Abstract
Purpose
Explore the retinoschisin (RS1) protein biochemical phenotype from an RS1 exon-5 deletion/insertion frame-shift mutation in an X-linked retinoschisis (XLRS) family and describe the clinical and electrophysiological features.
Methods
Six XLRS males underwent ophthalmologic examination and electroretinogram (ERG) recording. The RS1 gene was sequenced. Mutant RS1-RNA and protein expression were assessed by transfecting COS-7 cells with minigene constructs.
Results
All six males carried the RS1 c354del1-ins18 mutation in which an 18-bp insertion replaced nucleotide 354, duplicating the adjacent upstream intron-4-to-exon-5 junction and causing a premature termination codon downstream. Analysis indicated normal pre-mRNA splicing producing mRNA transcripts. Truncated RS1 protein was expressed transiently but was degraded rapidly by a proteasomal pathway rather than by nonsense mediated mRNA decay. Two boys, 1.5 and 5 years old (y/o), had foveal cysts and minimal peripheral schisis, and retained near-normal scotopic b-wave amplitude and normal ERG waveforms. The 5 y/o's ERG was reduced when repeated three years later. Four older XLRS relatives 32-45 y/o had substantial b-wave loss and strongly “electronegative” ERGs; three had overt macular atrophy. Cross-sectional family analysis showed the b/a-wave amplitude ratio as inversely related to age in the six males.
Conclusions
The c354del1-ins18 mutation causes an RS1 null biochemical phenotype and a progressive clinical phenotype in a 5-y/o male, while the older XLRS relatives had macular atrophy and marked ERG changes. The phenotypic heterogeneity with age by cross-sectional study of this family mutation argues that XLRS disease is not stationary and raises questions regarding factors involved in progression.
X-linked juvenile retinoschisis (XLRS) is a highly penetrant recessive retinal dystrophy characterized by early-onset central visual loss from bilateral foveo-macular cystic cavities involving the inner retina1 and additional retinal layers.2,3 Approximately half the affected males also exhibit peripheral retinoschisis.4 XLRS is one of the more common causes of juvenile macular degeneration in males, with estimated prevalence at 1:5,000 to 1:25,000.5–7
The RS1 gene contains six exons and encodes a 24-kDa secreted retinoschisin (RS1) protein, that includes a conserved discoidin domain8 homologous to proteins implicated in cell adhesion and cell–cell interactions.9 RS1 is highly expressed in photoreceptor cells and also in neurons of the inner retina.10–12 Over 150 mutations have been described across the small RS1 gene.13 The majority are missense mutations in exons 4–6; deletions, insertions, nonsense, and splice site mutations have also been reported.6–8,14–16
The XLRS phenotype is quite variable. The most common clinical finding is bilateral foveo-macular schisis.17 Retinal fundus examination and optical coherence tomography (OCT) scans of older affected males often demonstrate flattening and coalescence of the foveal schisis cavities.18 The retinal pigment epithelium (RPE) may show granularity and atrophy in later ages.18,19 The full-field electroretinogram (ERG) in XLRS generally shows a greater loss of the positive-going b-wave compared to the negative-going a-wave,20–23 although relative b-wave preservation has also been described.6,24 As the b-wave from bipolar cells lies post-synaptic to the photoreceptor a-wave, the ERG provides evidence of inner retinal dysfunction possibly involving the photoreceptor synapse.
Genotype–phenotype correlations can help to elucidate molecular genetic mechanisms underlying macular degeneration induced by RS1 mutations. We describe the clinical features of an XLRS family with an RS1 exon-5 deletion/insertion mutation (c354del1-ins18). The ERG changes observed across three years in a 5 year old (y/o) affected boy, and the ERG differences between the younger versus middle-age affected males raises questions about the nature of progression in XLRS disease.
Methods
Subjects
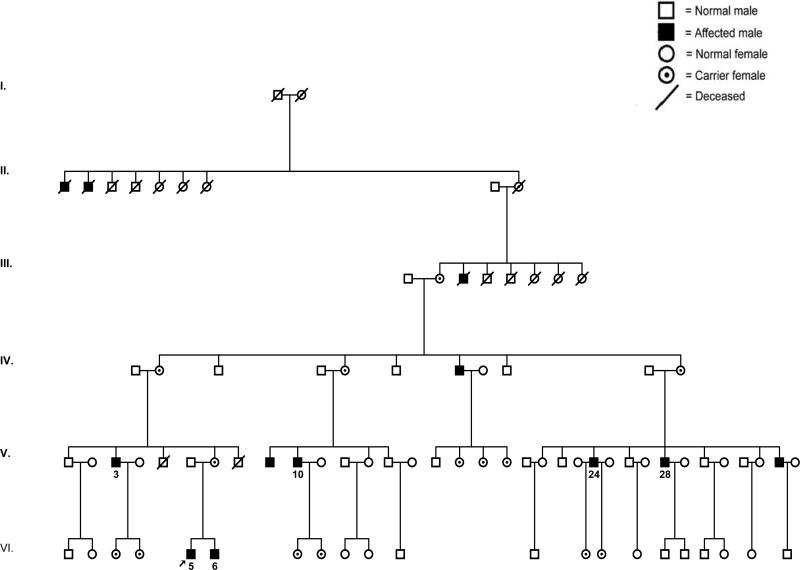
We studied six affected males in a multi-generation Caucasian Hispanic XLRS family (Fig. 1) including two young affected boys (VI.5, proband and brother VI.6), 5 and 1.5 y/o, and four older maternal male relatives, 32–45 y/o. The study protocols were approved by the National Institutes of Health IRB, consonant with the tenants of the Declaration of Helsinki, and the subjects gave informed consent.
Figure 1.
Pedigree of the XLRS family. The proband is indicated by an arrow.
Clinical Examination
Subjects were examined by fundus biomicroscopy and indirect ophthalmoscopy. Best-corrected Snellen visual acuity, Goldman kinetic perimetry (Haag-Streit, Bern, Switzerland) and optical coherence tomography (Stratus OCT 3, Carl Zeiss Meditec) were performed. The central visual absolute luminance threshold was measured after 30 minutes of dark adaptation with a Goldmann-Weekers adaptometer (Haag-Streit).
ERG Recording
ERG responses were elicited by full field flash stimuli (Utas 2000, LKC Technologies, Gaithersburg, MD) and Espion 1 (Diagnosys Inc., Lowell, MA) after pupil dilation (phenylephrine hydrochloride 2.5% and tropicamide 1%) and 30 minutes of dark adaptation. Burian-Allen bipolar corneal ERG electrodes (Hansen Ophthalmic Instruments, Iowa City, IA) were place after topical corneal anesthesia (propacaine hydrochloride 0.5%). Dark-adapted rod-mediated and combined rod-plus-cone ERG responses were recorded. Cone-mediated responses were then elicited after 10-minutes light adaption. ERG responses were amplified at 0.3–300 Hz, digitized, and stored. ERG a-wave and b-wave amplitudes (μV) and implicit times (milliseconds) were compared with 120 unaffected subjects.
PCR Amplification and Mutation Detection
Genomic DNA was isolated from peripheral blood leukocytes. Exons 1–6 and the intron/exon boundaries of RS1 were amplified by PCR using a combination of primer sets as previously described8 and were sequenced (ABI PRISM 310 Genetic Analyzer, Applied Biosystem, Foster City, CA).
