protein 4.1B can only be recruited to adhesion sites with microspheres via full length SynCAM1 (p < 0.001, n = 15). Error bars represent s.e.m. (E) Immunoprecipitation experiments confirm the specificity of the direct interaction between SynCAM1 and protein 4.1B. Recombinant and tagged versions of SynCAM1 (HA-SynCAM1) and 4.1B (GFP-4.1B) were immunoprecipitated using an antibody to the HA tag. Bound proteins and input proteins were visualized in Western blots using antibodies to the tags (anti-GFP and anti-HA). Deletion of the PDZ binding domain of SynCAM1 (HA-SynCAM1ΔPDZIIb) and the C-terminal domain of 4.1B (GFP-4.1B ΔCTD) did not affect the interaction of these two partners. In contrast, deletion of the FERM binding domain of SynCAM1 (HA-SynCAM1 ΔFERMb) and the FERM domain of protein 4.1B (GFP-4.1B ΔFERM) completely abolished the interaction of SynCAM1 and protein 4.1B.
As our assays show co-localization and not strictly a biochemical interaction, we performed immunoprecipitation experiments. Immobilization of HA-SynCAM1 on protein-A-sepharose beads led to the recovery of 4.1B-GFP from transfected COS7 cells (Fig.3E, lane 4, Bound). In contrast, HA-SynCAM1ΔFERMb failed to co-immunoprecipitate 4.1B-GFP (Fig.3E, lane 7, Bound), while deletion of the PDZ binding domain (HA-SynCAM1ΔPDZIIb) left the interaction with 4.1B intact (Fig.3, lane 8, Bound). Similarly, deletion of the FERM domain of 4.1B blocked recovery of 4.1B (Fig.3, lane 6, Bound), whereas deletion of the similar sized C-terminal domain (CTD; Fig.3B) did not affect interaction with HA-SynCAM1 (Fig.3E, lane 5, Bound). In conclusion, we have demonstrated that the interactions between SynCAM1 and protein 4.1B require the FERM binding domain of SynCAM1. Similarly, protein 4.1B’s localization to SynCAM1 at the membrane and its effect on NMDAR recruitment is dependent upon its FERM domain.
Protein 4.1B enhances synaptogenic properties of SynCAM1
Given the strong effect of protein 4.1B on the localization of NMDARs to sites of adhesion in the CAMRA, we decided to determine whether incorporation of protein 4.1B enhanced the development of a functional postsynaptic apparatus by using an HEK293 cell/neuronal co-culture assay (Fu et al., 2003). It was previously demonstrated that neurons co-cultured with non-neuronal cells expressing SynCAM1 formed functional presynaptic contacts onto those heterologous cells (Biederer et al., 2002; Sara et al., 2005). It is thought that SynCAM1 expressed in this fashion stabilizes contact with axon terminals through binding of its transynaptic partner located in those terminals. Activity from these stabilized axons can then be sensed at the HEK cell via co-transfected glutamate receptors. If a molecule such as 4.1B should have the ability to enhance the localization of functional glutamate receptors to the sites of contact with the neurons, then we reasoned that this effect would be reflected in the properties of the mEPSC’s measured in this assay.
First, we transfected HEK293 cells with SynCAM1 and NMDAR subunits (NR1 and NR2B) and co-cultured these cells with cerebellar granule neurons. Electrophysiological analysis verified the existence of synaptic currents resembling endogenous neuron-neuron synaptic responses (Fig.4A). Nearly 53% (52.6.± 2.8%) of HEK293 cells transfected with NMDAR subunits and SynCAM1 showed miniature excitatory postsynaptic currents (mEPSCs) in the presence of TTX (Fig.4B), while mEPSCs could only be detected in 9% (8.4± 1.2%) of HEK293 cells transfected with only the NMDARs and GFP. Due to the high affinity of NMDARs for glutamate (Hollmann and Heinemann, 1994), currents detected are not strictly synaptic. Thus, while this measure surely reflects the formation of functional presynaptic terminals onto HEK293 cells, further experiments are necessary to interpret whether NMDARs are specifically opposed to these terminals (see below).
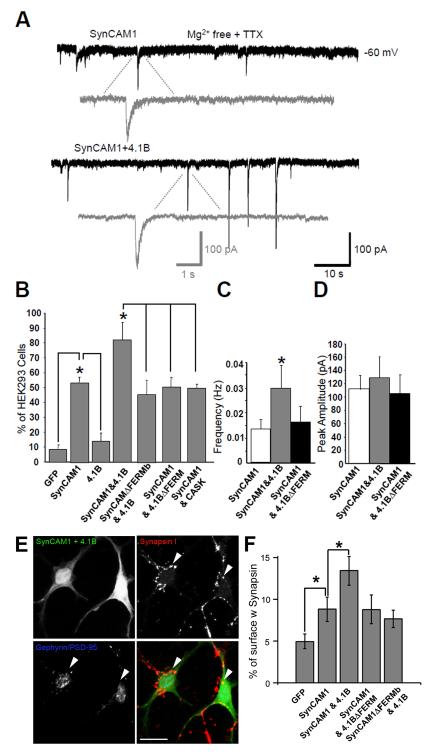
Fig. 4.
Protein 4.1B specifically enhances measures of NMDA-EPSCs in the HEK293 cell/neuronal co-culture assay. (A) Representative recordings of NMDA-mEPSCs from HEK293 cells transfected with either SynCAM1 and the NMDAR subunits (NR1/NR2B, top), or SynCAM1, 4.1B and the NMDAR subunits (bottom). The synaptic currents were measured in Mg2+-free extracellular solution with TTX. Gray traces are magnified individual NMDAR-mEPSCs. (B) Percentage of transfected HEK293 cells with recordable NMDA-mEPSCs in each experimental condition. SynCAM1 significantly enhanced recorded NMDA-mEPSCs over control conditions (p < 0.05, n >20). Protein 4.1B together with SynCAM1 significantly increased the number of cells with recordable currents over SynCAM1 alone conditions (p < 0.05, n > 20), while perturbing interactions mediated via the FERM domain canceled this increased activity. (C) Protein 4.1B significantly increased the frequency of NMDA-mEPSCs when compared to SynCAM1 alone conditions or the 4.1BΔFERM mutant (p < 0.05, n = 10). (D) Cotransfection of protein 4.1B did not significantly increase the amplitude of NMDA-mEPSCs (p = 0.09, n = 10). Error bars represent s.e.m. (E) Cotransfection of protein 4.1B significantly enhanced presynaptic stabilization. Immunolabeling of HEK293 cells transfected with SynCAM1 and 4.1B(upper left), co-cultured with neurons and labeled for synapsin I (upper right), PSD-95 and Gephryin (lower left). Arrows indicate regions of induced presynaptic contact where markers of postsynaptic structures are missing. Scale bar equals 20 μM. (F) Quantification of the percent surface area of transfected HEK293 cells covered with synapsin I, and not PSD-95 or Gephyrin, labeling under different transfection conditions. SynCAM1 significantly enhances percent surface covered with synapsin I over GFP conditions (* = p < 0.02, n = 17) and SynCAM1 plus 4.1B significantly increases percent surface area covered by synapsin I over SynCAM1 alone conditions (* = p < 0.02, n = 17). Error bars represent s.e.m.
The addition of protein 4.1B to SynCAM1 and NMDAR transfected HEK293 cells resulted in a 30% increase in the number of cells with recordable NMDAR-mEPSCs over SynCAM1 alone (82.2 ± 11.8%, p < 0.05, n = 20; Fig.4A and B). The increase in the percentage of cells with detectable mEPSCs was abolished by either removing the FERM binding domain in SynCAM1 (SynCAM1ΔFERMb) or the FERM domain from protein 4.1B (4.1BΔFERM; Fig. 4B). Similarly, addition of CASK, an effector molecule that interacts with SynCAM1 (Fig.2C and Supplemental Fig.1), but that did not significantly recruit NMDARs in the CAMRA (Fig.2B), did not increase the number of cells with detectable NMDAR mEPSCs above SynCAM1 alone (Fig.4B). These results suggest that 4.1B may either be increasing the localization of NMDARs to synaptic sites, as suggested by the CAMRA, or that protein 4.1B may act to increase the adhesive nature of SynCAM1 and thereby improve the formation of presynaptic terminals onto HEK293 cells.
Detailed analysis of the NMDAR-mediated mEPSC events revealed that the addition of 4.1B enhanced mEPSC frequency onto HEK293 cells (over 200% vs. SynCAM1 alone, p < 0.05; n = 14; Fig.4A and C). This effect was abrogated by deletion of the SynCAM-binding FERM domain (Fig.4C) and was not elicited by CASK (data not shown). Despite these strong effects on mEPSC frequency, it was surprising that we did not observe a significant increase in the peak amplitude of NMDA-mEPSCs as we expected given the CAMRA results (Fig.4D). However, we noticed that in about 35% of HEK293 cells co-expressing SynCAM1 and protein 4.1B a proportion of NMDAR-mEPSC events were larger than any observed in other experimental groups (vs. SynCAM1 only, p = 0.085, n = 10; Fig.4A, lower panel, and D). One possibility for the lack of the anticipated effect in this assay is that it is extremely difficult to normalize the size of miniature events relative to total receptor pool in a way that adequately addresses cell to cell variability in expression levels of receptors. Therefore, we consider this feature a limitation to determining the levels of NMDARs at synaptic sites in transfected HEK293 cells. However, this limitation is addressed if the other measures obtained in the assay are interpreted in light of the direct recruitment measurements calculated in the CAMRA, where we can more specifically measure localization and levels of NMDARs at sites of adhesion.
As mentioned, NMDARs have a very high affinity for glutamate and may well be detecting presynaptically-released glutamate at non-synaptic sites in addition to those closely juxtaposed to axon terminals. The results of the CAMRA (Fig.2) strongly support that protein 4.1B facilitates an increase in the concentration of NMDARs at sites of contact, however, it does not exclude the possibility that 4.1B may also enhance the stabilization of functional presynaptic terminals trans-synaptically via SynCAM1. To test whether 4.1B’s effects on the frequency of mEPSCs in the HEK293 co-culture study was at least partially due to a change in the number of stabilized presynaptic terminals, we labeled the co-cultured HEK293 cells with antibodies to SynapsinI, PSD-95 and Gephyrin. We then quantified the area of SynapsinI-positive regions located on transfected HEK293 cells, but eliminated regions that colocalized with the postsynaptic markers PSD-95 or Gephyrin to exclude endogenous neuron-neuron synapses (Biederer and Scheiffele, 2007). We found that SynCAM1 expressed in HEK293 cells significantly increased the proportion of SynapsinI positive surface area as compared to GFP transfected only (8.9±1.5% vs. 4.9±0.9%, p < 0.02, n=17; Fig.4F) confirming previous studies of SynCAM1’s ability to induce presynaptic differentiation on its own (Biederer et al., 2002; Sara et al., 2005). Surprisingly, the addition of 4.1B to SynCAM1 transfected HEK293 cells caused a significant increase in SynapsinI labeling relative to SynCAM1 only cells (8.9±1.5% vs. 13.9±1.7%, p < .02, n = 17; Fig.4E&F). Experiments using either of the deletion mutants, SynCAM1ΔFERMb or 4.1BΔFERM, showed that the FERM binding interaction was necessary for the increase in presynaptic stabilization (SynCAM1 vs. SynCAM1ΔFERMb + 4.1B: p = 0.26, n = 14; SynCAM1 vs. SynCAM1 + 4.1BΔFERM: p = .49, n = 13; Fig.4F). This result suggests that cytosolic protein 4.1B acts to enhance the formation of presynaptic terminals onto SynCAM1-expressing HEK293 cells.
