Abstract
How naïve CD4+ T cells commit to the T helper type 2 (TH2) lineage is poorly understood. Here we show that the basic helix-loop-helix transcription factor Dec2 is selectively expressed in TH2 cells. CD4+ T cells from Dec2-deficient mice exhibits defective TH2 differentiation in vitro and in vivo in an asthma model and in response to challenge with a parasite antigen. Dec2 promotes interleukin 4 (IL-4), IL-5 and IL-13 expression during early TH2 differentiation, and directly binds to and activates transcription of the Junb and Gata3 genes. As GATA3 induces Dec2 expression, these findings also indicate a feed-forward regulatory circuit during TH2 differentiation.
INTRODUCTION
CD4+ T helper (TH) cells are the central regulators of adaptive immunity and allergic diseases. Upon activation by antigen-presenting cells (APCs), naïve CD4+ precursors undergo clonal expansion and functional differentiation into cytokine-secreting effector cells. TH1 cells make interferon (IFN)-γ and promote antigen presentation and cellular immunity 1, 2. TH2 cells produce interleukin (IL)-4, -5 and -13, which together regulate anti-parasite and allergic responses 1, 2. TH-17 cells, distinct from TH1 and TH2 cells, secret IL-17, IL-17F, IL-22 and IL-21 and mediate tissue inflammation 3, 4.
TH effector differentiation is determined by the cytokine environment, which ultimately directs the expression of lineage-specific transcription factors 1, 2. During TH2 cell polarization, IL-4 is essential, as it activates STAT6, which induces the expression of the transcription factor GATA3 5. Naive CD4+ T cells start to express IL-4 mRNA 24 hours after stimulation of the T cell receptor (TCR) and costimulatory receptors; IL-4 mRNA expression increases even further at 48 hours post-stimulation6. The costimulatory receptor ICOS and the cytokine IL-25 are also important for early IL-4 production, as they promote expression of the transcription factors NFATc1 and JunB (http://www.signaling-gateway.org/molecule/query?afcsid=A001301)7–9. In addition to IL-4, IL-2 is involved in the initiation of TH2 differentiation 10, 11. When stimulated with low concentrations of antigenic peptide, TH cells produced IL-4, and this IL-4 production is regulated by IL-2–dependent STAT5 phosphorylation and IL-4–independent early GATA-3 expression 11. IL-2 also facilitates early TH2 differentiation by inducing expression of the IL-4 receptor α-chain in a manner dependent on STAT5 but independent of IL-4 12.
However, the precise mechanism controlling initial TH2 lineage commitment is not well understood. Here we show that Dec2 (also called Sharp1, Bhlhb3; Bhlhb2l or Bhlhe41) 13, 14, a basic helix-loop-helix transcription factor previously implicated in regulating circadian rhythm 15 and differentiation of a range of cell types 16, 17, is selectively expressed in the TH2 subset among all tested T helper cell subsets. We found that Dec2 promotes expression of GATA3 and JunB, the latter of which induces expression of IL-4 5, 18, 19 and IL-2 20–22 to promote TH2 differentiation.
Results
Regulation of Dec2 expression in TH2 cells
In a gene expression microarray (B.S. and Z.L., data not shown, and Z. Li, K. Mao, J. Zou, Y. Wang, Z. Tao, G. Lin, L.Tian, Y. Ji, X. Wu, X. Zhu, S. Sun, C. Xiang, and W. Chen, personal communication), expression of Dec2 mRNA was greatly elevated in TH2 cells compared to TH1 cells (13.5 fold). We thus further assessed the expression of Dec2 mRNA in TH1, TH2, TH-17, and inducible regulatory T (iTreg) cell subsets as well as naïve CD4+ and CD8+ T cells. CD4+ T cells purified from OT-II TCR transgenic mice were cultured in TH1, TH2, TH-17 or iTreg cell-polarizing conditions in vitro. As measured via quantitative real-time RT-PCR, Dec2 mRNA was highly expressed in TH2 cells but not in the other types of T cells (Fig. 1a). Dec2 mRNA was also expressed in non-T lineage immune cells, including plasmacytoid dendritic cells (DCs) and eosinophils (Supplementary Fig. 1).
