Abstract
Background and Purpose
Ischemic stroke (IS) is a multifactorial disorder with strong evidence from twin, family, and animal model studies suggesting a genetic influence on risk and prognosis. Several candidate genes for IS have been proposed, but few have been replicated. We investigated the contribution of 67 candidate genes (369 single nucleotide polymorphisms [SNPs]) on the risk of IS in a North American population of European descent.
Methods
Two independent studies were performed. In the first, 342 SNPs from 52 candidate genes were genotyped in 307 IS cases and 324 control subjects. The SNPs significantly associated with IS were tested for replication in another cohort of 583 IS cases and 270 control subjects. In the second study, 212 SNPs from 62 candidate genes were analyzed in 710 IS cases with subtyping available and 3751 control subjects.
Results
None of the candidate genes (SNPs) were significantly associated with IS risk independent of known stroke risk factors after correction for multiple hypotheses testing.
Conclusion
These results are consistent with previous meta-analyses that demonstrate an absence of genetic association of variants in plausible candidate genes with IS risk. Our study suggests that the effect of the investigated SNPs may be weak or restricted to specific populations or IS subtypes.
Keywords: candidate genes, genetics, ischemic stroke
Stroke is a multifactorial disorder or complex trait for which it is not possible to demonstrate classical patterns of inheritance. Ischemic stroke (IS) is a common neurological disease and a leading cause of severe disability and death in Western countries.1 Approximately 85% to 90% of strokes are ischemic.2,3 Strong evidence from twin, family, and animal model studies, however, have consistently suggested a genetic influence on stroke risk and prognosis.4 The genetic etiology of IS is likely to be complex with many loci modulating pathophysiological processes and conferring a small to moderate risk.
The most practical and widely used approach to identify common alleles influencing stroke has been candidate gene association analysis. The published genomewide association studies of IS5,6 have provided candidate loci of interest, although independent replication is required to establish the most convincing of these (12p13.33) as a definite risk locus for disease. Although several candidate genes for IS have been investigated by case–control analysis, few associations have been consistently replicated. We performed a case–control study in which cases (participants with an ischemic stroke) were compared with control subjects (participants free of a history of vascular events). We tested a total of 67 IS candidate genes (369 single nucleotide polymorphisms [SNPs]) that have been considered as potential genetic risk factors in vascular diseases for a role in stroke risk.
Methods
All samples used for Study 1 and Study 2 are of European descent.
Study 1: Candidate Genes and Replication (342 SNPs and 52 genes)
Samples (stroke cases and nonstroke control subjects) used in the discovery phase of Study 1 were collected from the Ischemic Stroke Genetics Study (ISGS), a case–control design. The replication phase consisted of samples from ISGS, Siblings with Ischemic Stroke Study (SWISS), and the National Institute of Neurological Disorders and Stroke-funded Neurogenetics Repository (Coriell Institute, http://ccr.coriell.org/ninds/). The SWISS-contributed cases (probands) were part of an affected sibpair (family study) design. Only one affected sibling of each family was included in the study.
Stroke cases from ISGS and SWISS included individuals diagnosed with IS according to the World Health Organization definition.7 A neurologist or a specialist in diagnosis classified all stroke and stroke subtype cases. The protocol for ISGS and SWISS has been reported previously.8,9 All cases included complete medical history and IS subtype based on the prespecified Trial of Org 10172 in Acute Stroke Treatment.10
In the replication phase, unrelated European descent neurologically normal control samples from different sites within the United States were from the Coriell repository. Each subject underwent a detailed medical history interview that included family history. Samples were not included if a first-degree relative had a history of the following neurological diseases: Alzheimer disease, amyotrophic lateral sclerosis, ataxia, autism, bipolar disorder, brain aneurysm, dementia, dystonia, and Parkinson disease. Sum scores on the Folstein Mini Mental State Examination11 ranged from 26 to 30. For more details, see http://ccr.coriell.org/ninds/controls/controls.html. Table 1 shows the demographic and clinical data in stroke subjects and control subjects in each phase of Study 1.
Table 1.