Expression Vectors
RS1 cDNA (NM 000330) was amplified from human retinal total RNA using RT-PCR and cloned into the pCR II-TOPO vector (Invitrogen, Carlsbad, CA).25 The entire RS1 cDNA coding sequence was further PCR-amplified using Not1 primed sense primer (5′-GCGGCCGCGCCACCATGTCACGCAAGATAGAAGGCTT-3′) and Xho1 primed antisense primer (5′-CTCGAGTCAGGCACACTTGCTGACGCAC-3′). The PCR product was digested by Not1 and Xho1, and subcloned in pCMV-Tag4A mammalian expression vector (Stratagene, La Jolla, CA) between the Not1/Xho1 enzyme sites (Tag4A-RS1). The sense primer design included the Kozak sequence (GCCACC) immediately upstream of the AUG start codon for efficient initiation of translation.
Site Directed Mutagenesis
For downstream cloning, EcoR1 site was introduced by four silent point mutations at NT-309–314 in exon 4 of RS1 cDNA. Sense and antisense oligomers that differed from the wild-type sequence at four sites (C309G; C312T; A313T; G314C) were chemically synthesized. These synthesized fragments were introduced into a Tag4A-RS1 vector using a Quick Change Site Directed Mutagenesis kit (Stratagene). The clones were verified by DNA sequencing. The creation of EcoR1 site altered neither the RS1-protein sequence nor the expression and localization.
RS1-Minigene Expression Vectors
The RS1 minigene included RS1 genomic introns 4 and 5 in frame with exons 1–6 of RS1 cDNA in the pCMV Tag 4A expression vector. Genomic DNA from normal control males and XLRS affected males of this family was PCR amplified to generate fragments encoding intron-4 and -5 sequences and the corresponding flanking exon sequences. Primers sequences were: Intron 4 with flanking sequences: Forward: 5′-CTGAATTCTCAAGGCTTTGGGTAAGCAGG-3′; Reverse: 5′-GGGTGCGAGCTGAAGTTGG-3′; Intron 5 with flanking sequences Forward: 5′-GAGAGCCAGCACCTGCGG-3′; Reverse: 5′-GCTCGAGTCAGGCACACTTGCTGACGCAC-3′.
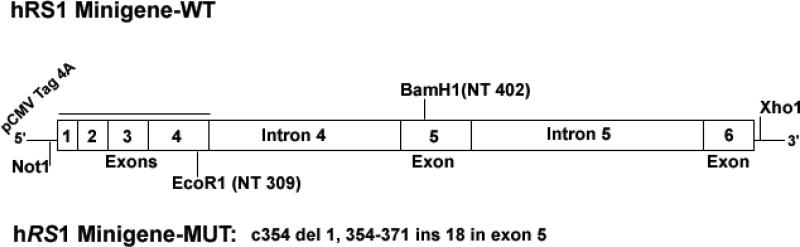
A central segment corresponding to intron 4 with the flanking exon-4 and -5 sequences was introduced between the EcoR1 and BamH1 sites. Likewise, intron 5 and its flanking exon-5 and -6 sequences were cloned between the BamH1 and Xho1 sites. The mutant minigene contained the exon-5 deletion/insertion mutation at NT-354. DNA sequence of all constructs was verified at each step.
Cell Culture and Transfection
Functionality of the wild-type (WT) and mutant (MUT) [c354del1-ins18] RS1 minigenes was studied. COS-7 cells were grown in DMEM medium with 10 % FBS. A day before transfection the cells were plated at 400,000 cells per 10 cm culture dishes. These were transiently transfected the next day using 18 μl of FuGENE 6 reagent (Roche-Applied Science, Indianapolis, IN) and 6 μg of pCMVTag 4A control plasmid (without insert) or with minigenes expressing RS1-WT or RS1-MUT. Whole-cell lysates were prepared 72 hours later by the freeze–thaw method in a lysis buffer (10 mM Tris-HCl [pH 7.4] 150 mM NaCl, 1% Triton X-100, 1 mM EDTA, 1 mM EGTA, and 0.5% Igepal CA 630, plus protease inhibitor cocktail). Transfected cell-culture medium was also collected and analyzed for RS1 protein. To investigate the protein degradation pathway, proteasome inhibitor MG132 (Z-Leu-Leu-Leu-al, Sigma Aldrich Corp, Saint Louis, MO) was added to cultures at 48 hours post-minigene transfections. After 6 hours, the cells were collected by harvesting for downstream applications.
RT-PCR and Subcloning
Total RNA was isolated from COS-7 cells expressing the RS1-WT and RS1-MUT minigenes using TRIZOL reagent (Invitrogen). DNA contamination was removed (DNA-free kit; Applied Biosystems). RT-PCR was performed in one step using SuperScript One Step RT-PCR with Platinum Taq (Invitrogen) with the following primers: (1) RS1 Exon 4-Forward: NT-206–227: 5′-TGGGTTTCGAGTCAGGGGGAGGTC-3′; (2) RS1 Exon 5-Reverse: NT-463–441: 5′-ACTGCACGCTGTACTTGGTCATC-3′; (3) RS1 Exon 1-Forward: NT-1–23: 5′-ATGTCACGCAAGATAGAAGGCTT-3′); (4) RS1 Exon 6-Reverse: NT-675–654: 5′-TCAGGCACACTTGCTGACGCAC-3′. The absence of genomic DNA in RNA was also verified by omitting the enzyme mix and substituting 2 units of Platinum Taq DNA Polymerase high fidelity in the reaction. The RT-PCR products were separated on a 1% agarose gel and visualized with ethidium bromide.
The intronless cDNA derived from RT-PCR analysis of transcripts encoded by the minigenes were cloned in pCR-Blunt II-TOPO vector using Zero Blunt TOPO PCR cloning kit (Invitrogen), and DNA was isolated. After confirming the authenticity of DNA sequences, the WT and MUT coding sequences were subcloned into pCMA Tag 4A expression vector. cDNA from full length WT and MUT mRNAs were later transiently transfected into COS-7 cells to analyze RS1 mRNA and protein expression.
Real Time RT-PCR
Real Time RT-PCR was performed using DNase treated total RNA, RS1 RT2 q PCR primer set (SA Biosciences, Frederick, MD), the house keeping GAPDH gene primer set and SYBR GreenER q PCR supermix according to the manufacturer's protocol (Invitrogen). The PCR amplifications were carried out in an I-cycler (BioRad Laboratories, Hercules, CA).
SDS-PAGE and Immunoblot Analysis
To analyze RS1-protein expression, the cells were lysed with IP buffer (50 mM Tris at pH 8, 150 mM NaCl, 1% Nonidet P40, 0.1% deoxycholate, 0.1% SDS, 1mM PMSF, 5 mM Na3VO4, 50 mM protease inhibitors for 30 minutes at 4°C). Total cell lysates were clarified by centrifugation at 14,000 rpm for 30 minutes. Aliquots of cell extracts containing an equivalent amount of proteins were resolved by SDS-polyacrylamide gel electrophoresis on 10% gels (SDS-PAGE) and transferred to polyvinylidene difluoride (PVDF) membranes (BioRad). The blots were probed with the anti-RS1 polyclonal antibody raised against amino acids 24–37 at the N-terminus.12 Horseradish peroxidase-conjugated goat anti-rabbit was the secondary antibody. Protein was visualized using an enhanced chemiluminescence kit (SuperSignal West Dura, Pierce Biotechnology, Rockford, IL).
