Abstract
Reliable identification of cancer-related mutations in TP53 is often problematic as these mutations can be randomly distributed throughout numerous codons and their relative abundance in clinical samples can fall below the sensitivity limits of conventional sequencing. To ensure the highest sensitivity in mutation detection, we adapted the recently described COLD-PCR method to employ two consecutive rounds of COLD-PCR followed by Sanger sequencing. Using this highly sensitive approach we screened 48 non-microdissected lung-adenocarcinoma samples for TP53 mutations. Twenty-four missense/frameshift TP53 mutations throughout exons 5–8 were identified in 23 of 48 (48%) lung-adenocarcinoma samples examined, including 8 low-level mutations at an abundance of ~1–17%, most of which would have been missed using conventional methodologies. The identified alterations include two rare lung-adenocarcinoma mutations, one of which is a ‘disruptive’ mutation currently undocumented in the lung cancer mutation-databases. A sample harboring a low-level mutation (~2% abundance) concurrently with a clonal mutation (80%-abundance) revealed intra-tumoral TP53 mutation heterogeneity. The ability to identify and sequence low-level mutations in the absence of elaborate micro-dissection, via COLD-PCR-based Sanger-sequencing, provides a platform for accurate mutation profiling in clinical specimens and the use of TP53 as a prognostic/predictive biomarker, evaluation of cancer risk, recurrence, and further understanding of cancer biology.
Keywords: lung cancer, TP53, low-level mutation, intra-tumor heterogeneity, COLD-PCR
Introduction
Lung adenocarcinoma represents ~48% non-small cell lung cancer (NSCLC), a major type of lung cancer (Herbst et al. 2008). The tumor suppressor gene-tumor protein p53 (TP53; MIM# 191170) is a multifunctional transcriptional factor that plays regulatory roles in cell cycle, apoptosis, and DNA repair (Foulkes 2007; Vousden and Lane 2007) and is the most frequently mutated gene in lung adenocarcinoma (Ding et al. 2008). Eighty percent of the reported TP53 mutations in lung adenocarcinoma are scattered throughout exons 5 to 8, a region encoding the DNA binding domain of the TP53 protein (Olivier et al. 2002). Tumor-encountered TP53 mutations have also been associated with poor prognosis for a range of cancers including lung cancer (Ahrendt et al. 2003; Huang et al. 1998) and loss of TP53 function is correlated with the onset of multi-drug resistance (Wallace-Brodeur and Lowe 1999). The type and position of TP53 mutations often defines their prognostic value (Huang et al. 1998) and can reveal whether they are driver or passenger mutations based on functional studies. Accordingly, reliable identification and sequencing of TP53 mutations in tumor biopsies or surgical samples is imperative. However, unless the mutation exists at or exceeds ~20–25% abundance relative to wild-type alleles, conventional Sanger sequencing does not possess adequate sensitivity for reliable detection in a clinical setting. Yet, mutations as low as 1–5% can have major clinical significance; for example epidermal growth factor receptor (EGFR; MIM# 131550) T790M mutations in lung adenocarcinoma can result in drug resistance (Engelman et al. 2006). Unlike Sanger sequencing, mutation scanning methods have the sensitivity to detect the presence of a mutation down to ~5% abundance (Bastien et al. 2008; Janne et al. 2006; Xiao and Oefner 2001). However, using mutation scanning alone does not allow for specific identification of the mutation type and position, hence their prognostic significance is difficult to assess (Huang et al. 1998; Milbury et al. 2009).
It is often perceived that low-level mutations in clinical samples occur due to a mixture of tumor cells within an excess of normal cells. This is frequently true for infiltrating tumor types, biopsies, tumor margins, lymph nodes and bodily fluids (Behn et al. 1998; Harden et al. 2004; Jenkins et al. 1998); however, tumors are known to be genetically heterogeneous, and low-level mutations may also exist in samples comprising abundant tumor cells (Janne et al. 2006). Although intra-tumor heterogeneity of TP53 mutations has been observed in certain type of skin cancers (Backvall et al. 2005), there has been no report of intra-tumor heterogeneity regarding TP53 mutations in lung adenocarcinoma to date. Heterogeneity over multiple genes is a potential consequence of cancer mutator phenotypes or genetic mosaicism, both of which can occur in lung cancer (Bielas et al. 2006; Sachidanandam et al. 2001). Whether a result of stromal contamination or intra-tumor heterogeneity, low-level TP53 mutations are important to identify and sequence, if they are to be used as cancer biomarkers.
Recently, we developed a novel PCR technology, co-amplification at lower denaturation temperature-PCR (COLD-PCR) (Li et al. 2009; Li et al. 2008b) that uses a critical denaturation temperature (Tc) to preferentially enrich low-level mutations within mixtures of wild-type and mutation containing sequences, irrespective of where an unknown mutation lies. Consequently, COLD-PCR amplification from genomic DNA yields PCR products containing high percentages of mutant alleles, thus permitting their detection. In brief, we observed that, (i) for each DNA sequence there is a critical denaturation temperature (Tc) that is typically 0.5–1.5°C lower than the amplicon melting temperature (Tm), below which PCR efficiency drops abruptly; and (ii) DNA amplicons differing by a single nucleotide reproducibly result in different amplification efficiencies relative to wild-type when the PCR denaturation temperature is set to the Tc. These new observations can be exploited during PCR amplification for the selective enrichment of minority alleles differing by one or more nucleotides at any position of a given sequence. Mutation-enrichment via COLD-PCR can be achieved via two formats: full COLD-PCR and fast COLD-PCR. In full COLD-PCR, an intermediate annealing temperature is used during PCR cycling to allow for the cross-hybridization of mutant and wild-type alleles to form heteroduplexes, which generally melt at lower denaturation temperatures than homoduplexes. Denaturation at the selected Tc preferentially denatures mismatch-containing heteroduplexes over homoduplexes, resulting to the enrichment of the mutant alleles. Mutant-to-wild type ratio can be increased up to 50% when full COLD-PCR is functioning at maximum efficiency. Fast COLD-PCR is another manifestation of COLD-PCR, which only applies for Tm-reducing mutations (e.g. G:C>A:T or G:C>T:A). In this case the cross-hybridization step is not necessary because denaturing at the Tc will preferentially denature the Tm-reducing mutant-containing sequences while wild-type DNA sequences remain double-stranded. When fast COLD-PCR is optimized to amplify at maximum efficiency, it can increase mutant prevalence up to 100%, i.e. the wild-type diminishes (Li and Makrigiorgos 2009). The choice between full and fast COLD-PCR depends on the intended use. Full COLD-PCR is used to identify all possible mutations; however the enrichment may be modest compared to fast COLD-PCR. Fast COLD-PCR can be used to provide a strong enrichment of Tm-reducing mutations, which comprise the majority of mutations in cancer (Greenman et al. 2007) and is simpler and convenient because the amplification is completed in less time.
In order to ensure the accurate identification and sequencing of low-level TP53 mutations abundant at as low as 1%, here we adapted the COLD-PCR method to employ two consecutive rounds of fast COLD-PCR followed by Sanger sequencing. We have used this highly sensitive new approach to screen 48 non-microdissected lung adenocarcinoma samples for the presence of TP53 mutations.
