Abstract
Spliceosomes catalyze the maturation of precursor mRNAs from yeast to humans. Their catalytic core comprises three small nuclear RNAs (U2, U5 and U6) involved in substrate positioning and catalysis. It has been postulated, but never shown experimentally, that the U2/U6 complex adopts at least two conformations that reflect different activation states. We have used single-molecule fluorescence to probe the structural dynamics of a protein-free RNA complex modeling U2/U6 from yeast and mutants of highly conserved regions. Our data show the presence of at least three distinct conformations in equilibrium. The minimal folding pathway consists of a two-step process with an obligatory intermediate. The first step is strongly magnesium dependent and we provide evidence suggesting the second corresponds to the formation of the genetically conserved helix IB. Site-specific mutations in the highly conserved AGC triad and the U80 base in U6 suggest that the observed conformational dynamics correlate with residues that play an important role in splicing.
Keywords: Splicing, RNA folding, Structural dynamics, Single-Molecule Fluorescence
INTRODUCTION
Splicing is an essential step in eukaryotic precursor messenger RNA (pre-mRNA) maturation. Single nucleotide errors can be lethal to the cell, and anomalous pre-mRNA splicing has been linked to numerous cancers and neurodegenerative disorders.1,2 The spliceosome is a multi-megadalton protein-RNA complex consisting of five small nuclear RNAs (snRNAs U1, U2, U4, U5 and U6) and over 150 proteins.3 It catalyzes splicing and is a critical center for alternative splicing regulation.4 During spliceosomal assembly and catalysis, snRNAs associate with proteins to form ribonucleoprotein complexes (snRNPs), which bind to and dissociate from the pre-mRNA substrate in a highly orchestrated process yielding catalytically competent spliceosomes.5 Splicing consists of two sequential transesterification reactions that produce free lariat intron and ligate two exons into a mature mRNA.
Only U2, U5 and U6 are present in active spliceosomes, but evidence suggests U2 and U6 are directly involved in the first catalytic step. U2 and U6 form an extensive base pair network and directly bind the 5' splice site and the branch point, positioning them for the first reaction (Figure 1a).6,7 A highly conserved element of U5 has been shown to be dispensable for the first splicing step in vitro and in mammalian cell extracts.8,9 Splicing related catalysis has been reported in vitro using a protein-free human U2/U6 complex.10,11 Parallels between the structure and catalytic mechanism of the spliceosome and self-splicing group II introns support the idea of a common molecular ancestor and that the spliceosome is a ribozyme.12–14 However, recent crystal structures of a Prp8 fragment suggest that both protein and RNA are involved in catalysis.15
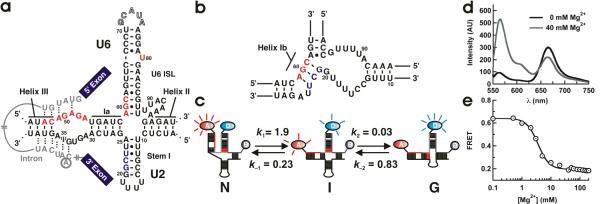
Figure 1.
(a) Secondary structure model of the spliceosomal snRNAs U2/U6 from S. cerevisiae with an intron bound.19 Highly conserved residues in U6 (ACAGAGA loop, AGC triad and U80) are highlighted in red. U2 residues involved in helix IB formation are highlighted in grey. The U6 ISL pentaloop (outlined residues G71–A75) was deleted to facilitate construct labeling. (b) Proposed structure of helix IB. (c) Proposed folding reaction pathway for the U2/U6 complex (rates measured at 40 mM Mg2+, units are s−1). At least three distinct conformations are observed for the U2/U6 snRNA complex: a high FRET conformation that resembles the 4-helix structure (N), a low FRET conformation that resembles the 3-helix structure with helix IB (G), and a previously unobserved folding intermediate (I). (d) Mg2+-induced conformational change in the U2/U6 spliceosomal complex. Fluorescence emission spectra of the fluorophore labeled U2/U6 complex, in the absence (black) and presence (grey) of Mg2+ ions. The donor fluorophore (Cy3) emits at 565 nm and the acceptor (Cy5) at 665 nm. Mg2+ ions decrease the acceptor emission and increase the donor emission, revealing the presence of a conformational change. (e) Calculated FRET ratio as a function of [Mg2+]. The line is a fit to a modified Hill equation (KMg = 3.0 ± 0.2 mM and n = 2.2 ± 0.2).
The U2/U6 structure has been a matter of debate over recent years. Early in vivo genetic studies support a 3-helix structure where the highly conserved AGC triad in U6 forms three base pairs with U2 (Figure 1b).16 Believed to form during catalytic activation, this structure has been proposed as a requirement for both splicing steps.17,18 More recently, NMR studies have shown that in absence of Mg2+ ions and proteins, the AGC triad forms intramolecular base pairs that extend the U6 internal stem loop (ISL, Figure 1a), forming a 4-helix structure.19,20 To reconcile these two structures, it has been postulated that each corresponds to a different spliceosomal activation state,21 yet any conformational change has remained undefined. To address this issue, we have used Förster Resonance Energy Transfer (FRET) and single-molecule fluorescence to characterize the structure and dynamics of a protein-free U2/U6 complex from yeast. Our results show that Mg2+ ions trigger a large amplitude conformational change that separates the ISL and the ACAGAGA loop (Figure 1). This two-step conformational change contains a previously unobserved obligatory intermediate, where only the first step is strongly Mg2+ dependent. Mutations in the highly conserved AGC triad show that helix IB forms only in the lowest FRET conformation, and correlate the observed structural dynamics in vitro with previously published mutations linked to activation of the second step in vivo.
