Abstract
During neurogenesis, expression of the basic Helix-Loop-Helix NeuroD6/Nex1/MATH-2 transcription factor parallels neuronal differentiation, and is maintained in differentiated neurons in the adult brain. To further dissect NeuroD6 differentiation properties, we previously generated a NeuroD6-overexpressing stable PC12 cell line, PC12-ND6, which displays a neuronal phenotype characterized by spontaneous neuritogenesis, accelerated NGF-induced differentiation, and increased regenerative capacity. Furthermore, we reported that NeuroD6 promotes long-term neuronal survival upon serum deprivation. In this study, we identified the NeuroD6-mediated transcriptional regulatory pathways linking neuronal differentiation to survival, by conducting a genome-wide microarray analysis using PC12-ND6 cells and serum deprivation as a stress paradigm. Through a series of filtering steps and a gene-ontology analysis, we found that NeuroD6 promotes distinct but overlapping gene networks, consistent with the differentiation, regeneration, and survival properties of PC12-ND6 cells. Using a gene set enrichment analysis, we provide the first evidence of a compelling link between NeuroD6 and a set of heat shock proteins in the absence of stress, which may be instrumental to confer stress tolerance to PC12-ND6 cells. Immunocytochemistry results showed that HSP27 and HSP70 interact with cytoskeletal elements, consistent with their roles in neuritogenesis and preserving cellular integrity. HSP70 also colocalizes with mitochondria located in the soma, growing neurites and growth cones of PC12-ND6 cells prior to and upon stress stimulus, consistent with its neuroprotective functions. Collectively, our findings support the notion that NeuroD6 links neuronal differentiation to survival via the network of molecular chaperones and endows the cells with increased stress tolerance.
Keywords: Heat shock proteins, heat shock transcription factors, NeuroD family, basic helix-loop-helix transcription factors, stress tolerance
INTRODUCTION
During neural development, neurotrophic factors induce a variety of signaling cascades, ultimately resulting in modulation of gene expression promoting differentiation and/or survival of neurons (Oppenheim, 1991). A plethora of studies has demonstrated that survival of most immature neurons is dependent on some form of trophic support to actively repress the default pro-apoptotic pathway, while mature neurons have the default anti-apoptotic pathway actively turned on to respond to stress-induced signals (Benn and Woolf, 2004). However, the identity of the transcriptional regulators controlling this molecular switch remains elusive.
The neurogenic basic helix-loop-helix (bHLH) NeuroD6 transcription factor, a member of the NeuroD family, is a potential candidate based on its differentiation-inducing properties during brain development. During corticogenesis, NeuroD6, whose expression is triggered at embryonic day E11, contributes to the specification of multipotential progenitors toward a glutamatergic pyramidal fate and is believed to execute cell cycle exit and terminal differentiation based on our vitro cell culture studies (Bartholomae and Nave, 1994; Shimizu et al., 1995; Schwab et al., 1998; Uittenbogaard and Chiaramello, 2002; 2004; Wu et al., 2005).
The NeuroD family is composed of four members, NeuroD, NeuroD2, NeuroD6 (also known as Nex1/MATH-2), and NeuroD4 (NeuroM/MATH-3), which display overlapping patterns of expression during brain development, resulting in functional compensation in most knockout models (Schwab et al., 1998; 2000). Nevertheless, these knockout models did highlight the notion that they may promote survival of specific neuronal lineages, while executing neuronal differentiation as illustrated in the double knockout Nex1/NeuroD mice, which display increased cell death of immature granule neurons of the dentate gyrus combined with decreased hippocampal size (Schwab et al., 2000). These studies generated the global hypothesis that cell death may be the result of failure in proper cell cycle withdrawal and differentiation (Miyata et al., 1999; Pennesi et al., 2003) and that a potential link between neurotrophin-induced differentiation and the survival pathway may occur in certain lineages (Kim et al., 2001; Olson et al., 2001; Liu et al., 2004). More specifically, our previous studies suggested that NeuroD6 might play a central role in the switch from pro- to anti-apoptotic pathways in differentiating neurons (Uittenbogaard and Chiaramello, 2002; 2005).
To elucidate NeuroD6 intrinsic properties, we engineered a stable NeuroD6- overexpressing PC12 cell line, PC12-ND6, and showed that NeuroD6 mimics NGF effect in terms of neuronal differentiation and survival and is an important effector of the NGF pathway (Uittenbogaard and Chiaramello, 2002; 2004). More recently, we provided the first direct evidence of NeuroD6 neuroprotective properties using serum deprivation as a stress paradigm. PC12-ND6 cells remain viable and retain a neuronal morphology after 15 days of serum deprivation, while naïve PC12 cells undergo apoptosis within 48 hours. We identified the initial steps of the NeuroD6 intrinsic survival-promoting transcriptional program, which included expression of G1 phase inhibitors and the anti-apoptotic regulators Bcl-xL, Bcl-w, XIAP, and survivin (Uittenbogaard and Chiaramello, 2005).
The main objective of this study is to expand the underlying mechanism of NeuroD6 transcriptional network in promoting neuronal survival and to decipher the molecular switch linking neuronal differentiation to survival. We performed a genome-wide analysis by comparing the gene expression profile upon NeuroD6 overexpression and two days of serum deprivation. We validated our genomic findings with comprehensive protein analyses, which included two additional time points after stress induction, 6 and 15 days, to understand the long-term effects of chronic stress, often associated with neurodegenerative disorders. Our results provide the first demonstration that NeuroD6 triggers a molecular chaperone network through the expression of a multitude of heat shock proteins (HSPs) in the absence of stress. This correlation may be relevant to the link between differentiation and survival, as HSPs interface at different connective points of these neuronal pathways (Fitzgerald et al., 2007; Herbert et al., 2007).
MATERIALS AND METHODS
Cell Culture
Control rat pheochromocytoma PC12 cells and PC12-ND6 cells (previously referred to as Nex1/MATH-2-overexpressing PC12-Nex1 cells) were grown on collagen I-coated plates (Becton Dickinson Labware, San Jose, CA, USA) in F-12K modified medium (Kaighn’s modification) supplemented with 2.5% fetal bovine serum and 15% horse serum (Invitrogen, Carlsbad, CA, USA), as described in Uittenbogaard and Chiaramello, 2005. When indicated, cells were differentiated in the presence of NGF (100 ng/ml) (Roche Molecular Biochemicals, Nutley, NJ, USA). For trophic factor deprivation, cells were seeded at 70% confluency and then washed the following day three times with serum-free medium, as described in Uittenbogaard and Chiaramello, 2005. The cells were grown in the absence of serum for specific periods of time, as indicated. For long-term serum deprivation, the serum-free medium was changed three times a week without splitting cells, as they stopped proliferating.
Total RNA isolation
Total RNA was isolated from six replicates of serum-grown control PC12 and PC12-ND6 cells as well as serum-deprived PC12-ND6 cells for two days to ensure adequate reproducibility. Total RNA was isolated using the RiboPure™ kit (Applied Biosystem, Foster City, CA) according to the manufacturer’s recommendations and the quality was assessed with the Agilent 2100 BioAnalyzer (Agilent Technologies, Palo Alto, CA).
cDNA synthesis and biotin-labeling of antisense cRNA
First strand cDNA was synthesized using a T7-(dT)24 oligomer and Superscript III enzyme (Invitrogen, Carlsbad, CA). Second-strand cDNA synthesis was performed using T4 DNA polymerase (Invitrogen) followed by phenol/chloroform extraction. Amplification and biotin-labeling of antisense cRNA was performed using the Bioarray™ High Yield RNA transcript labeling kit (Affymetrix, Santa Clara, CA). Unincorporated NTPs were removed using RNeasy spin columns (Applied Biosystem) and labeled cRNA was fragmented by metal-induced hydrolysis.
Microarray and Data Analysis
Fragmented and labeled amplified cDNA from each cell type (control PC12 or PC12-ND6) and growing condition (in the presence or absence of serum) done in six replicates was hybridized to the rat genome 230 A array as described in the Affymetrix array processing manual. Arrays (18 total) were washed at low and high stringency before being scanned by an Agilent laser scanner. Signal intensities were calculated using Affymetrix GeneChip Microarray Suite software 5.0. Microarray data were generated using the Affymetrix GCOS program. CEL files were generated and data normalization was performed using GC-RMA and per gene normalization to median (without averaging probe sets associated with a specific gene, when applicable). Data from the three conditions (serum-grown control PC12, serum-grown PC12-ND6 and serum-deprived PC12-ND6) were analyzed using the GeneSpring software version 7.3.1 (Agilent, Santa Clara, CA).
To identify statistically relevant probe sets, we performed a 1-Way ANOVA with the parametric test, don’t assume equal variances, a false discovery rate of 0.05 (p< 0.05) and a multiple testing correction with Benjamini and Hochberg false discovery rate. To correlate differential expression upon NeuroD6 overexpression (fold change of 1.5) with statistical significance (p-value< 0.05), a volcano plot filter was carried out between control PC12 and PC12-ND6 samples. The expression profile of the 951 NeuroD6 up-regulated probe sets was examined upon serum removal by applying a second Volcano plot filter (fold change of 1.5 fold and p-value cut-off of 0.05) between serum-grown and serum-deprived PC12-ND6 samples. All microarray data have been deposited in the Gene Expression Omnibus (GEO) of the National Center for Biotechnology Information (NCBI) (Edgar et al., 2002) and are accessible through the accession number GSE15942 (http://www.ncbi.nim.nih.gov/geo). Transcripts with differential expression levels upon NeuroD6 overexpression (PC12-ND6 cells) and upon withdrawal of trophic factors were classified into functional categories using the GO annotation tool in GeneSpring. A gene set enrichment analysis (GSEA), using the algorithm described in Subramanan et al. in 2005, was conducted to determine the rate of enrichment for each relevant functional category by comparing the percent of genes represented on the GeneChip® with the percent of genes upregulated upon NeuroD6 overexpression.
Immunoblot Analysis
Serum-grown control PC12 and PC12-ND6 cells, as well as serum-deprived PC12-ND6 cells for specific time periods as indicated in the figure legend were lysed in M-per mammalian protein extraction buffer (Pierce, Rockford, IL, USA) in the presence of a cocktail of protease inhibitors (Roche Molecular Biochemicals), as described in Uittenbogaard and Chiaramello, 2005. Protein concentration was determined by the Bradford assay (Bio-Rad) and proteins (40 μg) were resolved on a 10% NuPAGE Bis-Tris gels (Invitrogen) with either MES-sodium dodecyl sulfate (SDS) or MOPS-SDS running buffer, as recommended by the manufacturer, and transferred onto the nitrocellulose. Nitrocellulose membranes were stained with Ponceau-S (Sigma, St Louis, MO) to confirm uniform transfer of proteins, and subsequently blocked using Superblock™ blocking buffer (Pierce Biotechnology, Rockford, IL, USA) in phosphate buffer saline containing 0.05% Tween-20. The membranes were probed with various antibodies described in Table S1 and corresponding secondary horseradish peroxidase-conjugated antibodies (Pierce). The antigen-antibody complex was detected using the Supersignal West Pico Chemiluminescent Substrate kit (Pierce). Blots were then stripped using Restore™ western blot stripping buffer (Pierce) according to the manufacturer’s recommendations, and re-probed with a monoclonal antibody against glyceraldehyde 3-phosphate dehydrogenase (GAPDH) to confirm equal protein loading. Protein levels visualized by immunoblot analysis were quantified by GelEval 1.21 image software (FrogDance Software, http://www.frogdance.dundee.ac.uk) and normalized against the GAPDH loading control.
Immunocytochemistry and Confocal Fluorescence Microscopy
For immunocytochemistry labeling, control PC12 and PC12-ND6 cells grown in the presence or absence of serum were seeded on poly-D-lysine–coated coverslips placed in a six-well plate (Nunc, Rochester, NY, USA). Mitochondria were labeled by incubating cells in the presence of 500 nM MitoTracker® Red (Molecular Probes) for 10 minutes at 37 °C. All samples were fixed in 4.0% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA) for 5 minutes, and permeabilized in 0.2% Triton X-100 for 5 minutes. To label polymerized actin filaments, control PC12 and PC12-ND6 cells were incubated with AlexaFluor® 488 phalloidin (Molecular Probes) diluted to a final concentration of 82.5 nM in PBS for 20 minutes at RT following fixation and permeabilization. Cells were then blocked in 10% goat serum (Invitrogen) for one hour at RT. All primary and secondary antibody incubations were performed at RT as follows: one hour incubation with primary antibodies (Table S1), diluted in PBS, followed by one hour incubation with the appropriate Alexa Fluor® conjugated secondary antibodies diluted 1:1000 in PBS (Table S1). When indicated, cells were incubated with the nuclear counterstain DAPI (Molecular Probes). All samples were then mounted with Fluoromount G (Electron Microscopy Sciences), allowed to dry at room temperature overnight, and then stored at 4 °C. Images were acquired using a Zeiss LSM 510 confocal system and Observer Z1 microscope equipped with a 63x planapo chromat (NA, 1.4) objective and Zen software (4.5, 2007) interface. DAPI was excited with a 30 mW diode laser emitting at 405 nm and Alexa Fluor® 488 was excited with a 30 mW argon 488 nm laser. Alexa Fluor® 568 and MitoTracker Red were excited with a 1.2 mW helium/neon 543 nm laser, while Alexa Fluor® 647 was excited with a 5 mW helium/neon 633 nm laser.
RESULTS
Identification of differentially expressed genes upon constitutive expression of NeuroD6
In this study, we investigated the NeuroD6 genomic “signature” bridging neuronal differentiation and survival by performing a comparative genome-wide microarray analysis using an Affymetrix-based GeneChip® platform. The relative change in gene expression was analyzed under two conditions: 1) the global effect of constitutive NeuroD6 overexpression by comparing control PC12 and PC12-ND6 cells, and 2) the effect of serum deprivation by comparing serum grown PC12-ND6 and serum-deprived PC12-ND6 cells (t= 48 hrs). Serum removal is a widely used experimental paradigm for cell death, as it elicits within 48 hours apoptosis of control PC12 cells due to inappropriate cell cycle progression (Greene, 1978; Rukenstein et al., 1991; Farinelli and Greene, 1996) and increased ROS production (Troy and Shelanski., 1994; Satoh et al., 1996). The rat 230A high-density oligonucleotide GeneChip® was simultaneously screened with total RNA isolated from six replicates of the above-mentioned samples to ensure adequate reproducibility and robust statistical analyses.