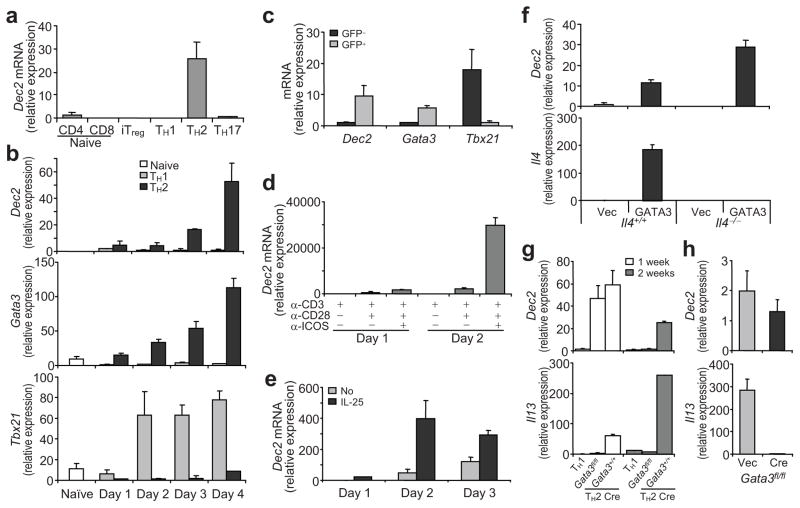
Fig. 1. Expression and regulation of Dec2.
(a) Dec2 mRNA expression in TH1, TH2, TH-17 or iTreg cells was measured by quantitative RT-PCR. Dec2 expression in naïve CD4+ T cells was set as 1. (b) Kinetic expression of Dec2, Gata3 and Tbx21 mRNA in TH1 or TH2 cells. (c) Lung-associated lymph node cells from 4-get mice with asthma were restimulated with OVA for 48 hr.and IL-4-GFP+ and GFP− cells were sorted on a CD4+CD44hi gate and mRNA expression was examined by real-time RT-PCR. (d, e) Naïve CD4+ T cells were activated with the indicated plate-bound antibodies (d) or with plate-bound anti-CD3 and anti-CD28 in the presence or absence of IL-25 (e) and Dec2 mRNA expression was analyzed daily. (f) Dec2 and Il4 mRNA expression in naïve CD4+ T cells from Il4−/− or Il4+/+ mice infected with retrovirus expressing GATA3-IRES-GFP (GATA3) or GFP only (Vec).(g) Dec2 and Il13 mRNA expression in naïve CD4+ T cells from Gata3fl/fl or Gata3+/+ mice polarized under TH2 conditions, infected with the bicistronic retrovirus hCre-IRES-GFP and further cultured under TH2 conditions for 1 or 2 weeks. (h) Dec2 and Il13 mRNA expression in naïve CD4+ T cells from Gata3fl/fl mice activated under TH2 conditions and infected with a Cre-encoding virus or a control virus. (f–h) GFP+ fractions were sorted and analyzed. Data were normalized to Actb (a–f, h) or Cd4 (g) and show mean and s.d. (b–h) The sample with lowest detectable expression was set as 1. All experiments were repeated 2–3 times with consistent results.
To understand the function of Dec2 in T cells, we tested the kinetics of Dec2 induction during TH2 differentiation. CD4+CD25−CD62LhiCD44lo naïve T cells from C57BL/6 (B6) mice purified by fluorescence-activated cell sorting (FACS) were activated with plate-bound anti-CD3 and anti-CD28 under TH1 or TH2-polarizing conditions for 1 to 4 days and gene expression was assessed daily. Similar to Gata3 mRNA, Dec2 mRNA expression increased with time in developing TH2 but not in TH1 cells; in contrast, TH1 but not TH2 cells expressed abundant Tbx21 mRNA (Fig. 1b). To test whether Dec2 is expressed in TH2 cells generated under physiological conditions, we induced experimental asthma in Il4-Gfp reporter (4-get) mice 23 and purified IL-4-GFP+ and GFP− cells by FACS from the CD4+CD44hi population among lung-associated lymph node cells. As expected, Dec2 was highly expressed in IL-4-GFP+ TH2 cells but not in GFP− non-TH2 cells (Fig. 1c). Thus, Dec2 is selectively expressed in TH2 cells generated in vitro and in vivo.