RESULTS
Mg2+ induces a large amplitude conformational change
We have developed a labeling strategy that enables us to study the U2/U6 conformational dynamics by FRET and single-molecule fluorescence (Figure 1c and Supplementary Figure 1). We have incorporated Cy5 (FRET acceptor) at the U6 5' end (nucleotides 45–70) and a biotin at the U6 3' end (nucleotides 76–100) for surface immobilization. To assess the dynamics between the ISL and the ACAGAGA loop, we have deleted the U6 pentaloop (Figure 1a, outlined) and labeled U70 with Cy3 (FRET donor). The RNAs were labeled and purified as described (Methods).22 To confirm three-strand complex formation, we used non-denaturing gel electrophoresis (Supplementary Figure 1, single bands in lanes 4, 7 10 and 11 are reliable indicators of homogeneity), fluorescence and single-molecule microscopy (Supplementary Figure 2). The 4- and 3-helix structures are expected to yield high and low FRET ratios, respectively.16,19
We first characterized the folding behavior of our fluorophore-labeled U2/U6 complex using bulk solution FRET (Figure 1d) in the presence and absence of Mg2+ in standard buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl). Absent Mg2+ (black), the donor intensity is lower than the acceptor (FRET = 0.64), indicating that U2/U6 adopts an initial conformation where the ACAGAGA loop and the ISL are in close proximity, as suggested by the 4-helix structure.19 In 40 mM MgCl2, the donor intensity increases and the acceptor intensity decreases (FRET = 0.19), indicating that Mg2+ ions induce a conformational change separating the ACAGAGA loop and the ISL, as expected for the 3-helix struture.16 Titrating the observed FRET efficiency as a function of [Mg2+] (Figure 1e) reveals a cooperative decrease in FRET efficiency with increasing [Mg2+]. This conformational change occurs with a dissociation constant KMg = 3.3 ± 0.7 mM, near the physiological range (~1 mM),23 and a cooperativity coefficient n = 2.2 ± 0.2. These data indicate that U2/U6 acts as a Mg2+-dependent conformational switch. The high cooperativity coefficient likely reflects the presence of multiple Mg2+ binding sites, in agreement with previous studies that have linked several coordination sites near the highly conserved base U80 in U6, the ACG triad and the ACAGAGA loop (Figure 1) with spliceosomal activation.24–28
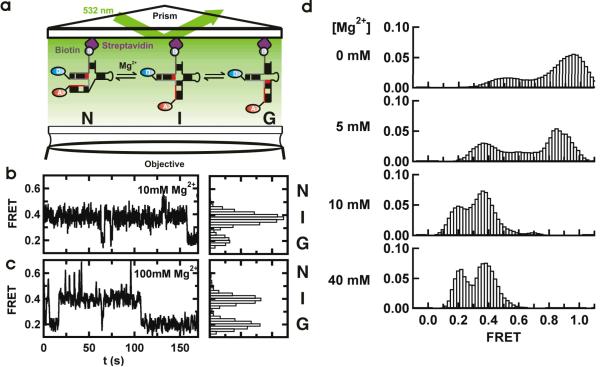
Single-molecule FRET reveals three dynamic conformations
Single-molecule FRET (smFRET) uncovers key structural and dynamic information otherwise hidden in ensemble-averaged experiments.29 Here, we have used smFRET to elucidate the folding reaction of the U2/U6 complex (Figure 2a). Characteristic FRET time trajectories of surface-immobilized U2/U6 in 10 and 100 mM Mg2+ are shown in Figures 2b and c. The observed FRET efficiency jumps randomly between three different values (~0.6, ~0.4 and ~0.2), revealing the presence of at least three distinct conformational states in dynamic equilibrium. Based on their FRET ratios, we initially assigned the high-FRET state to the 4-helix structure (N),19 the low-FRET state to the 3-helix structure (G) and the mid-FRET state to a previously unobserved folding intermediate (I, Figure 1c).
Figure 2.
Single-molecule FRET reveals a three-state folding pathway. (a) Schematic diagram of the single-molecule experiments. The RNA complex is surface-immobilized via a biotin-streptavidin bridge. The fluorophores are excited in a prism-based total internal reflection microscope. Fluorescence is collected by the objective and monitored with a CCD camera. (b and c). Single-molecule FRET time trajectories in 10 and 100 mM Mg2+. The different conformations (N, I and G) can be identified by their FRET ratios (~0.6, ~0.4 and 0.2) in the corresponding FRET histograms. (d) Mg2+ ions modulate the stability of the U2/U6 conformations. Each panel corresponds to an average smFRET histogram from >100 single-molecule trajectories as a function of [Mg2+], as indicated. In the absence of Mg2+, a high FRET state predominates the histogram. As [Mg2+] increases, the 0.4 and 0.2 FRET states become more populated.
FRET histograms from the trajectories (Figures 2b and c, right panels) reveal that I and G are the predominant states at high [Mg2+] (>10 mM), and N is only transiently populated. The high FRET state becomes more apparent at [Mg2+] < 10 mM (Figure 2d). Among the >3,200 transitions observed, only 40 (1.2%) go directly from N to G. In these instances, the dwell time in the I state may be shorter and than our 33 ms time resolution, which is direct evidence that I is an obligatory intermediate. Based on our single-molecule measurements, we propose a two-step folding pathway for the U2/U6 complex (Figure 1c).