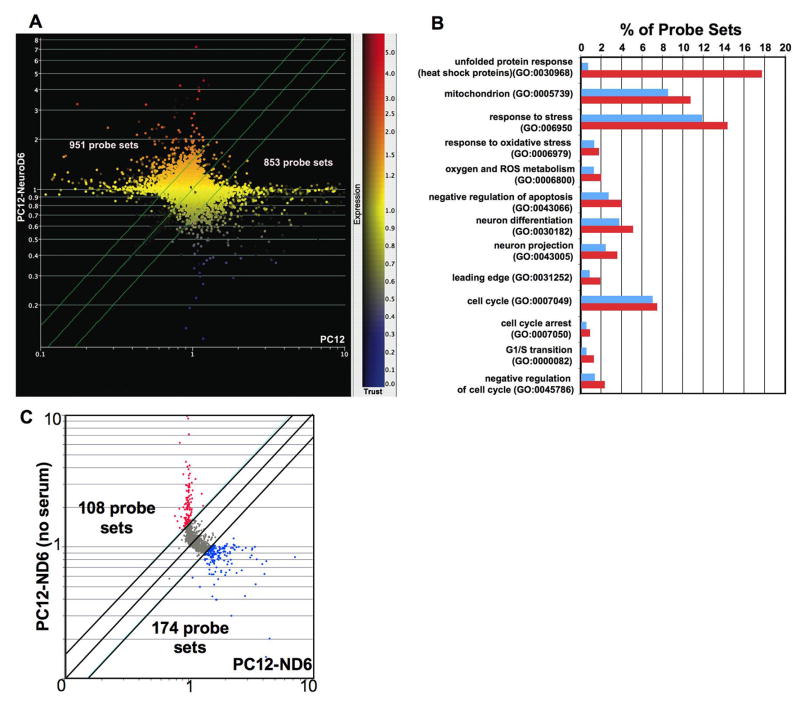
GC-RMA normalization was performed to eliminate all probe sets for which the expression data was not reproducible between the replicate GeneChips®. Upon a 1-way ANOVA statistical analysis combined with a Benjamini-Hochberg correction for false discovery rate (FDR) of 0.05, we generated a list of 6,059 statistically relevant probe sets from the 15,923 queried on the Affymetrix Rat 230 A high-density oligonucleotide GeneChip®. Differentially expressed probe sets upon NeuroD6 overexpression were then identified by performing a Volcano plot analysis with at least a 1.5 fold change and P-value cut-off of ≤0.05, as described in Materials and Methods. Figure 1A shows a scatterplot analysis of the differentially expressed probe sets upon NeuroD6 overexpression. Of the 6,059 statistically relevant probe sets, a total of 1,804 probe sets (30%) were differentially expressed in PC12-ND6 cells, with 951 transcripts up-regulated (16%) and 853 transcripts down-regulated (14%) (Table S2). We initially compared RNA levels of known NeuroD6 target genes from our previous restricted cDNA analysis and found that RNA levels of Gap43 (+6.21), p21CIP1 (Cdkn1a) (+4.31 and +2.63 for Affymetrix probe set 1387391_at and 1388674_at, respectively), neurofilament medium (nefm) (+3.43), and microtubule-associated protein-1A (Mtap1a) (+2.24) tightly correlated with our published studies (Uittenbogaard and Chiaramello, 2003; 2004), thus lending confidence in the genome-wide Affymetrix platform (Table S2).
Fig. 1.
Unbiased genome-wide microarray analysis upon NeuroD6 overexpression. (A) Scatterplot analysis showing differentially expressed probe sets upon NeuroD6 overexpression. Total RNA from control PC12 and PC12-ND6 cells were isolated in six replicates and hybridized to the rat 230A Affymetrix high density oligonucleotide GeneChip®. Differential expression was defined as at least a 1.5 fold change and a P-value cut-off of ≤0.05 after performing a 1-Way ANOVA and a multiple testing correction test with a Benjamini and Hochberg False Discovery Rate, followed by a Volcano analysis. Fluorescence intensities were graphed on a scatterplot, with control PC12 cells indicated on the horizontal axis and PC12-ND6 cells on the vertical axis (log10). The two green external lines on the scatterplot indicate boundaries for differential expression between 1 and ± 1.5 fold, while the green central line demarcates the lack of change in gene expression upon NeuroD6 overexpression. On the right side of the figure is shown the Colorbar representing expression levels on a continuous scale, with red indicating overexpression, yellow indicating average expression, and blue indicating underexpression in PC12-ND6 cells. The exact number of transcripts upregulated (951) and downregulated (853) by at least a 1.5 fold upon NeuroD6 overexpression are indicated on the graph. (B) Distribution of functional categories of differentially regulated transcripts upon NeuroD6 overexpression. The 951 probe sets upregulated by NeuroD6 were categorized in 13 functional groups using the gene ontology tool in GeneSpring, which were consistent with the reported differentiation and survival properties of PC12-ND6 cells. The percentage of upregulated transcripts in each category is indicated on the y axis with the corresponding functional group on the x axis. The percent of genes associated with each functional category on the 230A GeneChip® is indicated in blue, while the percent of genes upregulated in PC12-ND6 cells and associated with a specific functional category is indicated in red. The rate of enrichment was determined for each relevant functional category by comparing the percent of genes represented on the rat 230A GeneChip® with the percent of genes upregulated upon NeuroD6 overexpression. (C) Scatterplot analysis illustrating the expression profiles of the 951 NeuroD6-upregulated transcripts after two days of serum deprivation in PC12-ND6 cells. A Volcano analysis was performed with a minimum of a 1.5 fold change and P-value of ≤0.05 between serum-grown PC12-ND6 (x axis) and serum-deprived PC12-ND6 cells (y-axis). Fluorescence intensities were graphed on a log of 10. This analysis revealed 108 transcripts upregulated (red dots) and 174 transcripts downregulated (blue dots) upon serum deprivation of PC12-ND6 cells. The remaining 669 transcripts gray dots) did not display any significant changes in expression levels and thus were graphed between the two external diagonal lines representing the boundaries between ± 1.5 fold change in expression levels upon serum deprivation.
Overall, upregulated transcripts upon NeuroD6 overexpression belonged to canonical pathways associated with neurogenesis (Table S2), such as G1-phase inhibitors, including quiescin Q6 (Qscn6), E2F transcription factor 5 (E2f5), growth arrest specific-5 (gas5) and -6 (gas6), transcription factors cooperating with neurogenic bHLH regulators to promote neurogenesis and neuronal maturation, such as CCAAT/enhancer binding protein (C/EBP) (Nieto et al., 2001; Sun et al., 2001; Vetter, 2001; Ménard et al., 2002; Paquin et al., 2005), and the HMG-box protein SRY-box containing gene 4 (Sox4) (Bergsland et al., 2006), as well as enhancers of neural regeneration, such as neuronal regeneration related protein (Nrep) (Fujitani et al., 2004), arginosuccinate synthase (Ass) (Cai et al., 2002), small proline-rich repeat (SPRR1A) (Li and Strittmatter, 2003), matrix metallopeptidase 13 (Mmp13), a disintegrin and metallopeptidase domain 15 (Adam-15) and-23 (Adam-23) (Yong et al., 2001). As anticipated, significant down-regulation of key proliferating-promoting regulators, such as cyclin E (Ccne1), cyclin B1 (Ccnb1), cyclin B2 (Ccnb2), proliferating cell nuclear antigen (Pcna), and inhibitor of DNA binding 3 (Id3) was observed (Table S3), which is in keeping with previous gene expression profiling studies on NGF-treated PC12 cells using a serial analysis of gene expression (SAGE) approach (Angelastro et al., 2000; 2002) or a microarray platform (Dijkmans et al., 2008).
Functional gene categories regulated by NeuroD6
To gain further insight into the biological relevance of the NeuroD6-mediated gene network, we performed a gene ontology (GO) analysis on the 951 upregulated genes in PC12-ND6 cells, as described in Materials and Methods. This analysis employed the Gene Set Enrichment Analysis (GSEA) algorithm described in Subramanan et al. in 2005. These genes were distributed among 13 prevalent functional categories (Fig. 1B), which were consistent with the reported differentiation phenotype of PC12-ND6 cells, their long generation time (7 days) and survival properties (Uittenbogaard and Chiaramello, 2002; 2004; 2005). Genes showing increased expression belonged to functional groups involved in neuronal differentiation, such as neuronal projection (neurite extension), leading edge (formation of lamellipodium), and cell cycle (cell cycle arrest, G1/S phase transition and negative regulation of proliferation). Moreover, our GSEA analysis provided the first clue that overexpression of NeuroD6 resulted in increased expression of genes belonging to the heat shock response, mitochondrial bioenergetics, and stress response in the absence of stress stimulus (Fig. 1B).
Most of these functional groups displayed substantial gene enrichment, when compared to the percent of genes represented on the Rat 230 A high-density oligonucleotide GeneChip® (Fig. 1B). Interestingly, the category of heat shock proteins (HSPs) showed the most striking enrichment of genes upregulated in PC12-ND6 cells with a 18-fold increase in representation (Fig. 1B). Among the small HSPs members, HSP27 (hspb1) and HSP32, also called heme oxygenase-1 (hmox-1), displayed the highest increase in expression levels upon NeuroD6 overexpression with a 4.3 and 8.4 fold, respectively (Table 1). Several members of the DnaJ/Hsp40 family were also upregulated upon NeuroD6 overexpression. This observation is particularly relevant in the context of concomitant increased expression of several HSP-70 members in PC12-ND6 cells (Table 1), as DNAJ/HSP40 proteins interact with HSP70 proteins to stimulate their ATP hydrolysis activity for regulating protein folding (Qiu et al., 2006). We also observed increased expression of two organelle-specific members of the HSP70 family, the glucose-related proteins (Grps), Grp75 (Hspa9a) (1.5 fold) and Grp78 (hspa5) (2.3 fold) in PC12-ND6 cells (Table 1), which are known to possess protective functions against various stresses and insure proper protein folding in mitochondria and endoplasmic reticulum, respectively (Daugaard et al., 2007). In contrast, the constitutively expressed housekeeping Hsc70 (hspa8) member did not display differential expression upon NeuroD6 overexpression and upon serum deprivation (Table 1).
TABLE 1.
Differential expression levels of heat shock proteins in serum-grown and serum deprived PC12-ND6 cells for 48 hours.
| Members of HSP Families1 | Fold Change upon NeuroD6 | p-value* | Fold Change upon Serum Removal | p-value* |
|---|---|---|---|---|
| Small hsp family | ||||
| Hsp27 (hspb1) [24471] | 4.32 | 2.22 10−7 | −1.31 | 9.1310−5 |
| Hsp22 (hspb8) [113906] | 1.62 | 7.19 10−5 | 1.0 | 0.05 |
| Hsp32; HO-1 (hmox-1) [24451] | 8.4 | 4.82 10−6 | −1.28 | 0.00508 |
| Cpn10 (hspe1) [25462] | 1.3 | 5.79 10−5 | 1.0 | 0.05 |
| Hsp40 family | ||||
| 2 Dnajb1 predicted [361384] | 1.8 | 0.000354 | 1.0 | 0.005 |
| 3 Dnajb1 [361384] | 1.76 | 1.70 10−5 | 1.24 | 0.0136 |
| 4 Dnaja4 [300721 | 1.55 | 0.0131 | 1.26 | 0.0138 |
| 5 Dnaja4 [300721] | 1.51 | 0.00582 | 1.0 | 0.05 |
| 6 Dnaja1 [65028] | 1.94 | 0.000221 | 1.0 | 0.05 |
| 7 Dnaja1[65028] | 1.70 | 2.84 10−5 | 1.0 | 0.05 |
| Dnajb10 [501165] | 1.66 | 0.000792 | 1.0 | 0.05 |
| Dnajb9 [24908] | 2.0 | 0.000954 | 1.99 | 6.2210−5 |
| Dnajc3 [63880] | 1.97 | 0.00655 | −2.41 | 4.01510−5 |
| Hsp60 family | ||||
| Hsp60 (hspd1) [63868] | 1.3 | 8.16 10−5 | 1.0 | 0.05 |
| Hsp70 family | ||||
| Hsc70 (hspa8) [24468] | 1.3 | 0.015 | 1.0 | 0.05 |
| Hsp72; Hsp70-2 (hspa1b) [294254] | 2.8 | 0.0046 | 1.0 | 0.05 |
| BIP; GRP78 (hspa5) [25617] | 2.22 | 0.00608 | −2.44 | 2.3810−5 |
| mtHsp70; GRP75 (Hspa9a) [500372] | 1.56 | 0.0169 | −1.96 | 0.00531 |
| Hsp90 family | ||||
| Hsp90-1a (hsp90aa1) [299331] | 1.4 | 0.00131 | 1.25 | 0.05 |
| Hsp90-1b (hsp90ab1) [301252] | 1.4 | 0.0189 | 1.24 | 0.05 |
| Hsp110 family | ||||
| Hsp105 (hsph1)[288444] | 2.07 | 0.000729 | −1.1 | 0.0049 |
| Non-classical HSPs | ||||
| clusterin (clu) [24854] | 1.61 | 0.000145 | 2.18 | 3.34E-05 |
| Hsf members | ||||
| Hsf-1 [79245] | 1.0 | 0.05 | 1.0 | 0.05 |
| Hsf-2 [64441] | 1.0 | 0.05 | 1.0 | 0.05 |
| Hsf-4 [291960] | 1.73 | 4.75 10−5 | −1.46 | 0.00493 |
Hsf (heat shock factor), HSP (heat shock protein)
Members of the HSP family are listed by their known gene names (aliases) followed by their official or “interim” symbol in parentheses and their gene ID number in brackets, according to the NCBI nomenclature. Only members of the Hsp40 family are referred to as official symbols.
Corresponding Affymetrix probe set 1388722_at
Corresponding Affymetrix probe set 1383302_at
Corresponding Affymetrix probe set 1387780_at
Corresponding Affymetrix probe set 1378592_at
Corresponding Affymetrix probe set 1368852_at
Corresponding Affymetrix probe set 1398819_at
p-value calculated after a 1-Way ANOVA, a multiple testing correction with Benjamini and Hochberg with a false discovery rate of 0.05 (p< 0.05), followed by a Volcano analysis using the software GeneSpring version 7.3, as described in Materials and Methods.
Identification and functional distribution of NeuroD6 upregulated genes upon withdrawal of trophic factors
Since our recent studies revealed NeuroD6 neuroprotective function as an integral component of the differentiation program, we analyzed the expression profile of the 951-upregulated genes in the context of NeuroD6-mediated neuronal survival following withdrawal of trophic factors. Thus, we performed a second Volcano plot analysis with a minimum of a 1.5 fold change and P-value of ≤0.05 between serum-grown PC12-ND6 and serum-deprived PC12-ND6 cells (t=48 hrs). We found 108 genes (12%) to be significantly up-regulated upon serum removal and 174 genes (18%) to be down-regulated, while 301 genes (32%) were constant or moderately up-regulated (between 1.0 and +1.5), 358 genes (38%) were between 1.0 and −1.5, and 10 genes that were no longer statistically relevant under the serum deprivation conditions (Fig. 1C, Table S4).