We next asked how Dec2 expression is regulated during TH differentiation. Costimulatory signals especially those emanating from ICOS are important for TH2 differentiation 9 11. We thus sorted naïve CD4+ T cells from B6 mice and activated them with plate-bound anti-CD3 in the presence or absence of anti-CD28 or/and anti-ICOS for 1 or 2 days. On day 1, stimulation with anti-CD3 plus anti-CD28, but not with anti-CD3 alone, induced minimal upregulation of Dec2 mRNA expression (Fig. 1d). Addition of anti-ICOS further enhanced Dec2 expression. On day 2, Dec2 mRNA was much more abundant in cells receiving ICOS costimulation (Fig. 1d). These results indicate an important role of costimulation, particularly via ICOS, in regulation of Dec2 mRNA expression.
IL-25 facilitates the initiation of TH2 differentiation 7. Therefore, we also tested the effect of IL-25 on Dec2 mRNA expression. Naïve CD4+ T cells from B6 mice were activated with plate-bound anti-CD3 and anti-CD28 in the presence or absence of IL-25 for 1 to 3 days. Dec2 mRNA expression was strongly boosted in response to IL-25 stimulation (Fig. 1e).
As Dec2 mRNA is selectively expressed in TH2 cells, we assessed whether Dec2 expression is regulated by the transcription factor GATA3, which promotes TH2 differentiation. Naïve CD4+ T cells from Il4+/+ or Il4−/− mice were activated with plate-bound anti-CD3 and anti-CD28 for 36 hours and infected with bicistronic retroviruses encoding GATA3-IRES-GFP or GFP alone. Two days later, GFP+ cells were FACS sorted and Dec2 and Il4 mRNA expression was measured by quantitative RT-PCR. In both Il4+/+ and Il4−/− cells, over-expression of GATA3 increased Dec2 mRNA expression (Fig. 1f). In addition, we tested whether GATA3 is required for Dec2 expression. Established Gata3fl/fl or Gata3+/+ TH2 cells were infected with a bicistronic retrovirus encoding the Cre recombinase and GFP (hCre-IRES-GFP-RV)24. GFP+ cells were FACS-sorted after transduction and cultured under TH2-polarizing conditions for 1 or 2 weeks. Dec2 and Il13 mRNA expression was then measured. Unlike Il13 mRNA, the expression of which was quickly reduced after deletion of Gata3, Dec2 mRNA expression remained stable for 1 week after deletion of Gata3 but diminished after 2 weeks (Fig. 1g). Next, we tested the impact of acute GATA3 ablation on Dec2 mRNA expression during early TH2 differentiation. Naïve CD4+ T cells from Gata3fl/fl mice were activated under TH2-polarizing conditions and were infected with Cre-expressing or control retroviruses. Deletion of Gata3 at this early stage of TH2 differentiation did not substantially influence Dec2 mRNA expression, but it did markedly reduce Il13 mRNA expression (Fig. 1h). Taken together, these findings indicate that Dec2 mRNA expression is induced by costimulatory signals and by IL-25. GATA3 is sufficient to induce Dec2 mRNA expression but is not necessary for induction or maintenance of Dec2 mRNA expression.
Dec2 in TH2 differentiation in vitro
To assess the function of Dec2 in T cell differentiation, we generated Dec2-deficient mice. Two LoxP sites were introduced into the Dec2 locus, flanking the promoter region and exons 1 to 3. Germline Dec2-deficient mice were produced by breeding this floxed mouse with a CMV-Cre mouse (Supplementary Fig. 2a–b). Dec2+/− mice were backcrossed with B6 for 6–8 generations and were then interbred to generate Dec2−/− mice. In all experiments, Dec2−/− mice on a B6 background were used unless indicated. In Dec2−/− mouse, Dec2 messenger RNA in bone marrow was not detectable by RT-PCR (Supplementary Fig. 2c) and Dec2 protein expression in splenocytes was completely absent when tested by immunoblot (Supplementary Fig. 2d). The development of lymphoid populations in thymus and spleen appeared grossly normal in Dec2-deficient mice (Supplementary Fig. 3).