Materials and Methods
DNA and tumor samples
Genomic DNA from cell lines with defined TP53 mutations (T47D and SNU182, exon 6; DLD1, exon 7) was purchased from American Type Culture Collection (ATCC); the cell line SW480 (mutation in exon 8) was purchased from ATCC and genomic DNA was extracted from cultured cells. Forty-eight lung adenocarcinoma surgical samples (snap-frozen in liquid nitrogen within 1h following surgery) were obtained from the Massachusetts General Hospital Tumor Bank and used following Internal Review Board approval. Following manual macro-dissection, we isolated genomic DNA using the DNeasy™ tissue kit (Qiagen Inc.).
Determination of Critical Denaturation Temperature (Tc) of an amplicon
In order to determine Tc experimentally, the Tm of the amplicon was first identified. A real-time PCR of the target wild-type amplicon was performed in a Cepheid PCR machine in the presence of 0.1X LCGreen+ dye (Idaho Technologies Inc.) using conventional thermocycling conditions for the polymerase used (i.e. 98°C denaturation for Phusion™ DNA polymerase, New England Biolabs Inc), followed by a melting curve analysis. Next, a set of COLD-PCR reactions at graded temperatures below the Tm were performed, to identify the optimal critical denaturation temperature, Tc. In a Cepheid SmartCycler II, we set the following 10-second denaturation temperatures: a) a conventional temperature, 98°C, b) temperature equal to amplicon Tm, c) temperature equal to amplicon Tm − 0.5 °C, d) temperature equal to amplicon Tm− 1.0 °C, and e) temperature equal to amplicon Tm−1.5 °C. As validation systems we used wild-type DNA, and mutation-containing DNA diluted into wild-type DNA at a ratio of 1:10. We employed DNA with mutations that lower the melting temperature, such as G:C → A:T or G:C → T:A, to validate Tc for fast COLD-PCR. A RFLP assay, if the appropriate restriction endonuclease that discriminates mutant from wild-type is available, or direct sequencing was used post-PCR to determine the degree to which the mutant allele is enriched (Li et al. 2008b). The denaturation temperature that reproducibly produces robust PCR products combined with substantial enrichment of the mutations was selected as the Tc. By following this experimental procedure for all our amplicons, we observed that for a Cepheid PCR machine the empirical formula Tc = Tm −1 (10 seconds denaturation) typically produces high quality PCR amplicons combined with strong and reproducible mutation enrichment. This simple formula was validated and applied for all amplicons and samples tested in this investigation.
Two-round COLD-PCR
Two consecutive rounds of fast COLD-PCR, using nested primers for the second round, were performed prior to sequencing. Amplicons of 150–200 bp were designed for the first-round COLD-PCR, and shorter nested amplicons (100–150 bp) were amplified via second-round COLD-PCR. Both the first round and second round COLD-PCR reactions were performed and monitored in real-time in a Smart Cycler II thermocycler (Cepheid Inc.). Primer sequences, annealing temperatures (Ta), and critical denaturation temperatures (Tc) for all amplicons are presented in Supp. Table S1. COLD-PCR reactions contained final reagent concentrations as follows: 1X Phusion™ HF buffer (Finnzymes Inc.), 0.2 mM each dNTP, 0.2 µM forward and reverse primers (synthesized by Integrated DNA Technologies Inc), 0.1X LCGreen+ dye, 1X Phusion™ DNA polymerase and DNA template. Approximately 20–50 ng unamplified genomic DNA was used as template DNA in the first COLD-PCR for exons 5–8. Approximately 5,000 – 15,000 TP53 alleles are contained in 20–50 ng of genomic DNA, thus providing the required minimum number of mutant copies for detecting mutations with an abundance as low as 0.1%. Diluted product from the first COLD-PCR (dilution dependent upon amplification efficiency) was used as template for the second COLD-PCR using nested primers (Supp. Table S1). For amplicons with melting temperatures higher than 92 °C (e.g. 188 bp amplicon in exon 5), we added DMSO at a final concentration of 5% in order to lower its melting temperature and to increase COLD-PCR amplification efficiency. COLD-PCR cycling conditions for the first-round COLD-PCR was as follows: 98 °C, 30 s; 20–30 cycles of (98 °C, 10 s; Ta, fluorescence reading ON, 20 s; 72 °C, 20 s); then 20 cycles of (Tc, 10 s; Ta, fluorescence reading ON, 20 s; 72 °C, 20 s). PCR thermocycling conditions for the second round COLD-PCR were the same as that of the first round except that we used 5–10 cycles of conventional PCR and a Tc specific for the nested amplicon.
COLD-PCR amplification of mutation-positive samples was repeated at least three independent times starting from genomic DNA each time, and was always run in parallel with 3 to 4 wild-type DNA samples that served as negative controls. The starting DNA amount of the wild type controls was similar to the one used for the lung adenocarcinoma samples.
Sanger sequencing
PCR products were processed for Sanger di-deoxy sequencing at the molecular biology core facility of the Dana-Farber Cancer Institute. Two-round COLD-PCR products were sequenced using forward and reverse primers to achieve accurate and full coverage of each amplicon. To enable Sanger sequencing of PCR amplicons less than 100 bp in length, an M13 tail was added to the 5'-end of the forward or reverse primer and a long primer was used for sequencing as described (Diehl et al. 2008).
Pyrosequencing
Pyrosequencing was performed as described previously with the difference that we used universal biotinylated primers rather than amplicon-specific labeled primers. The design of the PCR and sequencing primers as well as the pyrosequencing reactions were carried out by EpigenDx Inc.
RFLP-based, independent verification of low-level mutations
To verify independently the low-level mutations identified via two-round COLD-PCR, a restriction fragment length polymorphism (PCR-RFLP)–based approach to achieve mutation enrichment (by using a native restriction site or an artificially-introduced site, AIRS-PCR) was used -see Supp. Methods. The digested products were examined via denaturing high performance liquid chromatography (dHPLC) to quantify the prevalence of low-level mutations.
Determination of loss of heterozygosity (LOH) at TP53 locus
The occurrence of loss of heterozygosity (LOH) was analyzed. Samples were analyzed via BstUI digestion of the polymorphism at codon 72 in Exon 4 or via a SNP array approach.
BstUI digestion of TP53 codon 72 polymorphism
To determine if TP53 loss of heterozygosity (LOH) occurred in certain lung tumor samples, an exon 4 segment (435 bp) was amplified from DNA obtained from matched normal and tumor tissue samples, using primers and PCR conditions described previously (Bastien et al. 2008). Amplified products were digested with BstUI (NEB Inc.) at 60 °C for 1 hour and analyzed via dHPLC.
SNP array
A SNP array-based approach was used to determine if TP53 loss of heterozygosity (LOH) is present in certain lung tumor tissue samples. Approximately 250 ng of genomic DNA from paired normal and lung adenocarcinoma samples were genotyped via the StyI 250K Affymetrix SNP arrays at the Dana-Farber Cancer Institute Microarray core facility. The analysis of LOH-call and gene copy number was performed via dCHIP software as described previously (Li 2008; Li et al. 2008a; Lin et al. 2004).