If the enhancement of mEPSC frequency that 4.1B elicits can be wholly accounted for by the enrichment of presynaptic terminals that we measured with SynapsinI labeling, then co-transfected glutamate receptors of the AMPA type could register similar changes in mEPSC frequency when 4.1B is present in conjunction with SynCAM1. To test this, the co-culture analysis was also performed by transfecting GluR1 in place of NR1/NR2B. In this case, SynCAM1 alone was again sufficient to cause a significant increase in the percentage of cells with recordable AMPAR currents over control transfection conditions (p < 0.05, n = 8). However, the co-expression of protein 4.1B with SynCAM1 did not further increase the proportion of HEK293 cells with recordable AMPAR-mEPSCs (data not shown). This suggests that the observed morphological increase in presynaptic contact due to the addition of protein 4.1B (Fig.4F) was not enough to significantly enhance mEPSC detection via AMPARs. Taken together, the HEK/neuron co-culture studies, in conjunction with the CAMRA experiments, suggest that SynCAM1interacts with 4.1B to facilitate enhanced localization of functional NMDARs to sites of contact and to enhance presynaptic stabilization.
Specific effect of protein 4.1 family members on glutamate receptor recruitment
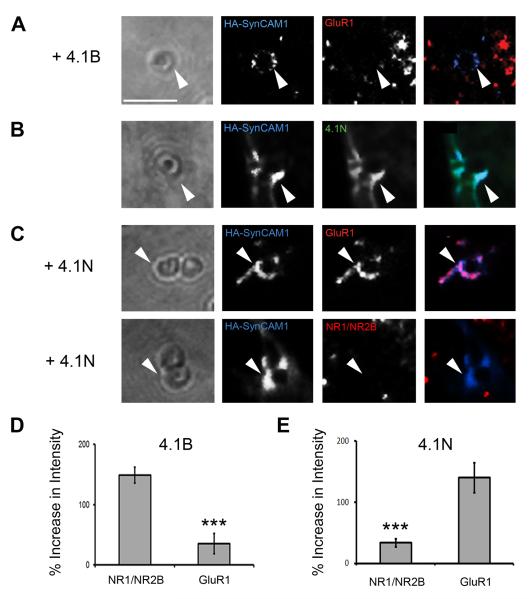
We decided to confirm the specificity of the SynCAM1-4.1B effect on NMDARs and not AMPARs using our CAMRA. When we co-expressed HA-SynCAM1, 4.1B and the GluR1 subunit in COS7 cells and applied microspheres directed to HA-SynCAM1, we found that there was significantly less clustering of GluR1 relative to the induced recruitment of NR1/NR2B described earlier (34.9 ± 16.8% vs. 148.7 ± 13.3%, p < 0.001, n = 15; Fig.5A & D). This suggests there are specific mechanisms by which protein 4.1B recruits only NMDARs (NR1/NR2B).
Fig. 5.
Specificity of 4.1 effector proteins to glutamate receptor recruitment. (A) Protein 4.1B did not significantly enhance recruitment of AMPA type receptors (GluR1) as compared to NR1/NR2B type receptors (arrowhead). Scale bar equals 2 μm. (B) Protein 4.1N was recruited to sites of HA-SynCAM1 accumulation in contact with microspheres (arrowheads). (C) Protein 4.1N induced significant recruitment of GluR1, but not NR1/NR2B, containing receptors to adhesion sites at microspheres (arrowheads). (D) Quantification of GluR1 recruitment vs. NR1/NR2B recruitment via protein 4.1B (34.9 ± 16.8% vs. 148.7 ± 13.3%, p < 0.001, n = 15). (E) Quantification of GluR1 recruitment vs. NR1/NR2B recruitment via protein 4.1N (139.8 ± 24.4% vs. 33.4 ± 6.95%, p < 0.001, n = 15). Error bars represent s.e.m.
To determine whether lack of recruitment of GluR1 is not a deficit of the CAMRA, we considered generating an alternative adhesion complex in COS7 cells that might recruit GluR1. Protein 4.1B belongs to a family of proteins that were first identified in erythrocytes (An et al., 1996) and later found to have a significant role in stabilizing the cytoskeleton by promoting the association of F-actin with tetrameric spectrin (Correas et al., 1986). The family members, coded for by distinct genes, are differentially expressed in distinct cell populations (Hoover and Bryant, 2000; Parra et al., 2000; Peters et al., 1998), but all may potentially interact with SynCAM1 via their conserved FERM domains. In particular, 4.1N is an additional family member highly expressed in neurons that localizes to postsynaptic specializations (Scott et al., 2001). Protein 4.1N interacts directly with GluR1 and promotes its recruitment to postsynaptic densities and to the plasma membrane (Shen et al., 2000). Therefore, we determined whether SynCAM1 / 4.1N could form an adhesion complex at microspheres and whether this would promote GluR1 clustering (Fig.5B, C and E). As predicted, protein 4.1N is recruited to HA-SynCAM1 clusters at microspheres (100 ± 12.7%, n = 15; Fig.5B). Additionally, GluR1 specific clustering was induced by this complex, while NR1/NR2B could not be recruited to HA-SynCAM1/4.1N adhesion sites (139.8 ± 24.4% vs. 33.4 ± 7%, p < 0.001, n = 15; Fig.5C & E). Thus, our experiments identify an additional SynCAM1 effector molecule, protein 4.1N. Our results further suggest that NMDAR and AMPAR recruitment specificity may be mediated by differential CAM / effector adhesion complexes.
Synaptic localization of protein 4.1B
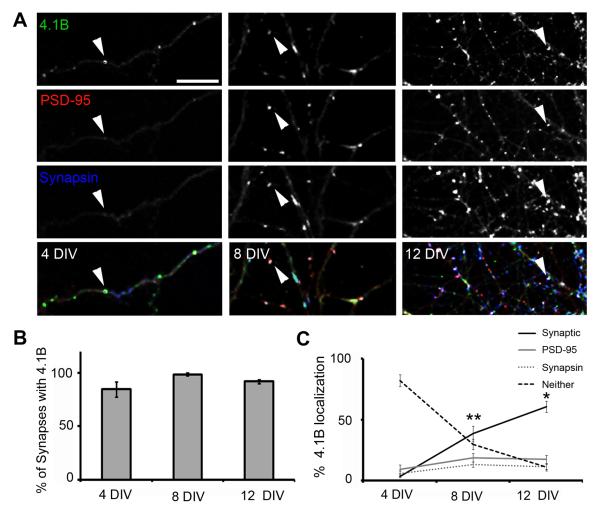
Intrigued by these initial observations in the COS7 and HEK293 cell culture assays, we expanded on these experiments to determine the relevance of protein 4.1B function in a neuronal context. Previous biochemical studies have reported that 4.1B is detected in PSD fractions prepared from rat forebrain, and could therefore be localized to excitatory synapses in vivo (Scott et al., 2001). To confirm synaptic localization of 4.1B, we immunolabeled cultured hippocampal neurons with antibodies to 4.1B and the markers Synapsin I and PSD-95 at 4, 8 and 12 days in vitro (DIV). Synapsin I is a protein that associates with synaptic vesicles and is thus located in presynaptic terminals, while PSD-95 is located in the glutamatergic postsynaptic density (Cho et al., 1992; Fletcher et al., 1991; Hunt et al., 1996). The immunolabeling of protein 4.1B revealed a highly punctate distribution at all ages examined (Fig.6A). At 4DIV, an age at which very few synapses have formed (Washbourne et al., 2002), protein 4.1B was localized in distinct puncta (3.7 ± 0.5 puncta/20μm, n = 10), some of which (3.4 ± 1.2% of total 4.1B puncta) were colocalized with the few synapses present, i.e. sites of colocalization of Synapsin I and PSD-95. The vast majority (81.9 ± 4.9 puncta/20μm, n = 10) were not colocalized with either Synapsin I or PSD-95 (Fig.6A,C) as 4.1B puncta far outweighed the presence of either Synapsin or PSD-95 puncta. However, almost all synapses present in the cultures at 4DIV (and at all other time points examined) demonstrated colocalization of protein 4.1B immunoreactivity (84.6 ± 7.7%, 98.5 ± 0.8%, 91.6 ± 2.1% of synapses at 4, 8 and 12 DIV, respectively: Fig.6B). Thus, as the number of synapses increased during development in vitro, the distribution of 4.1B puncta switched from not being associated with either pre- or postsynaptic markers to being colocalized with both Synapsin I and PSD-95 (Fig.6C). Interestingly, the intensity of the puncta decreased with age (Supplementary Figure 2), suggesting that the presence of protein 4.1B at synapses decreases with increasing maturity. Taken together, these results strongly suggest a function for 4.1B at synapses, and that protein 4.1B may play an important role during the early phases of synapse formation.
Fig. 6.
Localization of endogenous protein 4.1B in cultured hippocampal neurons. (A) Immunolabeling of protein 4.1B (green) with the synaptic markers PSD-95 (red) and synapsin I (blue) at 4, 8 and 12 DIV. Arrowheads indicate examples of protein 4.1B localization. Synaptic sites, areas enriched in all three proteins are white in the merged image (bottom row). Scale bar equals 10 μm. (B) Quantification of the percentage of synapses that show protein 4.1B localization over time in culture. Error bars represent s.e.m. (C) Quantification of the distribution of protein 4.1B puncta as determined by colocalization with synapsin I, PSD-95, both (Synaptic) or neither markers. Error bars represent s.e.m. (* p < 0.05, ** p < 0.01, n = 10).