Based on the neuroprotective properties of NeuroD6, we emphasized our analysis on functional categories associated with survival, such as negative regulation of apoptosis, mitochondrial bioenergetics, stress response, and heat shock response (Fig. S1). Remarkably, most of the HSPs remained expressed at unaltered levels upon serum deprivation (Fig. S1A, Table 1). Similarly, the majority of genes involved in mitochondrial bioenergetics and metabolism displayed constant RNA expression levels after 48 hours of serum deprivation in PC12-ND6 cells (Fig. S1B). As expected, most of the regulators repressing apoptosis remained expressed at constant levels in serum-deprived PC12-ND6 cells (Fig. S1C). Finally, other members of the stress response showed substantial variations in their expression levels upon serum removal (Fig. S1D, Table S4).
Thus, our microarray analysis provides the first clue of a correlation between the expression of NeuroD6 and HSP encoding genes in the absence of stress. This is particularly relevant to NeuroD6 functions, as the HSPs share pathways with NeuroD6, such as cell cycle control, cytoskeletal dynamics, and regulation of neuronal survival during neuronal differentiation (Richter-Landsberg, 2007). In addition, they participate in protein synthesis, folding, transport and translocalization via their chaperone activity and thereby preventing protein misfolding and aggregation upon stress stimuli (Liberek et al., 2008).
Validation and comparative analysis of HSP expression patterns between the NeuroD6 effect, serum removal and NGF-mediated differentiation
To validate the mRNA expression changes of the HSP members revealed by the microarray analysis, we selected HSP candidates with at least a 1.5 fold increase in mRNA expression levels and with well-established roles in neuronal differentiation and/or survival.
The protein expression levels of six HSP proteins belonging to the small HSP, HSP40, and HSP70 families were analyzed by immunoblot analysis and quantified as described in Materials and Methods, with the following objectives: first, to determine their expression levels upon NeuroD6 overexpression (PC12-ND6 cells); second, to determine their expression profile throughout serum deprivation of PC12-ND6 cells, to acquire a perspective on the dynamic aspect of stress response associated with long-term stress induction; and finally, to compare their levels of expression through NGF-mediated neuronal differentiation using control PC12 and PC12-ND6 cells, to compare the differentiation effect of NeuroD6 and versus NGF on the expression of distinct HSPs.
Overexpression of NeuroD6 resulted in increased expression levels of specific members of the small heat shock protein family
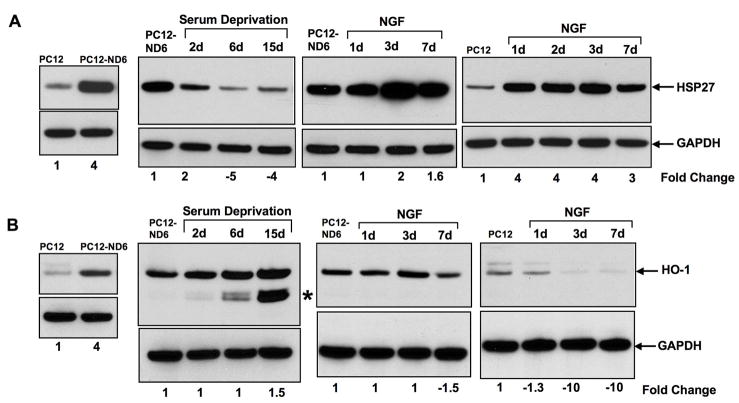
Based on the criteria aforementioned, we investigated the expression profile of two members of the small HSP family, HSP27 (hspb1) and HO-1 (hmox-1). We found a 4-fold increase in HSP27 protein levels upon NeuroD6 overexpression, which was in keeping with the 4.3-fold increased mRNA levels quantified by microarray analysis (Fig. 2A; Table 1). We further observed a tight correlation between decreased mRNA (−1.31-fold) and protein (−1.46-fold) levels after two days of serum deprivation in PC12-ND6 cells (Fig. 2A, Table 1). Surprisingly, expression levels of the HSP27 protein continued to decrease throughout serum deprivation (Fig. 2A). In contrast, HSP27 protein showed increased expression levels upon NGF exposure in both control PC12 and PC12-ND6 cells, with NGF-treated PC12-ND6 cells expressing higher levels of HSP27 protein than NGF-treated control PC12 cells (Fig. 2A).
Fig. 2.
Overexpression of NeuroD6 induces the expression of two members of the small heat shock protein family. Cells were grown and treated as described in Materials and Methods. Data shown are representative of at least three distinct experiments. Protein levels were quantified and normalized against the GAPDH loading control. Quantification results are expressed as fold change relative to the respective control cells, shown in lane 1 of each blot. Indicated quantification values are specific to the shown blot, with a standard deviation less than 10% when compared to the corresponding replicates. (A) Expression levels of HSP27 protein increase in serum-grown PC12-ND6, cells but collapse in serum-deprived PC12-ND6 cells. NGF treatment of both PC12-ND6 and control PC12 cells stimulates the expression levels of HSP27 protein. (B) Expression levels of HO-1 (HSP32) protein are upregulated upon NeuroD6 expression and further stimulated upon serum deprivation. An additional HO-1 isoform, indicated by an asterisk, was detected after six days of serum deprivation with maximal expression levels after 15 days of serum deprivation. NGF treatment failed to further increase expression levels of HO-1 protein in treated PC12-ND6 cells, whereas it failed to trigger expression of endogenous HO-1 protein in treated PC12 cells.
Immunoblot analysis confirmed increased expression levels of the HO-1 protein upon NeuroD6 overexpression in the absence of stress stimulus, albeit with a magnitude lower than that of the mRNA levels quantified by microarray analysis, 4-fold and 8.4-fold, respectively (Fig. 2B; Table 1). We further observed unaltered expression levels of the HO-1 protein during the first two days of serum deprivation in PC12-ND6 cells, which is in agreement with the HO-1 mRNA levels quantified by microarray analysis (Fig. 2B; Table 1). Interestingly, two additional HO-1 isoforms were detected after six days of serum deprivation in addition to the 32-kDa full-length HO-1 protein, which displayed increased expression levels (1.5 fold) after 15 days of serum deprivation (Fig. 2B). NGF treatment of PC12-ND6 cells did not alter HO-1 expression levels up to day seven, and induced the expression of the two potential HO-1 isoforms (Fig. 2B). As expected, NGF-treated PC12 cells failed to up-regulate expression of the endogenous HO-1 protein, since NGF exposure has been shown to trigger transient HO-1 expression only within the first two hours of exposure (Liu et al., 2003). Therefore, the NeuroD6 effect appears to diverge from the NGF pathway, as HO-1 expression was not sustained in NGF-treated PC12 cells, thus raising the possibility of a NeuroD6 direct regulation.
NeuroD6 upregulates the expression of members of the HSP70 chaperone system, independent of a stress stimulus
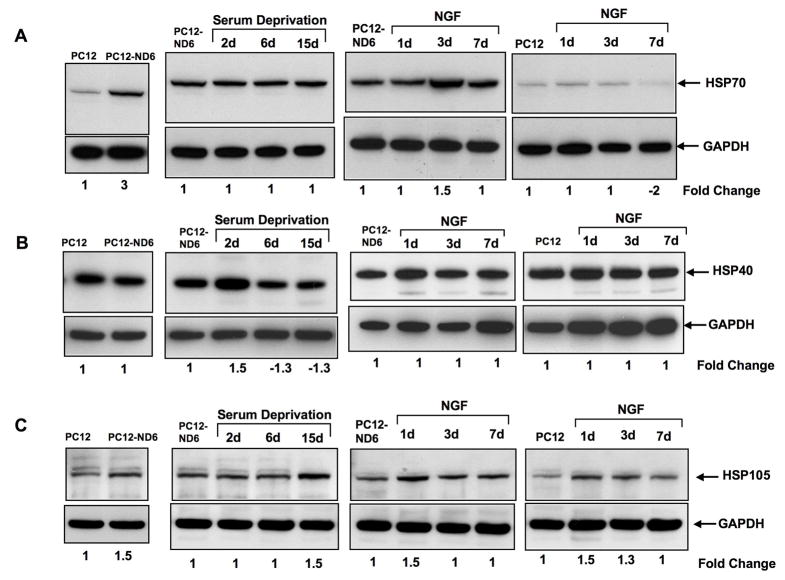
We examined the expression levels of the inducible form of Hsp70 (also called HSP72; hspa1b), as the microarray analysis revealed a 2.8-fold increase at the mRNA level upon NeuroD6 overexpression (Table 1). For the immunoblot analysis, we used an antibody specific to the inducible HSP70 form, which did not cross-react with the constitutive HSC70 form (Table S1). Strikingly, we found that PC12-ND6 cells expressed high levels of HSP70, while control PC12 cells only expressed barely detectable basal levels of HSP70 protein (Fig. 3A). Furthermore, NGF treatment failed to stimulate the expression levels of HSP70 protein in both cell types, implying that the hsp70 gene is not NGF responsive (Fig. 3A). Moreover, expression levels of HSP70 protein remained unaltered in serum-deprived PC12-ND6 cells (Fig. 3A). Collectively, these results suggest that the hsp70 gene may be a direct target gene of NeuroD6.
Fig. 3.
NeuroD6 and the HSP70 chaperone system. (A) Overexpression of NeuroD6 triggers expression of HSP70 protein, which remains at steady levels even upon serum deprivation and NGF treatment. In contrast, control PC12 cells express negligible expression levels of endogenous HSP70 protein, even upon NGF exposure. (B) PC12-ND6 cells express similar levels of endogenous HSP40 protein than control PC12 cells. However, expression levels of HSP40 protein remain at steady state in serum-deprived PC12-ND6 cells throughout the duration of the treatment. Expression levels of HSP40 protein fail to augment upon NGF exposure in both control PC12 and PC12-ND6 cells. (C) Overexpression of NeuroD6 stimulates the expression of HSP105 protein, which is further increased after 15 days of serum deprivation in PC12-ND6 cells. NGF treatment of control PC12 and PC12-ND6 cells have a modest impact on the expression levels of HSP105 protein.
Next, we focused on the HSP40 family, more specifically the Dnajb1 member, also known as HSP40 homolog, since it is a well-established co-chaperone for specific HSP70 proteins (Qiu et al., 2006). Although quantification of the microarray data revealed a 1.8 and 1.76-fold increase in DnaJb1 mRNA levels for probe set 1388722_at and 1383302_at, respectively upon NeuroD6 overexpression (Table 1), we failed to observe a similar increase at the protein levels by immunoblot analysis (Fig. 3B). However, after two days of serum deprivation, PC12-ND6 cells showed a 50% increased expression of the HSP40 protein, which was not maintained throughout the length of serum deprivation (Fig. 3B). In contrast, levels of DnaJb1 expression remained at steady levels throughout NGF treatment of either control PC12 or PC12-ND6 cells, implying that the DnaJb1 gene is not NGF-inducible (Fig. 3B).
We complemented the analysis of the Hsp70 chaperone system by focusing on the HSP105 (hsph1) chaperone for the following reasons: 1) our microarray analysis revealed a concomitant increase of HSP70 and HSP105 mRNA levels upon NeuroD6 overexpression (Table 1); and 2) HSP105 is known to cooperate with HSP70 in the disaggregation process of aggregated proteins (Zietkiewicz et al., 2004; 2006). Figure 3C shows a modest increase of the HSP105 protein in PC12-ND6 cells, consistent with the microarray data. Similarly, NGF treatment of PC12 and PC12-ND6 cells resulted in a slight but reproducible increase in HSP105 protein levels, although at higher levels in PC12-ND6 cells (Fig. 3C). Finally, the expression levels of HSP105 protein were only increased at 15 days of serum deprivation, suggesting that HSP105 may play a more prevalent role during the long-term phase of stress tolerance (Fig. 3C).
NeuroD6 stimulates the expression of organelle-specific members of the HSP70 family
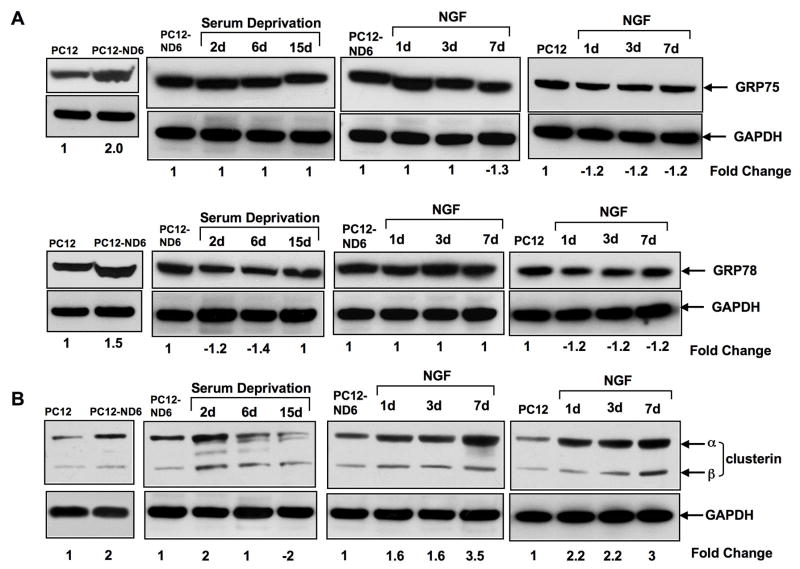
The microarray analysis revealed increased expression of two organelle-specific members of the Hsp70 family, GRP75 (hspa9a), also known as HSP70.9 or mtHSP70, and GRP78 (hspa5), also called Bip for binding protein), which are localized in mitochondria and endoplasmic reticulum, respectively (Table 2). We found that expression levels of both GRP75 and GRP78 proteins increased upon NeuroD6 overexpression in the absence of stimulus (Fig. 4A). Serum deprivation of PC12-ND6 cells did not alter GRP75 expression levels and only triggered a negligible decrease of GRP78 levels (Fig. 4A), all which are in accordance with the microarray results (Table 1). Finally, NGF-treated PC12-ND6 cells displayed sustained levels of both GRP75 and GRP78 proteins, whereas NGF treatment of control PC12 cells resulted in a slight but consistent decrease in GRP75 and GRP78 protein levels (Fig. 4A). Thus, these results imply that NeuroD6 upregulates GRP75 and GRP78 levels, independent NGF signaling.
Fig. 4.
NeuroD6 upregulates the expression of organelle-specific HSP70 members and the non-classical molecular chaperone clusterin. (A) NeuroD6 upregulates the expression of the mitochondrial-specific chaperone GRP-75 and the endoplasmic reticulum-specific chaperone GRP78. Serum deprivation does not impact expression levels of both GRP-75 and GRP78 in PC12-ND6 cells. NGF treatment of either control PC12 or PC12-ND6 cells did not result in further increased expression of GRP75 and GRP78 proteins. (B) Clusterin levels are increased upon NeuroD6 expression and after two days of serum deprivation in PC12-ND6 cells. NGF treatment stimulates endogenous expression levels of clusterin in both treated PC12 and PC12-ND6 cells, with an apparent synergistic effect between NGF and constitutive expression of NeuroD6.