To analyze the function of Dec2 in T cell function, we first examined CD4+ T cell activation in response to anti-CD3 and anti-CD28 stimulation. Naïve CD4+ T cells were FACS sorted from Dec2-deficient and control mice and activated with various concentrations of plate-bound anti-CD3 in the presence of a fixed amount anti-CD28. IL-2 production was measured by ELISA on day 2 and 3H-thymidine incorporation on day 3. Dec2-deficient CD4+ T cells exhibited reduced proliferation and IL-2 production compared with wild-type cells (Fig. 2a).
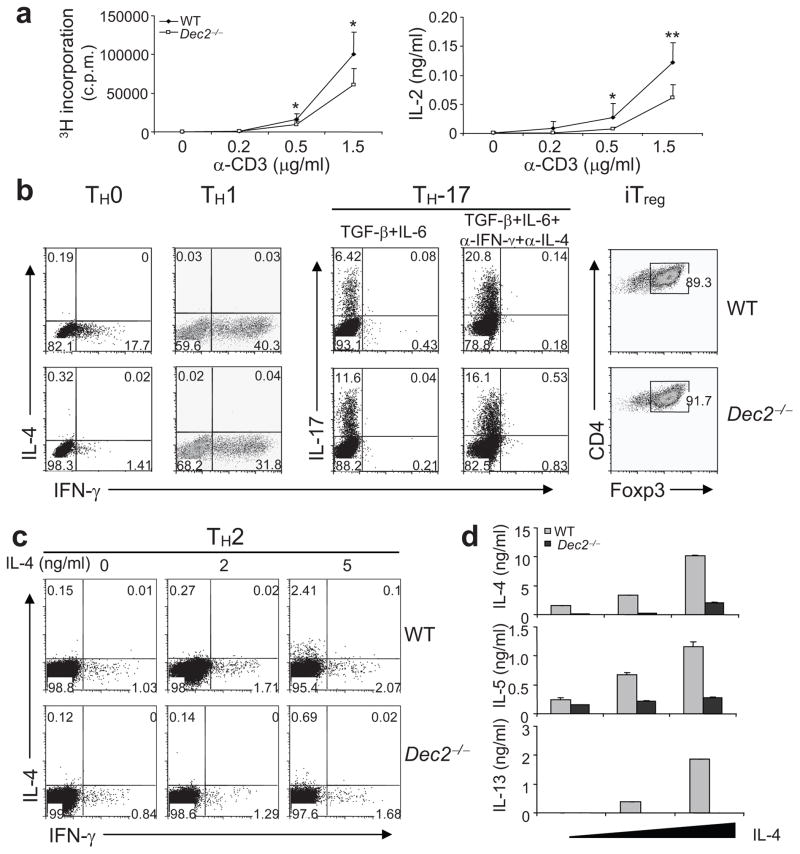
Fig. 2. Dec2 deficiency results in impaired TH2 differentiation in vitro.
(a) Naïve CD4+ T cells from the indicated mice (n = 3 per group, analyzed individually) were activated with indicated concentrations of anti-CD3 in the presence of anti-CD28. On day 3, proliferation was measured by 3H-thymidine incorporation (left). IL-2 protein expression was measured by ELISA on day 2 (right). (b) Naïve T cells from indicated mice were polarized under neutral (TH0), TH1, TH-17 and iTreg conditions for 4 days and cytokine production or Foxp3 expression was assessed by intracellular staining. (c). Naïve T cells from the indicated mice were polarized into TH2 cells with various concentrations of IL-4 for 4 days and were assessed by intracellular staining. (d) IL-4, IL-5 and IL-13 expression in the TH2 cells from (c) was measured by ELISA. All data are representative of 2 or 3 independent experiments with consistent results. (a, d) Values are means and s.d. Student t test, * P < 0.05; ** P < 0.005.