Protein 4.1B enhances synaptic localization of NMDARs in hippocampal neurons
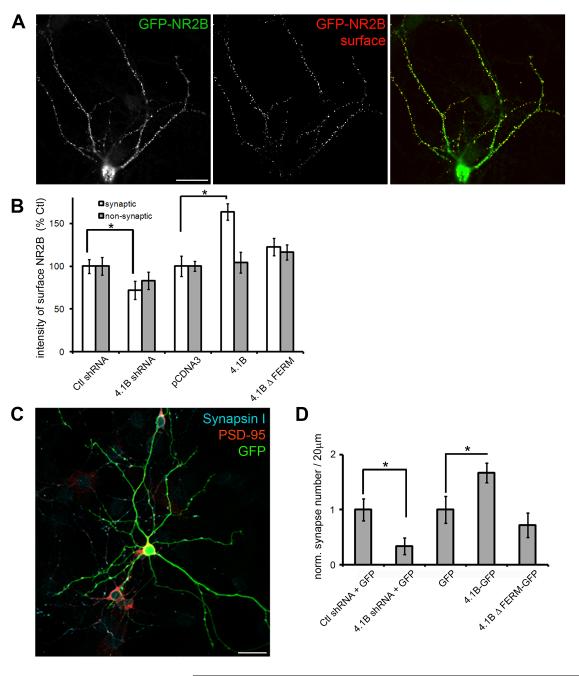
Because protein 4.1B is localized to synapses in hippocampal neurons (Fig.6), it was interesting to examine whether 4.1B could recruit NMDARs to “true” synapses, analogously to recruitment seen in the CAMRA and in HEK293 / neuron co-cultures (Fig.2 and 4). To specifically label surface exposed NMDARs, we transfected cultured hippocampal neurons at 7 DIV with a plasmid to express GFP-tagged NMDAR subunit 2B (GFP-NR2B). We also cotransfected expression plasmids for 4.1B or a short hairpin RNA (shRNA) construct to 4.1B, to manipulate the expression levels of protein 4.1B. 24 hours later, we labeled GFP-NR2B subunits present at the neuronal surface by exposing live, transfected neurons to GFP antibodies at 4°C (Washbourne et al., 2004b). Subsequently, neurons were fixed and permeabilized to visualize total GFP-NR2B (Fig.7A) and the presynaptic protein synaptophysin.
Fig. 7.
Protein 4.1B enhances NMDAR localization at synapses. (A) Transfected hippocampal neurons were incubated with antibodies to GFP (rb) at 4°C to label surface GFP-NR2B (surface, red) and subsequently fixed, permeabilized and reincubated with GFP antibodies (ms) to reveal total GFP-NR2B (total, green) and antibodies to synapsin I (not shown). Scale bar = 50 μm. (B) Quantification of the intensity of surface GFP-NR2B puncta at synaptic and non-synaptic sites in hippocampal neurons expressing shRNA to 4.1B (4.1B shRNA), control shRNA (Ctl shRNA), 4.1B and 4.1B ΔFERM and empty vector (pCDNA3) for 24 hours. Error bars represent s.e.m. (* p < 0.05). (C) Neurons expressing shRNA to 4.1B (4.1B shRNA) and GFP, control shRNA (Ctl shRNA) and GFP, 4.1B-GFP, 4.1B ΔFERM-GFP or GFP alone for 48 hours were immunolabeled with antibodies to synapsin I and PSD-95. Scale bar = 50 μm. (D) Quantification of the numbers of synapses per 20 μm of dendrite as determined by the colocalization of synapsin I and PSD-95. Synapse number was normalized to control conditions. Error bars represent s.e.m. (* p < 0.05, n = 11).
We first characterized the effectiveness of the shRNA. Expression of the 4.1B shRNA resulted in a 25% decrease in the intensity of 4.1B puncta 24 hours after transfection (from 9216 ± 621 a.u. to 6895 ± 423 a.u. for ctl and 4.1B shRNA, respectively, n = 11, p < 0.05). This may be an underestimation of knock-down as we did not take into account diffuse dendritic and cell body levels. Furthermore, knockdown was only performed for 24 hours, a relatively short time period. This short time period was important as we did not want high levels of GFP-NR2B expression to compromise trafficking pathways to synapses and we targeted a time period in development when 4.1B production would be high. Despite the relatively weak reduction in 4.1B levels with this shRNA protocol, we measured a significant reduction in the normalized intensity of punctate surface GFP-NR2B at synapses, as determined by colocalization with synaptophysin, when compared to mismatch (Ctl) shRNA (71.8% ± 10.6 of ctl, n = 11, p < 0.05; Fig. 7B). This reduction in NR2B surface intensity was not apparent at non-synaptic sites (83.1% ± 10.2 of ctl, n = 11, p = 0.3; Fig.7B). We also measured the ratio of surface to total GFP-NR2B to determine whether 4.1B was involved in delivery of NMDARs to the plasma membrane, as has been suggested for 4.1N (Shen et al., 2000). The ratio of surface-labeled to total GFP-NR2B subunits was unchanged in all conditions (n = 11 per condition, p > 0.05), suggesting that 4.1B specifically recruits NMDARs to synapses, including both internal and surface pools.
In contrast to knock-down with shRNA, overexpression of protein 4.1B resulted in an enhancement of the surface localization of GFP-NR2B at synapses (163.4% ± 9.7 of ctl, n = 11, p < 0.05; Fig.7B). The increase in surface localization of NR2B to synapses was abrogated by deletion of the FERM domain (ΔFERM, 122.4% ± 10.1, n = 11, p = 0.064; Fig. 7B). These results suggest that protein 4.1B plays an important role in the delivery of NMDAR subunits to synapses in hippocampal neurons, and that the FERM domain through which it may interact with SynCAM1 is necessary for this function.
Protein 4.1B enhances synaptogenesis in hippocampal neurons
In our co-culture experiments between CGCs and HEK293 cells (Fig.4), we measured a significant increase in the area occupied by presynaptic terminals on HEK293 cells expressing SynCAM1 and 4.1B, and that this was accompanied by an increase in NMDAR mEPSC frequency and the percentage of cells with recordable mEPSCs. This suggests a great facilitation of functional synapse formation between neurons and HEK293 cells. To test whether protein 4.1B exhibited a similar effect on functional synapse formation in neurons, we either increased or decreased 4.1B expression levels for two days (from 6 DIV to 8 DIV) and immunolabeled neurons with antibodies to the pre- and postsynaptic markers SynapsinI and PSD-95, respectively (Fig.7C). Quantification of the numbers of synapses, i.e. sites of colocalization of Synapsin I and PSD-95, revealed that a reduction in protein 4.1B expression levels due to shRNA expression results in a 3-fold decrease in synapse number (from 1.13 ± 0.2 synapses/20μm to 0.38 ± 0.06 synapses/20μm for ctl and 4.1B shRNA respectively, n = 11, p < 0.05; Fig. 7D). In contrast, increasing 4.1B expression levels by expressing 4.1B-GFP increased synapse number 1.7-fold (from 0.88 ± 0.2 synapses/20μm to 1.48 ± 0.3 synapses/20μm for GFP and 4.1B-GFP, respectively, n = 11, p < 0.05; Fig.7D). This increase in synapse number was completely abolished by deletion of the FERM domain (4.1B ΔFERM-GFP, 0.64 ± 0.1 synapses/20μm, n = 11, p > 0.05; Fig.7D). These results indicate that 4.1B is sufficient and necessary to drive the formation of synapses between hippocampal neurons in culture and that this activity is dependent on the FERM domain.
Protein 4.1B specifically enhances NMDAR currents in hippocampal neurons
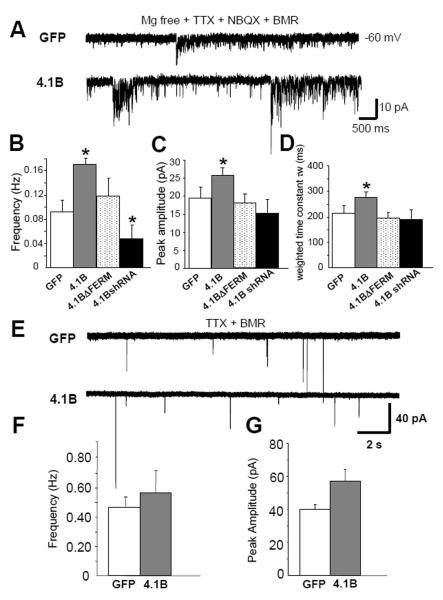
To further characterize the potential role of protein 4.1B on NMDAR recruitment in neurons, we analyzed the functional consequences of manipulating protein 4.1B expression levels in dissociated hippocampal neuronal culture. We were interested in whether protein 4.1B exacted effects analogous to those observed in our other assays. We recorded NMDA-mEPSCs in the presence of TTX, NBQX and BMR in a solution lacking Mg2+, as previously described in detail (Fu et al., 2005). To minimize variability due to cell heterogeneity and to measure synaptic currents with optimal space clamp and resolution, we selected hippocampal neurons with relatively small cell body size (15-20μm) and simple dendritic arborization. We compared recordings from neurons expressing either GFP, 4.1B-GFP, 4.1BΔFERM-GFP or 4.1B shRNA with GFP at 12-14 DIV (Fig.8). Protein 4.1B overexpression significantly increased both mean frequency (0.09 ± 0.02 Hz in GFP cells, 0.17 ± 0.01 Hz in 4.1B-GFP expressing cells, p < 0.05; n >30) and amplitude of NMDA-mEPSCs as compared to GFP expressing cells (19.2 ± 3.3 pA in GFP cells, 26.4 ± 2.7pA in 4.1B-GFP cells, p < 0.05; n >30; Fig.8A-C). Deletion of the FERM domain of 4.1B prevented the increase in frequency and amplitude (0.12 ± 0.03 Hz, 18.4 ± 2.3pA, respectively). The expression of 4.1B shRNA significantly decreased the frequency of NMDAR mEPSCs (0.05 ± 0.02 Hz), but not peak amplitude (15.6 ± 3.7 pA, p = 0.09, n > 30). It is possible that the knock-down by the 4.1B shRNA was not sufficient to significantly reduce the NMDAR mEPSC peak amplitude. However, our results are consistent with the idea that 4.1B plays a role in recruiting NMDARs to synapses in cultured hippocampal neurons.
Fig. 8.
Protein 4.1B enhances NMDAR mediated synaptic events in cultured hippocampal neurons. (A) Representative traces of isolated NMDA-mEPSCs recorded from hippocampal neurons (12-14 d.i.v) in Mg2+ free solution with TTX (0.5 μM), NBQX (5 μM) and BMR (50 μM) under different transfection conditions. (B) Overexpression of protein 4.1B increases and knock-down of 4.1B with shRNA decreases NMDA-mEPSC frequency (p < 0.05, n >30) (C) amplitude (p < 0.05, n >30) and (D) τw (p < 0.05, n >30) in hippocampal neurons. (E) Representative traces of isolated AMPA-mEPSC recordings in hippocampal neurons in presence of TTX (0.5 μM) and BMR (50 μM) in regular ECS with Mg2+ (F) 4.1B-GFP did not significantly affect the frequency of AMPA-mEPSCs (p = 0.79, n=16) or (G) amplitude (p = 0.51, n=16). Error bars represent S.E.M.
The NR2B subunit of the NMDAR has been shown to be more prevalent at synapses during development, with NR2A gradually taking over at mature synapses (Tovar and Westbrook, 1999). NR2B subunits cause a slow decay time component in NMDAR current kinetics (Kohr and Mody, 1994; Kohr and Seeburg, 1996; Monyer et al., 1994). Upon analysis of the decay time constant of NMDA-mEPSCs (τw) from our 4.1B-expressing cultured hippocampal neurons, we noticed that decay was significantly prolonged when compared to control transfected cells (212.5 ± 32.9 ms in GFP control cells, 275.6 ± 25.2 ms in 4.1B expressing cells, p < 0.05; n >30; Fig.8D). No significant effect on τw was seen in neurons expressing 4.1BΔFERM or 4.1B shRNA (198.6 ± 21.8 ms in 4.1BΔFERM cells, 188.3 ± 38.4 ms in 4.1B shRNA cells, p = 0.58; Fig.8D). The effect on τw indicates that protein 4.1B might specifically increase the recruitment of NR2B-containing NMDARs to synaptic sites.