Clusterin levels increase upon both NeuroD6 overexpression and serum deprivation
We analyzed the expression profile of clusterin (clu), also called apolipoprotein J, as the microarray analysis revealed a 1.61-fold and 2.18-fold increase in mRNA levels in PC12-ND6 and serum-deprived PC12-ND6 cells, respectively (Table 1). Furthermore, clusterin, which is structurally related to small HSPs, has recently been shown to possess neuroprotective properties (DeMattos et al., 2002; Poon et al., 2002; Carver et al., 2003). Interestingly, clusterin protein levels peaked at day two, but failed to sustain throughout the whole serum deprivation treatment, with a collapse at day 6 of serum deprivation (Fig. 4B). Finally, we observed that levels of clusterin protein were significantly upregulated upon NGF treatment of both control PC12 and PC12-ND6 cells (Fig. 4B), which is in keeping with a previous study reporting increased clusterin mRNA levels in NGF-treated PC12 cells (Gutacker et al., 1999). In summary, we found clusterin to be the only molecular chaperone displaying a transient increase in expression levels upon serum deprivation, which coincides with the transient increase in the expression of the anti-apoptotic Bcl-xL regulator, previously reported in our studies (Uittenbogaard and Chiaramello, 2005). This pattern of expression is consistent with the recently reported cytoprotective properties of clusterin (Björk and Sistonen, 2006; Loison et al., 2006).
Colocalization of actin and HSP27 in PC12-ND6 cells
We supplemented our analyses with immunocytochemistry studies to investigate whether specific heat shock proteins involved in cytoskeletal dynamics acquired appropriate subcellular localization upon NeuroD6 overexpression in the absence of extrinsic signaling. We focused on HSP27 (hspb1) and HSP70 (hspa1b), as they modulate key cytoskeletal elements during development of the nervous system (Herbert et al., 2007), which are also upregulated upon NeuroD6-mediated neuronal differentiation (Uittenbogaard and Chiaramello, 2002; 2004).
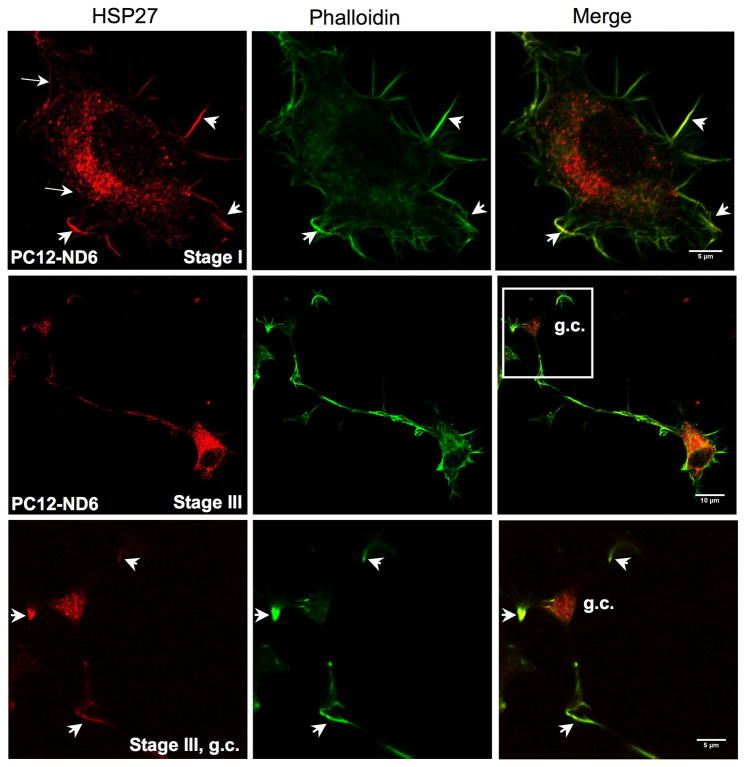
Since HSP27 influences neurite outgrowth of dorsal root ganglion neurons by modulating the actin network (Williams et al., 2005; 2006), we investigated this potential colocalization by immunocytochemistry using phalloidin to label actin and a rabbit polyclonal antibody against HSP27 (Table 1). One advantage of the PC12-ND6 cellular paradigm is that PC12-ND6 cells simultaneously recapitulate the first four stages of neuronal differentiation, as defined by Dotti et al., 1988 (Baxter et al., manuscript in preparation), thereby allowing the evaluation of the relationship between HSP27 and actin at distinct neuronal stages associated with intense actin remodeling, such as the early stage of lamellipodia formation (stage I) and the later stage of axonal outgrowth (stage III).
Figure 5A clearly shows that HSP27 colocalizes with actin at focal points of the leading edge of the lamellipodium (see arrows) as well as in filopodia (see arrowheads) of stage I PC12-ND6 cells. As expected, we also detected a significant amount of cytosolic HSP27 in the soma of PC12-ND6 cells. We further examined the distribution of the HSP27 protein at stage III of neuronal differentiation during which axonal outgrowth occurred, and found that HSP27 protein not only retained a robust cytosolic expression, but also colocalized with actin in the developing axon and filopodia extending from the growth cone. One interesting feature of HSP27 expression was its abundant accumulation in the growth cone, suggesting a potential role of HSP27 not only in neurite initiation, but also in axonal outgrowth via growth cone activity (Fig. 5A).
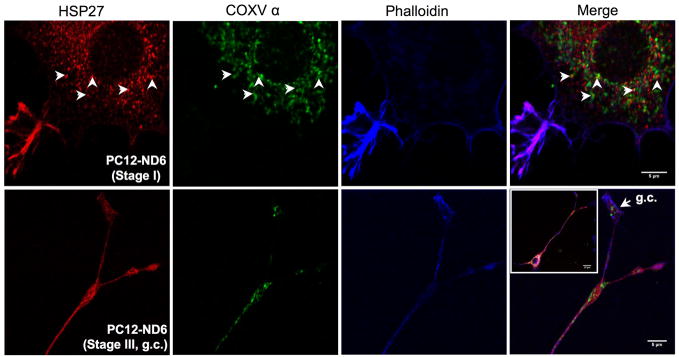
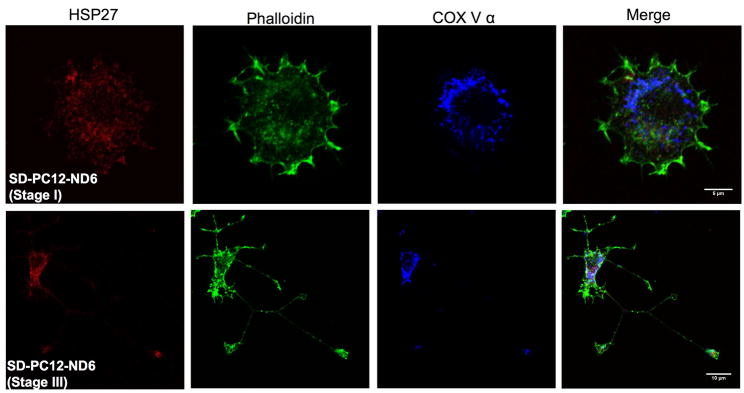
Fig. 5.
Subcellular localization of HSP27 protein in PC12-ND6 cells. (A) HSP27 protein colocalizes with actin and accumulates in growth cones of developing neurites. PC12-ND6 cells were seeded on poly-D-lysine-coated cover glass. To label polymerized actin filaments, PC12-ND6 cells were incubated with AlexaFluor® 488 phalloidin (82.5 nM) for 20 minutes at room temperature (RT), fixed and permeabilized as described in Materials and Methods. Cells were incubated with the anti-HSP27 primary antibody and then with the appropriate Alexa Fluor® 568 conjugated secondary antibody. Scale bars are indicated at the bottom right corner of merge images. The top row shows stained PC12-ND6 cells at stage I of neuronal differentiation, while the middle row displays stained stage III PC12-ND6 cells. The bottom row shows a magnification of the growth cone (GC) of stage III PC12-ND6 cell represented in the middle row. (B) Only a small fraction of HSP27 protein is located in the vicinity of mitochondria. PC12-ND6 cells were grown and processed for confocal fluorescence microscopy as described in panel A. Mitochondria were visualized with an antibody against the mitochondrial marker, the α subunit of complex V (COX V, also called ATP synthase). Mitochondria located in the vicinity of HSP27 protein are indicated with arrowheads. The top row shows stage I PC12-ND6 cells, while the bottom row illustrates a magnification of the growth cone (GC) of developing neurite from a stage III PC12-ND6 cell (see inset). Scale bars are indicated at the bottom right corner of merge images. (C) HSP27 expression not only decreases upon serum deprivation in PC12-ND6 cells, but also remains essentially cytosolic. The serum-deprived PC12-ND6 cells (SD-PC12-ND6) were processed for confocal fluorescence microscopy as described in Materials and Methods. The top row shows serum deprived (SD) stage I PC12-ND6 cells, while the bottom row shows serum-deprived stage III PC12-ND6 cells. Scale bars are indicated at the bottom right corner of merge images.
Although several studies using non-neuronal cells reported limited colocalization between HSP27 and mitochondria upon a stress stimulus (Bruey et al., 2000; Paul et al., 2002), we examined the HSP27 subcellular localization in relation to the mitochondria distribution prior to and during serum deprivation using PC12-ND6 cells. Mitochondria were visualized using an antibody against the α subunit of ATP synthase (COX V) (Table S1). Only a small fraction of cytosolic HSP27 was in the vicinity of mitochondria in the soma of serum-grown and serum-deprived PC12-ND6 cells (Fig. 5B and C, arrowheads). HSP27 did not colocalize with mitochondria located in the growth cones (Fig. 5B). Our immunocytochemistry results confirmed the decreased expression levels of HSP27 upon serum deprivation revealed by immunoblot analysis (Fig. 2). Thus, within the context of our neuronal differentiation/survival paradigm, these results imply that HSP27 essentially exerts its function through its colocalization with actin, rather than mitochondria, which is in accordance with the notion of HSP27 protecting non-neuronal HeLa cells by preserving actin integrity (Paul et al., 2002).
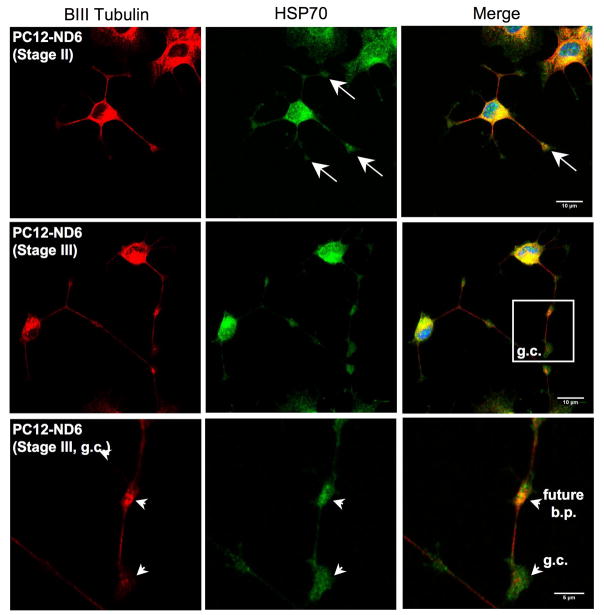
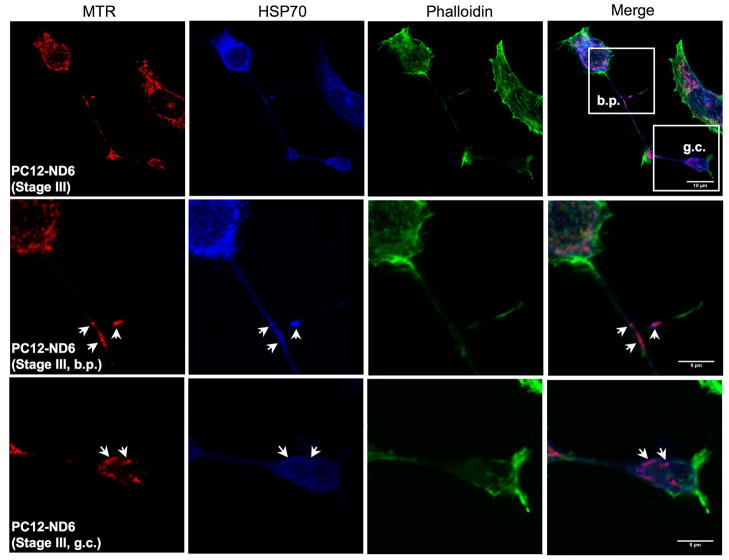
HSP70 protein colocalizes with tubulin and mitochondria in PC12-ND6 cells
We examined HSP70 (hspa1b) colocalization with cytoskeletal elements, as several studies have reported HSP70 associations with microtubules elements and the microtubule-binding protein Tau (Wallace et al., 1993; Kirby et al., 1994; Liang and MacRae, 1997). As expected, figure 6A shows strong colocalization of HSP70 and β-III tubulin in the soma and growing neurites of PC12-ND6 cells, with substantial accumulation of HSP70 protein at developing branch points (Fig. 6A, arrowheads). Furthermore, we observed accumulation of HSP70 in the neuritic tip of stage II PC12-ND6 cells (Fig. 6A, arrows). Interestingly, HSP70 protein remained present throughout the growth cone beyond the expression domain of β-III tubulin in stage III PC12-ND6 cells, but excluded from the leading edge of the growth cone and filopodia (Fig. 6A, arrowheads; Fig. 6B). This observation supports previous studies showing synaptic expression of HSP70 protein in rat brain upon hyperthermia and improvement of presynaptic performance at high temperature upon overexpression of HSP70 in Drosophila (Bechtold et al., 2000; Karunanithi et al., 2002). However, our results provide the first evidence of HSP70 expression in growth cones in the absence of heat shock or other type of stress.
Fig. 6.
HSP70 colocalizes with tubulin and mitochondria in PC12-ND6 cells. (A) HSP70 colocalizes with tubulin in the soma and extending neurites. However, it accumulates in growth cones in areas devoid of tubulin. PC12-ND6 cells were grown and processed for confocal microscopy as described in Materials and Methods. The top row shows stage II PC12-ND6 cells with neuritic tips indicated by arrows. The middle row illustrates stage III PC12-ND6 cells, which display growth cones (GC) and developing branching point (bp), both indicated by arrowheads. A magnification of the growth cone and neighboring developing branching point is shown in the bottom row. Scale bars are indicated at the bottom right corners of merge images. (B) HSP70 colocalizes with mitochondria located in the soma, while migrating in developing neurites and in growth cones. Mitochondria were stained with the MitoTracker® Red CMXRos dye (MTR). PC12-ND6 cells were stained with phalloidin to highlight the overall cellular morphology and to examine the HSP70-actin colocalization. The top row shows stage III PC12-ND6 cells, while the middle and bottom rows illustrate a magnification of a developing branching point (bp) and growth cone (GC), respectively. Arrowheads indicate colocalization of HSP70 and mitochondria. Scale bars are indicated at the bottom right corner of merge images. (C) HSP70 expression levels and subcellular localization remain unaltered by withdrawal of trophic factors in PC12-ND6 cells. Mitochondria were stained with MTR and the overall cellular morphology was revealed using an antibody against β-III tubulin. The top rows show a network of stage III SD-PC12-ND6 cells, while the bottom row illustrates the magnification of the neurite network indicated by a white box. Arrows indicate colocalization of mitochondria and HSP70 in growth cones of SD-PC12-ND6 cells. Scale bars are indicated at the bottom right corner of merge images.