Next, we assessed the function of Dec2 in TH differentiation. Naïve CD4+ T cells from wild-type and Dec2-deficient mice were differentiated under neutral (TH0), TH1, TH2, TH-17 and iTreg conditions for 4 days. As revealed by intracellular staining, no IL-4+ cells were detected under TH0 or TH1 conditions. Under the TH0 condition, Dec2 deficiency resulted in reduced numbers of IFN-γ-producing cells (Fig. 2b). Reduced percentages of IFN-γ-producing cells were also observed under the TH1 condition (Fig. 2b). TH-17-polarizing conditions (TGF-β plus IL-6) induced greater percentages of IL-17-producing cells in Dec2-deficient than wild-type populations; addition of anti-IFN-γ and anti-IL-4 diminished this difference (Fig. 2b), suggesting that the enhanced TH-17 differentiation in Dec2-deficient T cells may be due to their impaired production of IFN-γ and/or IL-4. Under the iTreg condition, Dec2-deficient T cells developed into Foxp3+ cells as efficiently as wild-type cells (Fig. 2b). During TH2 differentiation in the presence of increasing doses of exogenous IL-4, Dec2 deficiency led to greatly reduced percentages of IL-4-producing cells (Fig. 2c). As measured by ELISA, Dec2-deficient T cells in these experiments secreted reduced amounts of the TH2 cytokines IL-4, IL-5 and IL-13 (Fig. 2d). High doses of exogenous IL-4 partially restored IL-4 and IL-5 but not IL-13 expression in Dec2-deficient cells. In summary, our analysis of Dec2-deficient T cells indicates that Dec2 is needed for maximal IL-2 production and TH2 differentiation in vitro.
Dec2 promotes TH2 responses in vivo
To analyze the function of Dec2 in vivo, we immunized Dec2-deficient and B6 mice subcutaneously with chicken ovalbumin (OVA) protein in Complete Freund’s Adjuvant (CFA). Five days later, splenocytes were isolated and stimulated ex vivo with OVA protein. OVA immunization in CFA elicited a strong inflammatory response, and Dec2-deficient cells expressed comparable amounts of IFN-γ and IL-17 as wild-type cells, and neither Dec2-deficient nor wild-type cells expressed detectable amounts of IL-4 (Fig. 3a). These results indicate that Dec2 is dispensable for T cell activation and TH1 and TH-17 differentiation in vivo.
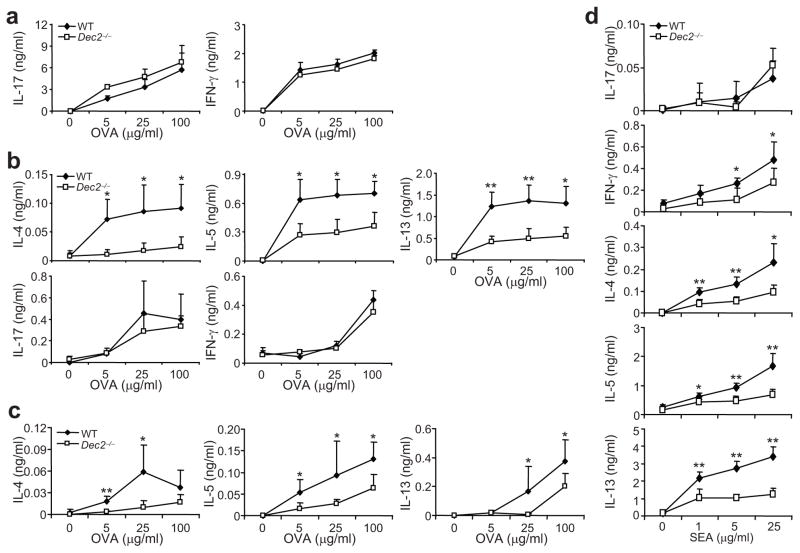
Fig. 3. Defective TH2 responses in vivo in the absence of Dec2.