Mg2+ ions influence the U2/U6 structural dynamics
To assess the effect of Mg2+ ions on the U2/U6 folding dynamics, we measured smFRET histograms between 0 and 40 mM [Mg2+] (Figure 2d). These histograms, built from over 100 trajectories each, show how Mg2+ ions modulate the folding dynamics of the U2/U6 complex. Absent Mg2+ (top panel), most molecules exist in the high FRET state, and only a small fraction adopts the intermediate FRET conformation. Increasing the [Mg2+] to 5 mM (second panel) shifts some of the high FRET population into the intermediate FRET. In 10 mM Mg2+ or higher (bottom panels), the high FRET state becomes more transiently populated and virtually disappears. Only brief excursions to the high FRET state are observed at these concentrations, and the U2/U6 molecules spend most of their time in the intermediate and low FRET states. The average FRET values of the single-molecule results overlay the ensemble-averaged results within the calculated standard deviation (Supplementary Figure 2f), indicating that surface-immobilization does not appreciably affect the U2/U6 folding dynamics.
Interestingly, the high FRET peak gradually shifts from ~0.95 at 0 mM Mg2+ to ~0.6 at 40 mM Mg2+. Such smooth changes in FRET distributions are usually an indication that multiple dynamic states are present in solution, but their dynamics are faster than our time resolution.30 This idea is further corroborated by the broad distributions with flat shoulders at 0 and 5 mM Mg2+ (Figure 2d). A similar behavior has been previously reported for the Diels-Alderase ribozyme.31 It is possible that in low [Mg2+] the AU-rich helix III is partially unwound (Figure 1a and Supplementary Figure 3), yielding the 0.95 FRET value. As [Mg2+] increases, helix III forms and effectively distances the U6 5' end from the ISL, yielding the 0.6 FRET value. The highly dynamic time trajectories observed below 5 mM Mg2+ (Supplementary Figure 3), as well as previously reported Mg2+ binding sites in this region,24,28 support this.
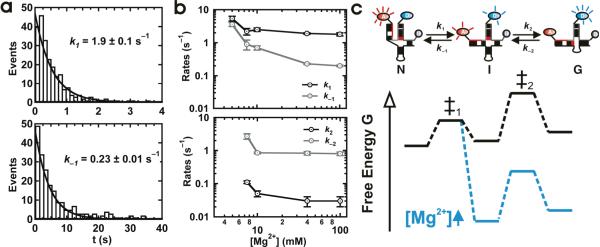
Dwell time analysis determines the rates of folding
We have determined the folding rate constants k1, k−1, k2 and k−2 (Figure 1c) for [Mg2+] = 5–100 mM. Below this range, dwell times were unidentifiable due to the highly dynamic time trajectories (see above). At 40 mM [Mg2+], the dwell time distributions can be readily fitted with single exponential decays to yield pseudo first order rate constants (Figure 3a, Supplementary Figure 4). Transitions out of the I state were divided into two categories depending on their final state: those that returned to N were used to determine k−1, while those that moved on to G were used to determine k2.13 The resulting rate constants and their Mg2+ dependence are shown in Figure 3b.
Figure 3.
Mg2+ dependence of the folding rate constants for the U2/U6 complex. (a) Dwell time distributions in the N (top) and I (bottom) conformations in 40 mM Mg2+. The distributions are fit to single exponential decays to yield the pseudo first order rates k1 and k−1, respectively. (b) Mg2+ dependence of k1 and k−1 (top) and k2 and k−2 (bottom). (c) Schematic diagram of the folding potential energy surface for the protein-free U2/U6 spliceosomal complex from yeast. In the absence of Mg2+ (black), the high FRET state (N) is the most stable. Upon addition of Mg2+ ions (green), the previously unobserved mid-FRET intermediate (I) becomes more stable, while N and the first transition state (‡1) remain approximately constant. This is evidenced by the large decrease in k−1 and not in k1. Meanwhile, the low FRET state (G) and the second transition state (‡2) are also similarly stabilized, as evidenced by the mild Mg2+ dependence of k2 and k−2.
These data explain how Mg2+ ions affect the U2/U6 folding dynamics and how to interpret them in terms of a folding potential energy surface comprising three minima (N, I and G) separated by two transition states (Figure 3c). Between 5 and 100 mM Mg2+, k1 decreases 3-fold while k−1 decreases 18-fold. The large decrease in k−1 indicates that Mg2+ ions preferentially stabilize I relative to the first transition state, leaving N approximately unchanged. Between 7 and 100 mM Mg2+, k2 and k−2 decrease only 4- and 3-fold, respectively, indicating that I, G and the second transition state are similarly stabilized by Mg2+ ions.
Helix IB forms in the low FRET state
To test for the presence of helix IB in the low FRET conformation (G), we designed a six-fold mutant that prevents helix IB formation while maintaining the stability of the extended ISL (Figure 4a), because the identity of junction-closing base pairs can affect the junction stability and dynamics.32 An ensemble-averaged Mg2+ titration shows that these mutations do not affect KMg, but the observed FRET ratio at high [Mg2+] is higher than the wild-type (Supplementary Figure 5). The absence of the 0.2 FRET state in the single-molecule trajectories and corresponding FRET histogram (Figure 4a) support our initial assignments. Dwell time analysis of this mutant shows that k1 and k−1 are within two-fold of the wild-type (Supplementary Figure 5c), showing that this mutation does not affect the first step of folding.
Figure 4.