Demographic and Clinical Data in Stroke Subjects and Control Subjects in Study 1
| ISGS |
|||
|---|---|---|---|
| Discovery Phase | Cases | Control Subjects | P Value |
| No. of samples | 307 | 324 | |
| Age, median (SD) | 68.32 (14.67) | 65.04 (14.71) | 0.0096 |
| Gender (male), n (%) | 183 (60) | 124 (40) | 0.1085 |
| Hypertension, n (%) | 196 (64) | 124 (38) | <0.001 |
| Diabetes mellitus, n (%) | 77 (25) | 36 (11) | <0.001 |
| Smoking, n (%) | 217 (71) | 170 (52) | <0.001 |
| Heart disease, n (%) | 84 (27) | 42 (13) | <0.001 |
| ISGS |
Coriell |
SWISS |
|||||
|---|---|---|---|---|---|---|---|
| Replication Phase | Cases | Control Subjects | Cases | Control Subjects | Cases | Control Subjects | P Value* |
| No. of samples | 92 | 2 | 398 | 267 | 93 | 1 | |
| Age, median | 68.34 | 74.34 | 68.00 | 69.00 | 70.00 | 56.00 | <0.001 |
| SD | 15.34 | 9.60 | 14.08 | 8.78 | 10.45 | NA | |
| Gender (male), n (%) | 34 (37) | 1 (50) | 223 (56) | 175 (44) | 48 (52) | 1 (100) | 0.0149 |
| Hypertension, n (%) | 50 (54) | 1 (50) | 232 (59) | 78 (29) | 67 (72) | 1 (100) | <0.001 |
| Diabetes mellitus, n (%) | 16 (17) | 1 (50) | 58 (15) | 21 (8) | 27 (29) | 0 (0) | <0.001 |
| Smoking, n (%) | 55 (60) | 2 (100) | 235 (61) | 108 (40) | 91 (98) | 1 (100) | <0.001 |
| Heart disease, n (%) | 29 (32) | 1 (50) | 17 (4) | 31 (12) | 20 (22) | 0 (0) | 0.8182 |
P value showing differences between cases and control subjects.
NA indicates not applicable.
Genotyping
Candidate genes were chosen based on evidence of association from previous studies of IS and according to biological plausibility, including the presence of genes involved in lipid metabolism, coagulation cascade, nitric oxide production, homocysteine metabolism, the renin–angiotensin system, and other stroke risk factors. Tagging SNPs were chosen for each candidate. Loci for the SNP mapping panel are described in the Supplemental Table (available at http://stroke.ahajournals.org).
In the discovery phase, genotyping was performed using the Illumina GoldenGate assay. DNA samples (5 μL) were genotyped according to the manufacturer's instructions on an Illumina Bead-Station 500G Golden Gate genotyping platform using a custom panel (GS0006623-OPA) of 364 candidate and ancestry informative SNPs.
In the replication phase, high-throughput SNP genotyping was performed using the 5′ nuclease allelic discrimination assay (TaqMan assay) on an ABI PRISM 7900HT Sequence Detection System. The assay includes the forward target-specific polymerase chain reaction primer, the reverse primer, and the TaqMan MGB probes labeled with 2 special dyes: FAM and VIC.
Cases and control subjects were pooled and distributed across plates. We also randomly selected 134 individuals from the discovery phase and genotyped them for these SNPs using the TaqMan assays to ensure the 2 methods used in the different phases generated the same genotypes for each individual (concordance rate 99.9%).
Genotypes were assigned in separate cluster files using Beadstudio Version 2.0 genotyping software. Genotypes with GenCall scores >0.25 were called. SNPs were excluded if SNPs had call rates <95% in cases or control subjects or the SNP genotyping was considered poor quality. Any samples with a call rate <95% of the SNPs were excluded from the analysis.
All genotype assignments were performed blind with regard to clinical data.
Statistical Analysis
Tests of significance between cases and control subjects were performed using 2-sample tests for binomial proportions (using a χ2 test of independence). Fisher exact test was used when appropriate. For comparison of age (continuous) among case–control groups, a 2-sample t test for independent samples was performed. For each SNP, tests of deviations from Hardy-Weinberg expectations were performed using the methods described by Wigginton et al.12 SNPs were dropped if the Hardy-Weinberg equilibrium probability value in the control group was <0.05 and minor allele frequency was <1% in the population.
For tests of association with individual SNPs between cases and control subjects, a series of generalized estimating equations were used that permitted inclusion of recognized stroke risk factors as covariates (age, sex, hypertension status, presence of atrial fibrillation, history of myocardial infarction, smoking status, presence of diabetes mellitus, and family history of stroke). Probability values were computed using the 2 degrees-of-freedom generalized test of association. When the generalized test of association was significant (P<0.05), additional models were tested that assumed an underlying mode of inheritance of risk (dominant, additive, recessive) with a one-degree-of-freedom test.
Tests for association with stroke risk were computed using the expectation–maximization algorithm. Statistical significance was assessed using a permutation test of the likelihood ratio statistic.
Study 2: Candidate Genes in 710 IS and 3751 Control Subject (212 SNPs, 62 genes)
At the time of completion of Study 1, genotyping from 710 IS cases with subtype data available and 3751 control subjects in >500 000 SNPs was obtained. We re-examined associations with the Study 1 candidate genes (SNPs) as well as others previously reported to be significantly associated with IS in published meta-analyses or studies with replication.13–21 We also examined SNPs contained in these genes or close to these genes (within 1 MB) that have not been reported in the literature but were included in the genotyping assays used here.