Results
Ophthalmological Examination
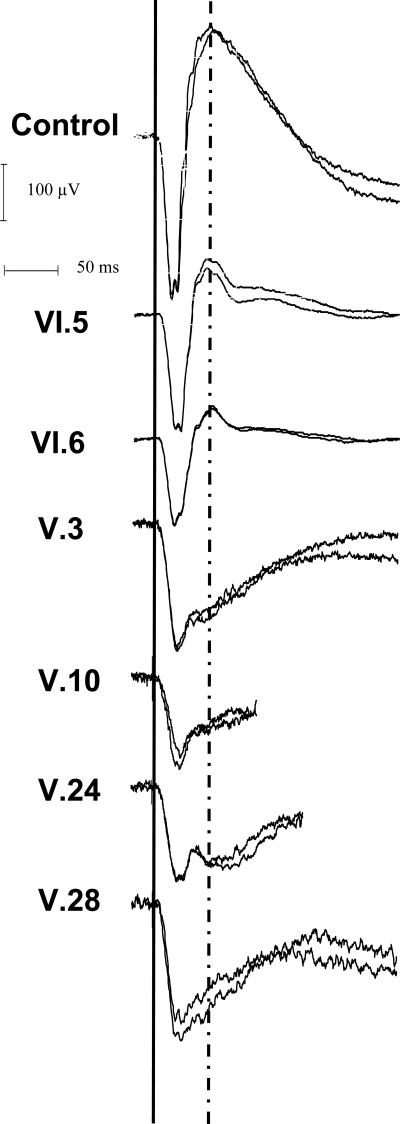
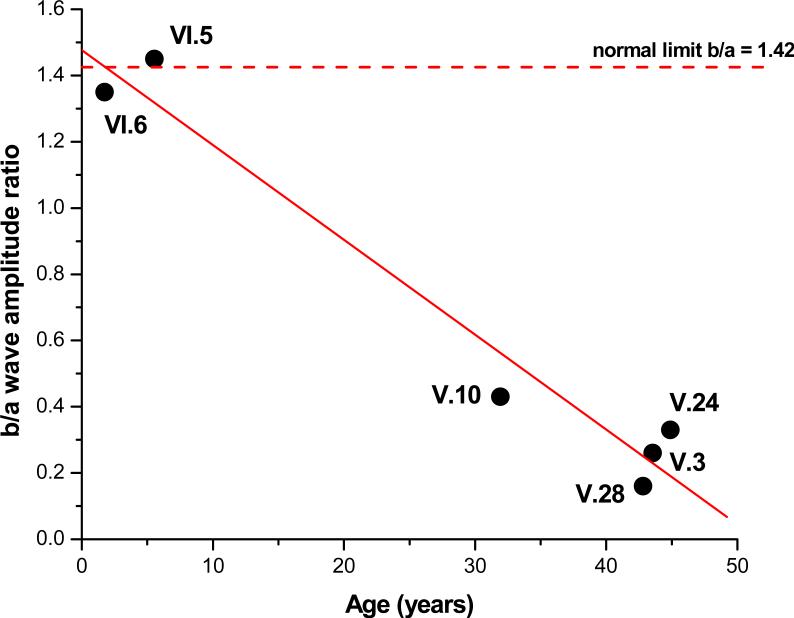
The six XLRS males showed heterogeneity and clinical and electroretinographic severity (Table 1). The two young boys had a cartwheel pattern of microcystic macular lesions typical of classic XLRS24 (Figs. 2–3). The four older relatives all had some degree of foveo-macular atrophy in one or both eyes and showed an advanced and typical “electronegative” ERG waveform. In contrast, the b-wave amplitude was normal for the two younger subjects (Fig. 4). The younger affected (VI.5 and VI.6) had nearly normal b/a-wave ratios (respectively 1.43 and 1.35), indicating that the synaptic connections between photoreceptors and bipolar cells were preserved across much of the retina. By contrast, the four older relatives all had a quite substantially lower b/a-wave ratio, indicating widespread retinal disturbances of synaptic and/or bipolar function. The difference with age is readily observed on the plot of age versus b/a-wave ratio (Fig. 5).
Table 1.
ERG and Visual Function Test Results
| SUBJECT | AGE | EYE | VISUAL ACUITY | SPHERICAL EQUIVALENT | VISUAL FIELD (both eyes similar) | a-WAVE AMPLITUDE (μV)* | ERG b/a-WAVE RATIO† | THRESHOLD LUMINANCE‡ (log cd/m2) |
|---|---|---|---|---|---|---|---|---|
| VI.5 | 5 | OD | 20/80 | +0.25 | normal V4 isopter other isopters constricted central scotoma | 212 | 1.45 | not measured |
| OS | 20/80 | +0.25 | not recorded | not recorded | ||||
| VI.6 | 1.5 | OD | too young | +7 | Not tested due to age | 159 | 1.35 | not measured |
| OS | too young | +7.5 | not recorded | not recorded | ||||
| V.3 | 43 | OD | 20/250 | +6 | normal V4 isopter other isopters reduced central scotoma | 235 | 0.26 | −3.9 |
| OS | 20/80 | +5.75 | 248 | 0.42 | −3.9 | |||
| V.10 | 32 | OD | 20/250 | +6.25 | normal V4 isopter other isopters reduced central scotoma | 169 | 0.43 | −4.4 |
| OS | 20/640 | +8 | 158 | 0.68 | −3.6 | |||
| V.24 | 45 | OD | 20/320 | +2 | normal V4 isopter other isopters reduced displaced central scotoma | 188 | 0.33 | −4.2 |
| OS | 20/400 | +2.25 | 182 | 0.31 | −3.9 | |||
| V.28 | 43 | OD | 20/125 | +10 | normal V4 isopter other isopters reduced central scotoma | 237 | 0.16 | −3.1 |
| OS | 20/160 | +9.5 | 275 | 0.20 | −3.4 | |||
Non-parametric reference values for a-wave amplitude (obtained from 96 normal volunteers)
Median: 297 μV Lower limit: 185 μV
Non-parametric reference values for b-wave/a-wave amplitude ratio (obtained from 96 normal volunteers)
Median: 1.89 Lower limit: 1.42
Reference values for absolute luminance threshold (obtained from 20 normal volunteers)
Mean: −4.89 log cd/m2 Upper limit: −4.13 log cd/m2
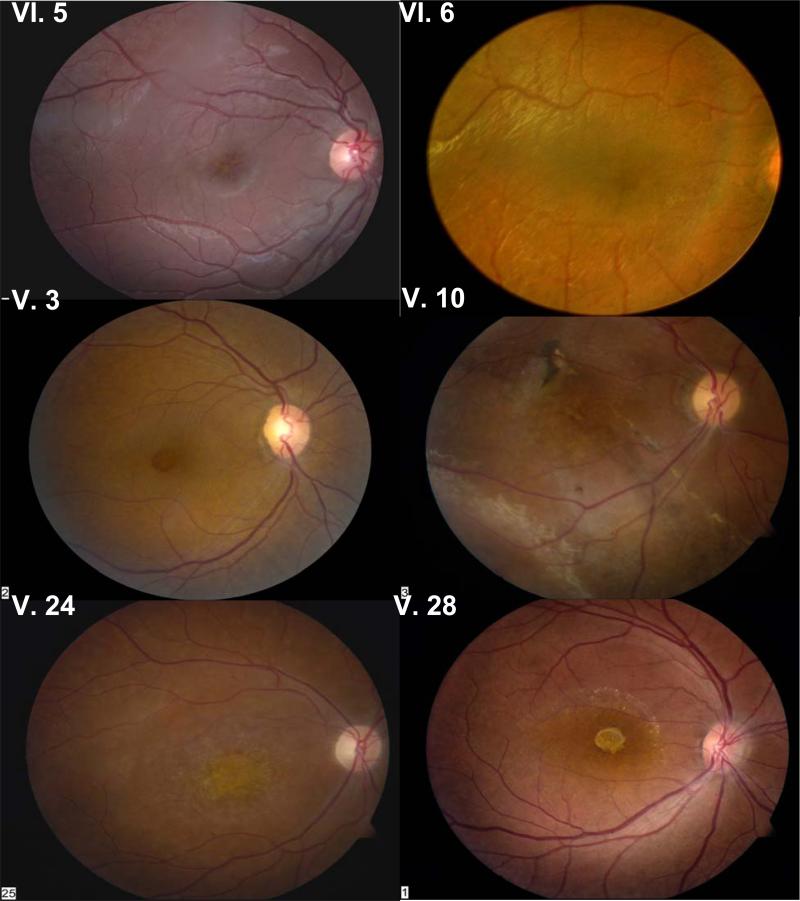
Figure 2.
