Abstract
Bacteria from nodules of the legume Acaciella angustissima native to the south of Mexico were characterized genetically and their nodulation and competitiveness were evaluated. Phylogenetic studies derived from rpoB gene sequences indicated that A. angustissima is nodulated by Sinorhizobium mexicanum, Rhizobium tropici, Mesorhizobium plurifarium and Agrobacterium tumefaciens and by bacteria related to Sinorhizobium americanum, Sinorhizobium terangae, Rhizobium etli and Rhizobium gallicum. A new lineage related to S. terangae is recognized based on the sequences of gyrA, nolR, recA, rpoB and rrs genes, DNA–DNA hybridization and phenotypic characteristics. The name for this new species is Sinorhizobium chiapanecum and its type strain is ITTG S70T. The symbiotic genes nodA and nifH were similar to those from S. mexicanum strains, which are Acaciella symbionts as well, with nodA gene sequences grouped within a cluster of nod genes from strains that nodulate plants from the Mimosoideae subfamily of the Leguminosae. Sinorhizobium isolates were the most frequently obtained from A. angustissima nodules and were among the best strains to promote plant growth in A. angustissima and to compete in interstrain nodule competition assays. Lateral transfer of symbiotic genes is not evident among the genera that nodulate A. angustissima (Rhizobium, Sinorhizobium and Mesorhizobium) but may occur among the sympatric and closely related sinorhizobia that nodulate Acaciella.
Keywords: Acaciella angustissima, N2 fixation, nodulation, rhizobial diversity, legume symbiosis
Introduction
Bacteria in the roots or the stems of legumes fix nitrogen and provide the plant with this nutrient. Symbiotic bacteria have been studied from only a small proportion of the extant legume species, and diverse genera such as Rhizobium, Sinorhizobium, Mesorhizobium, Bradyrhizobium, Devosia, Methylobacterium, Burkholderia and Cupriavidus have been reported to contain nodulating species (Young & Haukka, 1996; Sprent, 2001; Sy et al., 2001; Rivas et al., 2002; Chen et al., 2003; Vandamme & Coenye, 2004; Elliott et al., 2007a,b;). Mexico is a very diverse country and occupies the fourth place in plant diversity terms (Rzedowsky, 1978) with many endemic legumes. The genus Acaciella is found mainly in Mexico and has well-supported botanical differences to be recognized as a new species different from the Acacia genus where it was formerly classified (Rico-Arce & Bachean, 2006).
Nitrogen-fixing trees and shrubs are valuable to maintain forest fertility and N2 fixation allows their growth in infertile soils while enriching soil nitrogen. In Chiapas, Acaciella angustissima shrubs that can grow in poor soils are being used in agroforestry systems, due to their rapid growth rate, high capacity for nitrogen fixation and the quality of the tannins that accumulate in their bark (Rincón-Rosales & Gutiérrez-Miceli, 2008). Interestingly, these shrubs are the preferred hosts of Llaveia mexicanorum (Williams & MacVean, 1995), a native homeoptera scale insect, which is used by indigenous people of Chiapas and Mesoamerica to produce a fat for traditional lacquer wood handcrafts (Grillasca, 2007). We established nurseries to propagate A. angustissima plants and became aware that inoculants were required to attain good plant development. This prompted us to analyze and select strains for inoculation.
One of the sinorhizobial groups we encountered corresponded to a new species and we proposed the name Sinorhizobium mexicanum for this lineage (Lloret et al., 2007). However, modifications in the Sinorhizobium genus taxonomy have occurred (Young, 2003). The bacteria belonging to Sinorhizobium have been transferred to the genus Ensifer (Young, 2003) because according to judicial rules, Ensifer has priority over Sinorhizobium. This new Sinorhizobium species had to be named as Ensifer mexicanus (Lloret et al., 2007) instead of S. mexicanum. In this work, we chose to use the former name Sinorhizobium as used in many recently published papers.
Sinorhizobium mexicanum was not the only symbiont found in A. angustissima nodules. The objective of this study was to characterize the other symbionts of A. angustissima in Mexico (including a novel sinorhizobial species), their interstrain nodulation competitiveness and their plant growth promotion in A. angustissima.
Materials and methods
Sample sites
Isolates used in this study were obtained from root nodules of A. angustissima collected from the Sumidero Canyon National Park in Chiapas, Mexico, and from nodulated trap plants grown in pots containing soils collected from an ecological reserve area in Sierra de Huautla in Morelos, Mexico (Supporting Information, Fig. S1). The Chiapas and Morelos collecting sites were c. 1000 km apart and both are characterized by deciduous forest vegetation (Lloret et al., 2007).
Bacterial strains
The A. angustissima strains analyzed in this study are listed in Table 1. Bacteria were obtained as described by Vincent (1970) using peptone yeast agar (PY) as growth medium (Toledo et al., 2003). Plates were incubated aerobically at 28 °C for 3 days and the isolates were purified by streaking single colonies on fresh PY plates. Single colony formation and morphology were observed in yeast extract mannitol (YEM) and PY media at 28 °C as reported by Toledo et al. (2003). The acid/alkaline reaction was verified by spreading the inoculum on YEM plates (pH 7.0) containing 25 μg mL−1 bromothymol blue (Vincent, 1970).
Table 1.
Bacteria isolated from nodules of Acaciella angustissima
| Species and strains* | Geographical origin |
|---|---|
| Agrobacterium tumefaciens | |
| ITTG S2 | Chiapas, Mexico |
| ITTG S6 | Chiapas, Mexico |
| ITTG S9 | Chiapas, Mexico |
| ITTG S10 | Chiapas, Mexico |
| CFN ESH11 | Morelos, Mexico |
| CFN ESH16 | Morelos, Mexico |
| Mesorhizobium plurifarium | |
| CFN ESH5 | Morelos, Mexico |
| CFN ESH18 | Morelos, Mexico |
| CFN ESH19 | Morelos, Mexico |
| CFN ESH22 | Morelos, Mexico |
| CFN ESH26 | Morelos, Mexico |
| Rhizobium sp. (R. gallicum related) | |
| ITTG S11 | Chiapas, Mexico |
| Rhizobium sp. (R. leguminosarum/R. etli related) | |
| CFN ESH6 | Morelos, Mexico |
| CFN ESH7 | Morelos, Mexico |
| CFN ESH34 | Morelos, Mexico |
| Rhizobium tropici | |
| CFN ESH9 | Morelos, Mexico |
| CFN ESH10 | Morelos, Mexico |
| CFN ESH23 | Morelos, Mexico |
| CFN ESH25 | Morelos, Mexico |
| CFN ESH27 | Morelos, Mexico |
| CFN ESH29 | Morelos, Mexico |
| ITTG S7 | Chiapas, Mexico |
| Sinorhizobium sp. (S. americanum related) | |
| ITTG S8 | Chiapas, Mexico |
| Sinorhizobium chiapanecum sp. nov. | |
| ITTG R11 | Chiapas, Mexico |
| ITTG S1 | Chiapas, Mexico |
| ITTG S68 | Chiapas, Mexico |
| ITTG S70T | Chiapas, Mexico |
| ITTG S71 | Chiapas, Mexico |
| Sinorhizobium mexicanum | |
| CFN ESH1 | Morelos, Mexico |
| CFN ESH2 | Morelos, Mexico |
| CFN ESH3 | Morelos, Mexico |
| CFN ESH4 | Morelos, Mexico |
| ITTG R4 | Chiapas, Mexico |
| ITTG R7T | Chiapas, Mexico |
| ITTG S3 | Chiapas, Mexico |
| ITTG S4 | Chiapas, Mexico |
| ITTG S5 | Chiapas, Mexico |
| ITTG S64 | Chiapas, Mexico |
Identity according to the sequence analysis of the chromosomal gene rpoB.