To further test this hypothesis, we recorded mEPSCs in the presence of ifenprodil (10 μM), as it is a specific antagonist of NR1/NR2B-containing NMDARs (Mott et al., 1998). Changes in our measures in the presence of ifenprodil should then reflect the proportion of the NMDARs containing only NR1 and NR2B subunits. Application of ifenprodil to neurons overexpressing 4.1B resulted in a 73% (73 ± 5%, n = 12) decrease in mEPSC frequency compared to before ifenprodil application (Supplementary Fig. 3). This inhibition was only marginally decreased in the GFP only expressing neurons (69 ± 2%, n = 12, p > 0.05). This suggests a potentially small increase in the NR2B-only population of NMDARs when 4.1B is overexpressed. However, we did measure a significant difference between 4.1B overexpression and 4.1B knock-down with shRNA (60 ± 4%, p = 0.03). Taken together, the small changes in ifenprodil sensitivity might reflect the presence of heterotrimeric NR1/NR2A/NR2B containing NMDARs, which are not as sensitive to ifenprodil as NR1/NR2B NMDAR subtypes. Nevertheless, our results are consistent with the idea that protein 4.1B enables the recruitment of NMDARs containing NR2B subunits to synapses.
To test whether the effects of protein 4.1B were specific to NMDARs and not to AMPARs, we recorded from transfected hippocampal neurons in the presence of BMR and TTX. No significant effects were seen for AMPAR-mEPSC mean frequency (0.48 ± 0.08 Hz in GFP cells, 0.56± 0.14 Hz in 4.1B-GFP overexpressing cells, p = 0.79; n = 16) or amplitude of mEPSCs (40.1 ± 10.3 pA in GFP cells, 57.8 ± 13.7pA in 4.1B-GFP cells, p = 0.51; n = 16; Fig.8E-G). Taken together, these results demonstrate that protein 4.1B specifically recruits NMDARs, and not AMPARs, to synapses; the same glutamate receptor specificity as characterized in our CAMRA and HEK293 co-culture assays.
DISCUSSION
While a large number of proteins have been localized to the postsynaptic density and are thought to contribute towards its development and function (Reviewed in (Dillon and Goda, 2005; Feng and Zhang, 2009), it remains unclear which molecular interactions are sufficient to recruit glutamate receptors to synaptic sites early in development. In particular, the relatively novel synaptic cell adhesion molecule SynCAM1 has yet to be shown important for postsynaptic development and, thus, even less is known about its potential interactions leading to receptor recruitment. Here, we have used a microsphere-based assay, the CAMRA, and an HEK293 / neuronal co-culture assay as effective tools to identify SynCAM1 effector molecules sufficient to induce glutamate receptor recruitment. We identified protein 4.1B as a specific and potent effector of NMDAR recruitment to SynCAM1 adhesion sites. We also observed that other potential effectors could interact with SynCAM1 (CASK, Syntenin1), but these molecules did not impact NMDAR recruitment to the extent of 4.1B in our assays. Furthermore, in an attempt to determine the degree of specificity that SynCAM1 and protein 4.1B have on the recruitment of glutamate receptor types, we identified 4.1N as an additional SynCAM1 effector molecule sufficient to differentially recruit AMPA type receptors.
In this set of studies we were also able to demonstrate three key findings regarding the function of protein 4.1B. Specifically, protein 4.1B (1) facilitated the direct aggregation of NMDARs at sites of contact and adhesion, (2) enhanced morphological measures of presynaptic differentiation, and (3) specifically enhanced the frequency and amplitude of NMDAR, not AMPAR, mediated mEPSCs in neurons. Thus, this is the first report of a role for protein 4.1B at excitatory synapses and is the first demonstration of a potential mechanism by which SynCAM1 may directly participate in the early developmental process of postsynaptic differentiation.
The results obtained in COS7 cells and HEK293 cell/neuron co-culture help clarify that protein 4.1B largely promotes the development of functional postsynaptic structures in neurons containing NMDARs with a very specific composition. Overexpression of protein 4.1B in neurons enhanced surface presentation of NMDARs at synapses (Fig.7), and increased the peak amplitude of NMDAR-mediated mEPSCs (Fig.8), but not that of AMPAR-mediated mEPSCs. In keeping with this, knock-down of 4.1B with shRNA resulted in a decrease in synaptic localization of NMDARs (Fig.7). Knock-down did not affect the peak amplitude (Fig.8), however this may be due to insufficient knock-down in this assay. Nonetheless, our results suggest potent effects of protein 4.1B specifically on NMDAR recruitment at the postsynaptic specialization.
Interestingly, the combined data from our assays also strongly support that protein 4.1B possesses a role in the stabilization of functional presynaptic terminals via trans-synaptic interactions. Our studies using the HEK293 cell / neuronal co-culture assay and the overexpression studies in hippocampal neurons support that postsynaptically localized protein 4.1B exerts an effect on presynaptic differentiation or release in addition to its effects on receptor recruitment. For example, the increase in mean mEPSC frequency in hippocampal neurons and HEK293 cell/neuron co-culture support modulation of presynaptic properties. This physiological result is directly supported by the observed increase in labeling of presynaptic terminals in the HEK293cell / neuron co-culture study. However, we repeatedly fail to observe an increase in AMPA mediated mEPSCs in every assay. This strongly implies that protein 4.1B’s NMDAR-specific recruitment capabilities interact with its ability to enhance presynaptic differentiation to impact these measures of mEPSCs. Regardless, protein 4.1B acts to enhance the adhesive nature of SynCAM1 contact sites, resulting in more release sites, and may additionally modulate vesicle release properties in a retrograde fashion through SynCAM1 adhesion to mediate presynaptic differentiation. Thus, protein 4.1B may play a partial role in stabilization of presynaptic structures, but further experimentation will be needed to clarify its trans-synaptic role in neurons (Sara et al., 2005).
Given that protein 4.1B, a molecule most notably involved in cytoskeletal organization (Sun et al., 2002), exerted such a direct and strong effect on NMDAR recruitment to synapses, it is interesting to consider how it may perform its role at the postsynaptic density. 4.1 family proteins regulate actin dynamics via direct binding through their spectrin-actin binding (SAB) domain. Furthermore, NMDAR localization to synaptic sites requires significant actin stabilization, while non synaptic clusters of NMDARs can be maintained even when the actin cytoskeleton is destabilized (Allison et al., 1998). Moreover, spectrin has even been reported to directly bind NMDARs in the brain (Wechsler and Teichberg, 1998). As our assay revealed differential receptor recruitment activity of two 4.1 family members, 4.1B and 4.1N, their functions at the synapse are not simply explained by the possession of a single protein-protein binding region such as the SAB domain. It will be important in future studies to determine which domains differentially modulate NMDAR versus AMPAR recruitment and which processes are actin dependant and independent.
The specificity of the recruitment of NR2B subunit containing NMDARs, as measured by the weighted decay constant τw (Fig. 8), is particularly interesting, as recruitment of the NR2B subunit is especially relevant to early developmental processes. NR2B subunits are more prevalent at synapses than NR2A early during development (Stocca and Vicini, 1998; Tovar and Westbrook, 1999). Furthermore, scaffolding molecules such as SAP102 and PSD-95 appear to mediate the NR2B to NR2A switch that is a key feature of synaptic maturation (van Zundert et al., 2004). It is possible that 4.1B must now also be considered in playing a role in this process. However, consistent with the role for protein 4.1B at “young” developing synapses, we found that the endogenous localization of protein 4.1B to excitatory synapses in neuronal culture appeared very early (4 DIV) and the intensity at synapses dropped off with time (Fig. X). Previous in situ hybridization studies in newborn mouse brain show specific protein 4.1B expression in the Purkinje cell layer of the cerebellum, regions CA1 and CA3 of the hippocampus and throughout the cortex (Parra et al., 2000). Many of these regions are primarily the targets of major glutamatergic inputs and so it is interesting to speculate that protein 4.1B regulates excitatory postsynaptic differentiation in these areas before or near birth.
The protein structure of the 4.1 family proteins has been intensively studied, and it is clear that in addition to the FERM, CTD and SAB domains, critical Ca2+-sensitive and insensitive calmodulin binding domains regulate the association between 4.1 proteins and transmembrane proteins in erythrocyte membranes (Nunomura et al., 2000). Ca2+/calmodulin is known to be a key regulator of processes underlying synapse formation such as actin cytoskeleton dynamics (Konur and Ghosh, 2005; Oertner and Matus, 2005; Saneyoshi et al., 2008) and glutamate receptor activity and localization (Ehlers et al., 1996; Wyszynski et al., 1997). This suggests that a 4.1 family member’s activities and dynamics would be subject to the specific regulatory mechanisms known to affect glutamate receptor recruitment to the synapse and subsequent synaptic maturation. Such dynamics have not yet been reported for 4.1 molecules as they have been for other synaptic proteins (Sharma et al., 2006), but would further support the idea that protein 4.1B could act as a regulated developmental signal central to the specific localization of glutamate receptors.
Interestingly, we demonstrated that both family members (4.1B and 4.1N) were readily recruited to adhesion sites via SynCAM1. We assume that 4.1N also binds to SynCAM1through its FERM domain, as it is about 73% identical to the FERM domain of 4.1B (Parra et al., 2000). However, 4.1N and 4.1B show completely opposite specificities for glutamate receptor subtypes (Fig. 5). 4.1N is known to bind to GluR1 through the CTD (Shen et al., 2000), a domain that is also 73% identical to the CTD of 4.1B (Parra et al., 2000). This means that enough amino acids have changed between 4.1N and 4.1B in the CTD to switch binding from AMPAR subunits to NMDAR subunits. Alternatively, the ~30% amino acid difference between 4.1B an 4.1N can simply abrogate binding of 4.1B to AMPARs and other domains have acquired the ability to bind NMDARs directly or indirectly. 4.1B has at least three additional domains (U1, U2 and U3) that show significant sequence differences compared with other 4.1 proteins. These domains have no identified protein interaction or regulatory roles as yet. We hypothesize that these U domains might play a role in positively regulating the interaction of 4.1B with NMDARs but not with AMPARs. Future studies investigating the effects of 4.1B deletion constructs will provide insight into these possibilities.
We believe this study highlights the effectiveness of the CAMRA in identifying postsynaptic effector molecules of CAMs. However, in this study we have examined only tripartite complexes consisting of SynCAM1, effector and receptor. It is important to consider the possibility that some molecules may exert a strong effect on receptor recruitment, but only in the presence of additional effectors. For example, we observed no significant effect of CASK on NMDAR recruitment. Nevertheless, it was recently discovered that CASK plays a significant role in trafficking of NMDARs to the membrane surface and their localization to synapses in cultured hippocampal neurons (Jeyifous et al., 2009). However, this activity of CASK requires that it work in tandem with SAP97 and it remains unclear whether interactions with SynCAM1 or other CAMs at the final stages of synaptic targeting are relevant. Thus, further combinatorial studies may yet uncover additional effector molecules of SynCAMs that are capable of recruiting glutamate receptors or other synaptic components to nascent synaptic contacts.