Mutations in the AGC triad (red) show that helix IB forms only in the low FRET state. (a) Single-molecule trajectory of a six-fold mutant in the U6 snRNA with a base-flipped AGC triad that prevents formation of helix IB. The low FRET state is never observed in the trajectories or in the corresponding FRET histogram (right). (b) Single-molecule trajectory of an A91G mutant in the U6 snRNA previously shown to stabilize the formation of helix IB. Excursions to the low FRET state last longer, and the corresponding peak in the FRET histogram (right) is larger. (c) Single-molecule trajectory of an A59C mutant in the U6 snRNA that blocks the second step of splicing in vivo. Neither the 0.4 nor 0.2 FRET states are observed in the corresponding trajectories. The FRET histogram (right) reveals the presence of a new state with intermediate FRET (~0.3). (d) A U23G mutation in the U2 snRNA rescues formation of helix IB in the presence of the U6 A59C mutation. The single-molecule trajectories and corresponding histograms show the recovery of the 0.4 – 0.2 FRET dynamics in vitro. All measured in 50 mM Tris-HCl, pH 7.5, 100 mM NaCl and 40 mM MgCl2. The structures on the left are intended only to show the location of the mutations.
Helix IB formation can be favored by the U6 mutation A91G, extending helix II by one base pair and stabilizing it relative to stem I in U2 (Figures 1a and 3b, Halsig, J., Sashital, D.G. and Butcher, S.E., University of Wisconsin, Madison, personal communication). The trajectories clearly show that excursions to the low FRET state last longer than in the wild-type, and the corresponding smFRET histogram reveals a larger low FRET peak than the wild-type. Dwell time analysis shows that k1, k−1 and k−2 are very similar to the wild-type, but k2 is 60-fold faster, in agreement with these observations (Supplementary Figures 5d and e).
Taken together, these results show that the second folding step corresponds to a spontaneous intramolecular junction migration from I to G. The viability of the six-fold mutant has never been tested in vivo, but mutations that block helix IB formation can be lethal for the cell,16,17 thus raising the interesting possibility that the observed dynamics in vitro are linked to spliceosomal activation in vivo.
The structural dynamics correlate with splicing activation
To test the role of the observed dynamics in spliceosomal activation, we used previously published mutations. The U6 mutation A59C (Figure 3c) has been shown to prevent helix IB formation and block the spliceosome between the two splicing steps in vivo.17 The U2 compensatory mutation U23G (Figure 3d) restores base-pairing in helix IB and rescues the second step of splicing.
We have tested the effect of these mutations on the structural dynamics of U2/U6 in 40 mM Mg2+. The trajectories for A59C show an effect on both the junction structure and its dynamics (Figure 3c). The low FRET states (I and G) disappear, and a new state (~0.30 FRET) replaces them. Two possible scenarios likely explain these results. The junction mutation may disrupt the junction structure, trapping it in an intermediate conformation with the U6 5' end pointing in an intermediate direction, explaining the observed FRET ratio. Alternatively, the mutation may accelerate the junction dynamics faster than our time resolution, making the two FRET states appear as single state with an average FRET value between 0.2 and 0.4.
The trajectories for the compensatory mutation clearly show the rescue of the low FRET state and the structural dynamics (Figure 3d), in agreement with the in vivo experiments. Interestingly, the FRET histogram shows that the intermediate state FRET ratio is 0.32, indicating that the disrupted junction structure, rather than the averaged fast dynamics, is the most likely explanation for the A59C mutant intermediate FRET ratio. Additionally, bulk titrations show that KMg of A59C is nearly 4-fold higher than the wild-type, and the high [Mg2+] FRET ratio is higher than the wild-type (Supplementary Figure 5), supporting the single-molecule results. The compensatory mutant KMg and FRET ratio are in excellent agreement with the wild-type.
These results link the observed structural dynamics of the U2/U6 complex to a conformational rearrangement prior to the second splicing step, suggesting that junction dynamics may contribute to successful splicing in vivo.
U80 is involved in high FRET state stabilization
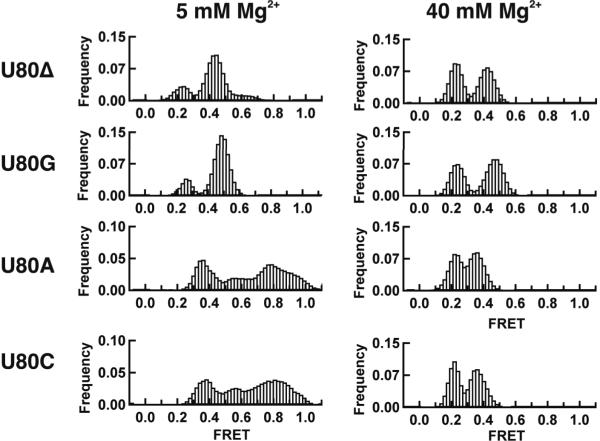
The U6 base U80 is highly conserved and has been involved in binding a metal ion important for catalysis.26 To assess its role in the U2/U6 folding dynamics, we deleted and mutated it to A, G or C (Figure 5 and Supplementary Figure 6). The U80G mutation and deletion are lethal in yeast, while U80A and C are viable.33 The Mg2+ binding affinity for U80A and C mutants shows excellent agreement with the wild-type, while the U80G mutation and deletion reveal two significant differences (Supplementary Figure 6). The initial, low [Mg2+], FRET value is lower, indicating the absence of the high FRET conformation (N), and the cooperativity coefficient decreases to 1.1 ± 0.1, indicating the loss of a Mg2+ ion binding site. All titrations converge to similar high [Mg2+], low FRET values, and all the KMg are within 2-fold of the wild-type. The corresponding 5 mM Mg2+ smFRET histograms (Figure 5) show that the broad distribution characteristic of the high FRET conformation disappears for the U80G mutation and deletion, while it remains present in both the U80A and C mutants. In 40 mM Mg2+, however, the histograms are similar for all mutants (Figure 5). The centers of the intermediate FRET distributions for the U80 deletion and U80G mutant shift to 0.43 and 0.47, respectively, indicating that the ISL bends differently in these mutants. A comparison of the NMR structures of the wild-type ISL and the U80G mutant shows a ~12 Å displacement of U70 (location of Cy3), supporting this idea.33,34
Figure 5.