Nodulation tests
Acaciella angustissima seeds were scarified with H2SO4 for 15 min and surface sterilized with 1% (v/v) sodium hypochlorite for 10 min. Treated seeds were germinated on 0.8% agar–water plates and then placed in glass tubes filled with vermiculite moistened with Fahraeus medium (Fahraeus, 1957). Inoculation tests were also performed with Acacia farnesiana, Leucaena leucocephala and Phaseolus vulgaris cv. Negro Jamapa as described (Lloret et al., 2007). Bacteria for inoculation were grown in individual PY plates and suspended in 1 mL sterile distilled water, serially diluted, and absorbance was determined at A600 nm. Final cell numbers were determined by plating on PY medium to count CFUs. Approximately 106 bacteria mL−1 were added to each plant rootlet and the plants were grown in a plant growth chamber at 28 °C (Räsänen et al., 2001). A negative control with uninoculated seedlings was included. After 30 days, surface-disinfected nodules were harvested and crushed in PY plates and single colonies were picked up and reinoculated for their authentication. Bacteria were conserved in 65% glycerol–PY broth and stored at −80 °C. Working cultures were maintained on YEM slants at 4 °C (Vincent, 1970).
DNA isolation, genomic fingerprinting and DNA–DNA hybridization
Isolates were grown overnight in 2 mL PY. Total DNA was isolated and purified using the Genomic Prep™ kit (Amersham). Enterobacterial repetitive intergenic consensus (ERIC) genomic fingerprinting was obtained by PCR using primers ERIC1R and ERIC2 as described by Versalovic et al. (1994). The fingerprints were visually analyzed after resolution of PCR products using electrophoresis in 1.5% agarose gels loaded with half the volume of the 25 μL PCR reaction. ERIC fingerprinting was used only to confirm that the isolates analyzed were not clones or siblings (Ormeño-Orrillo et al., 2006; Lloret et al., 2007;). Strains showing different patterns were considered for sequencing and phylogenetic analysis. The DNA relatedness was determined using DNA–DNA hybridization experiments using 32P-labelled DNA of the newly proposed species (described below) Sinorhizobium chiapanecum ITTG S70T as a probe. A filter hybridization method described previously was used (Martínez-Romero et al., 1991). The amounts of DNA were standardized using integrating gel fluorescence with the Eagle Eye II system (Stratagene). anova and t-tests were performed to compare the percentage of DNA–DNA hybridization values among species and within species using angular transformation of percentage data (Knudsen & Curtis, 1947; Martínez-Romero & Rosenblueth, 1990;).
PCR amplification and gene sequencing
An internal fragment of the chromosomal genes gyrA, nolR, recA, rpoB and 16S rRNA gene (rrs), and the symbiotic genes nifH and nodA were amplified using standard PCRs. Primers and annealing temperatures used for gyrA, nolR, recA, rrs, rpoB and nifH genes were performed as described in Lloret et al. (2007) and by Haukka et al. (1998) for nodA. Before sequencing, the amplification mixture was purified using the PCR product purification system of Roche™. The sequences generated were deposited in the GenBank public database and their accession numbers were included in the phylogenetic trees.
Phylogenetic analysis
The protein-coding sequences were aligned using the program clustal w (Thompson et al., 1994) and then aligned based on codons using dambe v4.2.13 (Xia & Xie, 2001). The alignments were edited with bioedit v5 (Hall, 1999). The best-fit evolutive models for each set of sequences were selected by the AKAIKE information criterion implemented in the modeltest v3.06 (Posada & Buckley, 2004). rpoB gene sequences were analyzed using the TrN+I+Γ model of evolution based on an alignment of 642 nucleotides from positions 3262 to 3903; nodA using the GTR+I+Γ model with 522 nucleotides from positions 67 to 588; nifH with the TrN+I+Γ model of evolution based on 474 nucleotides from positions 313 to 787; and for the gyrA, recA and nolR genes the model of evolution and alignment positions were as reported by Lloret et al. (2007). These positions were based on the rpoB, nolR, nodA and nifH genes of Sinorhizobium meliloti 1021 and recA and gyrA of Agrobacterium tumefaciens C58. The phylogenetic trees were inferred with the maximum-likelihood (ML) method using the program phyml v2.4.4 (Guindon & Gascuel, 2003) considering the α-parameter for the Gamma distribution and the proportion of invariable sites estimated by the program. For the inference of the rrs phylogenetic tree, Sinorhizobium type strains were analyzed by the neighbor-joining method (NJ) (Saitou & Nei, 1987) implemented in mega v3.1 (Kumar et al., 2004) using the TrN+G model with the α-parameter for the Gamma distribution estimated with modeltest. The rrs phylogenetic tree was constructed using an alignment of 1417 nucleotides from positions 28 to 1444 with respect to the rrs gene of S. meliloti 1021. The topology robustness was estimated by a nonparametric bootstrap test using 100 pseudoreplicates for ML and 1000 for NJ.
Competition assays
The nodulation capacity was evaluated in competition assays of S. mexicanum ITTG R7T or S. chiapanecum ITTG S70T against one randomly selected strain from each of the bacterial groups identified previously by rpoB gene sequence analysis. Twenty-one treatments resulted from the 12 combination mixtures plus each of the eight single strains as positive nodulation controls, and the negative control (uninoculated plants). Four replicates of inoculated plants were used per treatment. The plant growth conditions were as mentioned above for nodulation tests. The competitiveness was evaluated by the number of nodules obtained from each member of the mixture with respect to the total number of nodules. The identity of the reisolated strains was determined by plasmid patterns using the Eckhardt procedure (Eckhardt, 1978). The variation in nodule number was analyzed statistically by anova using sas software (SAS Institute Inc., 1989), followed by comparison of means by Tukey's test (P<0.05).
Plant inoculation assays
The strains with the best nodulation capacity and high competitiveness were used as inoculants. Germinated seedlings of A. angustissima were planted in vermiculite tubes with Fahraeus medium (Fahraeus, 1957) and inoculated as described above. Plants without inoculum, with or without 30 mg KNO3-N per plant, served as control (Hungria et al., 2001). Six replicate tubes were used per treatment and these were arranged in a completely randomized design. The plants were grown in a climate chamber at 28 °C for 90 days. At harvest, the shoot height, shoot dry weight, root dry weight and nodule number were determined, and total shoot nitrogen was assayed using the Kjeldahl method (Bremner & Mulvaney, 1982). The effect of the inoculation was analyzed statistically by anova, followed by comparison of means using Tukey's test (P<0.05).
Results
Strain identity, diversity and phylogeny
A total of 94 strains were obtained from A. angustissima root nodules in Chiapas and Morelos that were confirmed to form nodules in the original host. Thirty-eight strains that represented the different ERIC-PCR electrophoretic patterns were used for PCR amplification and sequencing. The taxonomic position of the selected strains from A. angustissima was determined according to the phylogenetic analysis performed with partial sequences of the chromosomal gene rpoB, which encodes the β-subunit of RNA polymerase (Fig. 1).
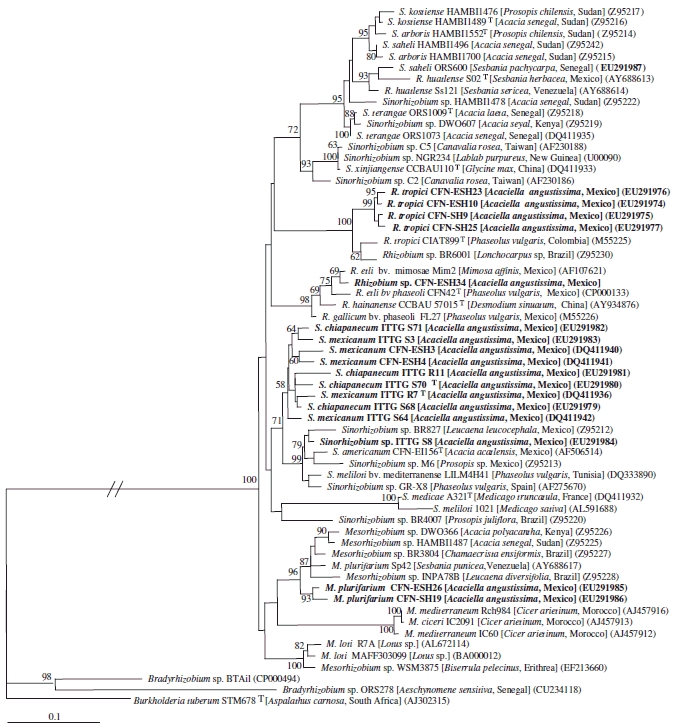
Fig. 1.