(a) The indicated mice (n = 4 in each group, analyzed individually) were subcutaneously immunized with OVA in CFA. Five days later, splenocytes were collected and restimulated with OVA protein for 2 days and IFN-γ and IL-17 were measured by ELISA. (b) The indicated mice (n = 4 in each group, analyzed individually) were i.p. immunized with OVA in alum. Sox days later, splenocytes were collected and restimulated with OVA protein for 3 days. IL-4, IL-5, IL-13, IFN-γ and IL-17 were measured by ELISA. (c) CD4+ T cells enriched from indicated naïve mice were mixed with B cells from B6 mice and transferred into Rag1−/− mice. Recipients were i.p. immunized with OVA in alum the next day. Five days later, the splenocytes were harvested and restimulated with OVA protein for 3 days. Cytokine expression was determined by ELISA. (d) Dec2-deficient (n = 5) and wild-type B6 (n = 4) mice were i.p. injected with inactivated Schistosoma mansoni eggs. Eight days later, splenocytes were collected and restimulated with soluble SEA for 3 days. IL-4, IL-5, IL-13, IFN-γ and IL-17 were measured by ELISA. Values are means and s.d. *, Student t test, P < 0.05.
To determine the role of Dec2 in TH2 responses in vivo, we immunized mice intraperitoneally (i.p.) with OVA and the adjuvant alum. Upon restimulation with OVA, T cells from Dec2-deficient animals exhibited significantly reduced IL-4, IL-5 and IL-13 expression and comparable IL-17 and IFN-γ expression compared with those from wild-type mice (Fig. 3b). Next we adoptively transferred CD4+ T cells from Dec2-deficient or wild-type mice together with wild-type B cells into Rag1−/− mice. Recipients of Dec2-deficient T cells produced less IL-4, IL-5 and IL-13 after immunization with OVA and alum, suggesting that the defective TH2 cytokine expression in Dec2-deficient mice was due to a CD4+ T cell-intrinsic defect (Fig. 3c).
TH2 cells play an important role in immunity against parasite infection. To characterize the role of Dec2 in type 2 immunity, we injected Dec2-deficient and wild-type mice i.p. with Schistosoma mansoni eggs, which elicit strong TH2 responses 25. In response to restimulation with soluble egg antigen (SEA), splenocytes from Dec2-deficient animals exhibited defective TH2 cytokine expression compared with those from wild-type mice (Fig. 3d). In addition, IFN-γ but not IL-17 expression was reduced in Dec2-deficient cells.
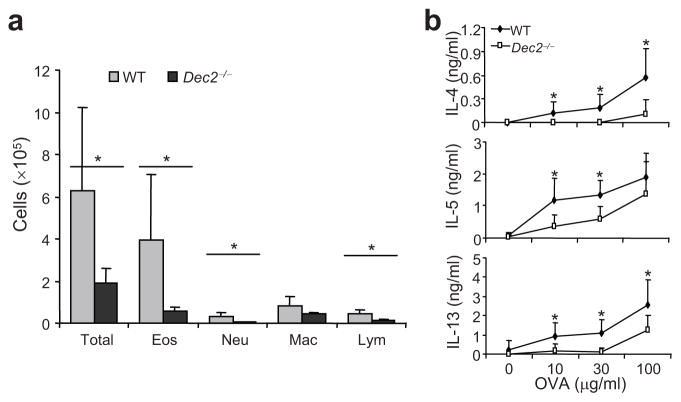
As TH2 cells are pathogenic in asthma, we induced experimental allergic asthma in Dec2-deficient and wild-type mice by using a standard protocol 26. Compared with wild-type mice, Dec2-deficient mice contained significantly fewer total cells, eosinophils, neutrophils and lymphocytes in bronchoalveolar lavage (BAL) fluid (Fig. a). Moreover, upon ex vivo restimulation with different doses of OVA protein, lung-associated lymph node cells from Dec2-deficient mice produced lower amounts of IL-4, IL-5 and IL-13 than did cells from wild-type mice (Fig. 4b). To rule out the possibility that Dec2 deficiency affects eosinophil development, we injected recombinant murine IL-5 i.p. into Dec2-deficient and wild-type mice and examined blood eosinophils with Wright-Giemsa staining. No significant difference was found in the numbers of blood eosinophils in wild-type and Dec2-deficient mice (Supplementary Fig. 4). Thus, the reduced eosinophila in asthmatic Dec2-deficient mice was not likely due to a defect in eosinophils or IL-5 signaling, but rather was caused by less T cell IL-5 expression in response to OVA + alum challenge. In summary, our data indicate that Dec2 is required for TH2 responses in vivo.