Analysis of TP53 mutations
The published mutation incidence, prediction of functional consequences, and yeast trans-activation assay results were obtained from the IARC TP53 database (Olivier et al. 2002).
Results
Overview of two-round COLD-PCR
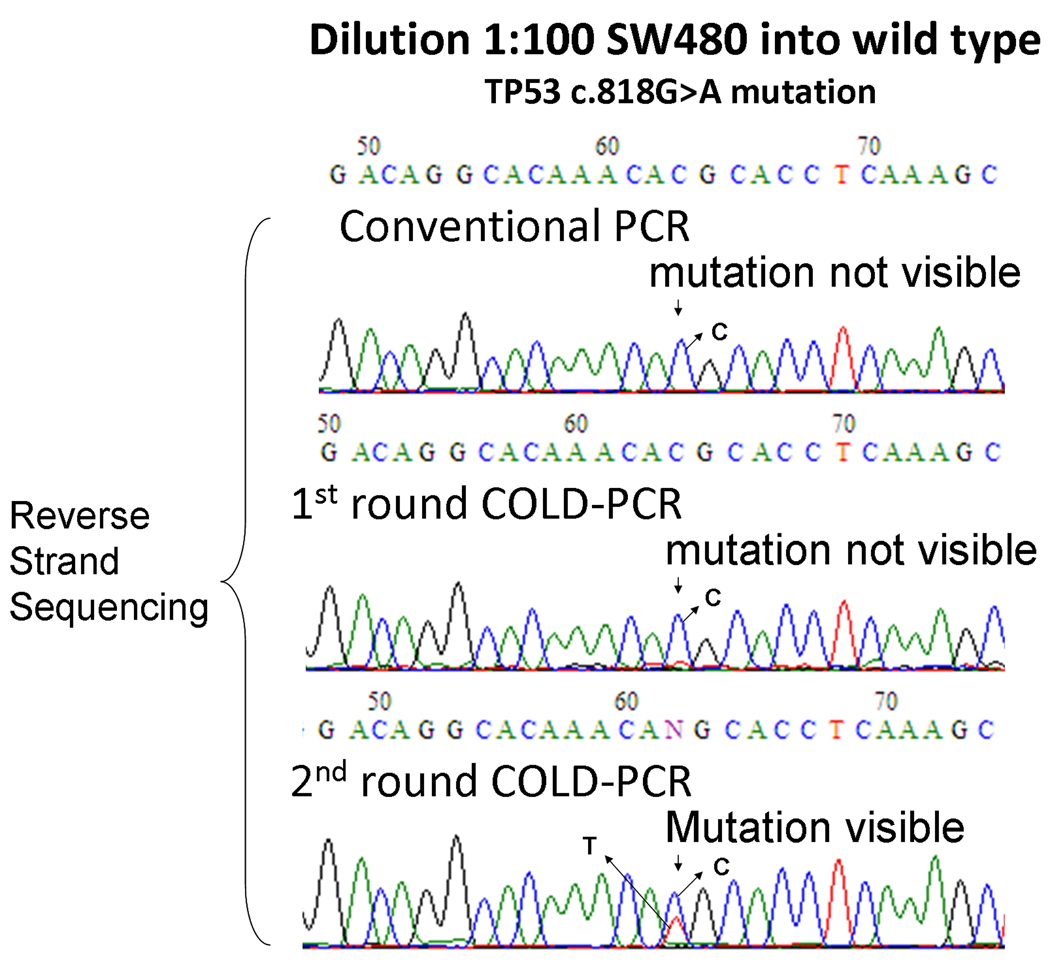
Both full and fast COLD-PCR produce substantial enrichment of mutations spanning the length of the amplicon and can be employed prior to Sanger sequencing (Delaney et al. 2009; Li and Makrigiorgos 2009), although enrichment via fast COLD-PCR is higher. The large majority (~70% (Olivier et al. 2002)) of TP53 mutations in lung adenocarcinoma exons 5–8 belongs to the Tm-reducing category. Considering this trend, as well as the desire to maximize mutation enrichment lead us to perform fast COLD-PCR in this investigation. To account for the possibility that TP53 mutations exist at such low-levels that a single round of COLD-PCR enrichment is not sufficient for identification via Sanger sequencing, two consecutive rounds of fast COLD-PCR were applied. Based on the length and position of the nested amplicon for each exon (Supp. Table S1), the sequence coverage of two-round cold PCR is determined to be 61% (c.376-c.487) in exon 5, 100% (c.560-c.672) in exon 6, 91% (c.684-c.782) in exon 7, and 74% (c.791-c.889) in exon 8. We used Phusion™ DNA polymerase (Finnzymes Inc.), a very high-fidelity DNA polymerase, to restrict the potential generation of PCR-introduced errors. In preliminary experiments, we diluted mutation-containing DNA by 100-fold into wild-type DNA and amplified each TP53 exon with two-round COLD-PCR. PCR products amplified by conventional PCR, one-round COLD-PCR, and two-round COLD-PCR were processed via Sanger sequencing to examine the degree of mutation enrichment and the potential presence of polymerase errors at different sequence positions. Results for SW480 cell line DNA (TP53 c.818G > A) are presented in Fig. 1. The SW480 mutation was not detectable in the conventional PCR and one-round COLD-PCR amplicons but was clearly detectable in two-round COLD-PCR amplicons. Furthermore, after two rounds of COLD-PCR enrichment, the background 'noise' in the sequencing chromatograms remains low, (i.e. polymerase misincorporations were not observed). Additionally, wild-type and clinical samples lacking the codon 273 mutations also demonstrate a lack of polymerase-introduced errors at all sequence positions for TP53 exon 8 (Supp. Figure S1).
Figure 1.
Two-round COLD-PCR improves the sensitivity of mutation detection over one-round COLD-PCR. Genomic DNA containing a mutation in TP53 exon 8 (SW480 cell line, TP53 c.818G > A) was diluted 100-fold in wild-type DNA and amplified by conventional, one-round and two-round COLD-PCR, followed with reverse strand sequencing. Arrows indicate the position of the G>A mutation.