smFRET histograms for the U80 mutations (U80Δ, U80G, U80A and U80C) in 5 and 40 mM Mg2+, as indicated. The U80 deletion and G mutation result in the disappearance of the high FRET conformation in the low [Mg2+] histogram, but do not affect the intermediate and low FRET conformations in the high [Mg2+] histogram. The smFRET histograms for the U80A U80C are similar to the wild-type. These results indicate that U80 is involved in a functionally important tertiary contact that stabilizes the high FRET conformation.
One possible explanation for these results is that U80 is involved in a key tertiary contact stabilizing the high FRET conformation and not the intermediate and low FRET states. U80 deletion prevents the formation of this contact, destabilizing the high FRET conformation and causing its disappearance from the histograms. Similarly, U80G results in the C67–G80 base pair, preventing the putative tertiary contact formation. Conversely, U80A and C do not disrupt the C67–A79 wobble pair, allowing the tertiary contact to take pace, and the high FRET distribution is recovered in the histograms. A likely partner for this interaction may be located in the ACAGAGA loop. To test this, we mutated the U2 bases 31–36 to replace the ACAGAGA loop with a double stranded helix (Supplementary Figure 7). The corresponding bulk titration and single-molecule experiments in 5 mM Mg2+ also show the disappearance of the high FRET conformation. These results show that the high FRET state (N) is stabilized by a tertiary interaction involving U80 and the ACAGAGA loop. The presence or absence of the high FRET conformation in all these mutants correlates well with viable or lethal mutations in yeast, supporting the idea that the in vitro dynamics may be important for splicing in vivo.
DISCUSSION
The U2/U6 complex lies at the heart of the catalytic core of the eukaryotic spliceosome, but its structure has been highly debated. Here, we have used smFRET to show that this important RNA complex acts as a Mg2+-dependent conformational switch that can adopt at least three distinct conformations. This supports the hypothesis that U2/U6 adopts multiple conformations at various splicing stages,21 which may or may not reflect the presence of unique active sites for each step.35
In the high FRET conformation (N), the AGC triad extends the ISL as predicted for the 4-helix structure,19 and the ACAGAGA loop and U80 in the ISL are brought in close proximity by a stabilizing tertiary contact. Because of the expected relationship between these three regions during the first splicing step, it is possible that this conformation resembles the active conformation. The recent Group II intron ribozyme crystal structure provides a framework for this scenario (Supplementary Figure 8)14. In this structure, C358G359C360 (proposed equivalents of the AGC triad) are clearly base paired with G383U384G385, extending domain 5 (proposed equivalent of the U6 ISL). The C360–G383 base pair forms a triple with C377 (proposed equivalent of U80), which in turn stacks on G288 from J2/3 (proposed equivalent of the ACAGAGA loop). G288 also forms a triple with G359 and U384. Our U80 mutant results support the idea that these interactions also take place in the high FRET conformation of the U2/U6 complex. In the crystal structure, O2 in U80 is a hydrogen bond acceptor in the base triple. The same O2 in U80C and N3 or N7 in U80A could play a similar role in the mutants.
In the mid FRET conformation, the AGC triad still forms the extended ISL (4-helix structure),19 but the tertiary contact between the ACAGAGA loop and U80 is no longer formed, as its stability does not depend on the U80 mutations. Our current data, however, do not allow us to conclude whether this conformation plays a more direct role in splicing.
In the low FRET conformation, a junction migration takes place, resulting in the formation of helix IB (3-helix junction).16 The U80 mutants do not affect its stability, indicating that the ACAGAGA loop and U80 are not in direct contact. A recent paper has shown that helix I is important for both steps of splicing and that a conformational rearrangement must precede each step.18 This result is in agreement with our mutational data that link the formation of the low FRET conformation to an activating step between the first and second splicing steps. It is tempting to hypothesize that the observed structural dynamics also play an activating role in the first splicing step, however the mutations tested here do not provide enough support for this conclusion.
The role of Mg2+ ions at different stages of spliceosomal activation and catalysis has been established by experiments using phosphorothioate substituted RNAs.25,26,36,37 A key sensitive nucleotide is the phylogenetically conserved U80, which suggests either that this base plays a direct role in catalysis or is at least involved in a closely related step.26 Phosphorothioate substitutions in the 5' and 3' splice sites also involve Mg2+ ions in both splicing steps by activating the nuc-leophilic attack and stabilizing the leaving groups,25,36 and suggest the presence of a Mg2+-dependent conformational change specific to the second step of splicing.36,37 We propose that this Mg2+-induced conformational change corresponds to the one observed here. Based on this, however, one might expect an inverse effect, whereby Mg2+ ions would preferentially stabilize the high FRET structure. In the presence of essentially a constant supply of Mg2+ ions in vivo, it is possible that the role of at least some of the spliceosomal proteins is to adjust the relative stability of these conformations to time the U2/U6 structural dynamics for accurate and efficient splicing.