Fundus photographs of the right eye of all subjects. VI.5 and VI.6 have classical foveal schisis; V.3 has a macular hole and abnormal RPE pigmentation is associated with atrophic macular changes in V.10, V.24, and V.28. See detailed description in text. Fundus photographs obtained using FF 450 Fundus Camera, Carl Zeiss Meditec, Dublin, CA.
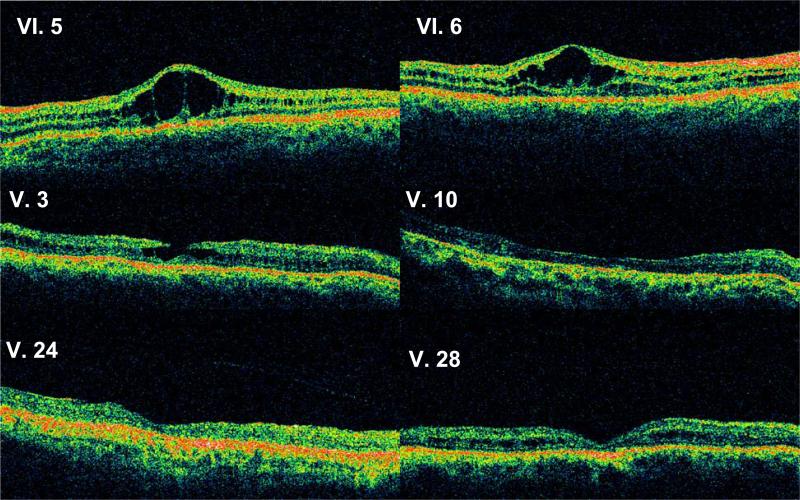
Figure 3.
Optical coherence tomograms through a horizontal section of the right eye of all subjects. VI.5 and VI.6 show classical foveal schisis. More severe abnormalities are seen in V.3 (pseudo-macular hole) and in V.10, V.24, and V.28 (atrophic macular thinning).
Figure 4.
Combined (rod and cone mediated) ERG response of the right eye of all subjects and of a representative normal control subject. Note the relative preservation of b-waves in VI.5 and VI.6, but the marked reduction in b-wave amplitude in V.3, V.10, V.24, and V.28, which gives these responses an electronegative configuration that is prototypical of XLRS disease.
Figure 5.
B/a-wave amplitude ratio (right eye) plotted as a function of age for the six XLRS subjects examined. Pearson correlation coefficient R = –0.979 with p = 0.0006.
The following is a brief description of findings (see Table 1).
The 5 y/o proband (VI.5) has macular radial cysts typical for XLRS in both eyes, confirmed by OCT (Fig. 3). The peripheral RPE had grayish pigmentation in the right-eye inferior quadrant and left-eye temporal quadrant. Vitreous veils were detected bilaterally in the posterior vitreous but no retinal bullous schisis was seen. Follow-up three years later at 8 y/o showed no new fundus changes, but the scotopic ERG a-wave and b-wave amplitudes were both reduced 25% compared to the first recording at 5 y/o.
The brother (VI.6), 1.5 y/o, had esotropia. Retinal exam under general anesthesia showed foveal cysts and parafoveal radial cysts in both eyes, with grayish RPE pigmentation of flat schisis in the temporal periphery. Vitreous veils were present in the superior temporal quadrant of both eyes but no bullous schisis. The ERG recording showed only a minimal technical reduction of a-wave and b-wave amplitudes. Clinical examination three years later showed no overt progression.
Subject V.3, 43 y/o, was a maternal uncle of the proband. He recalled central visual loss by 6–7 y/o. At 43 y/o, the right eye had a foveal pseudo-hole documented by OCT (Fig. 3). Parafoveal granular RPE pigmentation and mid peripheral white reticular areas and linear hyperpigmentation in the inferior temporal quadrant were seen. OCT of the left eye showed foveal atrophy. The scotopic ERG of both eyes was quite electronegative, although the photoreceptor a-wave remained normal.
Subject V.10 was diagnosed with XLRS at 8 y/o and had bilateral poor vision since childhood. At 32 y/o, both maculas had granular pigmentation and fibrotic scarring. The OCT showed foveal atrophy in the right eye. Bullous schisis was present in the inferotemporal periphery of the right eye. Flat schisis was observed in the inferotemporal periphery of the left eye. The scotopic ERG was electronegative.
Subject V.24 had exotropia and “macular dystrophy” since childhood. At 45 y/o, he had bilateral macular atrophy confirmed by OCT (Fig. 3). A vitreous band spanned the superotemporal periphery. The scotopic ERG was grossly electronegative.
Subject V.28 reported poor vision since childhood. At 43 y/o, the right-eye fovea was atrophic, and the left-eye fovea had multiple pigmented lesions. XLRS cysts could not be seen clinically, but were evident on OCT. The retinal periphery of both eyes had infero-temporal whitish deposits, but no overt schisis. The scotopic ERG was grossly electronegative.
Molecular Genetic Findings: Exon-5 Deletion/Insertion Mutation
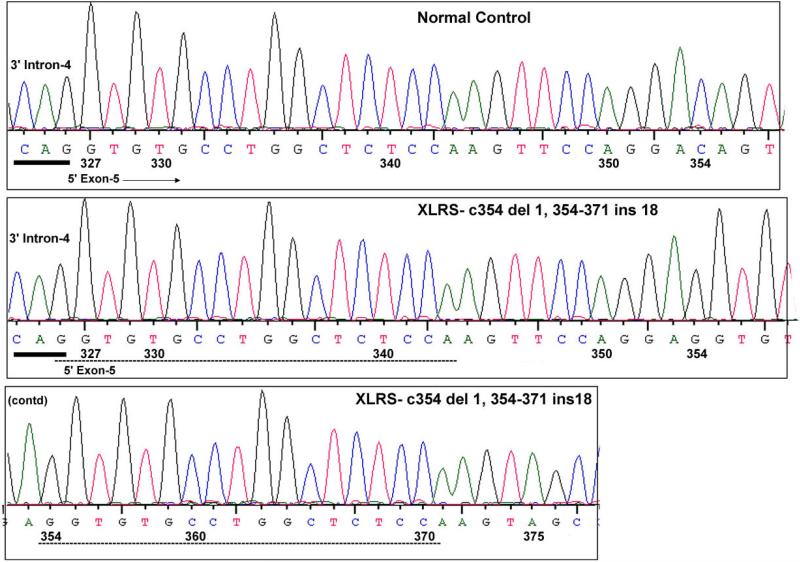
Sequencing RS1 along both strands showed a deletion/insertion mutation in all six affected subjects, with a 1-bp deletion replaced by an 18-bp insertion at the same nucleotide [354 del C, 354–371 Ins GGTGTGCCTGGCTCTCCA] (Fig. 6). The resulting frame shift creates a UAG termination signal six codons downstream. The inserted sequence duplicates the adjacent upstream sequence of both strands. The duplicated sequence spanned an intron–exon junction, from the last nucleotide of intron 4 through the first 17 nucleotides of exon 5. This duplication involves the RNA splice-donor and splice-acceptor sites surrounding the splice-donor site of intron 4 and generates an additional intron–exon junction 27 bp downstream of the original intron–exon junction. Consequently, we examined whether this mutation affects pre-RNA splicing.