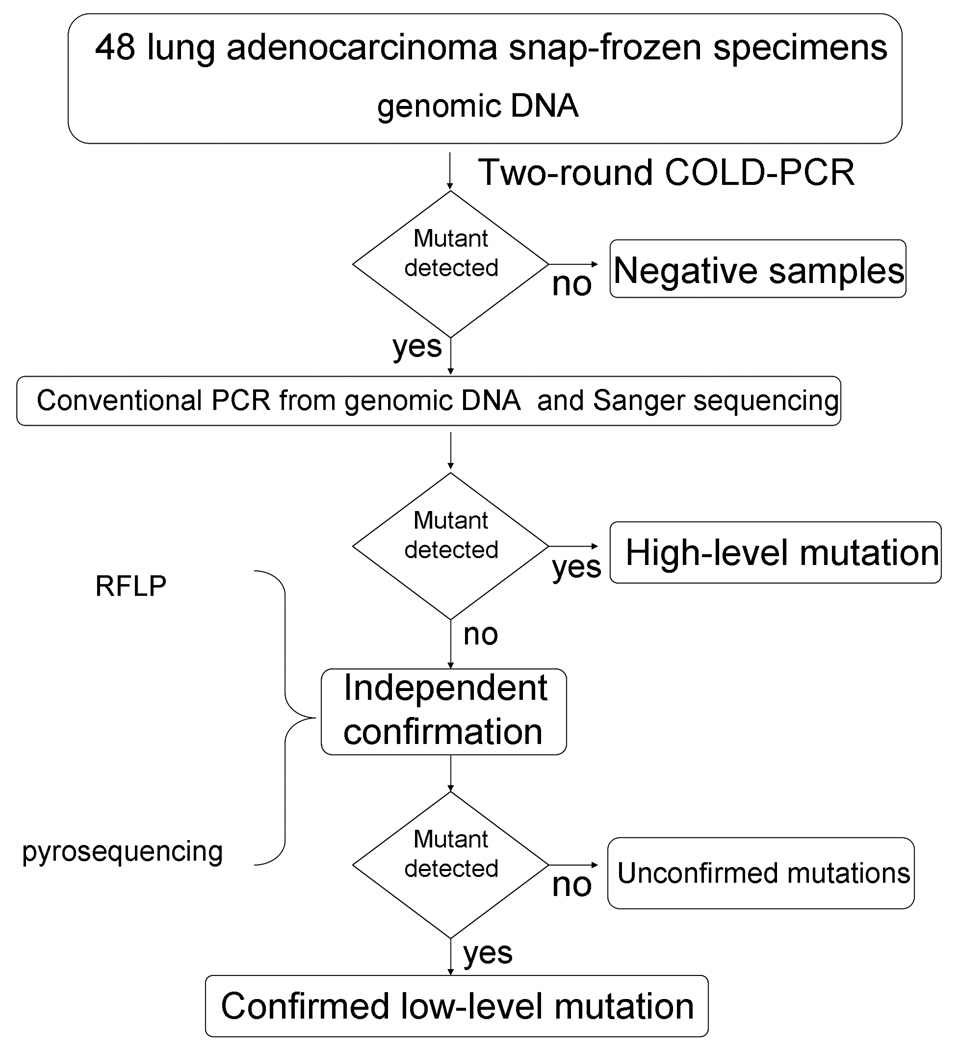
Strategy for two-round COLD-PCR-based deep-sequencing
As depicted in Fig. 2, genomic DNA, isolated from 48 surgical lung adenocarcinoma specimens, was amplified via two-round COLD-PCR and sequenced to identify mutations in TP53 exons 5 through 8. When a mutation was identified, an independent assessment via conventional PCR-sequencing directly from genomic DNA was also performed. If a mutation was detected clearly following conventional PCR-sequencing, this was deemed to be a high-level (clonal) mutation. If the mutation was not visible in the sequence chromatogram of conventional PCR amplicons, thus potentially suggesting a low-level mutation below the sensitivity of Sanger sequencing, an RFLP-based assay was designed to enrich the specific type of mutation at the sequence position that was indicated via COLD-PCR-sequencing. Following this conventional PCR-RFLP-based enrichment, Sanger sequencing on the enriched sequence was additionally used to independently confirm the low-level mutation. Alternatively, for some mutation-positive samples pyrosequencing was used as an independent verification method and also for quantifying the mutant allele frequency. Both the PCR-RFLP and the pyrosequencing-based verifications were performed starting from unamplified genomic DNA and were always run in parallel with multiple, similarly-treated wild-type DNA samples that served as negative controls.
Figure 2.
Procedure schematic used for COLD-PCR-based deep sequencing of TP53 mutations from lung adenocarcinoma samples.
Novel TP53 mutations identified via two-round COLD-PCR
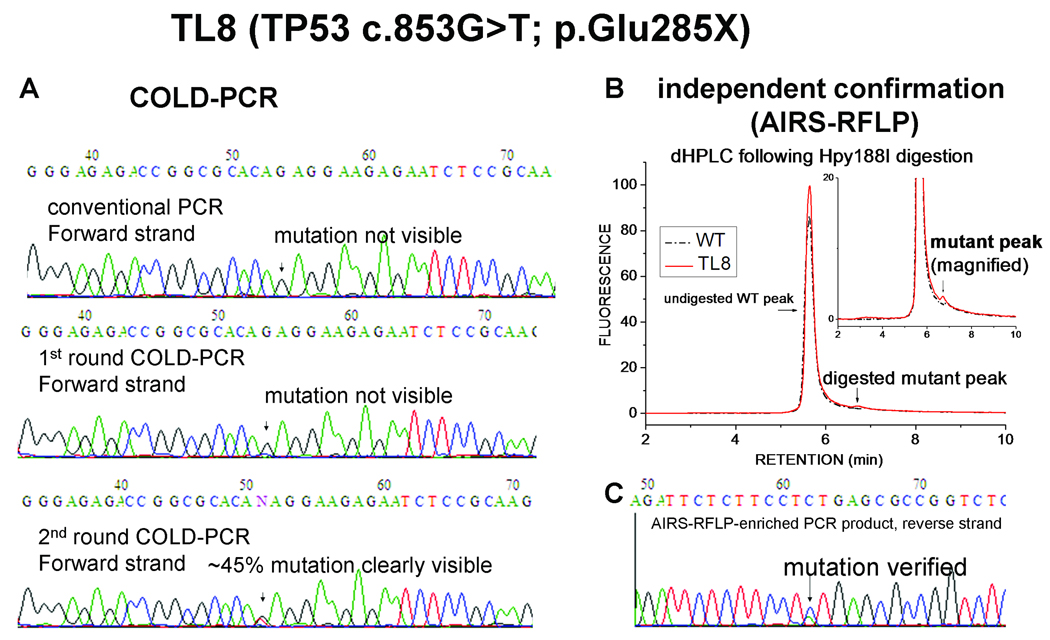
As depicted in Fig. 3 panel A, for lung tumor sample TL8, no mutation was detected in the sequence chromatograms following conventional PCR or single-round COLD-PCR amplicons. However, two-round COLD-PCR-sequencing chromatograms clearly and reproducibly exhibited a G>T mutation (Glu to STOP) at codon 285. For independent verification, the artificial introduction of a restriction site (AIRS-RFLP (Jenkins et al. 1998)) was used to introduce an Hpy188I restriction site during conventional PCR, which then enables selective post-PCR digestion of the wild-type DNA. Fig. 3 panel B presents a dHPLC chromatogram exhibiting a residual undigested PCR product (small peak) following amplification of DNA from sample TL8, while a wild-type sample treated in parallel depicts no PCR product. Comparing the area under the curves of the mutant and the wild type peaks revealed that the abundance of the mutation is approximately 1–2%. Following dHPLC analysis, the digested products were re-amplified using conventional PCR and sequenced, and the mutation was confirmed (Fig. 3 panel C). Although this mutation was reported as a rarely-encountered mutation in rectal and other cancers (Olivier et al. 2002), it has remained undocumented thus far for lung cancer in the IARC TP53, the COSMIC and the p53-Free databases.