SUMMARY
Using a cell adhesion molecule / receptor recruitment assay or CAMRA we have identified the 4.1 proteins, 4.1B and 4.1N, as postsynaptic effector molecules of SynCAM1. Our studies have revealed three important and novel findings: 1) SynCAM1 interacts with protein 4.1B to directly recruit NMDARs shortly after synaptic-like contact; 2) Postsynaptic protein 4.1B enhances presynaptic differentiation through SynCAM1; and 3) proteins 4.1B and 4.1N differentially regulate glutamate receptor recruitment to sites of adhesion. Studies in cultured hippocampal neurons suggest that 4.1B plays an important role in the recruitment of NR2B-containing NMDARs to synapses during development. Thus, these studies delineate a novel function for 4.1 proteins as SynCAM1 effectors in the recruitment of specific glutamate receptor types to synapses.
METHODS
Expression vectors and constructs
Human 4.1B cDNA was obtained from Irene Newsham (University of Texas, Houston, TX). Deletions of the FERM domain (ΔFERM; amino acids 106-302) and the C-terminal domain (ΔCTD; amino acids 894-1097) were performed by PCR. Full length and the deletion mutants were subcloned into pEGFP-N1 (Clontech, Mountain View, CA) and pCDNA3. To generate shRNA to mouse 4.1B, the following sequence of 4.1B was subcloned into the pSuper vector (Brummelkamp et al., 2002): 5′- CGTGACCGGCTTCGAATAA -3. For control shRNA, the following sequence was used: 5′ — GATCTGAAGGCGCCTATAC — 3′. HA-SynCAM1 was obtained by PCR from mouse cDNA using the following primers: gccgaagcttatggcgagtgctgtgctgccg and tcgggaattcctagatgaagt actctttctttcttcgg. An HA tag and SalI site were inserted using the megaprimer PCR technique: cttctccttcaatcactgtagcgtagtctgggacgtcgtatgggtagtcgactttag taaacagattctgtcc. Deletion of the FERM binding domain (ΔFERMb; amino acids 376-389) was performed by megaprimer PCR: ggccgctattttgccgccaaaggagccgatgac. The PDZ binding domain (ΔPDZIIb; amino acids 415-418) was generated by PCR. The resulting HA-tagged constructs were transferred to pCDNA3. 4.1N-GFP was a gift from Richard Huganir (Johns Hopkins University, Baltimore, MD). GFP-NR2B was provided by Anne Stephenson (University College London, UK). Syntenin1-GFP was provided by Jeremy Henley (University of Bristol, UK). Myc-CASK was a gift from Ben Margolis (University of Michigan, Ann Arbor, MI) and GRIP1-GFP was obtained from Casper Hoogenraad (Erasmus Medical Center, Rotterdam, NL). GluR1-GFP has been described previously (Washbourne et al., 2002).
COS7 cell culture and transfection
COS7 cells (ATCC® Manassas, VA) were plated at a density of 50,000 cells per ml onto poly-L-lysine (Sigma, St. Louis, MO) coated glass coverslips and maintained in DMEM, 10%FCS, 25 units Penicillin and 25 μg streptomycin/ml. 24 hours later, or approximately at 60-70% confluency, cells were transiently transfected with 3.5 μg total DNA per well of a 12 well plate using lipofectamine 2000 (Invitrogen). Transfection reagent was added to wells containing DMEM, 10% FCS without pen/strep and incubated for 4-5 hours at which time the media was replaced by fresh DMEM, 10%FCS and 2mM kynurenic acid. Microsphere clustering is performed on live cells 28-30 hours later.
Microsphere preparation
100μl of ProteinA coated microspheres (~1μM diameter; Bangs laboratory, Inc) were resuspended in 750 μl of protein A/G buffer (0.1 M TE and 0.15 M NaCl, pH 7.5) and then centrifuged at 4°C, 10,000×g for 5 minutes. The supernatant was discarded and microspheres were washed similarly two more times. After the final wash, microspheres were resuspended in 50 μl protein A/G buffer and 50 μl COS7 maintenance media with target IgG (10μg α-HA, rabbit polyclonal, Bethyl Laboratories Inc., Montgomery, TX). Microspheres were incubated with antibody solution for 1hr at 4°C with gentle agitation and vortexing every 5-10 minutes. After incubation microspheres, were washed three times in the same manner as before using protein A/G buffer.
Microsphere application and surface immunolabeling
6-9 μl of prepared microsphere suspension was added to transfected COS7 cells in 1 ml of fresh media and plates were swirled gently to distribute microspheres. Microspheres were incubated with cells for 35-45 minutes at 37 °C. Live cells were then rinsed 2× gently with warmed 1×PBS, then fixed for 8 minutes in 1.5% PFA, 4% sucrose at 4°C. After fixation, coverslips were blocked with 10%BSA, 1% blocking reagent (Roche) for 30 minutes at room temperature. Primary solution, antibodies against HA (anti-mouse IgG1, 1:1000, Covance, Emeryville, CA) and GFP (chicken polyclonal, 1:1000, Chemicon-Millipore, Billerica, MA) in 5% BSA, 0.5% blocking reagent, was added for 45 minutes (to not more than 1 hour) at room temperature. Coverslips were washed and incubated in secondary antibodies for 35-40 minutes at room temperature (Alexa Fluor® 633 goat α-mouse IgG1 1:800, Alexa Fluor® 546 goat α-chicken IgG 1:800 Molecular Probes Eugene, OR). A control stain on cells with only internal GFP (GFP-NR2B alone without NR1 co-transfection or a GFP tagged-effector) was always performed in parallel to the described experiments. In the experiments where surface receptor accumulation was measured, the effectors were not GFP tagged. Additionally, CAM - Effector colocalization experiments were conducted independent of CAM — Effect or - Receptor experiments.
Hippocampal cell culture and immunolabeling
Medium density hippocampal cultures were prepared from embryonic day 19 Sprague Dawley rat pups as described (Brewer et al., 1993), with minimal modifications. Briefly, neurons were plated in plating media (10% FCS, 20 mM dextrose, 25 units Penicillin and 25μg streptomycin in MEM (Invitrogen/GIBCO)) at a density of 40,000-60,000 cells/ml on 12 mm coverslips coated with poly-L-lysine (Sigma, St. Louis, MO) and incubated for 4-5 hours. This media was changed for the remainder of culturing to a maintenance medium (Neurobasal medium (Invitrogen, Carlsbad, CA), 1× B-27 (Invitrogen), 0.5 mM Glutamax-I (Invitrogen), 50 units Penicillin/ 50μg Streptomycin (Sigma) and 0 .07% beta-Mercaptoethanol). Neurons were fed fresh maintenance media in half changes every three days in culture. This protocol was slightly modified in the preparation of the hippocampal neurons for the electrophysiology where neurons where derived from P1 mice. Endogenous staining of protein 4.1B (goat polyclonal to EPB41L3/DAL-1, 1:500 Abcam Inc. Cambridge, MA), PSD-95 (mouse monoclonal IgG2A clone 28/43, 1:250, James Trimmer — UC Davis/NIMH NeuroMab Davis, CA) and SynapsinI (rabbit polyclonal 1:800, Chemicon-Milipore, Billerica, MA) were performed on hippocampal neurons from 4 to 12 d.i.v. Cells were fixed 10 minutes in 4% PFA + 4% sucrose at 4°C and then 5 minutes in 100% MeOH at −20 °C, permeablized in 0.25% Triton X-100 for 5 minutes at room temperature and blocked for 1hour in 10%BSA+ 1% Blocking reagent (BR; Roche). Cells were incubated in primary solution (antibodies + 3% BSA+ 0.3% BR) for 3 hours at room temperature and then in secondary antibodies for 45 minutes at room temperature (Alexa fluor® 546 Goat anti-mouse IgG2A and 633 Goat anti-rabbit 1:600 Molecular Probes® Eugene, OR and Cy™ 2 Donkey anti-Goat 1:600 Jackson Immuno Research, West Grove, PA.)
Surface immunolabeling of GFP-NR2B in Hippocampal cell culture
Hippocampal neuronal cultures were prepared as described previously, and then transfected at 7 d.i.v. with a 1:5 mixture of GFP-NR2B to untagged target plasmid: either control shRNA, 4.1BshRNA, 4.1B full length, 4.1B ΔFERMb or plasmid vector. A total of 4-5μg of plasmid DNA was transfected using the Calcium Phosphate transfection kit, ProFection® (Promega, Madison, WI). Neurons were incubated in precipitate for up to 1 hour and then placed back into fresh media with pen/strep and allowed to express for 24 hours. To label surface GFP-NR2B, total GFP-NR2B and presynaptic structures, transfected neurons were washed gently 2 times in fresh ACSF at room temperature and then fresh media without pen/strep plus anti-GFP antibody (rabbit polyclonal, 1:1000 Chemicon-Millipore, Billerica, MA) was added for 15-20 minutes at 4°C. Cells were gently washed afterwards 3× in 4°C PBS and then fixed with 4% PFA for 20 minutes at 4°C. Fixed cells were treated with 0.25% tritonX-100 for 5 minutes at room temperature, blocked in 10% BSA+1% Roche Blocking medium for 1 hour at room temperature. Secondary for the surface labeled GFP was added for 30 minutes at room temperature (Alexa fluor® 546 Goat anti-rabbit). After washing, primaries against GFP (chicken polyclonal, 1:1000, Chemicon-Millipore, Billerica, MA) and Synaptophysin (mouse monoclonal 1:1,500, SIGMA, Saint Louis, MO) were added for 1 hour at room temperature. Cells were washed again and then secondary against the primaries for total GFP and synaptophysin was applied for 45 minutes at room temperature (Alexa fluor® 488 Goat anti-chick and Goat anti-mouse IgG1,1:600 Molecular Probes® Eugene, OR). To confirm that we label surface GFP specifically in each experiment, we ensured that GFP only, and/ or GFP-4.1BFL controls did not have significant immune label in the 546 channel. Further all puncta that had signal in the 546 channel also contained 488 signal, confirming that we label both surface and total GFP.
All studies were conducted with approved protocols from both the University of Oregon Animal Care and Use Committee and the Georgetown University Animal Care and Use Committee, in compliance with NIH guidelines for the care and use of experimental animals.