Figure 6.
RS1 genomic sequence encompassing the c354del1-ins18 mutation in exon 5. The normal control and XLRS subject genomic sequences that are shown include a 3-bp upstream intron-4 sequence and downstream exon-5 sequence. The XLRS mutation is a 1-bp deletion and 18-bp insertion at NT-354. The inserted sequence at NT-354–371 is a direct copy of the upstream sequence (NT-327–343), both shown in dashed lines. The intron sequence is underlined/bold. The first G nucleotide of the inserted sequence probably originated from the terminal nucleotide of intron 4. The coding sequence derived from Homo sapiens retinoschisin (RS1), mRNA (Accession NM_000330) is numbered from 1–675.
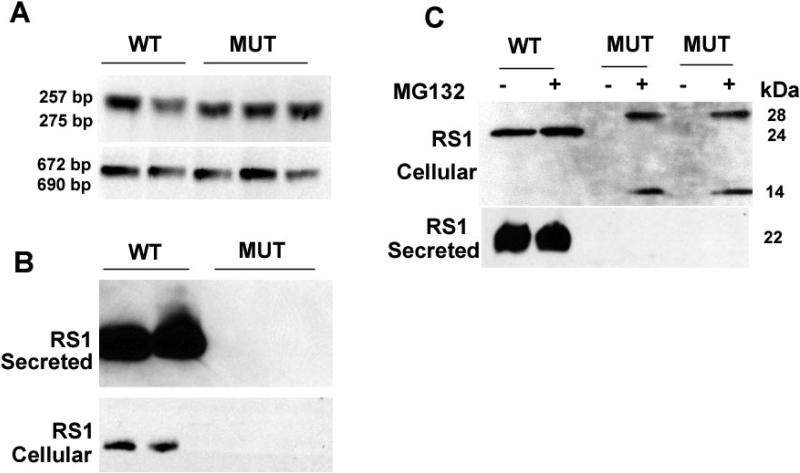
RS1-WT and RS1-MUT minigenes were constructed by PCR amplification of normal and the XLRS subject's genomic DNA [c354del1-ins18] (Fig.7), and COS-7 cells were transiently transfected to monitor the mutational effect on pre-mRNA splicing and RS1-protein expression. Total RNA isolated from COS-7 cells expressing the minigenes was used as templates for the RT-PCR with primers flanking the mutation (NT-206–463, exon 4–5 of RS1 cDNA). The RS1-WT minigene cells yielded the predicted 257-bp RT-PCR product. Cells expressing the RS1-MUT minigene produced a longer PCR product (Fig. 8A, top panel) that corresponded to the size of the 18-bp insertion. RT-PCR with primers spanning the CDS of RS1 gene (NT-1–675) produced the similar result, with longer PCR product in cells with RS1-MUT minigene (Fig. 8A, lower panel). No aberrantly spliced products were observed for the mutant mRNA, indicating that the duplication of the intron–exon junction 27 bp 3′ downstream of the original splice junction did not affect pre-RNA splicing. This finding was confirmed by subcloning the full length cDNA derived from RNA isolated from COS-7 cells expressing the minigenes. DNA sequence analysis of several of these subclones confirmed that the wild type and mutant RNAs are processed similarly.
Figure 7.
Schematic representation of the RS1-WT and RS1-MUT minigenes. RS1 genomic introns 4 and 5 were placed in frame with exons 1–6 of RS1 cDNA that was cloned into a mammalian expression vector pCMV Tag 4a. The RS1 genomic introns 4 and 5 were amplified from the genomic DNA obtained from the XLRS subjects and the normal controls. The DNA segments with intron 4 and 5 and the corresponding flanking exon sequences were placed between the EcoR1/BamH1 and BamH1/Xho1enzyme sites respectively. The mutant minigene contained the c354del1-ins18 mutation in exon 5.
Figure 8.
A: RNA analysis: RNA derived from cells expressing either WT or MUT (c354del1-ins18) minigene was subjected to RT-PCR using RS1 cDNA primers spanning (a) exon 4/exon 5 or (b) exon 1/exon 6. The products were resolved on agarose gel. The mutant DNA is longer than the WT as a consequence of insertion. Subcloning and sequencing of the RT-PCR product revealed that splicing was not disrupted in the mutant. However, the insertion caused frameshift and introduced premature termination codons in the RNA. This finding was confirmed on repetition. B: RS1-protein expression: Whole cell lysates derived from cells transfected with WT or MUT minigene were subjected to SDS-PAGE followed by immunoblotting with RS1 antibody. Cells transfected with WT minigene express RS1, which is predominantly detected in the secreted fraction. No RS1 expression was seen with the MUT minigene possibly because of premature termination of translation and rapid degradation of the truncated product. The findings were confirmed on repetition. C: At 48-hours post-transfection, cells expressing the intronless WT- and MUT-cDNA constructs were exposed to a proteasome inhibitor MG132 (50 μM for 6 hours) followed by SDS-PAGE and immunoblotting. Two protein bands (14 and 28 kDa) immunoreactive to RS1 antibody were detected in MG132 treated cells expressing the intronless MUT. Presumably they represent the monomer and dimer of the truncated RS1 species. RS1 was not detected in the medium. Cells with intronless WT-cDNA construct expressed both the precursor (24 kDa) and mature (22 kDa) forms of RS1. More than one mutant clone was tested.
Western-blot analysis of the media of cells transfected with the RS1-WT minigene showed a single ~22 kDa RS1-protein band (Fig. 8B), with very little detected in the cell lysates, consistent with the secreted nature of RS1 protein. However, mutant RS1 protein was not detected either in the medium or in the cellular fractions of RS1-MUT minigene cells. The 18-bp insertion introduces an amber stop codon UAG that prematurely terminates translation, i.e., a frame shift (fs) change with Asp 118 as the first affected amino acid changing to Glu and creating a new reading frame ending in a UAG stop codon at position 14 (pAsp118Glu fsX14). We were unable to detect any truncated RS1 protein in the western blots. It is unlikely from loss of the antibody epitope, as the anti-RS1 polyclonal antibody against the N-terminus amino acids 24–37 of RS1 would recognize the N-terminus epitope if truncated 131 amino-acid protein were present. The absence of any truncated RS1 mutant is consistent with rapid degradation of the protein. The loss of mutant RS1 could also be due to loss of mRNA triggered by NMD. Nonsense-mediated mRNA decay (NMD) in mammalian cells is a well-characterized mRNA surveillance mechanism by which aberrant mRNAs harboring premature translation termination codons (PTCs) are rapidly degraded.26,27 This is an intron-dependent regulatory mechanism that eliminates abnormal transcripts. To analyze whether the premature termination codon introduced by the c354del1-ins18 bp mutation interferes with RS1 mRNA expression levels, we performed real-time RT-PCR on total RNA isolated from COS-7 cells expressing the RS1-WT and RS1-MUT minigenes. Control and mutant cells showed no difference of RNA levels, which indicate that the loss of mutant protein was not due to NMD. We then used intronless mutant cDNA construct, which escape NMD-dependent RNA degradation, followed by Fugene transfection to amplify the mutant protein. However, truncated RS1-protein product was never observed in cells expressing the intronless mutant minigene.