Figure 3.
A novel lung cancer TP53 mutation in sample TL8 was identified by two-round COLD-PCR-sequencing and confirmed by AIRS-RFLP. (A) TP53 exon 8 in sample TL8 was screened via conventional PCR, one-round COLD-PCR, and two-round COLD-PCR, and the forward strand was Sanger-sequenced. (B) Genomic DNA from TL8 sample and wild-type DNA were amplified with modified primers to generate an Hpy188I restriction site to selectively digest wild-type DNA at codon 285. The Hpy188I digested product was analyzed using dHPLC. (C) The digested product was re-amplified and the reverse strand was Sanger sequenced. Arrows represent the position of mutated nucleotides in the sequence chromatograms.
A low-level, rare G>A mutation (Glu to Lys) at TP53 codon 271 in adenocarcinoma sample TL82 was also identified. This mutation was previously reported in a single case (NSCLC, NOS) out of 2710 lung cancer TP53 mutations (Olivier et al. 2002). The mutation is not apparent in sequence chromatograms following conventional PCR; however, it is clearly apparent after either one or two rounds of COLD-PCR (Supp. Figure S2A). We validated this mutation via RFLP (Supp. Figure S2B) and via pyrosequencing (Supp. Figure S3); an abundance of ~11% was demonstrated for this mutation.
Concurrent low-level and clonal TP53 mutations revealed via two-round COLD-PCR: intra-tumor TP53 heterogeneity
Using a similar approach as described above, we identified a low-level G>A mutation (Cys to Phe) at TP53 codon 277 in sample TL6. This mutation is not visible via Sanger sequencing of regular PCR amplicons; it becomes barely visible after one-round COLD-PCR and clearly evident (~70% abundance) after two-round COLD-PCR (Supp. Figure S4A). For independent verification, an AIRS-RFLP assay specific to the position and type of the putative mutation indicated by COLD-PCR was designed. An NlaIII site was introduced in the primer sequence to selectively digest the wild-type DNA following PCR amplification from genomic DNA, thereby enabling discrimination of mutant from wild-type. Supp. Figure S4B presents the dHPLC chromatogram following NlaIII digestion indicating the presence of a low-level mutation (small peak that is absent in the wild-type). Following NlaIII digestion a second PCR was used to amplify PCR fragments resisting digestion. Direct sequencing of the AIRS-RFLP-enriched PCR product further confirmed the presence and type of the mutation (Supp. Figure S4C). This G>A mutation exhibits a ~2–3% mutation abundance in sample TL6. Moreover, in this sample we identified a second, clonal G>A mutation (Val > Met) at codon 197 present at approximately 80% abundance (Supp. Figure S4D). The concurrent presence of a clonal mutation and a low-level mutation in the same sample indicates that the low-level mutations are likely a result of intra-tumor TP53 mutational heterogeneity, as opposed to dilution by admixture of normal cells in these lung adenocarcinoma surgical samples.
Range of TP53 mutations identified via two-round COLD-PCR
TP53 mutations throughout exons 5–8 were identified in 23 out of 48 (48%) lung adenocarcinoma samples examined, and 24 mutations were identified in total (listed in Table 1). The resulting mutation spectrum for these 48 samples includes mutational ‘hot spots’ common in lung cancer such as in codons 157, 158, 245, 248, 249, 273 and 282 (Table 1). All the mutations are either missense mutations or frameshift mutations caused by micro-deletions. Among the 24 mutations, 8 (33%) have an abundance below the sensitivity of Sanger sequencing and reliable identification using conventional-PCR is not possible (Table 1). Presence of the low-level mutations identified via COLD-PCR was confirmed via independent methods (Supp. Figure S5–10). Except mutations in TL6 and TL8, all other low-level mutations were quantified by pyrosequencing (Table 1). Among the eight low-level mutations identified, five are G>T transversions and three are G>A transitions.
Table 1.
TP53 mutations identified via two-round COLD-PCR-based Sanger sequencing
| Samples | Mutation (nt) |
Mutation (aa) |
Detectable via conventional PCR - Sanger sequencing |
Independent confirmation |
Mutation abundance |
|---|---|---|---|---|---|
| TL96 | c.747G>T | p.Arg249Ser | No | R, P | 6.7% |
| TL121 | c.733G>A | p.Gly245Ser | No | R, P | 7% |
| TL119 | c.730G>T | p.Gly244Cys | No | R, P | 7.5% |
| TL6a | c.830G>T | p.Cys277Phe | No | R | ~2–3% |
| TL8 | c.853G>T | p.Glu285X | No | R | ~1–2% |
| TL82 | c.811G>A | p.Glu271Lys | No | R, P | 11% |
| TL135 | c.646G>A | p.Val216Met | No | P | 17% |
| TL22 | c.469G>T | p.Val157Phe | No | P | 16.6% |
| TL134 | c.733G>T | p.Gly245Cys | Yes | - | ~45% |
| TL125 | c.742C>T | p.Arg248Trp | Yes | - | ~90% |
| TL15 | c.818G>A | p.Arg273His | Yes | - | ~40% |
| TL123 | c.810_819del | p.Phe270fs | Yes | - | ~50% |
| TL14 | c.844C>G | p.Arg282Gly | Yes | - | ~60% |
| TL18 | c.839G>A | p.Arg280Lys | Yes | - | ~70% |
| TL92 | c.845del | p.Arg282fs | Yes | - | ~50% |
| TL124 | c.809_818del | p.Phe270fs | Yes | - | ~60% |
| TL5 | c.830G>T | p.Cys277Phe | Yes | - | ~37% |
| TL136 | c.601_602del | p.Leu201fs | Yes | - | ~80% |
| TL106 | c.460del | p.Gly154fs | Yes | - | ~70% |
| TL107 | c.461G>T | p.Gly154Val | Yes | - | ~33% |
| TL112 | c.473G>T | p.Arg158Leu | Yes | - | ~50% |
| TL117 | c.388C>G | p.Leu130Val | Yes | - | ~80% |
| TL6a | c.589G>A | p.Val197Met | Yes | - | ~80% |
| TL97 | c.743G>T | p.Arg248Leu | Yes | - | ~40% |
Reference sequence used for the TP53 gene was RefSeq NM_000546.4. Nucleotide numbering reflects cDNA numbering with +1 corresponding to the A of the ATG translation initiation codon in the reference sequence. The initiation codon is codon 1
represents two concurrent mutations in a single sample. R and P represent independent confirmation by RFLP, pyrosequencing, respectively. For mutations detectable via conventional PCR-Sanger sequencing, no independent confirmation was provided.
Analyzing TP53 polymorphism at codon 72 and our SNP-array data (see Supp. Methods), we discovered concurrent TP53 LOH with low-level TP53 mutation in samples TL82 (Supp. Figure S2C) and TL22 (Supp. Figure S11). The concurrent low-level TP53 mutations with LOH of TP53 in TL22 and TL82 is consistent with a classical “two-hit” cancer genetic change with one TP53 allele mutated and the other allele lost (25, 27).