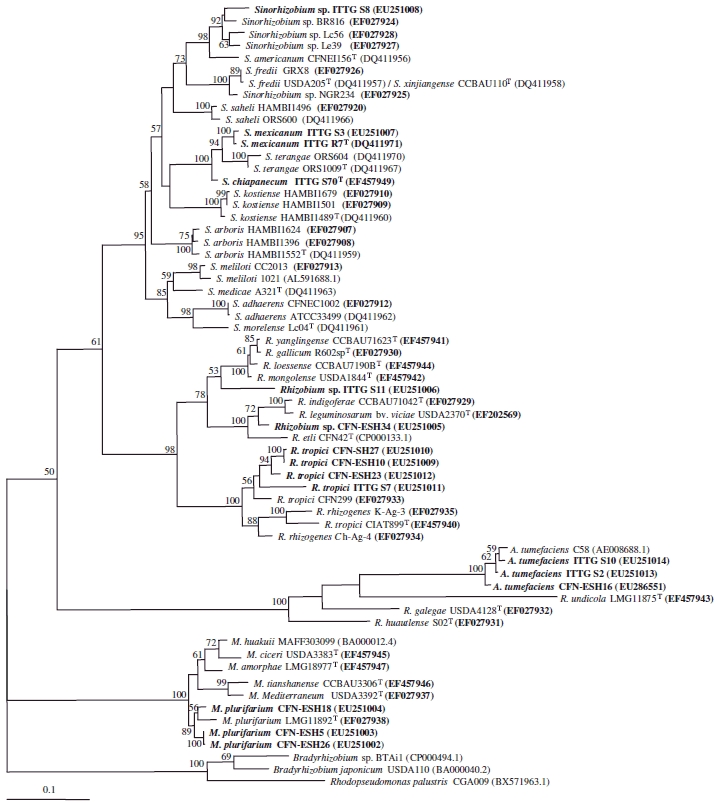
Phylogenetic tree estimated using the ML method with partial sequences of the chromosomal protein encoding gene rpoB using the phyml program. The alignment length was 642 nucleotides from positions 3262 to 3903 of the rpoB gene of Sinorhizobium meliloti 1021. Only bootstrap values ≥50% are shown. Type strains are indicated by superscript T. The Acaciella angustissima strains are shown in bold. Only haplotypes were included in each terminal branch. The accession numbers for the sequences are indicated within parentheses. Those generated in this work are shown in bold.
The largest percentage of isolates found at both sites corresponded to S. mexicanum (26.3%) while the lowest corresponded to bacteria related to S. americanum and Rhizobium gallicum, both with 2.6%. A new lineage related to S. mexicanum and Sinorhizobium terangae was isolated only in Chiapas while only the strains related to Rhizobium etli and Mesorhizobium plurifarium were found in Morelos. The largest percentage of the isolates in Chiapas corresponded to S. mexicanum (33.3%) and in Morelos Rhizobium tropici (30.0%).
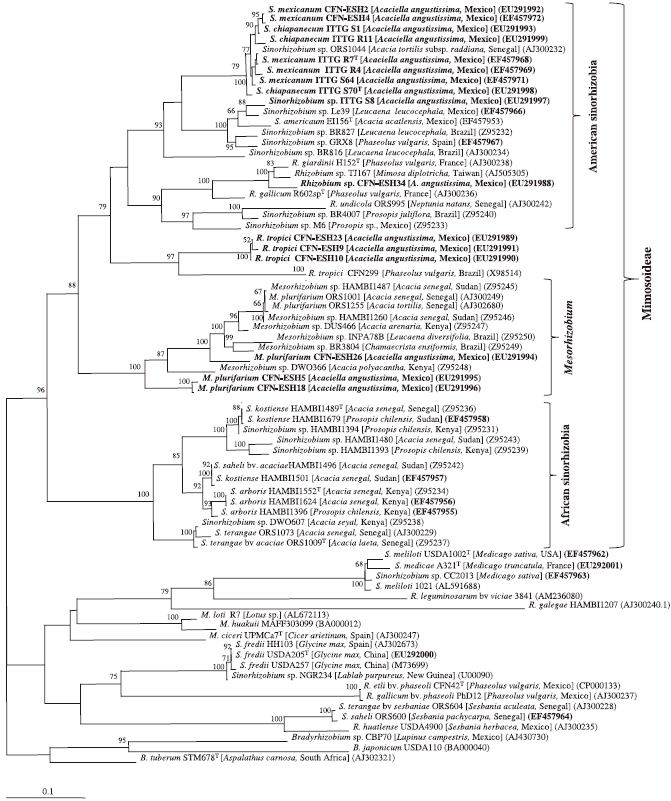
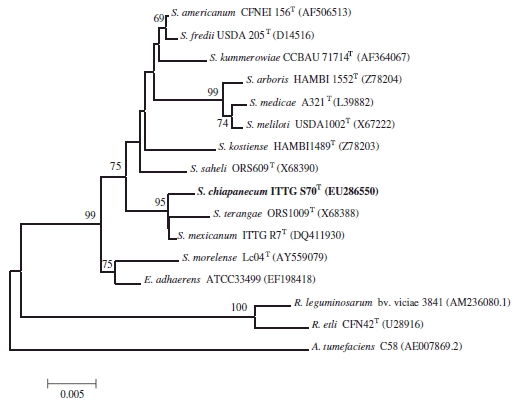
The Sinorhizobium sp. strain ITTG S70Trrs gene was found to be different from all sequences available in the GenBank database and had 99% identity to its closest relative S. mexicanum. The phylogenetic tree with the sequences of rrs genes (Fig. 2) and the phylogenetic trees with rpoB (Fig. 1), gyrA, nolR and recA genes (Fig. 3) were constructed including all of the type strains of Sinorhizobium species. The recA gene has been used previously in rhizobial phylogenetic studies (Gaunt et al., 2001; Vinuesa et al., 2005); gyrA, recA, nolR and rpoB were used previously to describe a new Sinorhizobium species (Lloret et al., 2007). gyrA encodes the alpha-subunit of DNA gyrase, nolR encodes a transcriptional regulator (Chen et al., 2000, 2005) and recA encodes the recombination protein RecA. In all phylogenetic trees, the position of strain ITTG S70T as a different lineage within the Sinorhizobium genus was well supported. Strains ITTG R11, ITTG S68 and ITTG S71 had sequences identical to those from ITTG S70T that was chosen to represent this new lineage.
Fig. 2.
Phylogenetic tree estimated by the NJ method with the sequences of rrs genes using the Tamura Nei model considering gamma (α-parameter=0.9271). The alignment length was 1416 nucleotides, from positions 28 to 1444 of the rrs gene of Sinorhizobium meliloti 1021. Only bootstrap values ≥50% are shown. Type strains are indicated by superscript T. Sinorhizobium chiapanecum is shown in bold.
Fig. 3.
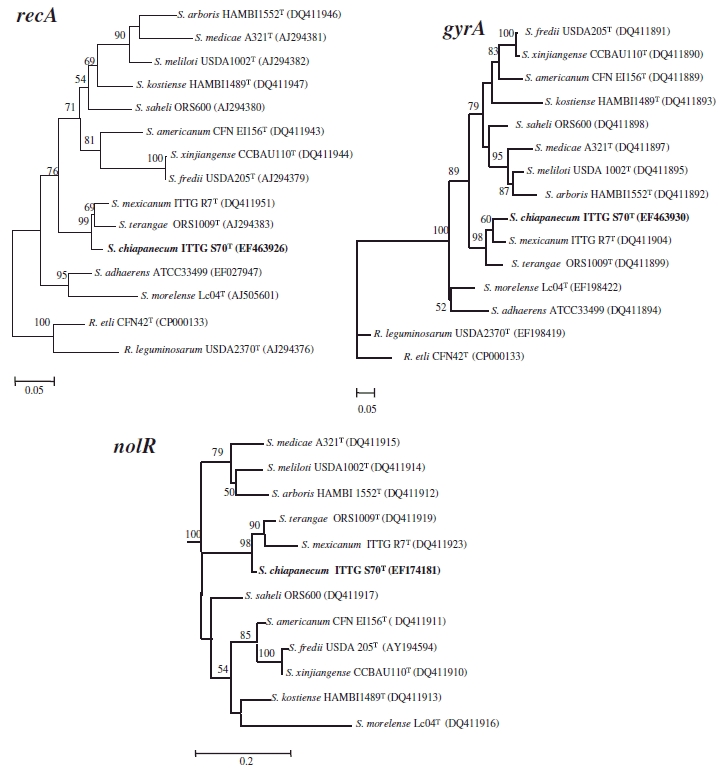
ML phylogenetic trees of partial sequences of the chromosomal protein encoding genes gyrA, recA and nolR. Type strains are indicated by superscript T. Only bootstrap values ≥50% are shown. The accession numbers for the sequences are indicated within parentheses. Those generated in this work for Sinorhizobium chiapanecum are shown in bold. The branches corresponding to the outgroup sequences Rhizobium leguminosarum USDA 2370T and Rhizobium etli CFN42T for the nolR tree are not shown because they were too divergent from the Sinorhizobium sequences.