Since HSP70 was detected in mitochondrial fractions of the human U937 leukaemic cell line (Bruey et al., 2000), we investigated the potential colocalization between HSP70 and mitochondria in PC12-ND6 cells and its time course in relation to the stress stimulus. Figure 6B shows that HSP70 colocalizes with mitochondria in the absence of stress stimulus and that such colocalization is not limited to the soma of serum-grown PC12-ND6 cells. More precisely, HSP70 colocalizes with migrating mitochondria (MTR, red) in growing neurites and at branching points (phalloidin, green) (Fig. 6B, middle row, arrows), and with mitochondria located in the growth cone (Fig. 6B, arrowheads). Interestingly, HSP70 was also present in areas of the growth cone devoid of mitochondria. HSP70 colocalization with mitochondria persisted in serum-deprived PC12-ND6 cells (Fig. 6C). It is worth noting that HSP70 protein remained present at high levels in developing neurites and growth cones as well as in the soma (Fig. 6C). As expected, HSP70 did not interact with actin (Fig. 6B). In conclusion, NeuroD6 not only triggers increased HSP70 expression, but also induces proper HSP70 trafficking in developing neurites and colocalization with microtubules and mitochondria.
Differential expression levels of the heat shock transcription factors HSF-1, HSF-2, and HSF-4 upon NeuroD6 overexpression
In view of the positive correlation between NeuroD6 and HSPs, we extended our analysis to the heat shock transcription factor (HSF) family, Hsf-1, Hsf-2, and Hsf-4, which bind to the Heat Shock Element (HSE) to regulate transcription of heat shock genes (Pirkkala et al., 2001; Morange, 2006; Åkerfelt et al., 2007). While Hsf1 mainly induces expression of HSPs in response to stress (Morimoto, 1998), Hsf-2 regulates not only the expression of Hsp genes in conjunction with Hsf-1 (Östling et al., 2007), but also the expression of target genes other than HSPs within the context of neuronal differentiation in the absence of stress (Chang et al., 2006). Much less is known on Hsf-4 mechanism of action within the context of HSP induction (Hu and Mivechi, 2006; Tu et al., 2006).
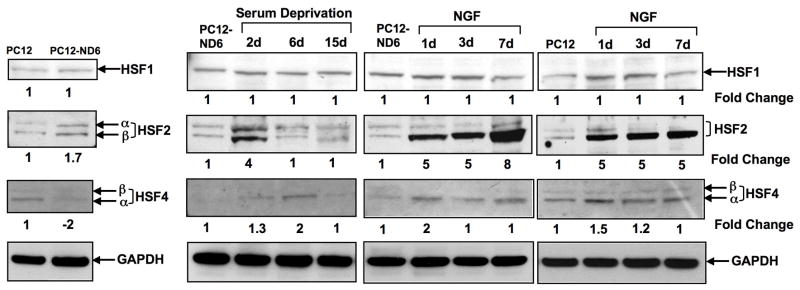
Our immunoblot analysis failed to detect any significant changes in expression levels of HSF-1 protein upon NeuroD6 overexpression, serum deprivation of PC12-ND6 cells, and NGF treatment (Fig. 7A). In contrast, HSF-2 protein expression levels were increased upon NeuroD6 overexpression, with both α and β isoforms being detected (Fig. 7A). HSF-2 expression levels were further increased upon two days of serum deprivation before returning to pre-stress levels (Fig. 7A). In keeping with the recently reported role of Hsf-2 during neuronal differentiation (Chang et al., 2006), we observed that NGF treatment induced expression of the HSF-2 β isoform (Fig. 7A), which is primarily expressed in the brain (Goodson et al., 1995). Strikingly, NGF-treated PC12-ND6 cells still displayed a stronger induction of HSF-2 β expression levels than NGF-treated PC12 cells, suggestive of a synergetic effect between NeuroD6 and NGF (Fig. 7A).
Fig. 7.
NeuroD6 and members of the heat shock transcription factors (HSFs). (A) Among the heat shock transcription factors, NeuroD6 only induces the expression of HSF2. Interestingly, NGF treatment only stimulates the expression levels of the β isoform of HSF2, in both the control PC12 and PC12-ND6 cells. Endogenous levels of HSF4 are repressed upon NeuroD6 overexpression. Serum deprivation results in a weak transient increase of HSF4 levels after 6 days, while NGF exposure triggers a modest but constant increase of HSF4 expression levels in both control PC12 and PC12-ND6 cells. (B) HSF2 protein is detected in growing neurites, branching points, and growth cones of PC12-ND6 cells. Control PC12 and PC12-ND6 cells were grown and processed for confocal microscopy, as described in Materials and Methods. Cells were stained with β-III tubulin to delineate their overall morphology. Scale bars are indicated at the bottom right corner of merge images. The immunocytochemistry results are in agreement with the immunoblot results shown in panel A. Growth cone (gc) and neighboring branching point (bp) in PC12-ND6 cells are indicated by arrowheads. (C) NGF treatment of PC12-ND6 cells results in increased expression of HSF2 in the nucleus, while sustaining HSF2 expression in developing neurites and growth cones. The top row shows PC12 cells exposed to NGF for 48 hours, while the middle row shows NGF-treated PC12-ND6 cells for 48 hours. The bottom row represents a magnification of the soma of NGF-treated PC12-ND6 cells to illustrate increased nuclear HSF2 expression, as indicated by an arrowhead. Scale bars are indicated at the bottom right corner of merge images.
On the other hand, PC12-ND6 cells expressed negligible levels of HSF-4 protein (Fig. 7A), which stands in contrast with the 1.73-fold increase in mRNA levels in PC12-ND6 cells (Table S2). The lack of concordance between mRNA and protein levels might be explained by the fact that: 1) the Affymetrix 230 A genechip® contains hsf-4 oligonucleotide probes designed from ETS sequences, therefore matching the 3′end of the predicted Hsf-4 mRNA; and 2) the existence of hsf-4 splice variants, the α and β isoforms (Nakai et al., 1997; Tanabe et al., 1999. Finally, HSF-4 protein levels were only marginally regulated upon seven days of NGF treatment in both control PC12 and PC12-ND6 cells, while serum deprivation of PC12-ND6 cells resulted in transient increase in HSF-4 expression levels on day 6 (Fig. 7A). Thus, among the HSF members, NeuroD6 only affects HSF2 expression levels regardless of serum-growth conditions or NGF exposure.
HSF-2 protein accumulates in branching points and growth cones of developing neurites in PC12-ND6 cells
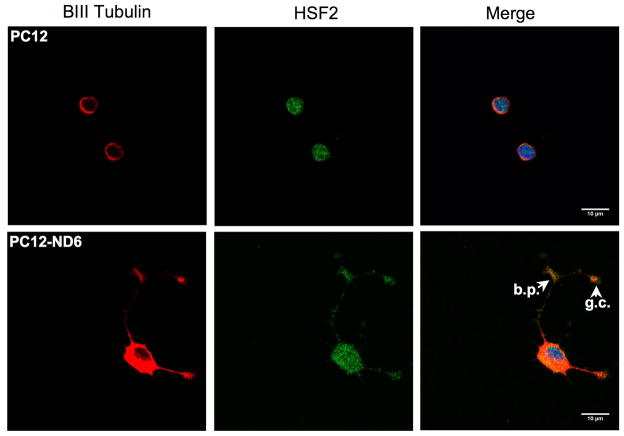
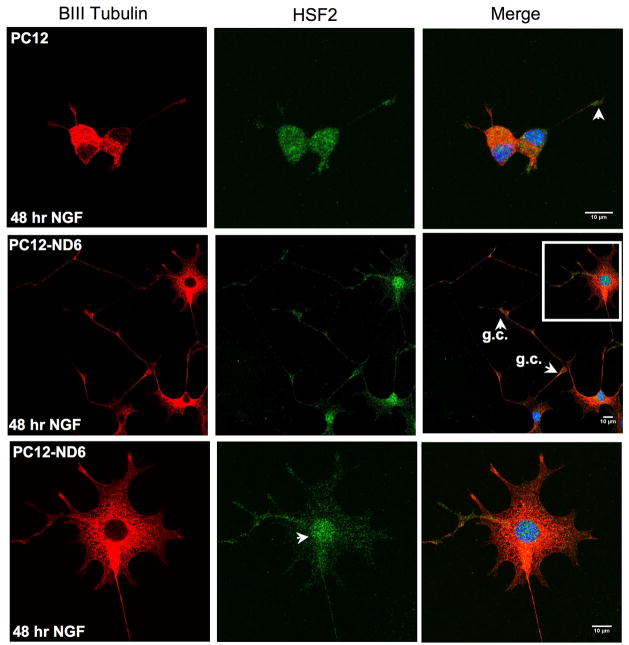
Since HSF-2 shuttles from the cytoplasm to nucleus to regulate gene expression, we assessed HSF-2 subcellular localization upon NeuroD6-or NGF-mediated differentiation.
Figure 7B shows that HSF-2 protein is surprisingly abundant in the developing neurites of PC12-ND6 cells with substantial accumulation in the branching point and growth cones (see arrowheads). HSF-2 protein was also detected in the nucleus, albeit at lower levels than in the cytoplasm (Fig. 7B), which is critical for HSF2 transcriptional activity.
We found that PC12 cells treated for two days with NGF displayed an HSF2 subcellular distribution similar to untreated PC12-ND6 cells, with the majority of HSF-2 staining in the cytoplasm, neurites and neuritic tip, accompanied by faint nuclear staining (Fig. 7C). NGF-treated PC12-ND6 cells, which displayed an extensive neurite network due to increased NGF responsiveness upon constitutive expression of NeuroD6 (Uittenbogaard and Chiaramello, 2002), exhibited HSF-2 staining in well developed neurites with accumulation of HSF-2 protein in growth cones and branching points (Fig. 7C, arrowheads). Intense HSF-2 nuclear localization tightly correlated with advanced neuronal differentiation.
Collectively, the immunocytochemistry results revealed an unexpected HSF2 staining pattern in the branching points and growth cones, suggestive of a potential emerging role in growth cone behavior and branching pattern, in addition to its well-established transcription activities.
DISCUSSION
The central issue of this study is to decipher the NeuroD6 transcriptional regulatory network linking neuronal differentiation to survival, as this molecular “bridge” is critical for proper neurogenesis and maintenance of the differentiated state. We took advantage of the long-term survival properties of PC12-ND6 cells to investigate how NeuroD6 promotes tolerance during prolonged stress mediated by serum deprivation, which may emulate chronic stress. We supplemented our analysis by comparing NeuroD6 effect with the NGF signaling cascade, since NeuroD6 is a critical effector of the NGF pathway (Uittenbogaard and Chiaramello, 2002).
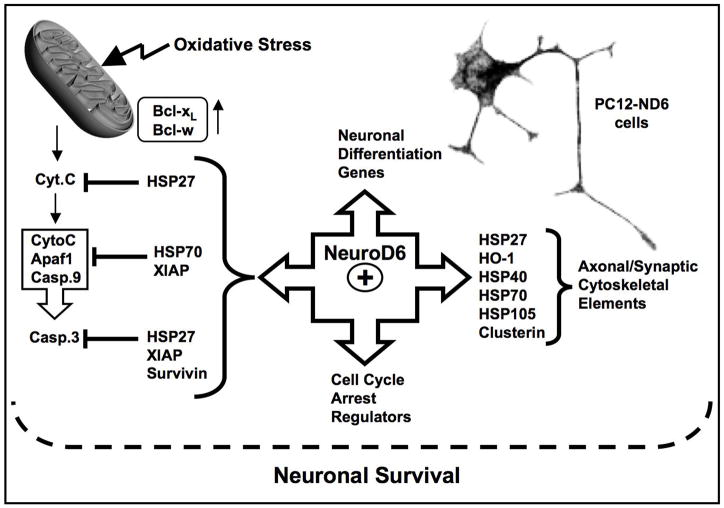
Our study provides the first demonstration that NeuroD6 regulates a molecular chaperone network in the absence of stress, with members displaying overlapping and distinct expression patterns throughout the different phases of stress tolerance. NeuroD6 induces expression of a set of heat shock proteins, including HSP27 (hspb1), the HSP70 chaperone system, Clusterin (clu), and heme-oxygenase-1 (hmox-1), known to either interface with cytoskeletal elements, the anti-apoptotic pathway, and/or to play a critical role in protein quality control (Fig. 8). The most striking finding is that the NeuroD6 effect on HSPs is not limited to their increased expression levels, but also to their precise spatial organization in PC12-ND6 cells vis-à-vis specific cytoskeletal elements involved in neuritogenesis (Liang and MacRae, 1997; Mounier and Arrigo, 2002; Dou et al., 2003).
Fig. 8.
Proposed model of the underlying mechanism by which NeuroD6 may bridge neuronal differentiation to survival. This model integrates the current results with our previously published studies, demonstrating that NeuroD6 upregulates the expression of G1 phase-promoting cell cycle regulators and several cytoskeletal elements in the absence of NGF (Uittenbogaard and Chiaramello, 2002; 2004), as well as maintain the expression levels throughout serum deprivation (Uittenbogaard and Chiaramello, 2005). In addition, NeuroD6 prevents caspase-3 activation upon serum deprivation by triggering expression of the anti-apoptotic Bcl2 members, Bcl-xL and Bcl-w, in conjunction with members of the Inhibitor of Apoptosis (IAP) family, such as XIAP and surviving (Uittenbogaard and Chiaramello, 2005). The NeuroD6-mediated pathways are illustrated by arrows with NeuroD6 located at the center of this nexus. The positive correlation between NeuroD6 and the heat shock network is shown as an integral component of the NeuroD6-mediated nexus, leading to the notion that NeuroD6 may bridge neuronal differentiation to survival via the expression of specific heat shock proteins, known to influence cytoskeletal dynamics and neuronal survival through the post-mitochondrial pathway depicted in the figure.
The differentiation properties of PC12-ND6 cells associated with increased HSP27 levels, colocalization with actin, and enrichment in branching points and growth cones are all consistent with the role of HSP27 in neuritogenesis (Williams et al., 2005; 2006) and neural regeneration (Costigan et al., 1998). Overexpression of HSP27 in cultured adult dorsal root ganglion (DRG) neurons stimulates neurite initiation resulting in increased neurite outgrowth accompanied by a complex branching pattern, while HSP27 silencing greatly diminishes neurite extension and branching pattern. HSP27 role in neuritogenesis is not limited to the DRG neuronal paradigm, as HSP27 expression is increased in NGF-treated PC12. Interestingly, constitutive expression of NeuroD6 alters the timing of maximal HSP27 expression, which occurs at day three of NGF exposure concomitantly with neurite branching pattern formation (Uittenbogaard and Chiaramello, 2002). This extensive branching architecture is specific to NGF-treated PC12-ND6 cells, as NGF-differentiated PC12 cells mainly extend thin poorly branched neurites. Thus, NeuroD6-mediated HSP27 expression essentially parallels major rearrangement and increase in neuronal cytoskeleton.