Discussion
Identification of the type and position of mutations by COLD-PCR-sequencing reveals their potential impact. For example, the novel low-level nonsense (Glu to STOP) mutation at codon 285 of sample TL8 results to a truncated protein. In squamous-cell head and neck carcinoma this mutation is known as a ‘disruptive mutation’ strongly associated with decreased overall survival (Poeta et al. 2007). Another interesting observation is in regard to the relative potential significance of the aforementioned concurrent mutations in sample TL6. The clonal mutation Val 197 Met has an 80% abundance while the low-level mutation Cys 277 Phe, has a ~2–3% abundance; yet, despite being a low-level mutation, the latter mutation is known to have a more prominent effect than the former. Functional assessment of the Val 197 Met clonal and the Cys 277 Phe low-level mutation in a yeast assay (Kato et al. 2003) showed that these two mutations belong to partially-functional and non-functional transactivation classes, respectively, with a ~125-fold difference in transactivation capability. Therefore identifying the type and position of mutation is of primary importance. It is noteworthy that identification of the type and position of mutations is a feature that established mutation scanning methods (HRM, SSCP) cannot provide unless followed by digital-PCR-based 'deep' sequencing. Additionally, because the inclusion of low-level mutations are important in accurately defining the mutational spectra (Fox et al. 2009), it is possible that large scale sequencing projects, such as The Cancer Genome Atlas (TCGA), that aim to catalogue cancer mutations may provide only a partial view of the lung cancer mutation spectra (Ding et al. 2008), thus missing low-level mutation spectra.
The concurrent presence of a clonal (G>A) and a low-level (G>T) mutation in the same tumor for sample TL6 is an interesting observation in the present study. This finding argues against substantial stromal admixture and indicates the presence of intra-tumor TP53 mutational heterogeneity in this sample. Lung adenocarcinoma is often a mixture of several histology subtypes of cancer, such as acinar, papillary, bronchio-alveolar, and solid adenocarcinoma with mucin formation (Mountain 1997). Therefore, the clonal and low-level TP53 mutations could reside in two different histological subtypes within the same sample. It is also possible that the low-level TP53 mutations maybe present in normal lung epithelial cells, due to a “field-cancerization” effect caused by tobacco carcinogen damage (Slaughter et al. 1953; Strong et al. 1984). Yet another possibility is that the two TL6 mutations represent a form of doublet mutations (two mutations in a single cell, possibly on the same allele) (Chen et al. 2008). Whatever the origin of the mutational heterogeneity in sample TL6, intra-tumor mutational heterogeneity may play an important role in tumor evolution and/or in personalized treatment and drug resistance (Engelman et al. 2006). Although there are reports showing intra-tumor heterogeneity of EGFR mutations in lung adenocarcinoma, to our knowledge this is the first documented study demonstrating TP53 mutational heterogeneity in this type of cancer.
One way to assess the importance of not missing TP53 mutations in cancer samples is to consider the extensive efforts applied towards association of TP53 mutations with prognosis. While earlier studies suggested that TP53 mutations are not significantly associated with poor prognosis (Olivier et al. 2002; Schiller et al. 2001), the current understanding is that TP53 mutations are associated with high grade lung tumors (Ding et al. 2008; Nakanishi et al. 2009) and that there is a significant correlation with poor prognosis in early stage lung cancer (Ahrendt et al. 2003; Huang et al. 1998). Investigations of different cancers, such as colon cancer, remain controversial in regards to the prognostic significance of TP53 mutations, with 12 studies showing positive association and 7 studies showing a lack of association (Olivier et al. 2002). While work in this area is ongoing, in view of the 8 low-level TP53 mutations of functional significance found in the 48 surgical lung samples examined, it seems unlikely that the controversy can be settled unless methods that accurately identify and sequence such low-level mutations are used, such as COLD-PCR-based sequencing. Arguably, if an elaborate micro-dissection scheme had been applied to the 8 samples found to contain low-level mutations, it is possible that more mutations might have been detected via Sanger sequencing or pyro-sequencing. However, micro-dissection can be quite laborious, is not always possible when infiltrating tumor types are studied, and it would not help with detection of low-level TP53 mutations in samples with intra-tumor heterogeneity such as TL6, since normal cell contamination was clearly not substantial in this sample.
We have successfully applied two-round COLD-PCR to enable thorough-sequencing in clinical lung adenocarcinoma samples and have demonstrated increased mutation enrichment relative to one-round COLD-PCR. However, the present study also has limitations as it was designed to maximize mutation enrichment and convenience by assessing only the Tm-reducing mutations (G:C>A:T and G:C>T:A), which comprise about 70% of all possible mutations in lung cancer (Olivier et al. 2002). In future investigations it would be advantageous to optimize the use of the general form of COLD-PCR (full-COLD-PCR) prior to sequencing in order to enrich and identify all types of mutations. At present, COLD-PCR has only been applied in the study of low-level mutations in specific genes rather than genome wide scanning. However, multiplex or highly parallel COLD-PCR can also be envisioned in view of the fact that amplicons of equal Tm are expected to also have the same Tc (Tc=Tm−1 under the experimental conditions presented here). Therefore, in principle, numerous iso-Tm amplicons could be enriched for mutations in a single COLD-PCR reaction.
Digital PCR-based approaches, including single-molecule PCR performed in the course of second generation sequencing are alternative powerful approaches to perform deep-sequencing in cancer samples (Rohlin et al. 2009; Thomas et al. 2006); these approaches require considerable oversampling (e.g. at least 2,500-fold for the detection of 2 mutant alleles in 100 copies of wild-type alleles in the 454 system, see http://www.454.com/downloads/protocols/5_AmpliconSequencing.pdf). Oversampling unavoidably reduces throughput, increases the cost, and is inefficient when a small number of sequences are examined. In contrast, COLD-PCR is simple, inexpensive, and does not require additional reagents or instrumentation. Alternatively, utilization of COLD-PCR to generate amplicons for examination via next-generation sequencing may potentially enable the simultaneous combination of deep-sequencing with high-throughput performance.
In summary, we have demonstrated an advantageous approach to discover and sequence low-level TP53 mutations in tumors via two-round COLD-PCR, and have identified intra-tumor TP53 heterogeneity as well as novel low-level TP53 mutations in lung adenocarcinoma. This technique may also potentially be applied in assessing tumor margins, detecting pre-cancerous genetic changes, screening DNA from bodily fluids, or when isolation of a relatively pure population of tumor cells is difficult, such as in pancreatic cancer or tissue from human airways (Park et al. 1999). Identification and determination of the consequences of low-level mutations may further advance our understanding of the origin and evolution of cancer, and assist in the evaluation of cancer risk, recurrence, and choice of treatment.
Supplementary Material
Acknowledgments
JL was supported by the JCRT Foundation and CM was supported by NIH training grant 5 T32 CA09078. This work was supported in part by NIH grants CA-138280 and CA-111994.