Imaging, Quantification and statistical analysis
COS7 cells were imaged on an inverted Nikon TU-2000 microscope with an EZ-C1 confocal system (Nikon) with a 100× oil-immersion objective (1.45 NA). Cells were imaged blind to specific co-transfection conditions. Cells were chosen for analysis if there were clear examples of microspheres with accumulations of the target CAM at site of contact (an average of a 110 ± 11.8% increase in intensity of HA-NLG1 of HA-SynCAM1 was measured at microspheres relative to background levels in the cell where visible accumulation was scored). 14-15 microspheres from 14-15 individual cells were chosen from three independent experiments in each condition. All channels were scanned sequentially and in the same plane of focus as apparent CAM aggregation. Images obtained in a given channel were obtained at constant laser intensities and the gain adjusted to just below intensity saturation. Images were converted to bitmaps and average intensity was analyzed surrounding an individual microsphere in each channel using Image Pro Plus® software. Briefly, average intensity increase at a given microsphere was calculated as follows: (Imicrosphere − Ibackground) / Ibackground * 100% = “% increase in intensity at microsphere” (Fig.1B). Imicrosphere is the average intensity in an annulus immediately surrounding the microsphere that is equal in width to the radius of the microsphere (annulus 1 in Fig.1B). Ibackground is the average intensity in an annulus surrounding the first around the microsphere (annulus 2 in Fig.1B) plus the average intensity in the area of the microsphere itself. The data are expressed in percent intensity above background, and given our conservative estimates of what is signal we are more likely to have underestimated total protein accumulation rather than overestimated. We did not assume a normal distribution of our CAMRA data and given the sample size in each experimental condition, we applied the non-parametric Mann Whitney test in all comparisons. Bonnferroni’s correction was applied to correct for multiple comparisons in the NMDAR and effector CAMRA experiments. Ten planned comparisons were made among the control and treatment groups. Given this number of multiple comparisons, comparison-level significance was tested at an alpha level of 0.0051 for an experimental-level Type I error rate of 0.05. Significance is depicted in graphs as asterisks: * is p < 0.05, ** is p < 0.01 and *** is p < 0.005. The non-parametric Mann Whitney-U was also used to test for significance in the comparisons of neuronal expression data unless otherwise noted. All data are expressed as mean ± standard error of the mean.
Immunoprecipitation and western blotting
COS7 cells were transfected with the plasmids of interest and cultured for 24 hours. Cells were lysed in 250μl of lysis buffer (150mM NaCl, 50mM Tris-HCl, 0.5mM EDTA, 0.2% Triton X-100 and protease inhibitor tablets, pH7.4) for 45 minutes at 4°C with agitation. This mixture was centrifuged at 14,000 × g for 5 minutes at 4°C and the supernatant collected. 20 μl of the supernatant was used as the input sample. 115 μl was incubated with 3 μg of anti-HA (mouse monoclonal; Bethyl Laboratories) overnight at 4°C with rotation, and 115μl was incubated without antibody. The following day, 50 μl of protein-A-sepharose beads in lysis buffer was added to both samples and incubated for 2 hours at 4°C with rotation. Protein-A-sepharose beads were collected by centrifugation and washed 3 times in lysis buffer. Bound proteins were eluted with 100 μl Laemmli sample buffer, boiled, separated on an SDS-PAGE gel, transferred to nitrocellulose membranes and probed with either anti -HA rabbit (1:1000; Bethyl Laboratories) or anti -GFP rabbit (1:1000; Invitrogen).
CGC and HEK293 cell co-culture and transfection
Primary cultures of mouse cerebellar granule cells (CGC) were prepared from postnatal day 5-7 (P5-7) from C57Bl6 mice. Mouse pups were sacrificed by decapitation in agreement with the guidelines of the Georgetown University Animal Care and Use Committee. The cerebella were dissociated as described in (Gallo et al., 1987). Cells were dispersed with trypsin (0.25 mg/ml, Sigma, St. Louis, MO) and plated at a density of 1.1×106 cells/ml on glass coverslips (Fisher Scientific, Pittsburgh, PA) coated with poly-L-lysine (10 μg/ml; Sigma) in 35 mm Nunc dishes. The cells were cultured in basal Eagle’s medium supplemented with 10% bovine calf serum, 2 mM glutamine, and 100 μg/ml gentamycin (all from Invitrogen Corporation Carlsbad, CA), and maintained at 37°C in 5% CO2. The final concentration of KCl in the culture medium was adjusted to 25 mM (high K+). To achieve functional synapse formation, at 5 d.i.v. the medium was replaced with the low (5 mM) potassium medium (MEM supplemented with 5 mg/ml glucose, 0.1 mg/ml transferrin, 0.025 mg/ml insulin, 2 mM glutamine, 20 μg/ml gentamicin, Invitrogen, and cytosine arabinofuranoside 10 μM, Sigma) as previously described (Chen et al., 2000; Prybylowski et al., 2002). Human embryonic kidney 293 cells (HEK293; American Type Culture Collection, Rockville MD, ATCC No. CRL1573) were grown in Minimal Essential Medium (Gibco BRL, Gaithersburg, MD), supplemented with 10% fetal bovine serum, 100 units /ml penicillin (Gibco BRL), and 100 units/ml streptomycin (Gibco BRL), in a 5% CO2 incubator. Exponentially growing cells were dispersed with trypsin, seeded at 2×105 cells/35-mm dish in 1.5 ml of culture medium and plated on 12 mm glass cover slips. HEK293 cells after transfection were dispersed with trypsin and plated on CGC cultures at a density of 1×104 cells/12-mm coverslip. HEK293 cells were transfected as described in (Vicini et al., 1998) using a modification of the calcium phosphate precipitation technique. Briefly, mixed plasmids (3 μg total) were added to the dish containing 1.5 ml MEM culture medium for 12-16 hours at 37oC under 5% CO2. Greater than 80% of cells expressed all the plasmids transfected as assessed independently with pEGFP, pDsRED2 and pECFP plasmids (not shown; Clontech).
HEK293cell/cerebellar co-culture for synaptic labeling
Human embryonic kidney 293 cells were grown in Minimal Essential Medium (Gibco), supplemented with 10% fetal bovine serum in a 5% CO2 incubator. Exponentially growing cells were dispersed with trypsin, seeded at 2 × 105 cells/35-mm dish in 1.5 ml of culture medium. HEK293 cells were then transfected within 24hrs after splitting as described Fu et al. (2003). Briefly, mixed plasmids (3 μg total) were added to the dish containing 1.5 ml MEM culture medium for 12–16 h at 37°C. HEK293 cells after transfection were dispersed and plated on CGCs cultures (DIV5) at a density of 2 ×104 cells/12-mm coverslip. 2 days later, live cultured HEK293-neuron co-culture was fixed with 4% paraformaldehyde, 4% sucrose in PBS for 10 min, and washed three times in PBS. Fixed neurons were permeabilized with 0.25% Triton X-100/PBS for 5 min, washed several times with PBS (5min per wash), and incubated in 10% bovine serum albumin in PBS for 1 hr to block non-specific staining. Cells were then incubated overnight (4°C) with the following primary antibodies: rabbit anti-synapsin1 antibody (1:1000; Chemicon, Temecula, CA), mouse anti-PSD-95 antibidy (1:100, abcam) and mouse anti-gephyrin (1:1000; mAb7α; Synaptic Systems). After washing with PBS, cells were incubated with goat anti-rabbit indocarbocyanine (Cy3)-conjugated secondary antibodies (1:1000; Jackson ImmunoResearch, West Grove, PA) and goat anti-mouse Alexa Fluor 647 —conjugated secondary antibodies for 1hr at RT. Prepared cells were imaged on an inverted Nikon TU-2000 microscope with an EZ-C1 confocal system (Nikon) with a 100× oil-immersion objective (1.45 NA). Cells were imaged blind to specific co-transfection conditions and specific criteria for chosen cells were set as per (Biederer and Scheiffele, 2007). Breifly, the measure taken and compared was percent of HEK293 cell surface area covered by synapsin I label where PSD-95 and Gephryin signal were absent. Each channel was scanned sequentially at constant laser intensities and gain was set just below saturation. Images were processed and analyzed using Image Pro Plus® software. Briefly, images were threshholded to subtract background in a semiautomatic fashion to obtain average intensity and area of labeled regions. Data is expressed as mean ± s.e.m. and a two-tailed unpaired Student’s t test was employed to determine significant differences between target comparisons (* = p < 0.05).
Electrophysiology
The recording chamber was continuously perfused at 5 ml/min with ECS composed of (in mM): NaCl (145), KCl (5), MgCl2 (1), CaCl2 (1), HEPES (5), glucose (5), sucrose (25), phenol red (0.25mg/l) and D-serine (5 μM) (all from Sigma) with pH adjusted to 7.4 with NaOH. All experiments were performed at room temperature (24-26°C). The recording solution contained (in mM): potassium gluconate (145), HEPES (10), ATP.Mg (5), GTP.Na (0.2), and BAPTA (10), adjusted to pH 7.2 with KOH. Electrodes were pulled in two stages on a vertical pipette puller from borosilicate glass capillaries (Wiretrol II, Drummond, Broomall, PA). Pipette resistance ranged from 5 to 7 MΩ. NMDA-mEPSCs were pharmacologically isolated using bicuculline methiobromide (BMR, 50 μM), TTX (0.5 μM) and 2,3-Dihydro-6-nitro- 1,2,3,4-tetrahydrobenzo[f]quinoxaline-7-sulfonamide (NBQX, 5 μM) (all from Sigma) in a Mg2+-free solution, while AMPA-mEPSCs were recorded in presence of BMR (50 μM) and TTX (0.5 μM) in regular extracellular solution (ECS) with Mg2+. Solutions and drugs were delivered locally with a Y-tube device (Murase et al., 1989). Whole-cell voltage-clamp recordings from neurons and HEK293 cells were made at −60 mV and performed at room temperature using an Axopatch 200 or an Axopatch-1D amplifier (Axon Instruments, Union City, CA). A transient current response to a hyperpolarizing 10 mV pulse was used to assess resistance and capacitance throughout the recordings. Currents were filtered at 2 kHz with a low-pass Bessel filter (Frequency Devices, Haverhill, MA), digitized at 5-10 kHz using an IBM-compatible microcomputer equipped with Digidata 1322A data acquisition board and pCLAMP9 software (both from Molecular Device Co., Sunnyvale CA). Off-line data analysis, curve fitting, and figure preparation were performed with Clampfit 9 (Molecular Device) software. NMDA-mEPSCs’ decay was fit using Clampfit 9 (Molecular Device) from averages of at least 20 events selected with Minianalysis (Synaptosoft Inc, Fort Lee). The decay phase of currents was fit using a simplex algorithm for least squares exponential fitting routine with a double exponential equation of the form I(t) = If X exp(−t/τf) + Is X exp(−t/τs), where Ix is the peak current amplitude of a decay component and τx is the corresponding decay time constant. To allow for easier comparison of decay times between experimental conditions, the two decay time components were combined into a weighted time constant τw = [If/(If+Is)]*τf + [Is /(If+Is)]*τs. All data are expressed as mean ± standard error of the mean, P-values represent the results of analysis of variance (ANOVA) for multiple comparisons or two-tailed unpaired Student’s t tests.
Supplementary Material
Supplemental Fig. 1 Recruitment of effector molecules to sites of HA-SynCAM1 mediated adhesion with microspheres. HA-SynCAM1 clustering via microspheres recruits protein 4.1B, CASK and Syntenin1, but not GRIP1 and PSD-95 (arrowheads).