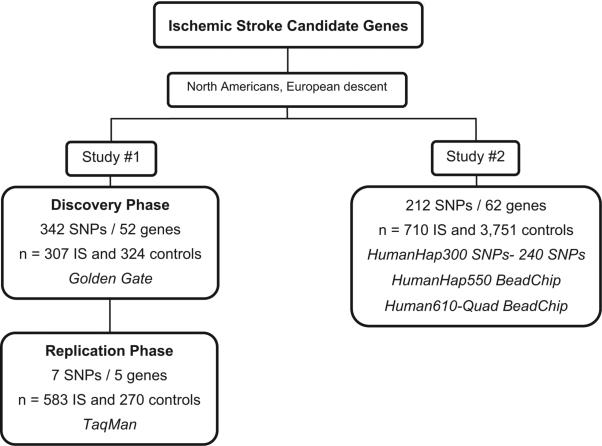
Ischemic Stroke samples came from ISGS, SWISS, and the Coriell repository. Control samples were selected from individuals who had participated in genomewide association studies performed by our group: 787 samples from the neurogenetics collection at the Coriell cell repository and 728 from the Baltimore Longitudinal Study of Aging (BLSA). In addition, 2236 samples were available from the Cancer Genetic Marker of Susceptibility Study (CGEMS). Details of CGEMS and BLSA collection of cohorts have been previously described.22,23 Briefly, CGEMS is a 3-year initiative of the National Cancer Institute that conducts genomewide association studies to identify common gene variations increasing the risk for cancer. On approval of a Data Access Request, we obtained raw genotype data from 1142 women from the Nurses Health Study and 1094 men from the Prostate, Lung, Colon and Ovarian Cancer Screening Trial. BLSA is a long-term study designed in 1958 to trace the effects of aging in humans. The BLSA study recruited individuals aged 17 to 96 years to participate in the assessment of health and physical and psychological performance. The average length of follow-up was 7.5 years with participants evaluated every 2 years in the Gerontology Research Center of the National Institutes of Health. Samples from individuals that developed IS, Alzheimer disease, Parkinson disease, or any other neurological conditions were not included. Table 2 shows the demographic and clinical data in stroke subjects and control subjects for each cohort used in Study 2. The Figure shows a study schematic.
Table 2.
Demographic and Clinical Characteristics of Each Cohort (212 SNPs, 62 Genes Study) for Study 2
| Stroke†, n (%) (n=710) | Coriell, n (%) (n=787) | BLSA, n (%) (n=728) | CGEMS, n (%) (n=2236) | P Value* | |
|---|---|---|---|---|---|
| Age, median (SD) | 67.42 (13.98) | 58.70 (16.40) | 69.61 (16.78) | 65.01 (5.94) | <0.001 |
| Gender, male | 409 (58) | 329 (42) | 402 (54) | 1094 (49) | <0.001 |
| Hypertension | 440 (60) | 176 (23) | 347 (48) | NA | <0.001 |
| Diabetes mellitus | 128 (18) | 51 (7) | 117 (16) | NA | <0.001 |
| Smoking | 452 (65) | 313 (42) | 390 (54) | NA | <0.001 |
| Heart disease | 106 (15) | 69 (9) | 69 (10) | NA | <0.001 |
P value showing differences between cases and control subjects.
From ISGS, SWISS, and Coriell cell repository.
NA indicates not available.
Figure.
Diagram of the 2 studies running independently to test for association in up to 62 candidate genes.
All DNA samples were obtained from individuals who were neurologically normal, of non-Hispanic European descent, had given signed informed consent, and for whom clinical phenotype data were available.
Statistical Analyses
Samples were excluded because demographic data or analysis of genetic background and/or pairwise identity-by-descent estimation examined by PLINK (http://pngu.mgh.harvard.edu/≈purcell/plink/) showed these samples not belonging to the European descent population or with a level of relatedness higher than expected. The source of the sample was also used as another covariate to correct for possible stratification.
A stepwise series of analyses was performed. Initially, the outcome (stroke case–control status) was a function solely of SNP and source of the sample (baseline model). This analysis was followed by models that included age, sex, hypertension, diabetes, heart disease, and smoking as covariates (independent predictors). Association between the investigated polymorphisms and risk of stroke were analyzed by means of logistic regression. The influence of multiple testing was evaluated using false discovery rate. All statistical analyses were done with PLINK.
Statistical Study Power
Power calculations were generated using a log-additive model of risk, 5% Type 1 error rate, and 80% power. For a minor allele frequency of 0.15, the detectable OR in the discovery phase of Study 1 was 1.51; the detectable OR in the replication phase of Study 1 was 1.47; and the detectable OR in Study 2 was 1.24.
Participants in both studies were enrolled prospectively under Institutional Review Board-approved protocols at participating institutions and all subjects gave written informed consent.