We removed the highly conserved U6 ISL loop, but in accordance with a previous genetic study, in which the ISL was extended by one base pair, we do not expect this loop to significantly affect the structure of U2/U6 in vitro.38 Nonetheless, we have tested the effect of this deletion on the U2/U6 structural dynamics using a two-strand construct (Supplementary Figure 9). Our results show that the two constructs behave almost identically. The three-strand construct, however, offers greater efficiency of synthesis, labeling and purification, and will facilitate future mutational studies.
We have recently elucidated the folding pathway of a self-splicing group II intron ribozyme, which has structural and catalytic similarities to the spliceosome.13 Its pathway also involves obligatory intermediates and is dominated by a Mg2+ capture step that activates junction dynamics for catalysis. The similarities between the U2/U6 structural dynamics and those of the group II intron ribozyme now expand the parallels between these two enzymes and suggest the existence of evolutionarily conserved structural dynamics.
ONLINE METHODS
RNA Preparation and Purification
There are 3 strands of RNA forming the U2-U6 complex in our construct (Supplementary Figure 1): U6-1 (5'-Cy5-AUA CAG AGA UGA UCA GCA GUU CCC CU-Cy3-3'), U6-2 (5'-AGG AUG AAC CGU UUU ACA AAG AGA UUU AU-Biotin-3') and U2 (5'-AUC UCU UUG CCU UUU GGC UUA GAU CAA GUG UAG UAU-3'). We purchased all the RNA samples from Keck Foundation Biotechnology Resource Laboratory at Yale University School of Medicine (New Haven, CT), and purified as described.39 These RNA samples have 2'OH protection groups that we removed by a 2'OH deprotection reaction according to the manufacture's protocol. We purified the deprotected RNA by denaturing gel electrophoresis (20% polyacrylamide and 8 M urea) and diffusion elution with elution buffer (0.5 M NH4OAc and 0.1 mM EDTA) at 4°C overnight. We further purified the RNA by chloroform extraction, ethanol precipitation, and C8 reverse-phase HPLC. The manufacturer labeled the 5' end of U6-1 with Cy5 during the RNA synthesis and attached a C7 amino linker to the 3' end, enabling the labeling of a Cy3 to 3' end for our FRET studies. During the 3'-labeling reaction, we mixed the Cy5-labeled RNA with Cy3 dye (GE Healthcare) in labeling buffer (100 mM Sodium tetraborate, pH 8.5). We kept this reaction mixture overnight at room temperature. We purified the doubly labeled samples by ethanol precipitation and reverse phase HPLC. We determined the RNA concentrations by UV-Vis absorbance at 260 nm.
Steady-State FRET
We carried out the steady-state FRET measurements of the doubly labeled U2–U6 complex in a spectrofluorometer (Cary Eclipse, Varian Inc., Palo Alto, CA). We excited Cy3 at 550nm (10 nm bandwidth) and measured the Cy3 and Cy5 emission intensities (ICy3 and ICy5) at 565 nm and 665 nm (5 nm bandwidth), respectively. We calculated the relative FRET efficiencies as FRET = ICy5/(ICy3 + ICy5). We first denatured a 130 μL solution with 250 nM U6-1, 500 nM U6-2 and 1 μM U2 in standard buffer (50 mM Tris-HCl, pH 7.5, 100mM NaCl and 25mM DTT) by heating at 90 °C for 2 min and then slowly annealed it at room temperature for 20 minutes. After this we scanned the RNA-only solution. We added manually 10 μL aliquots of varying concentrations of MgCl2 to the RNA. After 2 min equilibration, we measured the final FRET value. We obtained the Mg2+ dissociation constant (KMg) and cooperativity coefficients (n) by plotting FRET as a function of Mg2+ concentration and fitting to the modified Hill equation:
Single-molecule FRET
Single molecule experiments were performed as described.22 We annealed the three RNA strands (Fig. 1a) at 1 μM concentration in standard buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl) and variable [Mg2+]. We heated a 10 μL solution to 90°C for 45 sec before cooling at room temperature over 15 min. We diluted the annealed biotinylated and fluorophore-labeled complex to 25 – 50 pM concentration and bound the RNA to a streptavidin-coated quartz slide surface via the biotin-streptavidin bridge to generate a surface density of ~0.1 molecules per μm2. We The excited the donor fluorophore in a home-built total internal reflection microscope with a laser (532 nm, 3 mW, Spectra-Physics Excelsior, Mountain View, CA). We separated the donor and acceptor emission using appropriate dichroic mirrors (610DCXR, Chroma, Rockingham, VT), and detected it as two side-by-side images on a back-illuminated electron-multiplied CCD camera (Andor I-Xon, South Windsor, CT). We measured the individual donor (ID) and acceptor (IA) intensities by integration of their relative spot intensities. We used the donor (ID) and acceptor (IA) fluorescence signals of optically resolved single molecules (characterized by single-step photobleaching) to calculate the FRET ratio as IA/(IA + ID), and followed it in real time for each molecule. The resulting FRET time trajectories are essentially recorded movies of the molecular motion of the U2/U6 complex, which enable us to study its conformational dynamics. We performed measurements under variable [Mg2+] (0 – 100 mM) at 22°C, with an oxygen scavenging system consisting of 10% (wt/vol) glucose, 2% (vol/vol) 2-mercaptoethanol, 750 μg ml−1 glucose oxidase, and 90 μg ml−1 catalase to reduce photobleaching, with and without trolox (Sigma). We then calculated the dwell times of each folding event, constructed histograms, and determined the folding rate constants, as described.13,22
Fluorescence Based Gel Mobility Assay40
We performed 15% non-denaturing polyacrylamide (29:1acrylamide: bisacrylamide ratio) gel electrophoresis in 20 mM NaOAc, 20 mM Tris-acetic acid (pH6.5) and 40 mM Mg(OAc)2 using low-fluorescence glass plates. We heated 10 pmol doubly labeled RNA samples at 90 °C for 45 seconds and cooled down to room temperature in standard buffer. We equilibrated the acrylamide gel was for 15 minutes at 4 °C before loading. immediately after loading the samples on the gel, we applied a constant electric field (6 V cm–1).After electrophoresis for 8 hrs at 4 °C, we scanned the gel in a Typhoon 9210 Variable Model Imager (GE Healthcare), and analyzed with ImageQuant software (Amershan Bioscience). For color calibration purposes we used RNAs labeled with only Cy3 and Cy5. We used the Fluorsep software (Amershan Bioscience) to overlay the Cy3 and Cy5 gel images. Cy3 and Cy5 emissions appear as green and red, respectively.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Sam Butcher and David Brow for many helpful and stimulating discussions and for commenting the manuscript. This work was supported by the NIH (R01 GM085116) and an NSF CAREER award to DR (MCB-0747285).