Total DNA from the strain ITTG S70T showed low hybridization with the strains belonging to S. mexicanum (<42%) and S. terangae (<56%), while hybridization to three strains from its own group, ITTG S68, ITTG S71 and ITTG R11 (>74%), was higher than the limit proposed for new species (70%, Stackebrandt et al., 2002) (Table S2). DNA–DNA hybridization differences between S. chiapanecum strains and the closest species, S. mexicanum and S. terangae, were statistically significant with P<0.05 and P<0.10, respectively. It remains to be established whether similar plasmids in S. terangae and S. chiapanecum account for part of the DNA hybridization obtained. Description of species should be based on chromosomal and not plasmidic characteristics (Martínez-Romero & Jarvis, 1993). Also, phenotypic differences distinguishing S. chiapanecum, S. terangae and S. mexicanum are presented as Table S1.
The DNA–DNA hybridization, the phylogenetic position and phenotypic characteristics support that this Sinorhizobium lineage corresponds to a new species within the genus Sinorhizobium, and the proposed name is S. chiapanecum because it was isolated in Chiapas.
The nifH and nodA phylogenetic trees are shown in Figs 4 and 5, respectively. Symbiotic genes from the different rhizobia isolated from A. angustissima had affiliations with the corresponding genes from species in the genera Rhizobium, Sinorhizobium and Mesorhizobium. The phylogenies of these two symbiotic genes were incongruent with the phylogeny obtained with the chromosomal gene rpoB. The nodA sequences from Rhizobium sp. strains CFN ESH6 and CFN ESH34 (related to R. etli) isolated from A. angustissima were similar to the nodA gene of Rhizobium giardinii H152T isolated from common bean in France, but with the nifH gene analysis these strains were found to be related to the nifH gene of R. etli bv. mimosae Mim2 isolated from Mimosa affinis in Mexico. The nodA and nifH genes of R. tropici strains CFN ESH23, CFN ESH25, CFN ESH10, CFN ESH29 and CFN ESH9 were related but not identical to nodA and nifH gene sequences from R. tropici CFN299 isolated from P. vulgaris in Mexico. The symbiotic gene sequences of S. chiapanecum and S. mexicanum isolated from A. angustissima clustered together and were related to a different and well-supported group that included mainly sequences from Sinorhizobium isolated from American legumes, among them, the strains Sinorhizobium sp. BR827 and BR816 from L. leucocephala in Brazil and S. americanum CFN EI156 isolated from Acacia acatlensis in Mexico. The nodA and nifH gene sequences from S. terangae, the closest relative of S. mexicanum according to rpoB gene sequences, grouped in a far distant cluster. Mesorhizobium plurifarium isolated from A. angustissima has nodA and nifH gene sequences similar to several Mesorhizobium species isolated from American and African hosts, mainly with the strains Mesorhizobium sp. DWO366 isolated from Acacia polyacantha in Kenya and Mesorhizobium sp. INPA78b isolated from L. leucocephala in Brazil.
Fig. 4.
Phylogenetic tree estimated using the ML method with partial sequences of the symbiotic protein encoding nifH gene using the phyml program. The alignment length was 474 nucleotides from positions 313 to 786 of the nifH gene with respect to the nifH gene encoded on the pSymA of Sinorhizobium meliloti 1021. Only bootstrap values ≥50% are shown. Type strains are indicated by superscript T. The Acaciella angustissima strains are shown in bold. The accession numbers for the sequences are indicated within parentheses. Those generated in this work are shown in bold. Host and geographical origin are in parentheses.
Fig. 5.
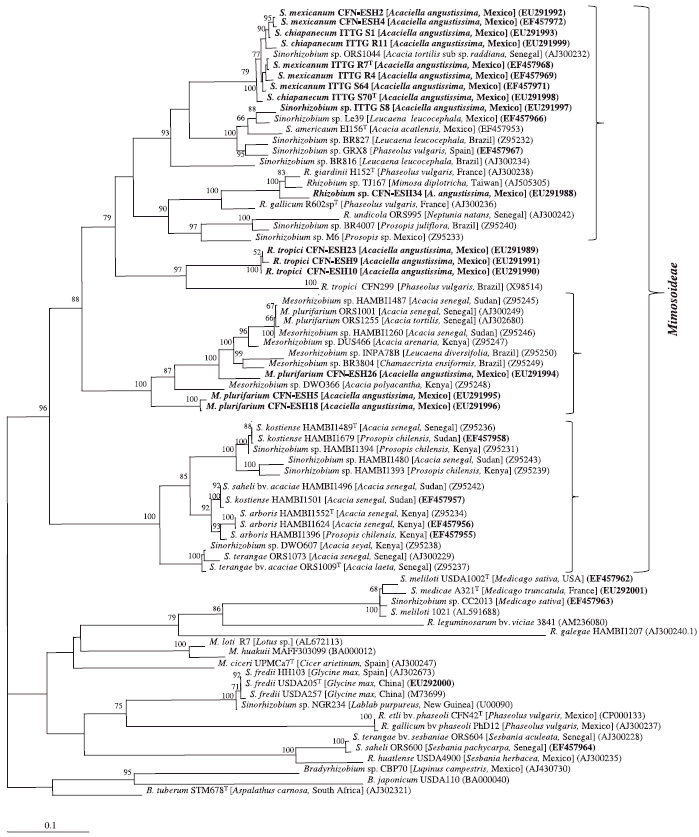
Phylogenetic tree estimated using the ML method with partial sequences of the symbiotic protein encoding the nodA gene using the phyml program. The alignment length was 522 nucleotides from positions 67 to 588 of the nodA gene with respect to the nodA encoded on the pSymA of Sinorhizobium meliloti 1021. Only bootstrap values ≥50% are shown. Type strains are indicated by superscript T. The Acaciella angustissima strains are shown in bold. The accession numbers for the sequences are indicated within parentheses. Those generated in this work are shown in bold. Host and geographical origin are in parentheses.
Nodulation and nodule occupancy in competition assays
Nodule occupancy evaluated from interstrain competition assays is shown in Table 2. The strains ITTG R7T, ITTG S70T, CFN ERSH34, CFN ERSH5 and ITTG S7 showed the best nodulation capacity and high competitiveness. Sinorhizobium mexicanum strain ITTG R7T always had a greater occupancy of the nodules than the respective competing strain, ranging from 65% when combined with Sinorhizobium sp. ITTG S8 to 100% when combined with Rhizobium sp. ITTG S11. The S. chiapanecum strain ITTG S70T did not always have a greater occupancy of the nodules than the competing strain, although this strain and S. mexicanum ITTG R7 were highly effective in inoculation assays with A. angustissima (Table 3). Mesorhizobium plurifarium CFN ESH5 and Rhizobium sp. CFN ESH34 had a greater occupancy than ITTG S70T (67% and 77%), respectively. Significantly lower numbers of nodules were obtained with M. plurifarium CFN ESH5, R. tropici ITTG S7, Sinorhizobium sp. ITTG S8 (related to S. americanum), Rhizobium sp. ITTG S11 (related to R. gallicum) and A. tumefaciens ITTG S2, with the latter showing the lowest number of nodules and very low nitrogen fixation. All reisolated strains showed colony morphology and plasmid patterns identical to the original inoculated strains (data not shown).
Table 2.