Footnotes
Supporting Information for this preprint is available from the Human Mutation editorial office upon request (humu@wiley.com)
References
- Ahrendt SA, Hu Y, Buta M, McDermott MP, Benoit N, Yang SC, Wu L, Sidransky D. p53 mutations and survival in stage I non-small-cell lung cancer: results of a prospective study. J Natl Cancer Inst. 2003;95:961–970. doi: 10.1093/jnci/95.13.961. [DOI] [PubMed] [Google Scholar]
- Backvall H, Asplund A, Gustafsson A, Sivertsson A, Lundeberg J, Ponten F. Genetic tumor archeology: microdissection and genetic heterogeneity in squamous and basal cell carcinoma. Mutat Res. 2005;571:65–79. doi: 10.1016/j.mrfmmm.2004.10.011. [DOI] [PubMed] [Google Scholar]
- Bastien R, Lewis TB, Hawkes JE, Quackenbush JF, Robbins TC, Palazzo J, Perou CM, Bernard PS. High-throughput amplicon scanning of the TP53 gene in breast cancer using high-resolution fluorescent melting curve analyses and automatic mutation calling. Hum Mutat. 2008;29:757–764. doi: 10.1002/humu.20726. [DOI] [PubMed] [Google Scholar]
- Behn M, Qun S, Pankow W, Havemann K, Schuermann M. Frequent detection of ras and p53 mutations in brush cytology samples from lung cancer patients by a restriction fragment length polymorphism-based "enriched PCR" technique. Clin Cancer Res. 1998;4:361–371. [PubMed] [Google Scholar]
- Bielas JH, Loeb KR, Rubin BP, True LD, Loeb LA. Human cancers express a mutator phenotype. Proc Natl Acad Sci U S A. 2006;103:18238–18242. doi: 10.1073/pnas.0607057103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Feng J, Buzin CH, Sommer SS. Epidemiology of doublet/multiplet mutations in lung cancers: evidence that a subset arises by chronocoordinate events. PLoS ONE. 2008;3:e3714. doi: 10.1371/journal.pone.0003714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney D, Diss TC, Presneau N, Hing S, Berisha F, Idowu BD, O'Donnell P, Skinner JA, Tirabosco R, Flanagan AM. GNAS1 mutations occur more commonly than previously thought in intramuscular myxoma. Mod Pathol. 2009;22:718–724. doi: 10.1038/modpathol.2009.32. [DOI] [PubMed] [Google Scholar]
- Diehl F, Li M, He Y, Kinzler KW, Vogelstein B, Dressman D. BEAMing: single-molecule PCR on microparticles in water-in-oil emulsions. Nat Methods. 2006;3:551–559. doi: 10.1038/nmeth898. [DOI] [PubMed] [Google Scholar]
- Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, Thornton K, Agrawal N, Sokoll L, Szabo SA, Kinzler KW, Vogelstein B, Diaz LA., Jr Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14:985–990. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan MB, Fulton L, Fulton RS, Zhang Q, Wendl MC, Lawrence MS, Larson DE, Chen K, Dooling DJ, Sabo A, Hawes AC, Shen H, Jhangiani SN, Lewis LR, Hall O, Zhu Y, Mathew T, Ren Y, Yao J, Scherer SE, Clerc K, Metcalf GA, Ng B, Milosavljevic A, Gonzalez-Garay ML, Osborne JR, Meyer R, Shi X, Tang Y, Koboldt DC, Lin L, Abbott R, Miner TL, Pohl C, Fewell G, Haipek C, Schmidt H, Dunford-Shore BH, Kraja A, Crosby SD, Sawyer CS, Vickery T, Sander S, Robinson J, Winckler W, Baldwin J, Chirieac LR, Dutt A, Fennell T, Hanna M, Johnson BE, Onofrio RC, Thomas RK, Tonon G, Weir BA, Zhao X, Ziaugra L, Zody MC, Giordano T, Orringer MB, Roth JA, Spitz MR, Wistuba B, II, Ozenberger PJ, Good AC, Chang DG, Beer MA, Watson M, Ladanyi S, Broderick A, Yoshizawa WD, Travis W, Pao MA, Province GM, Weinstock HE, Varmus SB, Gabriel ES, Lander RA, Gibbs M, Meyerson, Wilson RK. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman JA, Mukohara T, Zejnullahu K, Lifshits E, Borras AM, Gale CM, Naumov GN, Yeap BY, Jarrell E, Sun J, Tracy S, Zhao X, Heymach JV, Johnson BE, Cantley LC, Janne PA. Allelic dilution obscures detection of a biologically significant resistance mutation in EGFR-amplified lung cancer. J Clin Invest. 2006;116:2695–2706. doi: 10.1172/JCI28656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulkes WD. p53--master and commander. N Engl J Med. 2007;357:2539–2541. doi: 10.1056/NEJMp0707422. [DOI] [PubMed] [Google Scholar]
- Fox EJ, Salk JJ, Loeb LA. Cancer genome sequencing--an interim analysis. Cancer Res. 2009;69:4948–4950. doi: 10.1158/0008-5472.CAN-09-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, Edkins S, O'Meara S, Vastrik I, Schmidt EE, Avis T, Barthorpe S, Bhamra G, Buck G, Choudhury B, Clements J, Cole J, Dicks E, Forbes S, Gray K, Halliday K, Harrison R, Hills K, Hinton J, Jenkinson A, Jones D, Menzies A, Mironenko T, Perry J, Raine K, Richardson D, Shepherd R, Small A, Tofts C, Varian J, Webb T, West S, Widaa S, Yates A, Cahill DP, Louis DN, Goldstraw P, Nicholson AG, Brasseur F, Looijenga L, Weber BL, Chiew YE, DeFazio A, Greaves MF, Green AR, Campbell P, Birney E, Easton DF, Chenevix-Trench G, Tan MH, Khoo SK, Teh BT, Yuen ST, Leung SY, Wooster R, Futreal PA, Stratton MR. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harden SV, Thomas DC, Benoit N, Minhas K, Westra WH, Califano JA, Koch W, Sidransky D. Real-time gap ligase chain reaction: a rapid semiquantitative assay for detecting p53 mutation at low levels in surgical margins and lymph nodes from resected lung and head and neck tumors. Clin Cancer Res. 2004;10:2379–2385. doi: 10.1158/1078-0432.ccr-03-0405. [DOI] [PubMed] [Google Scholar]
- Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359:1367–1380. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Taki T, Adachi M, Konishi T, Higashiyama M, Miyake M. Mutations in exon 7 and 8 of p53 as poor prognostic factors in patients with non-small cell lung cancer. Oncogene. 1998;16:2469–2477. doi: 10.1038/sj.onc.1201776. [DOI] [PubMed] [Google Scholar]
- Janne PA, Borras AM, Kuang Y, Rogers AM, Joshi VA, Liyanage H, Lindeman N, Lee JC, Halmos B, Maher EA, Distel RJ, Meyerson M, Johnson BE. A rapid and sensitive enzymatic method for epidermal growth factor receptor mutation screening. Clin Cancer Res. 2006;12:751–758. doi: 10.1158/1078-0432.CCR-05-2047. [DOI] [PubMed] [Google Scholar]
- Jenkins GJ, Chaleshtori MH, Song H, Parry JM. Mutation analysis using the restriction site mutation (RSM) assay. Mutat Res. 1998;405:209–220. doi: 10.1016/s0027-5107(98)00138-9. [DOI] [PubMed] [Google Scholar]