REFERENCES
- 1.Kalnina Z, Zayakin P, Silina K, Line A. Alterations of pre-mRNA splicing in cancer. Genes Chromosomes Cancer. 2005;42:342–57. doi: 10.1002/gcc.20156. [DOI] [PubMed] [Google Scholar]
- 2.Licatalosi DD, Darnell RB. Splicing regulation in neurologic disease. Neuron. 2006;52:93–101. doi: 10.1016/j.neuron.2006.09.017. [DOI] [PubMed] [Google Scholar]
- 3.Stark H, Luhrmann R. Cryo-electron microscopy of spliceosomal components. Annu Rev Biophys Biomol Struct. 2006;35:435–57. doi: 10.1146/annurev.biophys.35.040405.101953. [DOI] [PubMed] [Google Scholar]
- 4.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 5.Brow DA. Allosteric cascade of spliceosome activation. Annu Rev Genet. 2002;36:333–60. doi: 10.1146/annurev.genet.36.043002.091635. [DOI] [PubMed] [Google Scholar]
- 6.Parker R, Siliciano PG, Guthrie C. Recognition of the TACTAAC box during mRNA splicing in yeast involves base pairing to the U2-like snRNA. Cell. 1987;49:229–39. doi: 10.1016/0092-8674(87)90564-2. [DOI] [PubMed] [Google Scholar]
- 7.Lesser CF, Guthrie C. Mutations in U6 snRNA that alter splice site specificity: implications for the active site. Science. 1993;262:1982–8. doi: 10.1126/science.8266093. [DOI] [PubMed] [Google Scholar]
- 8.O'Keefe RT, Norman C, Newman AJ. The invariant U5 snRNA loop 1 sequence is dispensable for the first catalytic step of pre-mRNA splicing in yeast. Cell. 1996;86:679–89. doi: 10.1016/s0092-8674(00)80140-3. [DOI] [PubMed] [Google Scholar]
- 9.Segault V, et al. Conserved loop I of U5 small nuclear RNA is dispensable for both catalytic steps of pre-mRNA splicing in HeLa nuclear extracts. Mol Cell Biol. 1999;19:2782–90. doi: 10.1128/mcb.19.4.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valadkhan S, Manley JL. Splicing-related catalysis by protein-free snRNAs. Nature. 2001;413:701–7. doi: 10.1038/35099500. [DOI] [PubMed] [Google Scholar]
- 11.Valadkhan S, Mohammadi A, Jaladat Y, Geisler S. Protein-free small nuclear RNAs catalyze a two-step splicing reaction. Proc Natl Acad Sci U S A. 2009;106:11901–6. doi: 10.1073/pnas.0902020106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sontheimer EJ, Gordon PM, Piccirilli JA. Metal ion catalysis during group II intron self-splicing: parallels with the spliceosome. Genes Dev. 1999;13:1729–41. doi: 10.1101/gad.13.13.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steiner M, Karunatilaka KS, Sigel RK, Rueda D. Single-molecule studies of group II intron ribozymes. Proc Natl Acad Sci U S A. 2008;105:13853–8. doi: 10.1073/pnas.0804034105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toor N, Keating KS, Taylor SD, Pyle AM. Crystal structure of a self-spliced group II intron. Science. 2008;320:77–82. doi: 10.1126/science.1153803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abelson J. Is the spliceosome a ribonucleoprotein enzyme? Nat Struct Mol Biol. 2008;15:1235–7. doi: 10.1038/nsmb1208-1235. [DOI] [PubMed] [Google Scholar]
- 16.Madhani HD, Guthrie C. A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell. 1992;71:803–17. doi: 10.1016/0092-8674(92)90556-r. [DOI] [PubMed] [Google Scholar]
- 17.Hilliker AK, Staley JP. Multiple functions for the invariant AGC triad of U6 snRNA. RNA. 2004;10:921–8. doi: 10.1261/rna.7310704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mefford MA, Staley JP. Evidence that U2/U6 helix I promotes both catalytic steps of pre-mRNA splicing and rearranges in between these steps. RNA. 2009;15:1386–97. doi: 10.1261/rna.1582609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sashital DG, Cornilescu G, McManus CJ, Brow DA, Butcher SE. U2-U6 RNA folding reveals a group II intron-like domain and a four-helix junction. Nat Struct Mol Biol. 2004;11:1237–42. doi: 10.1038/nsmb863. [DOI] [PubMed] [Google Scholar]
- 20.Sun JS, Manley JL. A novel U2-U6 snRNA structure is necessary for mammalian mRNA splicing. Genes Dev. 1995;9:843–54. doi: 10.1101/gad.9.7.843. [DOI] [PubMed] [Google Scholar]
- 21.Rhode BM, Hartmuth K, Westhof E, Luhrmann R. Proximity of conserved U6 and U2 snRNA elements to the 5' splice site region in activated spliceosomes. EMBO J. 2006;25:2475–86. doi: 10.1038/sj.emboj.7601134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao R, Rueda D. RNA folding dynamics by single-molecule fluorescence resonance energy transfer. Methods. 2009 doi: 10.1016/j.ymeth.2009.04.017. doi:10.1016/j.ymeth.2009.04.017. [DOI] [PubMed] [Google Scholar]