As this absence of mutant protein could result from reduced synthesis or accelerated degradation, we investigated whether the proteasome pathway is involved in degradation of the mutant protein. Cells expressing the intronless WT- and MUT-cDNA constructs were exposed to 50 μM MG 132 proteasome inhibitor for 6 hours and subjected immunoblot analysis. Protein bands immunoreactive to RS1 antibody were detected in MG132 treated cells with the intronless MUT (Fig. 8C). A protein band of ~14-kDa approximate mass was seen which corresponds to the size of the truncated RS1, along with a 28-kDa band presumed to be the dimer of the truncated species. However, no truncated RS1 was found in the medium. Untreated control cells did not exhibit these bands. The finding that truncated RS1 accumulates only in the presence of proteasome inhibitor demonstrates that the mechanism underlying the rapid degradation of the mutant protein involves the proteasomal pathway and that the c354del1-ins18 mutation gives a RS1-protein null phenotype.
Discussion
Molecular Biology of the Mutation
These findings implicate a putative mechanism that rapidly degrades truncated RS1 protein with premature termination codons (PTCs). Although the c354del1-ins18 mutation resulting in a PTC, we found no evidence for nonsense mediated decay of mRNA (NMD) as the underlying cause of the RS1-protein deficiency. Short deletion/insertion mutations with duplication of adjacent sequence have been described in other human diseases involving α2 globin and factor VIII genes,28,29 and in some of these mutations the inserted nucleotides duplicate the neighboring sequence on the same DNA strand, as we found in the XLRS family of this study. A combination of slipped mispairing during replication and intragenic recombination has been suggested as the mechanism for the deletion/insertion/duplication process.28,29 In our c354del1-ins18 XLRS family the inserted sequence duplicated the upstream intron–exon junction, but this had no effect on pre-RNA splicing. Nevertheless, the deletion/duplication mutation within the exon-5 coding sequence terminated the expression of RS1 protein.
The 1-bp deletion and 18-bp insertion creates a new reading frame that ends in a premature termination codon (UAG) at position 14. Eukaryotic mRNAs that contain PTCs are rapidly degraded by NMD.26,27 This conserved eukaryotic mRNA quality control system thereby prevents the accumulation of potentially detrimental truncated proteins. We investigated whether NMD is responsible for the deficiency of RS1 protein in the cells expressing mutant minigene. However, contrary to what NMD would predict, real-time RT-PCR analysis revealed no significant decrease in c354del1-ins18 mRNA levels in the cells expressing the RS1-MUT minigene. Although the stability of RS1 mRNA with PTC was surprising, several other reports have also indicated that not all mRNAs that contain PTCs are targeted for destruction by NMD27, and that a proteasome or proteases-dependent mechanism causes rapid degradation of truncated protein in some cases.30 This is consistent with the “rescue” of truncated RS1 we observed using the proteasome inhibitor MG132, confirming that rapid degradation of c354del1-ins18 mutant RS1 involves the proteasomal pathway.
Clinical Considerations
Because clinical changes from XLRS occur at most very slowly, it is difficult to document disease progression in single affected individuals, and XLRS has historically been described as a “stationary” condition.4,31,32 The present study, however, indicates that the condition is progressive in this RS1 mutation. The proband showed a 25% ERG reduction over a 3-year interval, between 5–8 y/o. Furthermore, the disease in this family varied considerably across 1.5–45 y/o and was substantially more severe in the older members. The four older males had major b-wave loss leading to quite classical XLRS electronegative ERGs, and three of them exhibited overt foveal atrophy. The two youngest had not progressed to that state and had more limited macular schisis and a nearly normal scotopic ERG. As all six affected males carried the same RS1 mutation, the cross-sectional analysis suggests progression with age.
This is consistent with the progression documented in natural history studies of the RS1 knockout-mouse model. Kjellstrom et al.33 found that the retinal cavities were greatest at 4 months age in the RS1-KO mouse and then collapsed and were progressively reduced at later ages. Progressive photoreceptor loss with age was also found, and by 16 months the outer nuclear layer showed major loss of cells in the mouse XLRS model. Both the RS1 mutation in this human family and the RS1-KO mouse model lack retinoschisin-protein expression, indicating that the absence of retinoschisin is deleterious and can cause progressive disease. (This study was conducted in accordance with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research.)
Some XLRS males exhibit severe disease even at very early age, including bilateral bullous schisis5,34–36 and vitreous hemorrhages and retinal detachment.35,37–39 By contrast, the two affected young boys in our family, despite the absence of RS1 protein with this mutation, had only macular schisis and retained nearly normal ERG response. The normal b/a-wave ratios indicate appropriate transmission through the rod synapse onto rod bipolar cells. The clinical findings in this family suggest that the absence of RS1 may initially cause less severe disease but that the retinopathy apparently is not stable, as the same mutation and absence of RS1 protein in the 32–45 y/o males caused considerably more dysfunction and vision loss. The central atrophy in the three older males implies a progressive death of the foveal cones. The atrophic maculopathy noted in three of the 32–45 y/o is not exclusive to a loss of RS1 protein, as macular atrophy has also been found in relatively young adults associated with RS1-missense mutations.40 Wu and Molday41 have shown in cell culture that some XLRS missense mutations fail to transport through the cell membrane.
Questions Raised
Environmental and genetic factors, including modifier genes, are predicted to account for some of the phenotypic variation in an XLRS mouse model,42 and hence, are the possible contribution to the wide phenotype heterogeneity found across the ages of members of this XLRS family. However, modifier genes have not been described for human XLRS. RS1 mutations can give rise to functional null biochemical phenotypes in at least two ways. Several discoidin domain missense mutations in exons 4–6 perturb RS1-protein folding and prevent secretion across the plasma membrane.40,43 Other missense mutations, including M1L in exon 144 and the deletion/insertion exon-5 mutation in this study, either abolish or severely reduce retinoschisin-protein expression. Whether these two biochemical phenotypes affect the clinical course of XLRS disease in the same or different ways remains unclear.
Retinoschisin has been described as a signaling protein, secreted by photoreceptor cells.45 Reid et al.46 suggested that retinoschisin interacts with bipolar and Müller cells and thereby normally contributes to the development of the cellular and synaptic architecture of the retina. Loss of retinoschisin disturbs retinal structure and causes loss of synaptic transmission to the retinal bipolar cells as reflected in the reduction of the scotopic ERG b-wave.11,12,45,47 Evidently, beyond the period of retinal development, RS1 is required for structural maintenance, and some XLRS subjects may have normal retinal development with substantial bipolar function at early age but subsequently exhibit a failure of maintenance with aging.24
These clinical and mutational considerations give rise to three questions:
Is the apparent progression of XLRS dysfunction due exclusively to age?