Results
Study 1: Candidate Genes and Replication (342 SNPs and 52 genes)
After quality filtering, 342 SNPs in 307 IS cases and 324 control subjects were evaluated. Each SNP was tested independently for association with IS. After adjustment for stroke risk factors (age, sex, race, and other stroke risk factors), 7 SNPs were significantly associated with IS risk (P<0.05). After Bonferroni correction, none of these SNPs remained significant. Because the Bonferroni correction is overly conservative, these 7 SNPs were genotyped in an independent cohort of 853 samples (583 patients with IS and 270 control subjects). After adjustment for stroke risk factors, none of the SNPs were significantly associated with IS. The summary statistics associated with these 7 SNPs are shown in Table 3. We tested these SNPs for association to the main subtypes of IS. After adjustment by Bonferroni correction, only the SNP rs1799983 was associated with the “other” IS subtype (additive model OR 1.56; 95% CI, 1.25 to 1.94; P=0.0003).
Table 3.
Summary Statistics of the 7 SNPs From 342 SNPs (52 Genes) That Were Initially Significantly Associated With IS Risk (P<0.05) and Replicated in an Independent Cohort
| Discovery (307 IS/324 Control Subjects) |
Replication (583 IS/270 Control Subjects) |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene | SNP | bp | C-HWE (pv) | 2DF (pv) | DOM (pv) | ADD (pv) | REC (pv) | C-HWE (pv) | 2DF (pv) |
| PPARG | rs10510419 | 12401936 | 0.400 | 0.026 | 0.007 | 0.008 | 0.616 | 0.515 | 0.061 |
| PPARG | rs709157 | 12437024 | 0.170 | 0.044 | 0.371 | 0.877 | 0.049 | 0.024 | 0.785 |
| F13A1 | rs3024388 | 6194644 | 0.623 | 0.009 | 0.010 | 0.002 | 0.019 | 0.871 | 0.756 |
| NOS3 | rs1799983 | 150133759 | 0.239 | 0.015 | 0.006 | 0.004 | 0.114 | 0.243 | 0.090 |
| MMP3 | rs520540 | 102214635 | 0.911 | 0.022 | 0.496 | 0.033 | 0.006 | 0.395 | 0.680 |
| MMP3 | rs679620 | 102218830 | 0.911 | 0.014 | 0.003 | 0.015 | 0.322 | 0.461 | 0.729 |
| LIPC | rs261336 | 56529710 | 0.239 | 0.013 | 0.004 | 0.010 | 0.977 | 0.228 | 0.354 |
C-HWE indicates Hardy-Weinberg equilibrium in control subjects; 2DF, 2-degrees-of-freedom generalized test of association; DOM, dominant model; ADD, additive model; REC, recessive model; bp, base pair; SNP, single nucleotide polymorphism; PPARG, peroxisome proliferator-activated receptor gamma; F13A1, coagulation factor XIII, A1 polypeptide; NOS3, nitric oxide synthase 3 (endothelial cell); MMP3, matrix metallopeptidase 3 (stromelysin 1, progelatinase); LIPC, lipase, hepatic.
Study 2: Candidate Genes in 710 IS and 3751 Control Subjects (212 SNPs and 62 genes)
The program PLINK was used to test for population stratification. All IS cases and control subjects were determined to have the same genetic structure.
Two hundred twelve SNPs from 62 genes that have been reported to be significantly associated with IS and were included in published meta-analyses or studies with replication, or SNPs investigated in our first study (using Golden Gate assay), were contained in the used Illumina Infinium BeadChips and passed our control quality criteria (call rate >95%, Hardy-Weinberg equilibrium P>1.0×10–7, minor allele frequency >1%). None of the SNPs in the candidate genes were significantly associated with IS after different measures of correction (Bonferroni, false discovery rate).
Analysis was performed in a subgroup of 710 IS and 1495 control subjects (samples from ISGS, SWISS, BLSA, and Coriell repository) that had complete stroke phenotypes. None of the SNPs in the candidate genes were significantly associated with IS after false discovery rate correction. We also examined other SNPs contained in these genes or within 1 MB of these genes not reported in the literature. None of the SNPs were significant after correcting for multiple tests. Given that samples coming from the BLSA study were followed up by at least 7.5 years, we can be more certain that these samples remained free of stroke. Thus, we further restricted these analyses to IS samples and control subjects from BLSA. None of the SNPs were significantly associated with risk for IS. The Supplemental Table shows SNPs that have been analyzed by our group or in published meta-analyses or studies with replication in European descent populations post-200513–21 and their results. Table 4 shows genetic common variants that have been found to be significantly associated with IS in meta-analyses or studies with replication. Table 5 shows details of the current study and other studies that did not find any association.
Table 4.