- 23.Hershey JWB, Merrick WC. The Pathway and Mechanism of Initiation of Protein Synthesis. In: Sonenberg N, Hershey JWB, Mathews MB, editors. Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 2000. [Google Scholar]
- 24.Fabrizio P, Abelson J. Thiophosphates in yeast U6 snRNA specifically affect pre-mRNA splicing in vitro. Nucleic Acids Res. 1992;20:3659–64. doi: 10.1093/nar/20.14.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sontheimer EJ, Sun S, Piccirilli JA. Metal ion catalysis during splicing of premessenger RNA. Nature. 1997;388:801–5. doi: 10.1038/42068. [DOI] [PubMed] [Google Scholar]
- 26.Yean SL, Wuenschell G, Termini J, Lin RJ. Metal-ion coordination by U6 small nuclear RNA contributes to catalysis in the spliceosome. Nature. 2000;408:881–4. doi: 10.1038/35048617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huppler A, Nikstad LJ, Allmann AM, Brow DA, Butcher SE. Metal binding and base ionization in the U6 RNA intramolecular stem-loop structure. Nat Struct Biol. 2002;9:431–5. doi: 10.1038/nsb800. [DOI] [PubMed] [Google Scholar]
- 28.Yuan F, et al. Use of a novel Forster resonance energy transfer method to identify locations of site-bound metal ions in the U2–U6 snRNA complex. Nucleic Acids Res. 2007;35:2833–45. doi: 10.1093/nar/gkm134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aleman EA, Lamichhane R, Rueda D. Exploring RNA folding one molecule at a time. Curr Opin Chem Biol. 2008 doi: 10.1016/j.cbpa.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 30.Nir E, et al. Shot-noise limited single-molecule FRET histograms: comparison between theory and experiments. J Phys Chem B. 2006;110:22103–24. doi: 10.1021/jp063483n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kobitski AY, Nierth A, Helm M, Jaschke A, Nienhaus GU. Mg2+−dependent folding of a Diels-Alderase ribozyme probed by single-molecule FRET analysis. Nucleic Acids Res. 2007;35:2047–59. doi: 10.1093/nar/gkm072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McKinney SA, Freeman AD, Lilley DM, Ha T. Observing spontaneous branch migration of Holliday junctions one step at a time. Proc Natl Acad Sci U S A. 2005;102:5715–20. doi: 10.1073/pnas.0409328102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sashital DG, Allmann AM, Van Doren SR, Butcher SE. Structural basis for a lethal mutation in U6 RNA. Biochemistry. 2003;42:1470–7. doi: 10.1021/bi027137h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reiter NJ, Nikstad LJ, Allmann AM, Johnson RJ, Butcher SE. Structure of the U6 RNA intramolecular stem-loop harboring an S(P)-phosphorothioate modification. RNA. 2003;9:533–42. doi: 10.1261/rna.2199103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moore MJ, Sharp PA. Evidence for two active sites in the spliceosome provided by stereochemistry of pre-mRNA splicing. Nature. 1993;365:364–8. doi: 10.1038/365364a0. [DOI] [PubMed] [Google Scholar]
- 36.Gordon PM, Sontheimer EJ, Piccirilli JA. Metal ion catalysis during the exon-ligation step of nuclear pre-mRNA splicing: extending the parallels between the spliceosome and group II introns. RNA. 2000;6:199–205. doi: 10.1017/s1355838200992069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sontheimer EJ. The spliceosome shows its metal. Nat Struct Biol. 2001;8:11–3. doi: 10.1038/82979. [DOI] [PubMed] [Google Scholar]
- 38.Sun JS, Manley JL. The human U6 snRNA intramolecular helix: structural constraints and lack of sequence specificity. RNA. 1997;3:514–26. [PMC free article] [PubMed] [Google Scholar]
- 39.Rueda D, Walter NG. Fluorescent energy transfer readout of an aptazyme-based biosensor. Methods Mol Biol. 2006;335:289–310. doi: 10.1385/1-59745-069-3:289. [DOI] [PubMed] [Google Scholar]
- 40.Rueda D, Wick K, McDowell SE, Walter NG. Diffusely bound Mg2+ ions slightly reorient stems I and II of the hammerhead ribozyme to increase the probability of formation of the catalytic core. Biochemistry. 2003;42:9924–36. doi: 10.1021/bi0347757. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.