Furthermore, it bears mentioning that increased HSP27 expression levels concord with increased expression of ATF3 and c-Jun in PC12-ND6 cells by 10-and nearly 2 folds, respectively (Table S2). PC12 cells infected with adenoviral vectors expressing ATF3 and c-Jun express HSP27 protein as a result of DNA-binding of the ATF3-c-Jun dimer and activation of the HSP27 promoter (Nakagomi et al., 2003), suggestive of a potential NeuroD6 regulation of the hsp27 gene via the ATF3-cJun pathway.
Several studies have demonstrated that HSP27 possesses survival properties in various neuronal paradigms (Costigan et al., 1998; Lewis et al., 1999; Benn et al., 2002; Nakagomi et al., 2003; Stetler et al., 2008). HSP27 promotes survival of injured sensory and motor neurons by binding to cytochrome c translocated from mitochondria to the cytosol, thereby preventing apoptosome formation and caspase-3 activation (Benn et al., 2002). In PC12 cells, this translocation event occurs within 24 hours of serum deprivation (Stefanis et al., 1996), during which naïve PC12 cells show negligible HSP27 levels, while PC12-ND6 cells display robust HSP27 levels without caspase-3 activation (Uittenbogaard and Chiaramello, 2005). Collectively, our results concur with the notion that HSP27 may be more relevant during the early rather than the late stages of long-term survival.
NeuroD6-mediated tolerance to prolonged stress may instead be acquired through HSP70 (hspa11b) expression, which contrary to HSP27 (hspb1) is maintained at steady levels up to 15 days of serum deprivation. Correlation between stress tolerance and HSP70 induction has been observed upon a variety of stressors, such as cerebral ischemia (Giffard et al., 2004; Brown, 2007), kainic acid-induced stress (Yenari et al., 1998), polyglutamine repeat-induced toxicity (Chai et al., 1999; Kobayashi et al., 2000; Muchowski and Walker, 2005), and beta-amyloid-mediated toxicity (Magrané et al., 2004; Smith et al., 2005). HSP70 may provide stress tolerance to PC12-ND6 cells by four distinct but complementary mechanisms of action, involving preservation of cytoskeletal integrity, synaptic protection, protein quality control, and translocation into mitochondria.
Here, we report the first evidence of HSP70 immunoreactivity in growth cones of developing neurites, suggesting that HSP70 may assist the folding of nascent polypeptides during local protein synthesis upon NeuroD6-mediated neuronal differentiation. This observation is in keeping with the reported local translation of HSP70 in regenerating axons and growth cones of injury-conditioned DRG neurons (Willis et al., 2005). In fact, such local translation is not limited to a single HSP, but rather to a group of HSPs, including αβ crystalline (hspb8), HSP27 and HSP60 proteins, with HSP70 levels being enriched in regenerating axons compared with other axonally synthesized proteins (Willis et al., 2005). Even though compelling evidence has demonstrated the importance of local protein translation in axonal growth cones in the context of neural development and regeneration (Koenig and Giuditta, 1999; Alvarez et al., 2000; Campenot and Eng, 2000; Zhang and Poo, 2002; Verma et al., 2005; Leung et al., 2006; Hengst and Jaffrey, 2007), the question of whether local protein translation is required for growth cone responses to guidance cues remains unresolved due to conflicting findings from different neuronal populations (Piper et al., 2006; Yao et al., 2006; Lin and Holt, 2007; Bouchard et al., 2008; Lang et al., 2008; Roche et al., 2009). In addition, our observation of sustained HSP70 and HSP27 expression in growth cones of serum-deprived PC12-ND6 cells suggests that they may promote neuronal survival by locally attenuating apoptotic signals upon stress and thereby protecting synapses, a notion consistent with the observed enhanced protection of pre-synaptic activity following heat stress in transgenic Hsp70 flies (Karunanithi et al., 2002). This argues for a potential cytoprotective role of HSP local translation beyond the reported role in axonal guidance, synaptic plasticity, and formation of long-term memory (reviewed by Twiss and van Minnen, 2006; Lin and Holt, 2007;van Horck and Holt, 2008). This is in keeping with the known neuro-protective effects of HSP27 or HSP70 overexpression to prevent excitotoxic neuronal cell death (Akbar et al., 2003; Kalwy et al., 2003).
HSP70 along with the co-chaperone HSP40 (Dnajb1), which is referred to as HSP70 chaperone system, may regulate protein quality control. While DnaJb1 protein regulates HSP70 ATPase activity essential for stable substrate interactions to prevent protein aggregation and misfolding (Qiu et al., 2006), HSP105 (hsph1) disaggregates aggregated proteins (Liberek et al., 2008). Moreover, HSP105 has been shown to confer neuroprotection in stably transfected PC12 cells upon stress stimulus, such as serum deprivation, heat shock, and JNK-induced apoptosis (Hatayama et al., 2001). In the case of staurosporine-treated HeLa cells, HSP105 overexpression prevents cell death by blocking Bax translocation to mitochondria and therefore cytochrome c release (Yamagishi et al., 2006).
Finally, HSP70 may in part confer stress tolerance of PC12-ND6 cells by translocating from the cytosol to mitochondria as assessed by confocal microscopy. In keeping with this notion, is the previously demonstrated correlation between HSP70 expression and heat shock-induced mitochondrial membrane depolarization in premonocytic U937 cells (Polla et al., 1996), which is consistent with HSP70 protein in the intermembrane space and matrix of mitochondria isolated from non-neuronal cells (Bruey et al., 2000). Thus, it is tempting to speculate that HSP70 may prevent mitochondrial dysfunction by maintaining proper folding of mitochondrial located proteins in conjunction with another member of the hsp70 family, the mitochondrial-specific GRP75, also simultaneously upregulated upon NeuroD6 expression. Moreover, HSP70 translocation to mitochondria may consequently preserve the functional and physical communication between endoplasmic reticulum (ER) and mitochondria, through the sustained expression of the endoplasmic reticulum chaperone GRP78. Furthermore, the dynamic ER-mitochondria interface, which is in part regulated by the chaperone machinery, plays a crucial role in cellular survival (Szabadkai et al., 2006; Hayashi and Su, 2007). Altogether, it suggests that HSP70 translocation to mitochondria may provide an additional mechanism to promote neuronal survival, and therefore further studies are warranted to establish its specific role in mitochondrial protection using a combination of biochemical and confocal microscopy approaches.
Finally, clusterin (clu), a chaperone molecule structurally related to small HSPs, may play a role in NeuroD6-mediated survival. Although its role as a survival-enhancing chaperone remains elusive, increased susceptibility to axotomy-induced cell death observed in clusterin null mice supports the notion of clusterin behaving as a modulator of cell death (Wicher and Aldskogius, 2005; Ohlsson and Havton, 2006). The fact that clusterin promotes neuritogenesis (Kang et al., 2005) makes it an attractive candidate in the NeuroD6-mediated HSP cascade to link neuronal differentiation to survival.
The positive correlation between NeuroD6 and HSPs combined with the presence of E-boxes in the proximal promoters of hsp27, Hmox-1, hspa1b, and clusterin bears the question of the transcriptional contribution of NeuroD6 in their regulation. It has been well established that HSFs are the main transcriptional regulators of the hsp genes, with HSF2 being the most relevant to brain development (Loones et al., 1997; Rallu et al., 1997; Brown and Rush, 1999; Kallio et al., 2002; Wang et al., 2003; Chang et al., 2006). Only recently, the role of HSF-2 upon stress has been resolved using non-neuronal cells. HSF-2 dimerizes with HSF-1 to modulate the activity of hsp promoters, such as hsp25, hsp40, hsp70, and hsp110 (Östling et al., 2007). This dimerization event is also relevant in the context of cellular differentiation. Clusterin is another hsp gene regulated by the HSF1-HSF2 complex in response to accumulation of aberrant proteins (Loison et al., 2006). NeuroD6 may participate to the transcriptional regulation of clusterin via E-boxes present in the proximal promoter (Gutacker et al., 1999). Similarly, NeuroD6 may contribute to the well-documented transcriptional regulation of the Hmox-1 gene involving multiple transcription factors belonging to distinct families, upon a variety of stress stimuli (Alam and Cook, 2007), as overexpression of NeuroD6 triggers a substantial increase in HMOX-1 expression in the absence of stress, possibly by binding to E-boxes located in the Hmox-1 promoter. Future studies will be required to address the question of whether NeuroD6 directly or indirectly stimulates the expression of these hsp genes.
Collectively, our findings support the concept that NeuroD6 may sustain a critical expression level of a set of HSPs to confer long-term stress tolerance to mature neurons. In conclusion, our study provides additional and compelling evidence that NeuroD6 harnesses an active survival pathway, which is an integral component of its differentiation-mediated network. Thus, NeuroD6 may constitute a new target to design therapeutic avenues for treatment of neurodegenerative diseases associated with cognitive deficits.
Supplementary Material
Acknowledgments
Contract grant sponsor: NIH; Contract grant number: R01-NS041391 grant to AC and P30HD40677.
We want to thank Dr. McCaffrey for his help with the microarray study. This work was supported in part by NIH grant R01-NS041391 to A.C. and by P30HD40677. Additional support was provided by the Catherine Birch McCormick Center and The St. Laurent Institute.
Footnotes
This work will be part of a dissertation presented to the graduate program of Molecular Medicine, The George Washington University Institute for Biomedical Sciences in partial fulfillment of the requirements for the Ph.D. degree.
References
- Akbar MT, Lundberg AM, Liu K, Vidyadaran S, Wells KE, Dolatshad H, Wynn S, Wells DJ, Latchman DS, de Belleroche J. The neuroprotective effects of heat shock protein 27 overexpression in transgenic animals against kainate-induced seizures and hippocampal cell death. J Biol Chem. 2003;278:19956–65. doi: 10.1074/jbc.M207073200. [DOI] [PubMed] [Google Scholar]
- Åkerfelt M, Trouillet D, Mezger V, Sistonen L. Heat shock factors at a crossroad between stress and development. Annals of the New York Academy of Sciences. 2007;1113:15–27. doi: 10.1196/annals.1391.005. [DOI] [PubMed] [Google Scholar]
- Alam J, Cook JL. How many transcription factors does it take to turn on the heme oxygenase-1 gene? Am J Respir Cell Mol Biol. 2007;36:166–174. doi: 10.1165/rcmb.2006-0340TR. [DOI] [PubMed] [Google Scholar]
- Alvarez J, Giuditta A, Koenig E. Protein synthesis in axons and terminals: significance for maintenance, plasticity and regulation of phenotype. With a critique of slow transport theory. Prog Neurobiol. 2000;62:1–62. doi: 10.1016/s0301-0082(99)00062-3. [DOI] [PubMed] [Google Scholar]
- Angelastro JM, Klimaschewski L, Tang S, Vitolo OV, Weissman TA, Donlin LT, Shelanski ML, Greene LA. Identification of diverse nerve growth factor-regulated genes by serial analysis of gene expression (SAGE) profiling. Proc Natl Acad Sci U S A. 2000;97:10424–9. doi: 10.1073/pnas.97.19.10424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelastro JM, Töröcsik B, Greene LA. Nerve growth factor selectively regulates expression of transcripts encoding ribosomal proteins. BMC Neurosci. 2002;3:3. doi: 10.1186/1471-2202-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholomae A, Nave KA. NEX-1: a novel brain-specific helix-loop-helix protein with autoregulation and sustained expression in mature cortical neurons. Mech Dev. 1994;48:217–228. doi: 10.1016/0925-4773(94)90061-2. [DOI] [PubMed] [Google Scholar]
- Bechtold DA, Rush SJ, Brown IR. Localization of the heat-shock protein Hsp70 to the synapse following hyperthermic stress in the brain. J Neurochem. 2000;74:641–6. doi: 10.1046/j.1471-4159.2000.740641.x. [DOI] [PubMed] [Google Scholar]
- Benn SC, Perrelet D, Kato AC, Scholz J, Decosterd I, Mannion RJ, Bakowska JC, Woolf CJ. Hsp27 upregulation and phosphorylation is required for injured sensory and motor neuron survival. Neuron. 2002;36:45–56. doi: 10.1016/s0896-6273(02)00941-8. [DOI] [PubMed] [Google Scholar]
- Benn SC, Woolf CJ. Adult neuron survival strategies- slamming on the brakes. Nature Rev Neurosci. 2004;5:686–700. doi: 10.1038/nrn1477. [DOI] [PubMed] [Google Scholar]
- Bergsland M, Werme M, Malewicz M, Perlmann T, Muhr J. The establishment of neuronal properties is controlled by Sox4 and Sox11. Genes and Development. 2006;20:3475–3486. doi: 10.1101/gad.403406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björk JK, Sistonen L. Clustering of heat-shock factors. Biochem J. 2006;395:e5–e6. doi: 10.1042/BJ20060071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown IR, Rush SJ. Cellular localization of the heat shock transcription factors HSF1 and HSF2 in the rat brain during postnatal development and following hyperthermia. Brain Res. 1999;821:333–40. doi: 10.1016/s0006-8993(99)01087-2. [DOI] [PubMed] [Google Scholar]
- Bouchard JF, Horn KE, Stroh T, Kennedy TE. Depolarization recruits DCC to the plasma membrane of embryonic cortical neurons and enhances axon extension in response to netrin-1. J Neurochem. 2008;107:398–417. doi: 10.1111/j.1471-4159.2008.05609.x. [DOI] [PubMed] [Google Scholar]
- Brown IR. Heat shock proteins and protection of the nervous system. Ann N Y Acad Sci. 2007;1113:147–58. doi: 10.1196/annals.1391.032. [DOI] [PubMed] [Google Scholar]