Genetic Common Variants and Characteristics of the Studies With Replication or Meta-Analyses Post-2005 That Have Found an Association With IS
| Reference No. | 16 | 16 | 14 | 13 | 21 | 15 | 15 | 15 | 15 |
|---|---|---|---|---|---|---|---|---|---|
| Gene | GP1BA | GP1BA | LTA* | NOS3 | PAI1 | F5 | MTHFR | F2 | ACE |
| Polymorphism | Thr(T)161 met (M) | Kozak variant | 252A> G* | glu298asp | 4G/5G | Arg506GIn | C677T | G20210A | I/D |
| Studies (W) | 6 (3) | 5 (3) | Consortium (6) | Original+replication | Original+meta-analysis (11) | 26 | 22 | 19 | 11 |
| IS | 893 | 1984 | 3550 | 2500 | 4588 | 3387 | 3028 | 2990 | |
| Control | 1313 | 1932 | 6560 | 3500 | 13 798 | 4597 | 7131 | 11 305 | |
| White samples (IS/C) | 358/615 | 2387/5089 | 1417/996 484/751 |
||||||
| Population | Mixed | Mixed | Mixed | European | Mixed | White | White | White | White |
| Analysis for each ethnic group | No | No | Yes | Germany | No | White | White | White | White |
| SNPs | 3 | 3 | 105 | 106 | 1 | 15 | 15 | 15 | 15 |
| P value | 0.004 | 4.54e-0.08 | 0 016 (W) | 0.05+0.04 | 0.002 | 0.001 | 0.002 | 0.006 | <0.001 |
| OR | (M/M versus T/T) 2.08 | (ADD) 1.28 | (ADD) 1.13–1.22 | (4G/4G versus 5G/5G) 0.89 | (DOM) 1.33 | (REC) 1.24 | (DOM) 1.44 | (REC) 1.21 | |
| 95% CI | 0.78–5.54 | 1.12–1.58 | 1.08–1.42 | 1.11–1.86 | 1.08–1.35 | ||||
| Heterogeneity in ORs | Yes | Extreme | Extreme | Yes | No | No | No | ||
| Use of covariates | No | No | Age, sex, HT, SM | Age, sex, Ch, D, HT | No | No | No | No | No |
| Multiple testing correction | FDR in all group | Permutations | |||||||
| Studies confirm association† | No | No | No | No | No | No | No | No | No |
| Studies not showing association‡ | Casas | Casas | Berger | Casas, Lalouschek, Wang | Casas, Lalouschek, Tsantes, Wang | Lalouschek, Wang | Berger, Lalouschek, Wang | Berger, Wang | Berger |
Significant in nonhypertensive IS sample.
Studies cited in references 13–16, 21.
Studies cited in references 17–20.
W indicates white; IS, ischemic stroke; C, control subjects; Ch, cholesterol; HT, hypertension; D, diabetes; SM, smoking; GP1BA, glycoprotein Ib (platelet), alpha polypeptide; LTA, lymphotoxin alpha (tumor necrosis factor superfamily, member 1); NOS3, nitric oxide synthase 3 (endothelial cell); PAI1, plasminogen activator inhibitor type 1; F5, coagulation factor V (proaccelerin, labile factor); MTHFR, 5,10-methylenetetrahydrofolate reductase (NADPH); F2, coagulation factor II (thrombin); ACE, angiotensin I converting enzyme (peptidyl-dipeptidase A) 1.
Table 5.
Details of the Current Study and Previously Published Studies With Replication or Meta-Analyses Post-2005 That Failed to Find an Association With IS
| Reference No. | Current Study | Current Study | 17 | 18 | 19 | 20 |
|---|---|---|---|---|---|---|
| Subjects (IS/C) | 307/324+583/270 | 710/3751 | 358/817 | 3104/4870 (2693/4320)* | 1606/2207 (845/1144)* | 774/1074 (661/1013)* |
| Studies | Original+replication | Original | Original | 15 (11)* | 8 (3)* | 7 (6)* |
| Population | European | European | European | Mix | Mix | Mix |
| Analysis for each ethnic group | European | European | European | Yes | Yes | Yes |
| SNPs (gene) | 342 (52 genes) | 212 (62 genes) | 60 (35 genes) | 1 (PAI-1) | 1 (TNFA) | 1 (ITGA2) |
| Heterogeneity in ORs | Yes | Yes | Yes | |||
| Use of covariates | Age, sex, AF, D, HT, MI, SM | Age, sex, D, HD, HT, SM | Age and sex | Age, sex, Ch, D, HT, SM | No | Age, sex, BMI, Ch, D, HT, SM |
| Use of multiple testing correction | No | FDR | Yes | |||
| Other features | Age range =45–57 | Association in child/Asian |
White samples.
C indicates control subjects; IS, ischemic stroke; AF, atrial fibrillation; BMI, body mass index; Ch, cholesterol; HD, heart disease; HT, hypertension; MI, myocardial infarction; SM, smoking; D, diabetes; FDR, false discovery rate.