How is the retina organized in the youngest two males in the absence of RS1 protein such that synaptic transmission can support a normal b/a-wave ratio?
Is the degree of ERG dysfunction and b/a-wave loss correlated with the extent of fundus involvement, as the pathology in the youngest two males appears relatively limited, whereas the older four males had considerable peripheral involvement in addition, suggesting that synaptic integrity might be impaired secondarily over a greater extent of the fundus?
We are currently exploring the possibility that the absence of RS1 protein results in up-regulation and over-expression of other adhesion proteins in the extracellular matrix and thereby provides a substitute that initially supports development and preserves synaptic integrity and retinal structure during the early stage of disease.
Acknowledgments
The authors thank Meira Meltzer, M.A., M.S., for coordinating subject visits and providing the pedigree; Gregory Short and John Rowan for performing the OCT scans; Leanne Reuter and Patrick Lopez for performing clinical and ERG testing; and Settara Chandrasekharappa, Ph.D. (NIH/NHGRI), for useful discussions.
Footnotes
Dislosure: C. Vijayasarathy, None; L. Ziccardi, None; Y. Zeng, None; N. Smaoui, None; R.C. Caruso, None; P.A. Sieving, None
References
- 1.Falcone PM, Brockhurst RJ. X-chromosome-linked juvenile retinoschisis: clinical aspects and genetics. Int Ophthalmol Clin. 1993;33:193–202. doi: 10.1097/00004397-199303320-00018. [DOI] [PubMed] [Google Scholar]
- 2.Prenner JL, Capone A, Jr., Ciaccia S, Takada Y, Sieving PA, Trese MT. Congenital X-linked retinoschisis classification system. Retina. 2006;26:S61–64. doi: 10.1097/01.iae.0000244290.09499.c1. [DOI] [PubMed] [Google Scholar]
- 3.Minami Y, Ishiko S, Takai Y, et al. Retinal changes in juvenile X linked retinoschisis using three dimensional optical coherence tomography. Br J Ophthalmol. 2005;89:1663–1664. doi: 10.1136/bjo.2005.075648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kellner U, Brummer S, Foerster MH, Wessing A. X-linked congenital retinoschisis. Graefes Arch Clin Exp Ophthalmol. 1990;228:432–437. doi: 10.1007/BF00927256. [DOI] [PubMed] [Google Scholar]
- 5.George ND, Yates JR, Moore AT. X linked retinoschisis. Br J Ophthalmol. 1995;79:697–702. doi: 10.1136/bjo.79.7.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sieving PA, Bingham EL, Kemp J, Richards J, Hiriyanna K. Juvenile X-linked retinoschisis from XLRS1 Arg213Trp mutation with preservation of the electroretinogram scotopic b-wave. Am J Ophthalmol. 1999;128:179–184. doi: 10.1016/s0002-9394(99)00144-0. [DOI] [PubMed] [Google Scholar]
- 7.Functional implications of the spectrum of mutations found in 234 cases with X-linked juvenile retinoschisis. The Retinoschisis Consortium. Hum Mol Genet. 1998;7:1185–1192. doi: 10.1093/hmg/7.7.1185. [DOI] [PubMed] [Google Scholar]
- 8.Sauer CG, Gehrig A, Warneke-Wittstock R, et al. Positional cloning of the gene associated with X-linked juvenile retinoschisis. Nat Genet. 1997;17:164–170. doi: 10.1038/ng1097-164. [DOI] [PubMed] [Google Scholar]
- 9.Vogel W. Discoidin domain receptors: structural relations and functional implications. FASEB J. 1999;13(Suppl):S77–82. doi: 10.1096/fasebj.13.9001.s77. [DOI] [PubMed] [Google Scholar]
- 10.Grayson C, Reid SN, Ellis JA, et al. Retinoschisin, the X-linked retinoschisis protein, is a secreted photoreceptor protein, and is expressed and released by Weri-Rb1 cells. Hum Mol Genet. 2000;9:1873–1879. doi: 10.1093/hmg/9.12.1873. [DOI] [PubMed] [Google Scholar]
- 11.Molday LL, Hicks D, Sauer CG, Weber BH, Molday RS. Expression of X-linked retinoschisis protein RS1 in photoreceptor and bipolar cells. Invest Ophthalmol Vis Sci. 2001;42:816–825. [PubMed] [Google Scholar]
- 12.Takada Y, Fariss RN, Tanikawa A, et al. A retinal neuronal developmental wave of retinoschisin expression begins in ganglion cells during layer formation. Invest Ophthalmol Vis Sci. 2004;45:3302–3312. doi: 10.1167/iovs.04-0156. [DOI] [PubMed] [Google Scholar]
- 13. Sequence variation database, http://www.dmd.nl/rs/
- 14.Huopaniemi L, Rantala A, Forsius H, Somer M, de la Chapelle A, Alitalo T. Three widespread founder mutations contribute to high incidence of X-linked juvenile retinoschisis in Finland. Eur J Hum Genet. 1999;7:368–376. doi: 10.1038/sj.ejhg.5200300. [DOI] [PubMed] [Google Scholar]
- 15.Huopaniemi L, Tyynismaa H, Rantala A, Rosenberg T, Alitalo T. Characterization of two unusual RS1 gene deletions segregating in Danish retinoschisis families. Hum Mutat. 2000;16:307–314. doi: 10.1002/1098-1004(200010)16:4<307::AID-HUMU3>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 16.Gehrig A, Weber BH, Lorenz B, Andrassi M. First molecular evidence for a de novo mutation in RS1 (XLRS1) associated with X linked juvenile retinoschisis. J Med Genet. 1999;36:932–934. [PMC free article] [PubMed] [Google Scholar]
- 17.Gregori NZ, Berrocal AM, Gregori G, et al. Macular spectral-domain optical coherence tomography in patients with X-linked retinoschisis. Br J Ophthalmol. 2009;93:373–378. doi: 10.1136/bjo.2007.136127. [DOI] [PubMed] [Google Scholar]
- 18.Tantri A, Vrabec TR, Cu-Unjieng A, Frost A, Annesley WH, Jr., Donoso LA. X-linked retinoschisis: a clinical and molecular genetic review. Surv Ophthalmol. 2004;49:214–230. doi: 10.1016/j.survophthal.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 19.Khan NW, Jamison JA, Kemp JA, Sieving PA. Analysis of photoreceptor function and inner retinal activity in juvenile X-linked retinoschisis. Vision Res. 2001;41:3931–3942. doi: 10.1016/s0042-6989(01)00188-2. [DOI] [PubMed] [Google Scholar]
- 20.Peachey NS, Fishman GA, Derlacki DJ, Brigell MG. Psychophysical and electroretinographic findings in X-linked juvenile retinoschisis. Arch Ophthalmol. 1987;105:513–516. doi: 10.1001/archopht.1987.01060040083038. [DOI] [PubMed] [Google Scholar]
- 21.Bradshaw K, George N, Moore A, Trump D. Mutations of the XLRS1 gene cause abnormalities of photoreceptor as well as inner retinal responses of the ERG. Doc Ophthalmol. 1999;98:153–173. doi: 10.1023/a:1002432919073. [DOI] [PubMed] [Google Scholar]