- Bruey J, Ducasse C, Bonniaud P, Ravagnan L, Susin SA, Diaz-Latoud C, Gurbuxani S, Arrigo A, Kroemer G, Solary E, Garrido C. Hsp27 negatively regulates cell death by interacting with cytochrome c. Nature Cell Biology. 2000;2:645–652. doi: 10.1038/35023595. [DOI] [PubMed] [Google Scholar]
- Cai D, Deng K, Mellado W, Lee J, Ratan RR, Filbin MT. Arginase I and polyamines act downstream from cyclic AMP in overcoming inhibition of axonal growth MAG and myelin in vitro. Neuron. 2002;35:711–716. doi: 10.1016/s0896-6273(02)00826-7. [DOI] [PubMed] [Google Scholar]
- Campenot RB, Eng H. Protein synthesis in axons and its possible functions. J Neurocytol. 2000;29(11–12):793–798. doi: 10.1023/a:1010939307434. [DOI] [PubMed] [Google Scholar]
- Carver JA, Rekas A, Thorn DC, Wilson MR. Small heat-shock proteins and clusterin: intra- and extracellular molecular chaperones with a common mechanism of action and function? Life. 200355:661–8. doi: 10.1080/15216540310001640498. Review. [DOI] [PubMed] [Google Scholar]
- Chai Y, Koppenhafer SL, Bonini NM, Paulson HL. Analysis of the role of heat shock protein (Hsp) molecular chaperones in polyglutamine disease. J Neurosci. 1999;19:10338–47. doi: 10.1523/JNEUROSCI.19-23-10338.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y, Östling P, Åkerfelt M, Trouillet D, Rallu M, Gitton Y, El Fatimy R, Fardeau V, Le Crom S, Morange M, Sistonen L, Mezger V. Role of heat-shock factor 2 in cerebral cortex formation and as a regulator of p35 expression. Genes and Development. 2006;20:836–847. doi: 10.1101/gad.366906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costigan M, Mannion RJ, Kendall G, Lewis SE, Campagna JA, Coggeshall RE, Meridith-Middleton J, Tate S, Woolf CJ. Heat shock protein 27: developmental regulation and expression after peripheral nerve injury. J Neurosci. 1998;18:5891–900. doi: 10.1523/JNEUROSCI.18-15-05891.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugaard M, Rohde M, Jäättelä M. The heat shock protein 70 family: Highly homologous proteins with overlapping and distinct functions. FEBS Letters. 2007;581:3702–3710. doi: 10.1016/j.febslet.2007.05.039. [DOI] [PubMed] [Google Scholar]
- DeMattos RB, O’dell MA, Parsadanian M, Taylor JW, Harmony JA, Bales KR, Paul SM, Aronow BJ, Holtzman DM. Clusterin promotes amyloid plaque formation and is critical for neuritic toxicity in a mouse model of Alzheimer’s disease. Proc Natl Acad Sci U S A. 2002;99:10843–8. doi: 10.1073/pnas.162228299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkmans TF, van Hooijdonk LW, Schouten TG, Kamphorst JT, Vellinga AC, Meerman JH, Fitzsimons CP, de Kloet ER, Vreugdenhil E. Temporal and functional dynamics of the transcriptome during nerve growth factor-induced differentiation. J Neurochem. 2008;105:2388–2403. doi: 10.1111/j.1471-4159.2008.05338.x. [DOI] [PubMed] [Google Scholar]
- Dotti CG, Sullivan CA, Banker GA. The establishment of polarity by hippocampal neurons in culture. J Neurosci. 1988;8:1454–1468. doi: 10.1523/JNEUROSCI.08-04-01454.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou F, Netzer WJ, Tanemura K, Li F, Hartl FU, Takashima A, Gouras GK, Greengard P, Xu H. Chaperones increase association of tau protein with microtubules. Proc Natl Acad Sci USA. 2003;100:721–726. doi: 10.1073/pnas.242720499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farinelli SE, Greene LA. Cell cycle blockers mimosine, ciclopirox, and deferoxamine prevent the death of PC12 cells and postmitotic sympathetic neurons after removal of trophic support. J Neurosci. 2006;16:1150–1162. doi: 10.1523/JNEUROSCI.16-03-01150.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald U, Gorman AM, Samali A. Heat chock proteins and the regulation of apoptosis. In: Richter-landsberg C, editor. Heat Shock Proteins in Neural cells. Neuroscience Intelligent Unit. Chapter 5. Springer Landes Bioscience; 2007. pp. 53–62. [Google Scholar]
- Fujitani M, Yamagishi S, Yong HC, Hata K, Kubo T, Ino H, Tohyama M, Yamashita T. P311 accelerates nerve regeneration of the axotomized facial nerve. J Neurochem. 2004;91:737–744. doi: 10.1111/j.1471-4159.2004.02738.x. [DOI] [PubMed] [Google Scholar]
- Giffard RG, Xu L, Zhao H, Carrico W, Ouyang Y, Qiao Y, Sapolsky R, Steinberg G, Hu B, Yenari MA. Chaperones, protein aggregation, and brain protection from hypoxic/ischemic injury. J Exp Biol. 2004;207:3213–20. doi: 10.1242/jeb.01034. [DOI] [PubMed] [Google Scholar]
- Goodson ML, Park-Sarge O, Sarge KD. Tissue-dependent expression of heat shock factor 2 isoforms with distinct transcriptional activities. Molecular and Cellular Biology. 1995;15:5288–5293. doi: 10.1128/mcb.15.10.5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene LA. Nerve growth factor prevents the death and stimulates the neuronal differentiation of clonal PC12 pheochromocytoma cells in serum-free medium. J Cell Biol. 1978;78:747–755. doi: 10.1083/jcb.78.3.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutacker C, Klock G, Diel P, Koch-Brandt C. Nerve growth factor and epidermal growth factor stimulate clusterin gene expression in PC12 cells. Biochem J. 1999;339:759–766. [PMC free article] [PubMed] [Google Scholar]
- Hatayama T, Yamagishi N, Minobe E, Sakai K. Role of hsp105 in protection against stress-induced apoptosis in neuronal PC12 cells. Biochem Biophys Res Commun. 2001;288:528–34. doi: 10.1006/bbrc.2001.5802. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Su TP. Sigma-1 receptor chaperones at the ER-mitochondrion interface regulate Ca(2+) signaling and cell survival. Cell. 2007;131:596–610. doi: 10.1016/j.cell.2007.08.036. [DOI] [PubMed] [Google Scholar]
- Hengst U, Jaffrey SR. Function and translational regulation of mRNA in developing axons. Semin Cell Dev Biol. 2007;18:209–215. doi: 10.1016/j.semcdb.2007.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert KR, Smali A, Gorman A. The role of Hsps in neuronal differentiation and development. In: Richter-landsberg C, editor. Heat Shock Proteins in Neural cells. Neuroscience Intelligent Unit. Chapter 3. Springer Landes Bioscience; 2007. pp. 25–37. [Google Scholar]
- Hu Y, Mivechi NF. Association and regulation of heat shock transcription factor 4b with both extracellular signal-regulated kinase mitogen-activated protein kinase and dual-specificity tyrosine phosphatase DUSP26. Molecular and Cellular Biology. 2006;26:3282–3294. doi: 10.1128/MCB.26.8.3282-3294.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallio M, Chang Y, Manuel M, Alastalo TP, Rallu M, Gitton Y, Pirkkala L, Loones MT, Paslaru L, Larney S, Hiard S, Morange M, Sistonen L, Mezger V. Brain abnormalities, defective meiotic chromosome synapsis and female subfertility in HSF2 null mice. EMBO J. 2002;21:2591–601. doi: 10.1093/emboj/21.11.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalwy SA, Akbar MT, Coffin RS, de Belleroche J, Latchman DS. Heat shock protein 27 delivered via a herpes simplex virus vector can protect neurons of the hippocampus against kainic-acid-induced cell loss. Brain Res Mol Brain Res. 2003;111:91–103. doi: 10.1016/s0169-328x(02)00692-7. [DOI] [PubMed] [Google Scholar]
- Kang SW, Shin YJ, Shim YJ, Jeong SY, Park IS, Min BH. Clusterin interacts with SCLIP (SCG10-like protein) and promotes neurite outgrowth of PC12 cells. Exp Cell Res. 2005;309:305–15. doi: 10.1016/j.yexcr.2005.06.012. [DOI] [PubMed] [Google Scholar]
- Karunanithi S, Barclay JW, Brown IR, Robertson RM, Atwood HL. Enhancement of presynaptic performance in transgenic Drosophila over-expressing heat shock protein Hsp70. Synapse. 2002;44:8–14. doi: 10.1002/syn.10048. [DOI] [PubMed] [Google Scholar]
- Kim W-Y, Fritzsch B, Seris A, Bakel LA, Huang EJ, Reichardt LF, Barth DS, Lee JE. NeuroD-null mice are deaf due to a severe loss of the inner ear sensory neurons during development. Development. 2001;128:417–426. doi: 10.1242/dev.128.3.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby BA, Merril CR, Ghanbari H, Wallace WC. Heat shock proteins protect against stress-related phosphorylation of tau in neuronal PC12 cells that have acquired thermotolerance. J Neurosci. 1994;14:5687–93. doi: 10.1523/JNEUROSCI.14-09-05687.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Kume A, Li M, Doyu M, Hata M, Ohtsuka K, Sobue G. Chaperones Hsp70 and Hsp40 suppress aggregate formation and apoptosis in cultured neuronal cells expressing truncated androgen receptor protein with expanded polyglutamine tract. J Biol Chem. 2000;275:8772–8728. doi: 10.1074/jbc.275.12.8772. [DOI] [PubMed] [Google Scholar]
- Koenig E, Giuditta A. Protein-synthesizing machinery in the axon compartment. Neuroscience. 1999;89:5–15. doi: 10.1016/s0306-4522(98)00282-6. [DOI] [PubMed] [Google Scholar]
- Lang S, von Philipsborn AC, Bernard A, Bonhoeffer F, Bastmeyer M. Growth cone response to ephrin gradients produced by microfluidic networks. Anal Bioanal Chem. 2008;390:809–816. doi: 10.1007/s00216-007-1363-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis SE, Mannion RJ, White FA, Coggeshall RE, Beggs S, Costigan M, Martin JL, Dillmann WH, Woolf CJ. A role for HSP27 in sensory neuron survival. J Neurosci. 1999;19:8945–8953. doi: 10.1523/JNEUROSCI.19-20-08945.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Strittmatter SM. Delayed systemic Nogo-66 receptor antagonist promotes recovery from spinal cord injury. J Neurosci. 2003;23:4219–4227. doi: 10.1523/JNEUROSCI.23-10-04219.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang P, MacRae TH. Molecular chaperones and the cytoskeleton. Journal of Cell Science. 1997;110:1431–1440. doi: 10.1242/jcs.110.13.1431. [DOI] [PubMed] [Google Scholar]
- Liberek K, Lewandowska A, Zietkiewicz S. Chaperones in control of protein disaggregation. EMBO J. 2008;27:328–335. doi: 10.1038/sj.emboj.7601970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin AC, Holt CE. Local translation and directional steering in axons. EMBO J. 2007;26:3729–3736. doi: 10.1038/sj.emboj.7601808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Nowak R, Chao W, Bloch KD. Nerve growth factor induces anti-apoptotic heme oxygenase-1 in rat pheochromocytoma PC12 cells. J Neurochem. 2003;86:1553–1563. doi: 10.1046/j.1471-4159.2003.01978.x. [DOI] [PubMed] [Google Scholar]
- Liu Y, Encinas M, Comella JX, Aldea M, Gallego C. Basic Helix-Loop-Helix proteins bind to TrkB and p21Cip1 promoters linking differentiation and cell cycle arrest in neuroblastoma cells. Mol Cell Biol. 2004;24:2662–2672. doi: 10.1128/MCB.24.7.2662-2672.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loison F, Debure L, Nizard P, Le Goff P, Michel D, Le Dréan Y. Up-regulation of the clusterin gene after proteotoxic stress: Implication of HSF1-HSF2 heterocomplexes. Biochem J. 2006;395:223–231. doi: 10.1042/BJ20051190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loones MT, Rallu M, Mezger V, Morange M. HSP gene expression and HSF2 in mouse development. Cell Mol Life Sci. 1997;53:179–90. doi: 10.1007/PL00000590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung KM, van Horck FP, Lin AC, Allison R, Standart N, Holt CE. Asymmetrical beta-actin mRNA translation in growth cones mediates attractive turning to netrin-1. Nat Neurosci. 2006;9:1247–1256. doi: 10.1038/nn1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magrané J, Smith RC, Walsh K, Querfurth HW. Heat shock protein 70 participates in the neuroprotective response to intracellularly expressed beta-amyloid in neurons. J Neurosci. 2004;24:1700–6. doi: 10.1523/JNEUROSCI.4330-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ménard C, Hein P, Paquin A, Savelson A, Yang XM, Lederfein D, Barnabé-Heider F, Mir AA, Sterneck E, Peterson AC, Johnson PF, Vinson C, Miller FD. An essential role for a MEK-C/EBP pathway during growth factor-regulated cortical neurogenesis. Neuron. 2002;36:597–610. doi: 10.1016/s0896-6273(02)01026-7. [DOI] [PubMed] [Google Scholar]
- Miyata T, Maeda T, Lee JE. NeuroD is required for differentiation of the granule cells in the cerebellum and hippocampus. Genes & Dev. 1999;13:1647–1652. doi: 10.1101/gad.13.13.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morange M. HSFs in development. Handbook of Experimental Pharmacology. 2006:153–169. doi: 10.1007/3-540-29717-0_7. [DOI] [PubMed] [Google Scholar]
- Morimoto RI. Regulation of the heat shock transcriptional response: Cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes and Development. 1998;12:3788–3796. doi: 10.1101/gad.12.24.3788. [DOI] [PubMed] [Google Scholar]
- Mounier N, Arrigo AP. Actin cytoskeleton and small heat shock proteins: how do they interact? Cell Stress Chaperones. 2002;7:167–176. doi: 10.1379/1466-1268(2002)007<0167:acashs>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchowski PJ, Wacker JL. Modulation of neurodegeneration by molecular chaperones. Nat Rev Neurosci. 2005;6:11–22. doi: 10.1038/nrn1587. [DOI] [PubMed] [Google Scholar]
- Nakagomi S, Suzuki Y, Namikawa K, Kiryu-Seo S, Kiyama H. Expression of the activating transcription factor 3 prevents c-Jun Nterminal kinase-induced neuronal death by promoting heat shock protein 27 expression and Akt activation. J Neurosci. 2003;23:5187–5196. doi: 10.1523/JNEUROSCI.23-12-05187.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai A, Tanabe M, Kawazoe Y, Inazawa J, Morimoto RI, Nagata K. HSF4, a new member of the human heat shock factor family which lacks properties of a transcriptional activator. Molecular and Cellular Biology. 1997;17:469–481. doi: 10.1128/mcb.17.1.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto M, Schuurmans C, Britz O, Guillemot F. Neural bHLH genes control the neuronal versus glial fate decision in cortical progenitors. Neuron. 2001;29:401–413. doi: 10.1016/s0896-6273(01)00214-8. [DOI] [PubMed] [Google Scholar]