Discussion
Association studies are becoming a common approach to mapping variants that affect IS. Although some studies, including some meta-analyses, indicate an association between the risk of stroke and certain SNPs, many other studies show lack of reproducibility. In this study, a series of highly plausible candidate genes were selected for intensive and comprehensive genetic study. No evidence for association with either IS or stroke subtype was observed.
The potential reasons that have been discussed for this lack of reproducibility are reduced to 3 potential causes: a false-positive association is correctly not replicated; a true association fails to be replicated in an underpowered follow-up study (false-negative); or a true association in one population is not true in a second population because of heterogeneity in genetic or environmental background.24 In the case of stroke, most studies that have identified polymorphisms related with IS are underpowered studies with a sample size not >300 patients. Until recently, the larger-scale studies came from meta-analysis. Casas et al performed meta-analyses of 120 stroke candidate gene case–control studies and a total of 51 polymorphisms in 32 genes. Statistically significant associations with IS were identified for factor V Leiden Arg506Gln, methylenetetrahydrofolate reductase C677T, prothrombin G20210A, and angiotensin-converting enzyme insertion/deletion.15
Wang et al evaluated the association between 105 polymorphisms in 64 inflammatory and cardiovascular system-related genes and IS. None of these SNPs remained statistically significant after false discovery rate correction. Only when the data were stratified on hypertension status, 2 polymorphisms on LTA were significantly associated with IS in nonhypertensive subjects. The data were not adjusted for other stroke risk factors such as diabetes or heart disease.14
Other meta-analyses restricted to one or more common variant from one gene reported an association with IS for GP1BA16 or a nonassociation for plasminogen activator inhibitor-1, tumor necrosis factor-α, and ITGA2.18–20 Although meta-analyses facilitate the overall interpretation of association, they also need to be interpreted with caution. Some meta-analyses do not include stroke risk factors as covariates and the sample sizes remain small when correctly taking into account differences in ethnicity and/or inclusion study criteria (inclusion of children and adults, patients with transient ischemic attack, and so on).
In an attempt to clarify these associations and avoid false-positive associations, we decided to conduct a case–control study and used an independent cohort as a replication study of the other. In uncorrected analysis for multiple testing, 7 polymorphisms were associated with IS in the first stage after adjustment for stroke risk factors. None of these polymorphisms were associated with IS in the replication study. In a second larger study, we investigated the same genes (192 SNPs in common) with the same results: modest, albeit nonsignificant, associations.
Berger et al investigated a total of 106 SNPs located in 63 candidate genes for potential associations with ischemic stroke in 2 independent case–control studies from a German population. All genes tested were related to pathways important in the pathophysiology of cardiovascular and inflammatory diseases. Only the glu298asp polymorphism in the nitric oxide synthase-3 gene was reported to be replicated in the second study. The association was independent of age, sex, hypertension, diabetes, and hypercholesterolemia in both studies. In the present study, a first association with the same SNP and IS was not replicated in the second cohort. Analysis of data of the genomewide association scan did not show an association either. Only in analysis by stroke subtype using the full sample did the glu298asp polymorphism become associated with increased risk of unknown stroke subtype independent of other stroke risk factors and while using Bonferroni correction.13
The study presented here also has some limitations. The size of the studied population in the first stage using the Golden Gate assay is relatively small and therefore underpowered to find genes with small effects influencing risk for IS. The inclusion of one second set of cases–control subjects as replication of the most significant SNPs rules out false-positive associations but does not rule out the possibility of false-negative or Type II error associations. However, analysis of many of these SNPs in a larger cohort and a review of the literature suggests that the majority of these SNPs have no significant effect for stroke risk in white populations.
The development and application of genomewide association studies represents a significant opportunity for stroke research and provides an opportunity to test the common disease common variant hypothesis in this disorder. Two genomewide association studies for stroke have been published thus far, a pilot study of modest size published by us in 20075 that failed to find any risk alleles of large effect and a recent study published by Ikram and colleagues that suggests a significant association between a locus at 12p13.33 with stroke. This locus was not present in the panel of loci selected for testing here; however, this locus clearly warrants replication in an independent study to confirm or refute this as an unequivocal risk factor for stroke.
In summary, 7 polymorphisms have been reported to be associated with IS after meta-analysis or replication; however, all of these SNPs have been investigated in at least one other large study in which no association with IS was reported. Differences in inclusion and exclusion criteria selection, type of statistical evaluation, covariates, correction for multiple testing, and inclusion of different patient populations may explain these discrepancies. This study indicates that any association that may exist is likely to be weak or perhaps restricted to specific populations or IS subtypes.
Supplementary Material
Supplemental Table. SNPs That Have Been Analyzed by Our Group or in Published Meta-Analyses or Studies With Replication in European Descent Populations Post-200513–21 and Their Results
Acknowledgments
We thank the subjects for participating in this research and the submitters for depositing samples at the National Institute of Neurological Disorders and Stroke Neurogenetics Repository, Coriell Institute for Medical Research, Camden, NJ.