- Kato S, Han SY, Liu W, Otsuka K, Shibata H, Kanamaru R, Ishioka C. Understanding the function-structure and function-mutation relationships of p53 tumor suppressor protein by high-resolution missense mutation analysis. Proc Natl Acad Sci U S A. 2003;100:8424–8429. doi: 10.1073/pnas.1431692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C. Automating dChip: toward reproducible sharing of microarray data analysis. BMC Bioinformatics. 2008;9:231. doi: 10.1186/1471-2105-9-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Beroukhim R, Weir BA, Winckler W, Garraway LA, Sellers WR, Meyerson M. Major copy proportion analysis of tumor samples using SNP arrays. BMC Bioinformatics. 2008a;9:204. doi: 10.1186/1471-2105-9-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Makrigiorgos GM. COLD-PCR: a new platform for highly improved mutation detection in cancer and genetic testing. Biochem Soc Trans. 2009;37:427–432. doi: 10.1042/BST0370427. [DOI] [PubMed] [Google Scholar]
- Li J, Wang L, Janne PA, Makrigiorgos GM. Coamplification at Lower Denaturation Temperature-PCR Increases Mutation-Detection Selectivity of TaqMan-Based Real-Time PCR. Clin Chem. 2009;55:748–756. doi: 10.1373/clinchem.2008.113381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wang L, Mamon H, Kulke MH, Berbeco R, Makrigiorgos GM. Replacing PCR with COLD-PCR enriches variant DNA sequences and redefines the sensitivity of genetic testing. Nat Med. 2008b;14:579–584. doi: 10.1038/nm1708. [DOI] [PubMed] [Google Scholar]
- Lin M, Wei LJ, Sellers WR, Lieberfarb M, Wong WH, Li C. dChipSNP: significance curve and clustering of SNP-array-based loss-of-heterozygosity data. Bioinformatics. 2004;20:1233–1240. doi: 10.1093/bioinformatics/bth069. [DOI] [PubMed] [Google Scholar]
- Milbury CA, Li J, Makrigiorgos GM. PCR-based methods for the enrichment of minority alleles and mutations. Clin Chem. 2009;55:632–640. doi: 10.1373/clinchem.2008.113035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mountain CF. Revisions in the International System for Staging Lung Cancer. Chest. 1997;111:1710–1717. doi: 10.1378/chest.111.6.1710. [DOI] [PubMed] [Google Scholar]
- Nakanishi H, Matsumoto S, Iwakawa R, Kohno T, Suzuki K, Tsuta K, Matsuno Y, Noguchi M, Shimizu E, Yokota J. Whole genome comparison of allelic imbalance between noninvasive and invasive small-sized lung adenocarcinomas. Cancer Res. 2009;69:1615–1623. doi: 10.1158/0008-5472.CAN-08-3218. [DOI] [PubMed] [Google Scholar]
- Olivier M, Eeles R, Hollstein M, Khan MA, Harris CC, Hainaut P. The IARC TP53 database: new online mutation analysis and recommendations to users. Hum Mutat. 2002;19:607–614. doi: 10.1002/humu.10081. [DOI] [PubMed] [Google Scholar]
- Park I, Wistuba A, II, Maitra S, Milchgrub AK, Virmani JD, Minna, Gazdar AF. Multiple clonal abnormalities in the bronchial epithelium of patients with lung cancer. J Natl Cancer Inst. 1999;91:1863–1868. doi: 10.1093/jnci/91.21.1863. [DOI] [PubMed] [Google Scholar]
- Poeta ML, Manola J, Goldwasser MA, Forastiere A, Benoit N, Califano JA, Ridge JA, Goodwin J, Kenady D, Saunders J, Westra W, Sidransky D, Koch WM. TP53 mutations and survival in squamous-cell carcinoma of the head and neck. N Engl J Med. 2007;357:2552–2561. doi: 10.1056/NEJMoa073770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohlin A, Wernersson J, Engwall Y, Wiklund L, Bjork J, Nordling M. Parallel sequencing used in detection of mosaic mutations: comparison with four diagnostic DNA screening techniques. Hum Mutat. 2009;30:1012–1020. doi: 10.1002/humu.20980. [DOI] [PubMed] [Google Scholar]
- Sachidanandam R, Weissman D, Schmidt SC, Kakol JM, Stein LD, Marth G, Sherry S, Mullikin JC, Mortimore BJ, Willey DL, Hunt SE, Cole CG, Coggill PC, Rice CM, Ning Z, Rogers J, Bentley DR, Kwok PY, Mardis ER, Yeh RT, Schultz B, Cook L, Davenport R, Dante M, Fulton L, Hillier L, Waterston RH, McPherson JD, Gilman B, Schaffner S, Van Etten WJ, Reich D, Higgins J, Daly MJ, Blumenstiel B, Baldwin J, Stange-Thomann N, Zody MC, Linton L, Lander ES, Atshuler D. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409:928–933. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- Schiller JH, Adak S, Feins RH, Keller SM, Fry WA, Livingston RB, Hammond ME, Wolf B, Sabatini L, Jett J, Kohman L, Johnson DH. Lack of prognostic significance of p53 and K-ras mutations in primary resected non-small-cell lung cancer on E4592: a Laboratory Ancillary Study on an Eastern Cooperative Oncology Group Prospective Randomized Trial of Postoperative Adjuvant Therapy. J Clin Oncol. 2001;19:448–457. doi: 10.1200/JCO.2001.19.2.448. [DOI] [PubMed] [Google Scholar]
- Slaughter DP, Southwick HW, Smejkal W. Field cancerization in oral stratified squamous epithelium; clinical implications of multicentric origin. Cancer. 1953;6:963–968. doi: 10.1002/1097-0142(195309)6:5<963::aid-cncr2820060515>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Strong MS, Incze J, Vaughan CW. Field cancerization in the aerodigestive tract--its etiology, manifestation, and significance. J Otolaryngol. 1984;13:1–6. [PubMed] [Google Scholar]
- Thomas RK, Nickerson E, Simons JF, Janne PA, Tengs T, Yuza Y, Garraway LA, LaFramboise T, Lee JC, Shah K, O'Neill K, Sasaki H, Lindeman N, Wong KK, Borras AM, Gutmann EJ, Dragnev KH, DeBiasi R, Chen TH, Glatt KA, Greulich H, Desany B, Lubeski CK, Brockman W, Alvarez P, Hutchison SK, Leamon JH, Ronan MT, Turenchalk GS, Egholm M, Sellers WR, Rothberg JM, Meyerson M. Sensitive mutation detection in heterogeneous cancer specimens by massively parallel picoliter reactor sequencing. Nat Med. 2006;12:852–855. doi: 10.1038/nm1437. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8:275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- Wallace-Brodeur RR, Lowe SW. Clinical implications of p53 mutations. Cell Mol Life Sci. 1999;55:64–75. doi: 10.1007/s000180050270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W, Oefner PJ. Denaturing high-performance liquid chromatography: A review. Hum Mutat. 2001;17:439–474. doi: 10.1002/humu.1130. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.