- Ohlsson M, Havton LA. Complement activation after lumbosacral ventral root avulsion injury. Neurosci Lett. 2006;394:179–83. doi: 10.1016/j.neulet.2005.10.037. [DOI] [PubMed] [Google Scholar]
- Olson JM, Asakura A, Snider L, Hawkes R, Strand A, Stoeck J, Hallahan A, Pritchard J, Tapscott SJ. NeuroD2 is necessary for development and survival of central nervous system neurons. Dev Biol. 2001;234:174–187. doi: 10.1006/dbio.2001.0245. [DOI] [PubMed] [Google Scholar]
- Oppenheim RW. Cell death during development of the nervous system. Annu Rev. 1991;14:453–501. doi: 10.1146/annurev.ne.14.030191.002321. [DOI] [PubMed] [Google Scholar]
- Östling P, Björk JK, Roos-Mattjus P, Mezger V, Sistonen L. Heat Shock Factor 2 (HSF2) contributes to inducible expression of hsp genes through interplay with HSF1. Journal of Biological Chemistry. 2007;282:7077–7086. doi: 10.1074/jbc.M607556200. [DOI] [PubMed] [Google Scholar]
- Paquin A, Barnabé-Heider F, Kageyama R, Miller FD. CCAAT/enhancer-binding protein phosphorylation biases cortical precursors to generate neurons rather than astrocytes in vivo. J Neurosci. 2005;25:10747–10758. doi: 10.1523/JNEUROSCI.2662-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul C, Manero F, Gonin S, Kretz-Remy C, Virot S, Arrigo A. Hsp27 as a negative regulator of cytochrome c release. Molecular and Cellular Biology. 2002;22:816–834. doi: 10.1128/MCB.22.3.816-834.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennesi ME, Cho J-H, Yang Z, Wu SH, Zhang J, Wu SM, Tsai M-J. Beta/NeuroD1 null mice: a new model for transcription factor-dependent photoreceptor degeneration. J Neurosci. 2003;23:453–461. doi: 10.1523/JNEUROSCI.23-02-00453.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper M, Anderson R, Dwivedy A, Weinl C, van Horck F, Leung KM, Cogill E, Holt C. Signaling mechanisms underlying Slit2-induced collapse of Xenopus retinal growth cones. Neuron. 2006;49:215–228. doi: 10.1016/j.neuron.2005.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polla BS, Kantengwa S, François D, Salvioli S, Franceschi C, Marsac C, Cossarizza A. Mitochondria are selective targets for the protective effects of heat shock against oxidative injury. Proc Natl Acad Sci U S A. 1996;93:6458–6463. doi: 10.1073/pnas.93.13.6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirkkala L, Nykänen P, Sistonen L. Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB Journal. 2001;15:1118–1131. doi: 10.1096/fj00-0294rev. [DOI] [PubMed] [Google Scholar]
- Poon S, Treweek TM, Wilson MR, Easterbrook-Smith SB, Carver JA. Clusterin is an extracellular chaperone that specifically interacts with slowly aggregating proteins on their off-folding pathway. FEBS Lett. 2002;513:259–66. doi: 10.1016/s0014-5793(02)02326-8. [DOI] [PubMed] [Google Scholar]
- Qiu X, Shao Y, Miao S, Wang L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cellular and Molecular Life Sciences. 2006;63:2560–2570. doi: 10.1007/s00018-006-6192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rallu M, Loones M-T, Lallemand Y, Morimoto R, Morange M, Mezger V. Function and regulation of heat shock factor 2 during mouse embryogenesis. Proc Natl Acad Sci U S A. 1997;94:2392–2397. doi: 10.1073/pnas.94.6.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter-landsberg C. Heat Shock Proteins in Neural cells. Neuroscience Intelligent Unit. Chapter 1. Springer Landes Bioscience; 2007. pp. 1–12. [Google Scholar]
- Roche FK, Marsick BM, Letourneau PC. Protein synthesis in distal axons is not required for growth cone responses to guidance cues. J Neurosci. 2009;29:638–652. doi: 10.1523/JNEUROSCI.3845-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rössler OG, Bauer I, Chung HY, Thiel G. Glutamate-induced cell death of immortalized murine hippocampal neurons: neuroprotective activity of heme oxygenase-1, heat shock protein 70, and sodium selenite. Neurosci Lett. 2004;362:253–7. doi: 10.1016/j.neulet.2004.03.033. [DOI] [PubMed] [Google Scholar]
- Rukenstein A, Rydel RE, Greene LA. Multiple agents rescue PC12 cells from serum-free cell death by translation- and transcription-independent mechanisms. J Neurosci. 1991;11:2552–63. doi: 10.1523/JNEUROSCI.11-08-02552.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas M, Diaz R, Abraham NG, Ruiz de Galarreta CM, Cuadrado A. Nerve Growth Factor Protects against 6-Hydroxydopamine-induced Oxidative Stress by Increasing Expression of Heme Oxygenase-1 in a Phosphatidylinositol 3-Kinase-dependent Manner. J Biol Chem. 2000;278:13898–13904. doi: 10.1074/jbc.M209164200. [DOI] [PubMed] [Google Scholar]
- Satoh T, Sakai N, Enokido Y, Uchiyama Y, Hatanaka H. Survival factor-insensitive generation of reactive oxygen species induced by serum deprivation in neuronal cells. Brain Res. 1996;733:9–14. doi: 10.1016/0006-8993(96)00527-6. [DOI] [PubMed] [Google Scholar]
- Schwab MH, Bartholomae A, Heimrich B, Feldmeyer D, Druffel-Augustin S, Goebbels S, Naya FJ, Zhao S, Frotscher M, Tsai MJ, Nave KA. Neuronal basic helix-loop-helix proteins (NEX and BETA2/Neuro D) regulate terminal granule cell differentiation in the hippocampus. J Neurosci. 2000;20:3714–3724. doi: 10.1523/JNEUROSCI.20-10-03714.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab MH, Druffel-Augustin S, Gass P, Jung M, Klugmann M, Bartholomae A, Rossner MJ, Nave KA. Neuronal basic helix-loop-helix proteins (NEX, neuroD, NDRF): spatiotemporal expression and targeted disruption of the NEX gene in transgenic mice. J Neurosci. 1998;18:1408–1418. doi: 10.1523/JNEUROSCI.18-04-01408.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu C, Akazawa C, Nakanishi S, Kageyama R. MATH- 2,a mammalian helix-loop-helix factor structurally related to the product of Drosophila proneural gene atonal, is specifically expressed in the nervous system. Eur J Biochem. 1995;229:239–248. doi: 10.1111/j.1432-1033.1995.tb20461.x. [DOI] [PubMed] [Google Scholar]
- Smith RC, Rosen KM, Pola R, Magrané J. Stress proteins in Alzheimer’s disease. Int J Hyperthermia. 2005;21:421–31. doi: 10.1080/02656730500133165. [DOI] [PubMed] [Google Scholar]
- Stefanis L, Park S, Yan C-YI, Farinelli SE, Troy CM, Shelanski ML, Greene LA. Induction of CPP-32-like activity in PC12 cells by withdrawal of trophic factor. J Biol Chem. 1996;271:663–30. 671. doi: 10.1074/jbc.271.48.30663. [DOI] [PubMed] [Google Scholar]
- Stetler RA, Cao G, Gao Y, Zhang F, Wang S, Weng Z, Vosler P, Zhang L, Signore A, Graham SH, Chen J. Hsp27 protects against ischemic brain injury via attenuation of a novel stress-response cascade upstream of mitochondrial cell death signaling. J Neurosci. 2008;28:13038–13055. doi: 10.1523/JNEUROSCI.4407-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Nadal-Vicens M, Misono S, Lin MZ, Zubiaga A, Hua X, Fan G, Greenberg ME. Neurogenin promotes neurogenesis and inhibits glial differentiation by independent mechanisms. Cell. 2001;104:365–376. doi: 10.1016/s0092-8674(01)00224-0. [DOI] [PubMed] [Google Scholar]
- Szabadkai G, Bianchi K, Várnai P, De Stefani D, Wieckowski MR, Cavagna D, Nagy AI, Balla T, Rizzuto R. Chaperone-mediated coupling of endoplasmic reticulum and mitochondrial Ca2+ channels. J Cell Biol. 2006;175:901–911. doi: 10.1083/jcb.200608073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe M, Sasai N, Nagata K, Liu X, Liu PCC, Thiele DJ, Nakai A. The mammalian HSF4 gene generates both an activator and a repressor of heat shock genes by alternative splicing. Journal of Biological Chemistry. 1999;274:27845–27856. doi: 10.1074/jbc.274.39.27845. [DOI] [PubMed] [Google Scholar]
- Troy CM, Shelanski ML. Down-regulation of copper/zinc superoxide dismutase causes apoptotic death in PC12 neuronal cells. Proc Natl Acad Sci U S A. 1994;91:6384–6387. doi: 10.1073/pnas.91.14.6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu N, Hu Y, Mivechi NF. Heat shock transcription factor (Hsf)-4br recruits Brg1 during the G1 phase of the cell cycle and regulates the expression of heat shock proteins. Journal of Cellular Biochemistry. 2006;98:1528–1542. doi: 10.1002/jcb.20865. [DOI] [PubMed] [Google Scholar]
- Twiss JL, van Minnen J. New insights into neuronal regeneration: the role of axonal protein synthesis in pathfinding and axonal extension. J Neurotrauma. 2006;23:295–308. doi: 10.1089/neu.2006.23.295. [DOI] [PubMed] [Google Scholar]
- Uittenbogaard M, Chiaramello A. Constitutive overexpression of the basic helix-loop-helix Nex1/MATH-2 transcription factor promotes neuronal differentiation of PC12 cells and neurite regeneration. J Neurosci Res. 2002;67:235–245. doi: 10.1002/jnr.10119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uittenbogaard M, Chiaramello A. Expression profiling upon Nex1/MATH-2-mediated neuritogenesis in PC12 cells and its implication in regeneration. J Neurochem. 2004;91:1332–1343. doi: 10.1111/j.1471-4159.2004.02814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uittenbogaard M, Chiaramello A. The basic helix-loop-helix transcription factor Nex-1/Math-2 promotes neuronal survival of PC12 cells by modulating the dynamic expression of anti-apoptotic and cell cycle regulators. J Neurochem. 2005;92:585–596. doi: 10.1111/j.1471-4159.2004.02886.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uittenbogaard M, Martinka DL, Chiaramello A. The basic helix-loop-helix differentiation factor Nex1/MATH-2 functions as a key activator of the GAP-43 gene. J Neurochem. 2003;84:678–688. doi: 10.1046/j.1471-4159.2003.01572.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Horck FP, Holt CE. A cytoskeletal platform for local translation in axons. Sci Signal 2008. 2008 Feb 26;1(8):pe11. doi: 10.1126/stke.18pe11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma P, Chierzi S, Codd AM, Campbell DS, Meyer RL, Holt CE, Fawcett JW. Axonal protein synthesis and degradation are necessary for efficient growth cone regeneration. J Neurosci. 2005;25:331–342. doi: 10.1523/JNEUROSCI.3073-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetter M. A turn of the helix: preventing the glial fate. Neuron. 2001;29:559–562. doi: 10.1016/s0896-6273(01)00231-8. [DOI] [PubMed] [Google Scholar]
- Wallace W, Johnson G, Sugar J, Merril CR, Refolo LM. Reversible phosphorylation of tau to form A68 in heat-shocked neuronal PC12 cells. Brain Res Mol Brain Res. 1993;19:149–55. doi: 10.1016/0169-328x(93)90160-q. [DOI] [PubMed] [Google Scholar]
- Wang G, Zhang J, Moskophidis D, Mivechi NF. Targeted disruption of the heat shock transcription factor (hsf)-2 gene results in increased embryonic lethality, neuronal defects, and reduced spermatogenesis. Genesis. 2003;36:48–61. doi: 10.1002/gene.10200. [DOI] [PubMed] [Google Scholar]
- Wicher GK, Aldskogius H. Adult motor neurons show increased susceptibility to axotomy-induced death in mice lacking clusterin. Eur J Neurosci. 200;21:2024–8. doi: 10.1111/j.1460-9568.2005.04008.x. [DOI] [PubMed] [Google Scholar]
- Williams KL, Rahimtula M, Mearow KM. Heat Shock Protein 27 Is Involved in Neurite Extension and Branching of Dorsal Root Ganglion Neurons In Vitro. J Neurosc Res. 2006;84:716–723. doi: 10.1002/jnr.20983. [DOI] [PubMed] [Google Scholar]
- Williams KL, Rahimtula M, Mearow KM. Hsp27 and axonal growth in adult sensory neurons in vitro. BMC Neuroscience. 2005:6. doi: 10.1186/1471-2202-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis D, Li KW, Zheng JQ, Chang JH, Smit A, Kelly T, Merianda TT, Sylvester J, van Minnen J, Twiss JL. Differential transport and local translation of cytoskeletal, injury-response, and neurodegeneration protein mRNAs in axons. J Neurosci. 2005;25:778–791. doi: 10.1523/JNEUROSCI.4235-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SX, Goebbels S, Nakamura K, Nakamura K, Kometani K, Minato N, Kaneko T, Nave KA, Tamamaki N. Pyramidal neurons of upper cortical layers generated by NEX-positive progenitor cells in the subventricular zone. Proc Natl Acad Sci U S A. 2005;102:17172–7. doi: 10.1073/pnas.0508560102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi N, Ishihara K, Saito Y, Hatayama T. Hsp105 family proteins suppress staurosporine-induced apoptosis by inhibiting the translocation of Bax to mitochondria in HeLa cells. Exp Cell Res. 2006;312:3215–23. doi: 10.1016/j.yexcr.2006.06.007. [DOI] [PubMed] [Google Scholar]
- Yao J, Sasaki Y, Wen Z, Bassell GJ, Zheng JQ. An essential role for beta-actin mRNA localization and translation in Ca2+-dependent growth cone guidance. Nat Neurosci. 2006;9:1265–1273. doi: 10.1038/nn1773. [DOI] [PubMed] [Google Scholar]
- Yenari MA, Fink SL, Sun GH, Chang LK, Patel MK, Kunis DM, Onley D, Ho DY, Sapolsky RM, Steinberg GK. Gene therapy with HSP72 is neuroprotective in rat models of stroke and epilepsy. Ann Neurol. 1998;44:584–91. doi: 10.1002/ana.410440403. [DOI] [PubMed] [Google Scholar]
- Yong VW, Power C, Forsyth P, Edwards DR. Metalloproteinases in biology and pathology of the nervous system. Nature Reviews Neuroscience. 2001;2:502–511. doi: 10.1038/35081571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Poo MM. Localized synaptic potentiation by BDNF requires local protein synthesis in the developing axon. Neuron. 2002;36:675–688. doi: 10.1016/s0896-6273(02)01023-1. [DOI] [PubMed] [Google Scholar]
- Zietkiewicz S, Krzewska J, Liberek K. Successive and synergistic action of the Hsp70 and Hsp100 chaperones in protein disaggregation. Journal of Biological Chemistry. 2004;279:44376–44383. doi: 10.1074/jbc.M402405200. [DOI] [PubMed] [Google Scholar]
- Zietkiewicz S, Lewandowska A, Stocki P, Liberek K. Hsp70 chaperone machine remodels protein aggregates at the initial step of Hsp70-Hsp100-dependent disaggregation. Journal of Biological Chemistry. 2006;281:7022–7029. doi: 10.1074/jbc.M507893200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.