Nodule occupancy by strains of Sinorhizobium mexicanum ITTG R7T and Sinorhizobium chiapanecum ITTG S70T and the coinoculated bacteria in competition assays in Acaciella angustissima
| Nodule occupancy (%) by |
|||
|---|---|---|---|
| Treatments | Nodule number (per plant) (±SD)* | First strain of the combination | Second strain of the combination |
| Uninoculated | 0h | ||
| A. tumefaciens ITTG S2 | 0.5 (±1.0)gh | ||
| M. plurifarium CFN ESH5 | 8.0 (±1.6)bcde | ||
| Rhizobium sp. ITTG S11 | 2.0 (±0.8)fgh | ||
| Rhizobium sp. CFN ESH34 | 10.5 (±2.5)abc | ||
| R. tropici ITTG S7 | 4.0 (±1.6)defgh | ||
| Sinorhizobium sp. ITTG S8 | 2.25 (±2.1)fgh | ||
| S. chiapanecum ITTG S70T | 10.0 (±1.6)abc | ||
| S. mexicanum ITTG R7T | 14.0 (±3.7)a | ||
| S. chiapanecum ITTG S70T+A. tumefaciens ITTG S2 | 4.5 (±1.3)defgh | 61 (18)† | 39 |
| S. chiapanecum ITTG S70T+M. plurifarium CFN ESH5 | 4.5 (±2.6)defgh | 33 (18) | 67 |
| S. chiapanecum ITTG S70T+Rhizobium sp. ITTG S11 | 4.25 (±1.7)defgh | 82 (17) | 18 |
| S. chiapanecum ITTG S70T+Rhizobium sp. CFN ESH34 | 3.25 (±1.9)efgh | 23 (13) | 77 |
| S. chiapanecum ITTG S70T+R. tropici ITTG S7 | 2.5 (±1.3)fgh | 80 (10) | 20 |
| S. chiapanecum ITTG S70T+Sinorhizobium sp. ITTG S8 | 3.75 (±2.1)defgh | 67 (15) | 33 |
| S. mexicanum ITTG R7T+A. tumefaciens ITTG S2 | 6.0 (±1.4)cdef | 75 (24) | 25 |
| S. mexicanum ITTG R7T+M. plurifarium CFN ESH5 | 8.75 (±5.1)abcd | 89 (35) | 11 |
| S. mexicanum ITTG R7T+Rhizobium sp. ITTG S11 | 5.5 (±1.9)cdefg | 100 (22) | 0 |
| S. mexicanum ITTG R7T+Rhizobium sp. CFN ESH34 | 12.0 (±1.4)ab | 71 (48) | 29 |
| S. mexicanum ITTG R7 T+R. tropici ITTG S7 | 3.75 (±1.3)defgh | 67 (15) | 33 |
| S. mexicanum ITTG R7T+Sinorhizobium sp. ITTG S8 | 6.5 (±3.4)cdef | 65 (26) | 35 |
Mean values of four replicates. The means followed by the same letter are not significantly different (P<0.05).
In parentheses, total number of nodules analyzed.
Table 3.
Effect of inoculation by the strains with high competitivity and nodulation capacity on the growth, nodulation and nitrogen fixation of Acaciella angustissima
| Strains | Shoot height (cm) | Shoot dry weight (mg) | Root dry weight (mg) | Nodule number | Total shoot N (mg per plant) |
|---|---|---|---|---|---|
| Uninoculated | 15.0 c* | 76.1 b | 46.3 b | 0 b | 30.4 c |
| M. plurifarium CFN ESH5 | 16.5 bc | 95.0 b | 40.3 a | 2.1 b | 39.9 c |
| Rhizobium sp. CFN ESH34 | 20.1 b | 101.0 b | 44.9 a | 2.3 b | 51.5 c |
| R. tropici ITTG S7 | 17.0 bc | 96.4 b | 42.9 a | 2.3 b | 44.3 c |
| S. chiapanecum ITTG S70T | 24.8 a | 112.9 a | 44.0 a | 5.3 a | 101.6 b |
| S. mexicanum ITTG R7T | 25.1 a | 134.7 a | 52.6 a | 5.8 a | 158.9 a |
| KNO3-N (30 mg per plant) | 15.3 c | 56.1 c | 25.3 b | 0 b | 22.4 c |
Mean values of six replicates. The means followed by the same letter are not significantly different (P<0.05).
Plant growth, nodulation and nitrogen fixation of A. angustissima inoculated with selected strains
The inoculation using the selected rhizobia strains had a significant effect on the growth of A. angustissima (Table 3). Rhizobium sp. CFN ESH34, S. mexicanum ITTG R7T and S. chiapanecum ITTG S70T had a positive effect on shoot height, shoot dry weight and root dry weight compared with the uninoculated control plants and those with added KNO3. Plants inoculated with these strains were on average 8.3 cm taller and weighted 109 mg more than noninoculated plants 90 days postinoculation. The number of nodules obtained with S. mexicanum ITTG R7T and S. chiapanecum ITTG S70T was significantly different (P<0.05) compared with the rest of the treatments. None of the noninoculated plants formed nodules. The plants inoculated with ITTG R7T showed a significantly higher total shoot nitrogen compared with other treatments (P<0.05). ITTG R7T was found to be the most effective strain in terms of plant growth promotion as indicated by total plant nitrogen content.
Characteristics of S. chiapanecum sp. nov
Sinorhizobium chiapanecum (chia.pa.ne'cum. N.L. neut. adj. chiapanecum of Chiapas, the name of a state in Mexico where the bacterium was isolated). Gram-negative, aerobic, motile and nonspore-forming rods. Strains are fast growing and acid producers in YEM medium. The generation time for ITTG S70T in YEM broth is 2.33 h at 28 °C. Colonies on PY or YEM are circular, pearly, slightly translucent and produce copious amounts of polysaccharides. Colonies are normally more than 2–4 mm in diameter within 2 days of incubation at 28 °C. The strains are resistant to nalidixic acid (120 μg mL−1) but not to carbenicillin (20 μg mL−1), ampicillin (10 μg mL−1) or chloramphenicol (10 μg mL−1). They grow in media containing 0.5%, 1.0% and 2.0% NaCl but not with 3.0% NaCl. Total DNA from strain ITTG S70T showed low hybridization values with the strains belonging to S. terangae ORS1009T (<48%) and with S. mexicanum ITTGR7T (<33%). This species can be differentiated from other described Sinorhizobium species on the basis of the phylogenetic analysis of the chromosomal genes rrs, gyrA, recA, rpoB and nolR. The type strain ITTG S70T was isolated from nodules of A. angustissima collected in the Sumidero Canyon National Park, Chiapas, Mexico. Sinorhizobium chiapanecum ITTG S70T nodulated and fixed nitrogen in A. angustissima, Acaciella cochliancantha, Acaciella farnesiana, Acaciella pennatula, Dolichos lablab, P. vulgaris, L. leucocephala and Lysiloma acapulcensis and tolerated salinity and acidity (data not shown). ITTG S70T has characteristics of the species.
Discussion
Tropical forests in Mexico harbor many endemic plants and a high richness of species (Rzedowsky, 1978). Forests have abiotic and biotic characteristics that allow such diversity to exist. Plant speciation in Mexico seems to be driven by geographical isolation due to the complex topography of the country. The tropics have a large diversity of rhizobia (Wang et al., 1999; Mohamed et al., 2000; Räsänen et al., 2001; Toledo et al., 2003; Wolde-Meskel et al., 2004;). Sinorhizobia seem to have radiated in Mexico in relation to the geographical isolation and diversity of climates, conditions and plants (Toledo et al., 2003; Lloret et al., 2007;). The sinorhizobia-nodulating legumes in Africa and in the Americas are considered to have had a long period of diverging evolution (Haukka et al., 1998; Toledo et al., 2003; Lloret et al., 2007;). Our results showed that A. angustissima was preferentially nodulated by closely related members of the Alphaproteobacteria, especially sinorhizobia. Differences in symbiotic efficiency and competitiveness were found among the isolates, with S. mexicanum and S. chiapanecum strains being highly effective symbionts and good competitors. In contrast to acacias, no bradyrhizobia or Betaproteobacteria strains were found nodulating this legume. Acaciella angustissima was among the legume hosts of Latin American origin that formed nitrogen-fixing nodules with the African sinorhizobial strains Sinorhizobium arboris HAMBI 1552T, Sinorhizobium kostiense HAMBI 1489T and S. terangae bv. acaciae ORS 1058 (Räsänen et al., 2001). Tropical legumes seem to have a mild specificity when associating with nodulating bacteria (Moreira et al., 1998), although under natural conditions predominant rhizobial species may be preferentially encountered in promiscuous plants (Bala & Giller, 2001; Bala et al., 2003; Martínez-Romero, 2003;) as shown here.