Supplemental Fig. 2 Protein 4.1 puncta decrease in intensity during development. (A) Immunolabeling for protein 4.1B, synapsin I and PSD-95 at 4 and 12DIV demonstrates an increase in the number of overall 4.1B puncta and decrease in their intensity. (B) Quantification of the mean puncta intensity for protein 4.1B immunolabeling over time in culture. Errors bars are s.e.m. (n = 11, **p < 0.01).
Supplemental Fig. 3 Analysis of mEPSCs after ifenprodil application. We measured mEPSC frequency in transfected hippocampal neurons before and during application of the NR1/NR2B containing NMDAR specific antagonist ifenprodil (10 μM). The only significant difference seen was between 4.1B overexpression and 4.1B knock-down with shRNA (p = 0.02, n =12).
ACKNOWLEDGEMENTS
We thank Irene Newsham for providing protein 4.1B cDNA, Richard Huganir for 4.1N cDNA, Casper Hoogenraad for GRIP1, Ben Margolis for CASK, and Jeremy Henley for Syntenin1 cDNAs. Thanks to Dan Pak for the hippocampal neuronal culture used in the electrophysiology experiments. We thank Leigh Ann Starcevich for advice on statistical analysis. We thank Sean Sweeney, Elva Diaz and members of the lab for comments on previous versions of this manuscript. We also thank Jimmy Giang, Shawn Brown and Keith Beadle for assistance with biochemical experiments, cell culture and general lab management, respectively. This work was supported by R01NS065795 from National Institute of Neurological Disorders, a grant from Autism Speaks to P.W., and R03MH085224 to Z.F. J.L.H. is an American Psychological Association DPN fellow: NIH APA DPN T32 MH18882-22
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Allison DW, Gelfand VI, Spector I, Craig AM. Role of actin in anchoring postsynaptic receptors in cultured hippocampal neurons: differential attachment of NMDA versus AMPA receptors. J. Neurosci. 1998;18:2423–2436. doi: 10.1523/JNEUROSCI.18-07-02423.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An XL, Takakuwa Y, Nunomura W, Manno S, Mohandas N. Modulation of band 3-ankyrin interaction by protein 4.1. Functional implications in regulation of erythrocyte membrane mechanical properties. J Biol Chem. 1996;271:33187–33191. doi: 10.1074/jbc.271.52.33187. [DOI] [PubMed] [Google Scholar]
- Barrow SL, Constable JR, Clark E, El-Sabeawy F, McAllister AK, Washbourne P. Neuroligin1: a cell adhesion molecule that recruits PSD-95 and NMDA receptors by distinct mechanisms during synaptogenesis. Neural Dev. 2009;4:17. doi: 10.1186/1749-8104-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biederer T. Bioinformatic characterization of the SynCAM family of immunoglobulin-like domain-containing adhesion molecules. Genomics. 2005a doi: 10.1016/j.ygeno.2005.08.017. [DOI] [PubMed] [Google Scholar]
- Biederer T. Progress from the postsynaptic side: signaling in synaptic differentiation. Sci STKE. 2005b;2005:pe9. doi: 10.1126/stke.2742005pe9. [DOI] [PubMed] [Google Scholar]
- Biederer T, Sara Y, Mozhayeva M, Atasoy D, Liu X, Kavalali ET, Sudhof TC. SynCAM, a synaptic adhesion molecule that drives synapse assembly. Science. 2002;297:1525–1531. doi: 10.1126/science.1072356. [DOI] [PubMed] [Google Scholar]
- Biederer T, Scheiffele P. Mixed-culture assays for analyzing neuronal synapse formation. Nat Protoc. 2007;2:670–676. doi: 10.1038/nprot.2007.92. [DOI] [PubMed] [Google Scholar]
- Bloch RJ. Cell-to-cell interactions during synaptogenesis. Curr Opin Cell Biol. 1989;1:940–946. doi: 10.1016/0955-0674(89)90062-8. [DOI] [PubMed] [Google Scholar]
- Brewer GJ, Torricelli JR, Evege EK, Price PJ. Optimized survival of hippocampal neurons in B27-supplemented Neurobasal, a new serum-free medium combination. J Neurosci Res. 1993;35:567–576. doi: 10.1002/jnr.490350513. [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- Chen L, Chetkovich DM, Petralia RS, Sweeney NT, Kawasaki Y, Wenthold RJ, Bredt DS, Nicoll RA. Stargazin regulates synaptic targeting of AMPA receptors by two distinct mechanisms. Nature. 2000;408:936–943. doi: 10.1038/35050030. [DOI] [PubMed] [Google Scholar]
- Cho KO, Hunt CA, Kennedy MB. The rat brain postsynaptic density fraction contains a homolog of the Drosophila discs-large tumor suppressor protein. Neuron. 1992;9:929–942. doi: 10.1016/0896-6273(92)90245-9. [DOI] [PubMed] [Google Scholar]
- Chow I, Poo MM. Release of acetylcholine from embryonic neurons upon contact with muscle cell. J Neurosci. 1985;5:1076–1082. doi: 10.1523/JNEUROSCI.05-04-01076.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correas I, Leto TL, Speicher DW, Marchesi VT. Identification of the functional site of erythrocyte protein 4.1 involved in spectrin-actin associations. J Biol Chem. 1986;261:3310–3315. [PubMed] [Google Scholar]
- Dean C, Scholl FG, Choih J, DeMaria S, Berger J, Isacoff E, Scheiffele P. Neurexin mediates the assembly of presynaptic terminals. Nat Neurosci. 2003;6:708–716. doi: 10.1038/nn1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon C, Goda Y. The actin cytoskeleton: integrating form and function at the synapse. Annu Rev Neurosci. 2005;28:25–55. doi: 10.1146/annurev.neuro.28.061604.135757. [DOI] [PubMed] [Google Scholar]
- Durand GM, Konnerth A. Long-term potentiation as a mechanism of functional synapse induction in the developing hippocampus. J Physiol Paris. 1996;90:313–315. doi: 10.1016/s0928-4257(97)87905-3. [DOI] [PubMed] [Google Scholar]
- Ehlers MD, Zhang S, Bernhadt JP, Huganir RL. Inactivation of NMDA receptors by direct interaction of calmodulin with the NR1 subunit. Cell. 1996;84:745–755. doi: 10.1016/s0092-8674(00)81052-1. [DOI] [PubMed] [Google Scholar]
- Feng W, Zhang M. Organization and dynamics of PDZ-domain-related supramodules in the postsynaptic density. Nat Rev Neurosci. 2009;10:87–99. doi: 10.1038/nrn2540. [DOI] [PubMed] [Google Scholar]
- Fletcher TL, Cameron P, De Camilli P, Banker G. The distribution of synapsin I and synaptophysin in hippocampal neurons developing in culture. J Neurosci. 1991;11:1617–1626. doi: 10.1523/JNEUROSCI.11-06-01617.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z, Logan SM, Vicini S. Deletion of the NR2A subunit prevents developmental changes of NMDA-mEPSCs in cultured mouse cerebellar granule neurones. J Physiol. 2005;563:867–881. doi: 10.1113/jphysiol.2004.079467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z, Washbourne P, Ortinski P, Vicini S. Functional excitatory synapses in HEK293 cells expressing neuroligin and glutamate receptors. J Neurophysiol. 2003;90:3950–3957. doi: 10.1152/jn.00647.2003. [DOI] [PubMed] [Google Scholar]
- Funke L, Dakoji S, Bredt DS. Membrane-associated guanylate kinases regulate adhesion and plasticity at cell junctions. Annu Rev Biochem. 2005;74:219–245. doi: 10.1146/annurev.biochem.74.082803.133339. [DOI] [PubMed] [Google Scholar]
- Gallo V, Kingsbury A, Balazs R, Jorgensen OS. The role of depolarization in the survival and differentiation of cerebellar granule cells in culture. J Neurosci. 1987;7:2203–2213. doi: 10.1523/JNEUROSCI.07-07-02203.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollmann M, Heinemann S. Cloned glutamate receptors. Annu Rev Neurosci. 1994;17:31–108. doi: 10.1146/annurev.ne.17.030194.000335. [DOI] [PubMed] [Google Scholar]
- Hoover KB, Bryant PJ. The genetics of the protein 4.1 family: organizers of the membrane and cytoskeleton. Curr Opin Cell Biol. 2000;12:229–234. doi: 10.1016/s0955-0674(99)00080-0. [DOI] [PubMed] [Google Scholar]
- Hunt CA, Schenker LJ, Kennedy MB. PSD-95 is associated with the postsynaptic density and not with the presynaptic membrane at forebrain synapses. J Neurosci. 1996;16:1380–1388. doi: 10.1523/JNEUROSCI.16-04-01380.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida J, Hirabayashi S, Sato Y, Hata Y. Synaptic scaffolding molecule is involved in the synaptic clustering of neuroligin. Mol Cell Neurosci. 2004;27:497–508. doi: 10.1016/j.mcn.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Irie M, Hata Y, Takeuchi M, Ichtchenko K, Toyoda A, Hirao K, Takai Y, Rosahl TW, Sudhof TC. Binding of neuroligins to PSD-95. Science. 1997;277:1511–1515. doi: 10.1126/science.277.5331.1511. [DOI] [PubMed] [Google Scholar]
- Isaac JT, Crair MC, Nicoll RA, Malenka RC. Silent synapses during development of thalamocortical inputs. Neuron. 1997;18:269–280. doi: 10.1016/s0896-6273(00)80267-6. [DOI] [PubMed] [Google Scholar]
- Jeyifous O, Waites CL, Specht CG, Fujisawa S, Schubert M, Lin EI, Marshall J, Aoki C, de Silva T, Montgomery JM, Garner CC, Green WN. SAP97 and CASK mediate sorting of NMDA receptors through a previously unknown secretory pathway. Nat Neurosci. 2009;12:1011–1019. doi: 10.1038/nn.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Burette A, Chung HS, Kwon SK, Woo J, Lee HW, Kim K, Kim H, Weinberg RJ, Kim E. NGL family PSD-95-interacting adhesion molecules regulate excitatory synapse formation. Nat Neurosci. 2006;9:1294–1301. doi: 10.1038/nn1763. [DOI] [PubMed] [Google Scholar]
- Ko J, Kim S, Chung HS, Kim K, Han K, Kim H, Jun H, Kaang BK, Kim E. SALM synaptic cell adhesion-like molecules regulate the differentiation of excitatory synapses. Neuron. 2006;50:233–245. doi: 10.1016/j.neuron.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Kohr G, Mody I. Kindling increases N-methyl-D-aspartate potency at single N-methyl-D-aspartate channels in dentate gyrus granule cells. Neuroscience. 1994;62:975–981. doi: 10.1016/0306-4522(94)90336-0. [DOI] [PubMed] [Google Scholar]