Sources of Funding
This work was supported in part by the Intramural Research Program of the National Institute on Aging, National Institutes of Health, Department of Health and Human Services, project Z01 AG000954-06. SWISS is supported by a grant from the National Institute of Neurological Disorders and Stroke (R01 NS39987).
Footnotes
Disclosures
None.
References
- 1.Bonita R. Epidemiology of stroke. Lancet. 1992;339:342–344. doi: 10.1016/0140-6736(92)91658-u. [DOI] [PubMed] [Google Scholar]
- 2.Flossmann E, Schulz UG, Rothwell PM. Systematic review of methods and results of studies of the genetic epidemiology of ischemic stroke. Stroke. 2004;35:212–227. doi: 10.1161/01.STR.0000107187.84390.AA. [DOI] [PubMed] [Google Scholar]
- 3.Brown RD, Whisnant JP, Sicks JD, O'Fallon WM, Wiebers DO. Stroke incidence, prevalence, and survival: secular trends in Rochester, Minnesota, through 1989. Stroke. 1996;27:373–380. [PubMed] [Google Scholar]
- 4.Wang XL, Mahaney MC, Sim AS, Wang J, Wang J, Blangero J, Almasy L, Badenhop RB, Wilcken DE. Genetic contribution of the endothelial constitutive nitric oxide synthase gene to plasma nitric oxide levels. Arterioscler Thromb Vasc Biol. 1997;17:3147–3153. doi: 10.1161/01.atv.17.11.3147. [DOI] [PubMed] [Google Scholar]
- 5.Matarin M, Brown WM, Scholz S, Simon-Sanchez J, Fung HC, Hernandez D, Gibbs JR, De Vrieze FW, Crews C, Britton A, Langefeld CD, Brott TG, Brown RD, Jr, Worrall BB, Frankel M, Silliman S, Case LD, Singleton A, Hardy JA, Rich SS, Meschia JF. A genome-wide genotyping study in patients with ischaemic stroke: initial analysis and data release. Lancet Neurol. 2007;6:414–420. doi: 10.1016/S1474-4422(07)70081-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ikram MA, Seshadri S, Bis JC, Fornage M, DeStefano AL, Aulchenko YS, Debette S, Lumley T, Folsom AR, van den Herik EG, Bos MJ, Beiser A, Cushman M, Launer LJ, Shahar E, Struchalin M, Du Y, Glazer NL, Rosamond WD, Rivadeneira F, Kelly-Hayes M, Lopez OL, Coresh J, Hofman A, DeCarli C, Heckbert SR, Koudstaal PJ, Yang Q, Smith NL, Kase CS, Rice K, Haritunias T, Roks G, de Kort PL, Taylor KD, de Lau LM, Oostra BA, Uitterlinen AG, Rotter JI, Boerwinkle E, Psaty BM, Mosley TH, van Duijn CM, Breteler MM, Longstreth WT, Jr, Wolf PA. Genomewide association studies of stroke. N Engl J Med. 2009;360:1718–1728. doi: 10.1056/NEJMoa0900094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.WHO MONICA Project Principal Investigators The World Health Organization MONICA Project (monitoring trends and determinants in cardiovascular disease): a major international collaboration. J Clin Epidemiol. 1988;41:105–114. doi: 10.1016/0895-4356(88)90084-4. [DOI] [PubMed] [Google Scholar]
- 8.Meschia JF, Brott TG, Brown RD, Jr, Crook RJ, Frankel M, Hardy J, Merino JG, Rich SS, Silliman S, Worrall BB, Ischemic Stroke Genetics Study The Ischemic Stroke Genetics Study (ISGS) Protocol. BMC Neurol. 2003;3:4. doi: 10.1186/1471-2377-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meschia JF, Brown RD, Jr, Brott TG, Chukwudelunzu FE, Hardy J, Rich SS. The Siblings With Ischemic Stroke Study (SWISS) Protocol. BMC Med Genet. 2002;3:1. doi: 10.1186/1471-2350-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meschia JF, Barrett KM, Chukwudelunzu F, Brown WM, Case LD, Kissela BM, Brown RD, Jr, Brott TG, Olson TS, Rich SS, Silliman S, Worrall BB, Siblings with Ischemic Stroke Study (SWISS) Investigators Interobserver agreement in the Trial of Org 10172 in Acute Stroke Treatment classification of stroke based on retrospective medical record review. J Stroke Cerebrovasc Dis. 2006;15:266–272. doi: 10.1016/j.jstrokecerebrovasdis.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Folstein MF, Folstein SE, McHugh PR. ‘Mini-mental state.’ A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975;12:189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- 12.Wigginton JE, Cutler DJ, Abecasis GR. A note on exact tests of Hardy-Weinberg equilibrium. Am J Hum Genet. 2005;76:887–893. doi: 10.1086/429864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berger K, Stogbauer F, Stoll M, Wellmann J, Huge A, Cheng S, Kessler C, John U, Assmann G, Ringelstein EB, Funke H. The glu298asp polymorphism in the nitric oxide synthase 3 gene is associated with the risk of ischemic stroke in two large independent case–control studies. Hum Genet. 2007;121:169–178. doi: 10.1007/s00439-006-0302-2. [DOI] [PubMed] [Google Scholar]