Mesorhizobium plurifarium strains originally isolated from Acacia senegal (de Lajudie et al., 1998) encompass a set of diverging strains. In this study, M. plurifarium strains were found in A. angustissima only in Morelos. In Mexico, M. plurifarium were found nodulating Sesbania (Papilioniodeae) trees (Wang et al., 1999) and L. leucocephala plants grown in Morelos soils. Acaciella and Leucaena belong to the Mimosoideae subfamily of the Leguminosae. Plant traps with soils from Morelos were used to collect the bacteria and it has been shown that by doing so a larger diversity of bacteria may be obtained nodulating a single legume (Hungria et al., 2001), and so we predicted that Mesorhizobium strains were the less adapted to nodulate A. angustissima. This turned out to be true.
We found seven isolates of Rhizobium similar to R. tropici type A, with nod genes more closely related (but not identical) to nodA of R. tropici than to other nodA genes. Rhizobium tropici strains are common in tropical soils and nodulate some trees from the Mimosoideae subfamily of the Leguminosae such as L. leucocephala (Martínez-Romero et al., 1991) as well as A. angustissima (not shown).
Rhizobium etli is commonly isolated from P. vulgaris (Segovia et al., 1993), but has also been isolated from other shrub legumes in Kenya (Odee et al., 2002). Biovars that refer to host specificity have been described in R. etli. Nodulation of Mimosoidea plants such as M. affinis and Leucaena spp. is the characteristic of biovar mimosae (Wang et al., 1999). It is probable that the Rhizobium sp. strains (related to R. etli and R. leguminosarum) from A. angustissima correspond to biovar mimosae. Strain ITTG S11 was found to be related to R. gallicum (Amarger et al., 1997). Rhizobium gallicum bv. gallicum was isolated from common bean and can also nodulate L. leucocephala (Amarger et al., 1997; Silva et al., 2005;) and other species from the Mimosoideae subfamily of the Leguminosae (Zurdo-Piñeiro et al., 2004) but it was not known that it nodulated Acaciella.
In addition, species of Agrobacterium were also found in this study. Bala & Giller (2001) reported that the legumes Acacia auriculiformis, L. leucocephala, Gliricidia sepium, P. vulgaris and Sesbania sesban formed effective nodules with one or more isolates that resembled A. tumefaciens. Agrobacterium strains have been isolated previously from nodules of Acacia mellifera, A. polyacantha, Acacia nilotica and S. sesban (Khbaya et al., 1998; de Lajudie et al., 1999;) and shrubs growing in the semi-arid and arid climates of north-western China (Tan et al., 1999). Odee et al. (2002) indicated that agrobacteria were often found in association with root nodules as a co-occupant with rhizobia. The Agrobacterium strains described here were capable of forming nodules on A. angustissima, but the nitrogen fixation was very low. Agrobacterium sp. ITTG S2 (similar to A. tumefaciens) showed a low level of competitiveness when inoculated in competition assays. Recently, some Agrobacterium strains were found to be capable of forming tumors on plants as well as nodulating (Rivas et al., 2004). In additional experiments, we evaluated the pathogenicity of the strains ITTG S2, ITTG S6 and ITTG S10 (all similar to A. tumefaciens) on sunflower plants (Helianthus annus) and found that these strains are not tumorogenic (not shown).
We showed that the phylogenies of the symbiotic genes were incongruent with the chromosomal genes as has been reported previously (Haukka et al., 1998; Wernegreen & Riley, 1999; Laguerre et al., 2001; Toledo et al., 2003; Lloret et al., 2007;). Symbiotic genes on elements such as plasmids and symbiotic islands are prone to lateral gene transfer (Sullivan & Ronson, 1998; Ochman & Moran, 2001;) and may be selected by hosts (Ueda et al., 1995; Haukka et al., 1998; Wernegreen & Riley, 1999;), as observed here, because the two species S. mexicanum and S. chiapanecum nodulating Acaciella have the same nodA genes. The three main groups described based on nod gene sequences (Haukka et al., 1998) corresponding to African and Latin–American sinorhizobia and some Mesorhizobium spp. were observed in the trees presented here with several more sequences included (Fig. 5). A large group was distinguished that corresponds to nod genes of symbionts with the capacity to nodulate many plants from the Mimosoideae subfamily of the Leguminosae (Fig. 5); it is worth noticing that within this group, R. giardinii and R. gallicum nod gene sequences were included.
Sinorhizobium sp. ITTG S8, a strain related to S. americanum, clustered in the rpoB tree with some American strains isolated from L. leucephala in Mexico (Wang et al., 1999) and with strain BR816 (van Rhijn et al., 1994) from Brazil. This group constitutes a sister clade to S. americanum and could have been identified as belonging to the same species, but unpublished DNA–DNA hybridization results from our lab showed that BR816 was not a member of S. americanum. A new biovar has been proposed (mediterranense) (Mnasri et al., 2007) to account for sinorhizobia closely related to Sinorhizobium fredii and with specificity for L. leucocephala and P. vulgaris. This biovar includes strain BR816 and some other strains that, despite being closely related to S. fredii, do not form nodules on soybean. In spite of the close relatedness of the symbiotic genes of bv. mediterranense and S. americanum to those from S. mexicanum and S. chiapanecum, we consider that A. angustissima symbionts would not correspond to biovar mediterranense because the isolates that we found to be closely related to biovar mediterranense were not efficient to nodulate A. angustissima, comprised only 2.6% of the original isolates and were outcompeted by S. mexicanum or S. chiapanecum.
Within the enlarged set of sequence data presented here, we observed that the nodA gene sequence from Rhizobium huautlense (not reported previously) forms a clade with other Sinorhizobium species nodulating Sesbania (Fig. 5), indicating the strong specificity for Sesbania nodulation and evidencing lateral transfer of symbiotic genes between Rhizobium and Sinorhizobium. The genetic coherence among symbiotic and chromosomal genes has been considered to be characteristic of rhizobia nodulating wild legumes (Wernegreen & Riley, 1999), but S. chiapanecum and S. mexicanum as well as R. huautlense and sinorhizobia from biovar sesbaniae, all from noncultivated hosts, do not follow this observation and show evidence of horizontal transfer of symbiotic genes.
Acknowledgments
We thank Posgrado of Ciencias Biologicas-UNAM and CONAcyT for a fellowship to R.R.-R. We also thank Aryana Chávez and Paola Espinosa for field work at the Sumidero Canyon National Park in Chiapas, Ivonne Toledo, M.A. Rogel and J.C. Martínez-Romero for their excellent technical assistance or sampling field work at the Sierra de Huautla, Morelos. We thank W.X. Chen, P. de Lajudie, E.T. Wang and K. Lindström for providing bacterial strains. We also wish to thank Lourdes Rico for her valuable contribution in the reclassification of the Acaciella genus. We thank Michael Dunn for reading this manuscript. Financial support was from DGAPA IN201106-3.
Authors' contribution
R.R.-R. and L.L. contributed equally to this work.
Statement
Reuse of this article is permitted in accordance with the Creative Commons Deed, Attribution 2.5, which does not permit commercial exploitation.