- 22.Shinoda K, Ohde H, Ishida S, Inoue M, Oguchi Y, Mashima Y. Novel 473-bp deletion in XLRS1 gene in a Japanese family with X-linked juvenile retinoschisis. Graefes Arch Clin Exp Ophthalmol. 2004;242:561–565. doi: 10.1007/s00417-004-0878-y. [DOI] [PubMed] [Google Scholar]
- 23.Lesch B, Szabo V, Kanya M, et al. Truncation of retinoschisin protein associated with a novel splice site mutation in the RS1 gene. Mol Vis. 2008;14:1549–1558. [PMC free article] [PubMed] [Google Scholar]
- 24.Eksandh L, Andreasson S, Abrahamson M. Juvenile X-linked retinoschisis with normal scotopic b-wave in the electroretinogram at an early stage of the disease. Ophthalmic Genet. 2005;26:111–117. doi: 10.1080/13816810500228688. [DOI] [PubMed] [Google Scholar]
- 25.Zeng Y, Takada Y, Kjellstrom S, et al. RS-1 gene delivery to an adult RS1h knockout mouse model restores ERG b-wave with reversal of the electronegative waveform of X-linked retinoschisis. Invest Ophthalmol Vis Sci. 2004;45:3279–3285. doi: 10.1167/iovs.04-0576. [DOI] [PubMed] [Google Scholar]
- 26.Behm-Ansmant I, Kashima I, Rehwinkel J, Sauliere J, Wittkopp N, Izaurralde E. mRNA quality control: an ancient machinery recognizes and degrades mRNAs with nonsense codons. FEBS Lett. 2007;581:2845–2853. doi: 10.1016/j.febslet.2007.05.027. [DOI] [PubMed] [Google Scholar]
- 27.Byers PH. Killing the messenger: new insights into nonsense-mediated mRNA decay. J Clin Invest. 2002;109:3–6. doi: 10.1172/JCI14841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oron-Karni V, Filon D, Rund D, Oppenheim A. A novel mechanism generating short deletion/insertions following slippage is suggested by a mutation in the human alpha2-globin gene. Hum Mol Genet. 1997;6:881–885. doi: 10.1093/hmg/6.6.881. [DOI] [PubMed] [Google Scholar]
- 29.Tavassoli K, Eigel A, Horst J. A deletion/insertion leading to the generation of a direct repeat as a result of slipped mispairing and intragenic recombination in the factor VIII gene. Hum Genet. 1999;104:435–437. doi: 10.1007/s004390050981. [DOI] [PubMed] [Google Scholar]
- 30.Zhang X, Dudek EJ, Liu B, et al. Degradation of C-terminal truncated αA-crystallins by the Ubiquitin–Proteasome Pathway. Invest Ophthalmol Vis Sci. 2007;48:4200–4208. doi: 10.1167/iovs.07-0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ewing CC, Ives EJ. Juvenile hereditary retinoschisis. Trans Ophthalmol Soc UK. 1970;89:29–39. [PubMed] [Google Scholar]
- 32.Forsius H, Krause U, Helve J, et al. Visual acuity in 183 cases of X-chromosomal retinoschisis. Can J Ophthalmol. 1973;8:385–393. [PubMed] [Google Scholar]
- 33.Kjellstrom S, Bush RA, Zeng Y, Takada Y, Sieving PA. Retinoschisin gene therapy and natural history in the Rs1h-KO mouse: long-term rescue from retinal degeneration. Invest Ophthalmol Vis Sci. 2007;48:3837–3845. doi: 10.1167/iovs.07-0203. [DOI] [PubMed] [Google Scholar]
- 34.Brockhurst RJ. Retinoschisis. Complication of peripheral uveitis. Arch Ophthalmol. 1981;99:1998–1999. doi: 10.1001/archopht.1981.03930020874012. [DOI] [PubMed] [Google Scholar]
- 35.Greven CM, Moreno RJ, Tasman W. Unusual manifestations of X-linked retinoschisis. Trans Am Ophthalmol Soc. 1990;88:211–225. discussion 226–218. [PMC free article] [PubMed] [Google Scholar]
- 36.Kawano K, Tanaka K, Murakami F, Ohba N. Congenital hereditary retinoschisis: evolution at the initial stage. Albrecht Von Graefes Arch Klin Exp Ophthalmol. 1981;217:315–323. doi: 10.1007/BF00429292. [DOI] [PubMed] [Google Scholar]
- 37.Conway BP, Welch RB. X-chromosone-linked juvenile retinoschisis with hemorrhagic retinal cyst. Am J Ophthalmol. 1977;83:853–855. doi: 10.1016/0002-9394(77)90913-8. [DOI] [PubMed] [Google Scholar]
- 38.Regillo CD, Tasman WS, Brown GC. Surgical management of complications associated with X-linked retinoschisis. Arch Ophthalmol. 1993;111:1080–1086. doi: 10.1001/archopht.1993.01090080076021. [DOI] [PubMed] [Google Scholar]
- 39.Roesch MT, Ewing CC, Gibson AE, Weber BH. The natural history of X-linked retinoschisis. Can J Ophthalmol. 1998;33:149–158. [PubMed] [Google Scholar]
- 40.Lesch B, Szabo V, Kanya M, et al. Clinical and genetic findings in Hungarian patients with X-linked juvenile retinoschisis. Mol Vis. 2008;14:2321–2332. [PMC free article] [PubMed] [Google Scholar]
- 41.Wu WW, Molday RS. Defective discoidin domain structure, subunit assembly, and endoplasmic reticulum processing of retinoschisin are primary mechanisms responsible for X-linked retinoschisis. J Biol Chem. 2003;278:28139–28146. doi: 10.1074/jbc.M302464200. [DOI] [PubMed] [Google Scholar]
- 42.Johnson BA, Aoyama N, Friedell NH, Ikeda S, Ikeda A. Genetic modification of the schisis phenotype in a mouse model of X-linked retinoschisis. Genetics. 2008;178:1785–1794. doi: 10.1534/genetics.107.084905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang T, Zhou A, Waters CT, O'Connor E, Read RJ, Trump D. Molecular pathology of X linked retinoschisis: mutations interfere with retinoschisin secretion and oligomerisation. Br J Ophthalmol. 2006;90:81–86. doi: 10.1136/bjo.2005.078048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim DY, Neely KA, Sassani JW, et al. X-linked retinoschisis: novel mutation in the initiation codon of the XLRS1 gene in a large family. Retina. 2006;26:940–946. doi: 10.1097/01.iae.0000224321.93502.a3. [DOI] [PubMed] [Google Scholar]
- 45.Reid SN, Yamashita C, Farber DB. Retinoschisin, a photoreceptor-secreted protein, and its interaction with bipolar and muller cells. J Neurosci. 2003;23:6030–6040. doi: 10.1523/JNEUROSCI.23-14-06030.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reid SN, Farber DB. Glial transcytosis of a photoreceptor-secreted signaling protein, retinoschisin. Glia. 2005;49:397–406. doi: 10.1002/glia.20131. [DOI] [PubMed] [Google Scholar]
- 47.Takada Y, Vijayasarathy C, Zeng Y, Kjellstrom S, Bush RA, Sieving PA. Synaptic pathology in retinoschisis knockout (Rs1-/y) mouse retina and modification by rAAV-Rs1 gene delivery. Invest Ophthalmol Vis Sci. 2008;49:3677–3686. doi: 10.1167/iovs.07-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]