- Kohr G, Seeburg PH. Subtype-specific regulation of recombinant NMDA receptor-channels by protein tyrosine kinases of the src family. J Physiol. 1996;492(Pt 2):445–452. doi: 10.1113/jphysiol.1996.sp021320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konur S, Ghosh A. Calcium signaling and the control of dendritic development. Neuron. 2005;46:401–405. doi: 10.1016/j.neuron.2005.04.022. [DOI] [PubMed] [Google Scholar]
- Liao D, Zhang X, O’Brien R, Ehlers MD, Huganir RL. Regulation of morphological postsynaptic silent synapses in developing hippocampal neurons. Nat Neurosci. 1999;2:37–43. doi: 10.1038/4540. [DOI] [PubMed] [Google Scholar]
- McAllister AK. Dynamic Aspects of CNS Synapse Formation. Annu Rev Neurosci. 2007 doi: 10.1146/annurev.neuro.29.051605.112830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer G, Varoqueaux F, Neeb A, Oschlies M, Brose N. The complexity of PDZ domain-mediated interactions at glutamatergic synapses: a case study on neuroligin. Neuropharmacology. 2004;47:724–733. doi: 10.1016/j.neuropharm.2004.06.023. [DOI] [PubMed] [Google Scholar]
- Monyer H, Burnashev N, Laurie DJ, Sakmann B, Seeburg PH. Developmental and regional expression in the rat brain and functional properties of four NMDA receptors. Neuron. 1994;12:529–540. doi: 10.1016/0896-6273(94)90210-0. [DOI] [PubMed] [Google Scholar]
- Mott DD, Doherty JJ, Zhang S, Washburn MS, Fendley MJ, Lyuboslavsky P, Traynelis SF, Dingledine R. Phenylethanolamines inhibit NMDA receptors by enhancing proton inhibition. Nat Neurosci. 1998;1:659–667. doi: 10.1038/3661. [DOI] [PubMed] [Google Scholar]
- Murase K, Ryu PD, Randic M. Excitatory and inhibitory amino acids and peptide-induced responses in acutely isolated rat spinal dorsal horn neurons. Neurosci Lett. 1989;103:56–63. doi: 10.1016/0304-3940(89)90485-0. [DOI] [PubMed] [Google Scholar]
- Nam CI, Chen L. Postsynaptic assembly induced by neurexin-neuroligin interaction and neurotransmitter. Proc Natl Acad Sci U S A. 2005;102:6137–6142. doi: 10.1073/pnas.0502038102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunomura W, Takakuwa Y, Parra M, Conboy JG, Mohandas N. Ca(2+)-dependent and Ca(2+)-independent calmodulin binding sites in erythrocyte protein 4.1. Implications for regulation of protein 4.1 interactions with transmembrane proteins. J Biol Chem. 2000;275:6360–6367. doi: 10.1074/jbc.275.9.6360. [DOI] [PubMed] [Google Scholar]
- Oertner TG, Matus A. Calcium regulation of actin dynamics in dendritic spines. Cell Calcium. 2005;37:477–482. doi: 10.1016/j.ceca.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Parra M, Gascard P, Walensky LD, Gimm JA, Blackshaw S, Chan N, Takakuwa Y, Berger T, Lee G, Chasis JA, Snyder SH, Mohandas N, Conboy JG. Molecular and functional characterization of protein 4.1B, a novel member of the protein 4.1 family with high level, focal expression in brain. J Biol Chem. 2000;275:3247–3255. doi: 10.1074/jbc.275.5.3247. [DOI] [PubMed] [Google Scholar]
- Peters LL, Weier HU, Walensky LD, Snyder SH, Parra M, Mohandas N, Conboy JG. Four paralogous protein 4.1 genes map to distinct chromosomes in mouse and human. Genomics. 1998;54:348–350. doi: 10.1006/geno.1998.5537. [DOI] [PubMed] [Google Scholar]
- Petralia RS, Esteban JA, Wang YX, Partridge JG, Zhao HM, Wenthold RJ, Malinow R. Selective acquisition of AMPA receptors over postnatal development suggests a molecular basis for silent synapses. Nat Neurosci. 1999;2:31–36. doi: 10.1038/4532. [DOI] [PubMed] [Google Scholar]
- Prybylowski K, Fu Z, Losi G, Hawkins LM, Luo J, Chang K, Wenthold RJ, Vicini S. Relationship between availability of NMDA receptor subunits and their expression at the synapse. J Neurosci. 2002;22:8902–8910. doi: 10.1523/JNEUROSCI.22-20-08902.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saneyoshi T, Wayman G, Fortin D, Davare M, Hoshi N, Nozaki N, Natsume T, Soderling TR. Activity-dependent synaptogenesis: regulation by a CaM-kinase kinase/CaM-kinase I/betaPIX signaling complex. Neuron. 2008;57:94–107. doi: 10.1016/j.neuron.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sara Y, Biederer T, Atasoy D, Chubykin A, Mozhayeva MG, Sudhof TC, Kavalali ET. Selective capability of SynCAM and neuroligin for functional synapse assembly. J Neurosci. 2005;25:260–270. doi: 10.1523/JNEUROSCI.3165-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheiffele P, Fan J, Choih J, Fetter R, Serafini T. Neuroligin expressed in nonneuronal cells triggers presynaptic development in contacting axons. Cell. 2000;101:657–669. doi: 10.1016/s0092-8674(00)80877-6. [DOI] [PubMed] [Google Scholar]
- Scott C, Keating L, Bellamy M, Baines AJ. Protein 4.1 in forebrain postsynaptic density preparations: enrichment of 4.1 gene products and detection of 4.1R binding proteins. Eur J Biochem. 2001;268:1084–1094. doi: 10.1046/j.1432-1327.2001.01968.x. [DOI] [PubMed] [Google Scholar]
- Sharma K, Fong DK, Craig AM. Postsynaptic protein mobility in dendritic spines: long-term regulation by synaptic NMDA receptor activation. Mol Cell Neurosci. 2006;31:702–712. doi: 10.1016/j.mcn.2006.01.010. [DOI] [PubMed] [Google Scholar]
- Shen L, Liang F, Walensky LD, Huganir RL. Regulation of AMPA receptor GluR1 subunit surface expression by a 4. 1N-linked actin cytoskeletal association. J Neurosci. 2000;20:7932–7940. doi: 10.1523/JNEUROSCI.20-21-07932.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocca G, Vicini S. Increased contribution of NR2A subunit to synaptic NMDA receptors in developing rat cortical neurons. J Physiol. 1998;507(Pt 1):13–24. doi: 10.1111/j.1469-7793.1998.013bu.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun CX, Robb VA, Gutmann DH. Protein 4.1 tumor suppressors: getting a FERM grip on growth regulation. J Cell Sci. 2002;115:3991–4000. doi: 10.1242/jcs.00094. [DOI] [PubMed] [Google Scholar]
- Sytnyk V, Leshchyns’ka I, Delling M, Dityateva G, Dityatev A, Schachner M. Neural cell adhesion molecule promotes accumulation of TGN organelles at sites of neuron-to-neuron contacts. J Cell Biol. 2002;159:649–661. doi: 10.1083/jcb.200205098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovar KR, Westbrook GL. The incorporation of NMDA receptors with a distinct subunit composition at nascent hippocampal synapses in vitro. J Neurosci. 1999;19:4180–4188. doi: 10.1523/JNEUROSCI.19-10-04180.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zundert B, Yoshii A, Constantine-Paton M. Receptor compartmentalization and trafficking at glutamate synapses: a developmental proposal. Trends Neurosci. 2004;27:428–437. doi: 10.1016/j.tins.2004.05.010. [DOI] [PubMed] [Google Scholar]
- Vicini S, Wang JF, Li JH, Zhu WJ, Wang YH, Luo JH, Wolfe BB, Grayson DR. Functional and pharmacological differences between recombinant N-methyl-D-aspartate receptors. J Neurophysiol. 1998;79:555–566. doi: 10.1152/jn.1998.79.2.555. [DOI] [PubMed] [Google Scholar]
- Wang CY, Chang K, Petralia RS, Wang YX, Seabold GK, Wenthold RJ. A novel family of adhesion-like molecules that interacts with the NMDA receptor. J Neurosci. 2006;26:2174–2183. doi: 10.1523/JNEUROSCI.3799-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washbourne P, Bennett JE, McAllister AK. Rapid recruitment of NMDA receptor transport packets to nascent synapses. Nat Neurosci. 2002;5:751–759. doi: 10.1038/nn883. [DOI] [PubMed] [Google Scholar]
- Washbourne P, Dityatev A, Scheiffele P, Biederer T, Weiner JA, Christopherson KS, El-Husseini A. Cell adhesion molecules in synapse formation. J Neurosci. 2004a;24:9244–9249. doi: 10.1523/JNEUROSCI.3339-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washbourne P, Liu XB, Jones EG, McAllister AK. Cycling of NMDA receptors during trafficking in neurons before synapse formation. J Neurosci. 2004b;24:8253–8264. doi: 10.1523/JNEUROSCI.2555-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler A, Teichberg VI. Brain spectrin binding to the NMDA receptor is regulated by phosphorylation, calcium and calmodulin. Embo J. 1998;17:3931–3939. doi: 10.1093/emboj/17.14.3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Malinow R, Cline HT. Maturation of a central glutamatergic synapse. Science. 1996;274:972–976. doi: 10.1126/science.274.5289.972. [DOI] [PubMed] [Google Scholar]
- Wyszynski M, Lin J, Rao A, Nigh E, Beggs AH, Craig AM, Sheng M. Competitive binding of alpha-actinin and calmodulin to the NMDA receptor. Nature. 1997;385:439–442. doi: 10.1038/385439a0. [DOI] [PubMed] [Google Scholar]
- Yageta M, Kuramochi M, Masuda M, Fukami T, Fukuhara H, Maruyama T, Shibuya M, Murakami Y. Direct association of TSLC1 and DAL-1, two distinct tumor suppressor proteins in lung cancer. Cancer Res. 2002;62:5129–5133. [PubMed] [Google Scholar]
- Zoghbi HY. Postnatal neurodevelopmental disorders: meeting at the synapse? Science. 2003;302:826–830. doi: 10.1126/science.1089071. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Fig. 1 Recruitment of effector molecules to sites of HA-SynCAM1 mediated adhesion with microspheres. HA-SynCAM1 clustering via microspheres recruits protein 4.1B, CASK and Syntenin1, but not GRIP1 and PSD-95 (arrowheads).
Supplemental Fig. 2 Protein 4.1 puncta decrease in intensity during development. (A) Immunolabeling for protein 4.1B, synapsin I and PSD-95 at 4 and 12DIV demonstrates an increase in the number of overall 4.1B puncta and decrease in their intensity. (B) Quantification of the mean puncta intensity for protein 4.1B immunolabeling over time in culture. Errors bars are s.e.m. (n = 11, **p < 0.01).
Supplemental Fig. 3 Analysis of mEPSCs after ifenprodil application. We measured mEPSC frequency in transfected hippocampal neurons before and during application of the NR1/NR2B containing NMDAR specific antagonist ifenprodil (10 μM). The only significant difference seen was between 4.1B overexpression and 4.1B knock-down with shRNA (p = 0.02, n =12).