References
- Amarger N, Macheret V, Laguerre G. Rhizobium gallicum sp. nov. and Rhizobium giardinii sp. nov. from Phaseolus vulgaris nodules. Int J Syst Bacteriol. 1997;47:996–1006. doi: 10.1099/00207713-47-4-996. [DOI] [PubMed] [Google Scholar]
- Bala A, Giller KE. Symbiotic specificity of tropical tree rhizobia for host legumes. New Phytol. 2001;149:495–507. doi: 10.1046/j.1469-8137.2001.00059.x. [DOI] [PubMed] [Google Scholar]
- Bala A, Murphy P, Giller KE. Distribution and diversity of rhizobia nodulating agroforestry legumes in soils from three continents in the tropics. Mol Ecol. 2003;12:917–929. doi: 10.1046/j.1365-294x.2003.01754.x. [DOI] [PubMed] [Google Scholar]
- Bremner JM, Mulvaney CS. Nitrogen-total: methods of soil and plant analysis. J Am Soc Agron. 1982;9:595–624. [Google Scholar]
- Chen H, Higgins J, Kondorosi E, Kondorosi A, Djordjevic MA, Weinman JJ, Rolfe BG. Identification of nolR-regulated proteins in Sinorhizobium meliloti using proteome analysis. Electrophoresis. 2000;21:3823–3832. doi: 10.1002/1522-2683(200011)21:17<3823::AID-ELPS3823>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Chen H, Gao K, Kondorosi E, Kondorosi A, Rolfe BG. Functional genomic analysis of global regulator NolR in Sinorhizobium meliloti. Mol Plant–Microbe Interact. 2005;18:1340–1352. doi: 10.1094/MPMI-18-1340. [DOI] [PubMed] [Google Scholar]
- Chen WM, James EK, Prescott AR, Klerans M, Sprent JI. Nodulation of Mimosa spp. by the β-Proteobacterium Ralstonia taiwanensis. Mol Plant–Microbe Interact. 2003;16:1051–1061. doi: 10.1094/MPMI.2003.16.12.1051. [DOI] [PubMed] [Google Scholar]
- De Lajudie P, Willems A, Nick G, et al. Characterization of tropical tree rhizobia and description of Mesorhizobium plurifarium sp. nov. Int J Syst Bacteriol. 1998;48:369–382. doi: 10.1099/00207713-48-2-369. [DOI] [PubMed] [Google Scholar]
- De Lajudie P, Willems A, Nick G, et al. Agrobacterium bv. 1 strains isolated from nodules of tropical legumes. Syst Appl Microbiol. 1999;22:119–132. [Google Scholar]
- Eckhardt T. A rapid method for the identification of plasmid desoxyribonucleic acid in bacteria. Plasmid. 1978;1:584–585. doi: 10.1016/0147-619x(78)90016-1. [DOI] [PubMed] [Google Scholar]
- Elliott GN, Chen WM, Chou JH, et al. Burkholderia phymatum is a highly effective nitrogen-fixing symbiont of Mimosa spp. and fixes nitrogen ex planta. New Phytol. 2007a;173:168–180. doi: 10.1111/j.1469-8137.2006.01894.x. [DOI] [PubMed] [Google Scholar]
- Elliott GN, Chen WM, Bontemps C, Chou JH, Young JPW, Sprent JI, James EK. Nodulation of Cyclopia spp. (Leguminosae, Papilionoideae) by Burkholderia tuberum. Ann Bot. 2007b;100:1403–1411. doi: 10.1093/aob/mcm227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahraeus G. The infection of clover root hair by nodule bacteria studied by a single glass slide technique. J Gen Microbiol. 1957;16:374–381. doi: 10.1099/00221287-16-2-374. [DOI] [PubMed] [Google Scholar]
- Gaunt MW, Turner SL, Rigottier-Gois L, Lloyd-Macgilp SA, Young JPW. Phylogenies of atpD and recA support the small subunit rRNA-based classification of rhizobia. Int J Syst Evol Microbiol. 2001;51:2037–2048. doi: 10.1099/00207713-51-6-2037. [DOI] [PubMed] [Google Scholar]
- Grillasca MMA. Laca Chiapaneca: Ensayo de una singular aventura. Consejo Estatal para las Culturas y las Artes de Chiapas. Mexico: CONECULTA; 2007. p. 127. [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hall TA. Bioedit a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Haukka K, Lindström K, Young JPW. Tree phylogenetic groups of nodA and nifH genes in Sinorhizobium and Mesorhizobium isolates from leguminous trees growing in Africa and Latin America. Appl Environ Microbiol. 1998;64:419–426. doi: 10.1128/aem.64.2.419-426.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hungria MA, De O, Chueire LM, Coca RG, Megias M. Preliminary characterization of fast growing rhizobial strains isolated from soybean nodules in Brazil. Soil Biol Biochem. 2001;33:1349–1361. [Google Scholar]
- Khbaya B, Neyra M, Normand P, Zerhari K, Filali-Matouf A. Genetic diversity and phylogeny of rhizobia that nodulate Acacia spp. in Morocco assessed by analysis of rRNA gene. Appl Environ Microbiol. 1998;64:4912–4917. doi: 10.1128/aem.64.12.4912-4917.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen L, Curtis JM. The use of the angular transformation in biological assays. J Am Stat Assoc. 1947;42:282–296. doi: 10.1080/01621459.1947.10501927. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Laguerre G, Nour SM, Macheret V, Sanjuan J, Drouin P, Amarger N. Classification of rhizobia based on nodC and nifH gene analysis reveals a close phylogenetic relationship among Phaseolus vulgaris symbionts. Microbiology. 2001;147:981–993. doi: 10.1099/00221287-147-4-981. [DOI] [PubMed] [Google Scholar]
- Lloret L, Ormeño-Orrillo E, Rincón R, Martínez-Romero J, Rogel-Hernandez MA, Martínez-Romero E. Ensifer mexicanus sp. nov. a new species nodulating Acacia angustissima (Mill.) Kuntze in Mexico. Syst Appl Microbiol. 2007;30:280–290. doi: 10.1016/j.syapm.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Martínez-Romero E. Diversity of Rhizobium-Phaseolus symbiosis: overview and perspectives. Plant Soil. 2003;252:11–23. [Google Scholar]
- Martínez-Romero E, Rosenblueth M. Increased bean (Phaseolus vulgaris L.) nodulation competitiveness of genetically modified Rhizobium strains. Appl Environ Microbiol. 1990;56:2384–2388. doi: 10.1128/aem.56.8.2384-2388.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Romero E, Jarvis BDW. International committee on systematic bacteriology, subcommittee of the taxonomy of Agrobacterium and Rhizobium. Int J Syst Bacteriol. 1993;43:622. [Google Scholar]
- Martínez-Romero E, Segovia L, Mercante FM, Franco AA, Graham P, Pardo MA. Rhizobium tropici, a novel species nodulating Phaseolus vulgaris L. beans and Leucaena sp. trees. Int J Syst Bacteriol. 1991;41:417–426. doi: 10.1099/00207713-41-3-417. [DOI] [PubMed] [Google Scholar]
- Mnasri B, Mrabet M, Laguerre G, Aouani ME, Mhamdi R. Salt-tolerant rhizobia isolated from a Tunisian oasis that are highly effective for symbiotic N2-fixation with Phaseolus vulgaris constitute a novel biovar (bv. mediterranense) of Sinorhizobium meliloti. Arch Microbiol. 2007;187:79–85. doi: 10.1007/s00203-006-0173-x. [DOI] [PubMed] [Google Scholar]
- Mohamed SH, Smouni A, Neyra M, Kharchaf D, Filali-Maltouf A. Phenotypic characteristics of root-nodulating bacteria isolated from Acacia spp. grown in Libya. Plant Soil. 2000;224:171–183. [Google Scholar]
- Moreira FMS, Haukka K, Young JPW. Biodiversity of rhizobia isolated from a wide range of forest legumes in Brazil. Mol Ecol. 1998;7:889–895. doi: 10.1046/j.1365-294x.1998.00411.x. [DOI] [PubMed] [Google Scholar]
- Ochman H, Moran N. Genes lost and genes found: evolution of bacterial pathogenesis and symbiosis. Science. 2001;292:1096–1098. doi: 10.1126/science.1058543. [DOI] [PubMed] [Google Scholar]
- Odee DW, Haukka K, McInroy SG, Sprent JI, Sutherland JM, Young JPW. Genetic and symbiotic characterization of rhizobia isolated from tree and herbaceous legumes grown in soils from ecologically diverse sites in Kenya. Soil Biol Biochem. 2002;34:801–811. [Google Scholar]