- 14.Wang X, Cheng S, Brophy VH, Erlich HA, Mannhalter C, Berger K, Lalouschek W, Browner WS, Shi Y, Ringelstein EB, Kessler C, Luedemann J, Lindpaintner K, Liu L, Ridker PM, Zee RY, Cook NR, RMS Stroke SNP Consortium A meta-analysis of candidate gene polymorphisms and ischemic stroke in 6 study populations: association of lymphotoxin-alpha in nonhypertensive patients. Stroke. 2009;40:683–695. doi: 10.1161/STROKEAHA.108.524587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Casas JP, Hingorani AD, Bautista LE, Sharma P. Meta-analysis of genetic studies in ischemic stroke: thirty-two genes involving approximately 18 000 cases and 58 000 controls. Arch Neurol. 2004;61:1652–1661. doi: 10.1001/archneur.61.11.1652. [DOI] [PubMed] [Google Scholar]
- 16.Maguire JM, Thakkinstian A, Sturm J, Levi C, Lincz L, Parsons M, Whyte S, Attia J. Polymorphisms in platelet glycoprotein 1balpha and factor VII and risk of ischemic stroke: a meta-analysis. Stroke. 2008;39:1710–1716. doi: 10.1161/STROKEAHA.107.507228. [DOI] [PubMed] [Google Scholar]
- 17.Lalouschek W, Endler G, Schillinger M, Hsieh K, Lang W, Cheng S, Bauer P, Wagner O, Mannhalter C. Candidate genetic risk factors of stroke: results of a multilocus genotyping assay. Clin Chem. 2007;53:600–605. doi: 10.1373/clinchem.2006.073494. [DOI] [PubMed] [Google Scholar]
- 18.Tsantes AE, Nikolopoulos GK, Bagos PG, Tsiara CG, Kapsimali V, Travlou A, Vaiopoulos G. Plasminogen activator inhibitor-1 4G/5G polymorphism and risk of ischemic stroke: a meta-analysis. Blood Coagul Fibrinolysis. 2007;18:497–504. doi: 10.1097/MBC.0b013e3281ec4eee. [DOI] [PubMed] [Google Scholar]
- 19.Pereira TV, Rudnicki M, Franco RF, Pereira AC, Krieger JE. Effect of the G-308A polymorphism of the tumor necrosis factor alpha gene on the risk of ischemic heart disease and ischemic stroke: a meta-analysis. Am Heart J. 2007;153:821–830. doi: 10.1016/j.ahj.2007.02.031. [DOI] [PubMed] [Google Scholar]
- 20.Nikolopoulos GK, Tsantes AE, Bagos PG, Travlou A, Vaiopoulos G. Integrin, alpha 2 gene C807T polymorphism and risk of ischemic stroke: a meta-analysis. Thromb Res. 2007;119:501–510. doi: 10.1016/j.thromres.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 21.Attia J, Thakkinstian A, Wang Y, Lincz L, Parsons M, Sturm J, McGettigan P, Scott R, Meldrum C, Levi C. The PAI-1 4G/5G gene polymorphism and ischemic stroke: an association study and meta-analysis. J Stroke Cerebrovasc Dis. 2007;16:173–179. doi: 10.1016/j.jstrokecerebrovasdis.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 22.Kawas C, Gray S, Brookmeyer R, Fozard J, Zonderman A. Age-specific incidence rates of Alzheimer's disease: the Baltimore Longitudinal Study of Aging. Neurology. 2000;54:2072–2077. doi: 10.1212/wnl.54.11.2072. [DOI] [PubMed] [Google Scholar]
- 23.Hunter DJ, Kraft P, Jacobs KB, Cox DG, Yeager M, Hankinson SE, Wacholder S, Wang Z, Welch R, Hutchinson A, Wang J, Yu K, Chatterjee N, Orr N, Willett WC, Colditz GA, Ziegler RG, Berg CD, Buys SS, McCarty CA, Feigelson HS, Calle EE, Thun MJ, Hayes RB, Tucker M, Gerhard DS, Fraumeni JF, Jr, Hoover RN, Thomas G, Chanock SJ. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39:870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newton-Cheh C, Hirschhorn JN. Genetic association studies of complex traits: design and analysis issues. Mutat Res. 2005;573:54–69. doi: 10.1016/j.mrfmmm.2005.01.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table. SNPs That Have Been Analyzed by Our Group or in Published Meta-Analyses or Studies With Replication in European Descent Populations Post-200513–21 and Their Results