- Ormeño-Orrillo E, Vinuesa P, Zúñiga-Dávila D, Martínez-Romero E. Molecular diversity of native bradyrhizobia isolated from lima bean (Phaseolus lunatus L.) in Peru. Syst Appl Microbiol. 2006;29:253–262. doi: 10.1016/j.syapm.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Posada D, Buckley TR. Model selection and model averaging in phylogenetics: advantages of AIC and Bayesian approaches over likelihood ratio tests. Syst Biol. 2004;53:793–808. doi: 10.1080/10635150490522304. [DOI] [PubMed] [Google Scholar]
- Räsänen AL, Sprent JI, Lindström K. Symbiotic properties of sinorhizobia isolated from Acacia and Prosopis nodules in Sudan and Senegal. Plant Soil. 2001;235:193–210. [Google Scholar]
- Rico-Arce ML, Bachean S. A taxonomic revision of Acaciella (Leguminosae, Mimosoideae) Anales del Jardín Bótanico de Madrid. 2006;63:189–244. [Google Scholar]
- Rincón-Rosales R, Gutiérrez-Miceli FA. Características biológicas de Acaciella angustissima (Mill.) Britton, Rose, en su hábitat natural y evaluación de su potencial cortical en Chiapas, México. Agrociencia. 2008;42:129–137. [Google Scholar]
- Rivas R, Velázquez E, Willems A, Vizcaino N, Subba-Rao NS, Mateos PF, Gillis M, Dazzo FB, Martínez-Molina E. A new species of Devosia that forms a unique nitrogen-fixing root-nodule symbiosis with the aquatic legume Neptunia natans (L.f.) Druce. Appl Environ Microb. 2002;68:5217–5222. doi: 10.1128/AEM.68.11.5217-5222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas R, Willems A, Palomo JL, García-Benavides P, Mateos PF, Martinez-Molina E, Gillis M, Velázquez E. Bradyrhizobium betae sp. nov., isolated from roots of Beta vulgaris affected by tumour-like deformations. Int J Syst Evol Microbiol. 2004;54:1271–1275. doi: 10.1099/ijs.0.02971-0. [DOI] [PubMed] [Google Scholar]
- Rzedowsky J. La vegetación de México. México: Editorial Limusa; 1978. p. 432. [Google Scholar]
- Saitou N, Nei M. The Neighbor-Joining method: a new method for reconstructing phylogenetics trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- SAS Institute Inc. SAS/STAT User's Guide. Version 6.0. 4th edn. Cary, NC: SAS Institute Inc.; 1989. [Google Scholar]
- Segovia L, Young JPW, Martínez-Romero E. Reclassification of American Rhizobium leguminosarum biovar phaseoli type I strains as Rhizobium etli sp. nov. Int J System Bacteriol. 1993;43:374–377. doi: 10.1099/00207713-43-2-374. [DOI] [PubMed] [Google Scholar]
- Silva C, Vinuesa P, Eguiarte LE, Souza V, Martínez-Romero E. Evolutionary genetics and biogeographic structure of Rhizobium gallicum sensu lato, a widely distributed bacterial symbiont of diverse legumes. Mol Ecol. 2005;14:4033–4050. doi: 10.1111/j.1365-294X.2005.02721.x. [DOI] [PubMed] [Google Scholar]
- Sprent JI. Nodulation in Legumes. London, UK: Royal Botanic Gardens, Kew; 2001. [Google Scholar]
- Stackebrandt E, Frederiksen W, Garrity GM, et al. Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int J Syst Evol Microbiol. 2002;52:1043–1047. doi: 10.1099/00207713-52-3-1043. [DOI] [PubMed] [Google Scholar]
- Sullivan JT, Ronson CW. Evolution of rhizobia by acquisition of a 500-kb symbiosis island that integrates into a phe-tRNA gene. Proc Natl Acad Sci. 1998;95:8985–8989. doi: 10.1073/pnas.95.9.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sy A, Giraud E, Jourand P, et al. Methylotrophic Methylobacterium bacteria nodulate and fix nitrogen in symbiosis with legumes. J Bacteriol. 2001;183:214–220. doi: 10.1128/JB.183.1.214-220.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan ZY, Wang ET, Peng GX, Zhu ME, Martínez-Romero E, Chen WX. Characterization of bacteria isolated from wild legumes in the north-western regions of China. Int J Syst Bacteriol. 1999;49:1457–1469. doi: 10.1099/00207713-49-4-1457. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo I, Lloret L, Martínez-Romero E. Sinorhizobium americanum sp. nov., a new Sinorhizobium species nodulating native Acacia spp. in Mexico. Syst Appl Microbiol. 2003;26:54–64. doi: 10.1078/072320203322337317. [DOI] [PubMed] [Google Scholar]
- Ueda T, Suga Y, Yahiro N, Matsuguchi T. Phylogeny of Sym plasmids of rhizobia by PCR-based sequencing of a nodC segment. J Bacteriol. 1995;177:468–472. doi: 10.1128/jb.177.2.468-472.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandamme P, Coenye T. Taxonomy of the genus Cupriavidus: a tale of lost and found. Int J Syst Evol Microbiol. 2004;54:2285–2289. doi: 10.1099/ijs.0.63247-0. [DOI] [PubMed] [Google Scholar]
- Van Rhijn P, Desair J, Vlassak K, Vanderleyden J. The NodD proteins of Rhizobium sp. strain BR816 differ in their interactions with coinducers and in their activities for nodulation of different host plants. Appl Environ Microbiol. 1994;60:3615–3623. doi: 10.1128/aem.60.10.3615-3623.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versalovic J, Schneider M, Brujin JF, Lupski JR. Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol Cell Biol. 1994;5:25–40. [Google Scholar]
- Vincent JM. A Manual for the Practical Study of Root Nodule Bacteria. Oxford: Blackwell Scientific; 1970. [Google Scholar]
- Vinuesa P, Leon-Barrios M, Silva C, Willems A, Jarabo-Lorenzo A, Perez-Galdona R, Werner D, Martinez-Romero E. Bradyrhizobium canariense sp. nov., an acid-tolerant endosymbiont that nodulates endemic genistoid legumes (Papilionoideae: Genisteae) from the Canary Islands, along with Bradyrhizobium japonicum bv. genistearum, Bradyrhizobium genospecies alpha and Bradyrhizobium genospecies beta. Int J Syst Evol Microbiol. 2005;55:569–575. doi: 10.1099/ijs.0.63292-0. [DOI] [PubMed] [Google Scholar]
- Wang ET, Martínez-Romero JM, Martínez-Romero E. Genetic diversity of rhizobia from Leucaena leucocephala nodules in Mexican soils. Mol Ecol. 1999;8:711–724. [Google Scholar]
- Wernegreen JJ, Riley MA. Comparison of the evolutionary dynamics of symbiotic and housekeeping loci: a case for the genetic coherence of rhizobial lineages. Mol Biol Evol. 1999;16:98–113. doi: 10.1093/oxfordjournals.molbev.a026041. [DOI] [PubMed] [Google Scholar]
- Williams ML, MacVean CM. Ethnococcidology: use of the giant margarodid, Llaveia spp. (Homoptera: Coccoidea: Margarodidae), by indigenous peoples of Mesoamerica in their culture, medicine and arts. Isr J Entomol. 1995;29:147–148. [Google Scholar]
- Wolde-Meskel E, Terefework Z, Lindström K, Frostegard A. Rhizobia nodulating African Acacia spp. and Sesbania sesban trees in Southern Ethiopian soils are metabolically and genomically diverse. Soil Biol Biochem. 2004;36:2013–2025. [Google Scholar]
- Xia X, Xie Z. DAMBE: software package for data analysis in molecular biology and evolution. J Hered. 2001;92:371–373. doi: 10.1093/jhered/92.4.371. [DOI] [PubMed] [Google Scholar]
- Young JM. The genus name Ensifer Casida 1982 takes priority over Sinorhizobium Chen et al. 1988, and Sinorhizobium morelense Wang et al. 2002 is a later synonym of Ensifer adhaerens Casida 1982. Is the combination “Sinorhizobium adhaerens” (Casida 1982) Willems et al. 2003 legitimate? Request for an opinion. Int J Syst Evol Microbiol. 2003;53:2107–2110. doi: 10.1099/ijs.0.02665-0. [DOI] [PubMed] [Google Scholar]
- Young JPW, Haukka KE. Diversity and phylogeny of rhizobia. New Phytol. 1996;133:87–94. [Google Scholar]
- Zurdo-Piñeiro JL, Velázquez E, Lorite MJ, Brelles-Mariño G, Schröder EC, Bedmar EJ, Mateos PF, Martinez-Molina E. Identification of fast-growing rhizobia nodulating tropical legumes from Puerto Rico as Rhizobium gallicum and Rhizobium tropici. Syst Appl Microbiol. 2004;4:469–477. doi: 10.1078/0723202041438437. [DOI] [PubMed] [Google Scholar]