Fig. 4. Dec2 is required in allergic asthma disease.
Allergic asthma was induced in Dec2−/− and wild-type (WT) mice (n = 5 in each group, analyzed individually) by i.p. injection of OVA in alum followed by intranasal challenges with OVA protein. (a) Cells in BAL fluid. Eos, eosinophil; Neu, neutrophil; Mac, macrophage; Lym, lymphocyte. (b) TH2 cytokine expression in lung-associated lymph node cells, which were harvested and stimulated with OVA protein for 3 days. Cytokine production were measured by ELISA. Values are means and s.d. *, Student t test, P < 0.05. Data shown represent two independent experiments with consistent results.
Dec2 in early TH2 lineage commitment
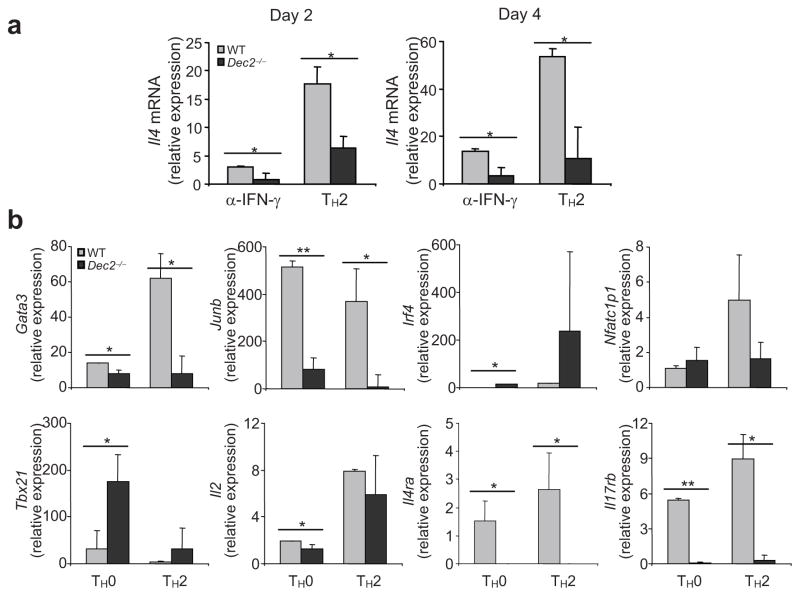
We next addressed how Dec2 regulates TH2 differentiation by asking whether Dec2 is required for TH2-specific gene expression during early TH2 development. Naïve CD4+ T cells from Dec2-deficient or wild-type mice were activated under the TH2-polarizing condition or with only anti-IFN-γ for 2 or 4 days. As measured by quantitative RT-PCR, Il4 mRNA expression was reduced in Dec2-deficient cells under both conditions on days 2 and 4 (Fig. 5a). These results indicate a critical role for Dec2 in early IL-4 expression in activated T cells. To further understand the function of Dec2, we differentiated naïve CD4+ T cells from Dec2-deficient or wild-type mice under TH0 or TH2 conditions for 2 days and analyzed their gene expression profiles. Dec2-deficient cells failed to up-regulate Gata3, Junb, Il4ra and Il17rb mRNA, but expressed wild-type amounts of Irf4, Nfatc1 and Tbx21 mRNA (Fig. 5b). Notably, IL-2 expression was reduced in Dec2-deficient cells under the TH0 but not TH2 conditions (Fig. 5b). These results indicate that Dec2 plays an important role in early TH2 differentiation and is required for GATA3 and JunB expression.
Fig. 5. Dec2 is required for early TH2 differentiation.
(a) Naïve CD4+ T cells from Dec2−/− and wild-type (WT) mice were differentiated with anti-IFN-γ or under TH2-polarizing conditions for 2 or 4 days and Il4 mRNA expression was assessed by real-time RT-PCR. (b) Naïve CD4+ T cells from Dec2−/− and WT mice were differentiated under TH0 and TH2 conditions for 2 days and mRNA expression was assessed by real-time RT-PCR. Values are means of 2–3 independent experiments, and s.d. are indicated. The results were normalized to a reference Actb and presented in arbitrary units. *, Student t test, P < 0.05.
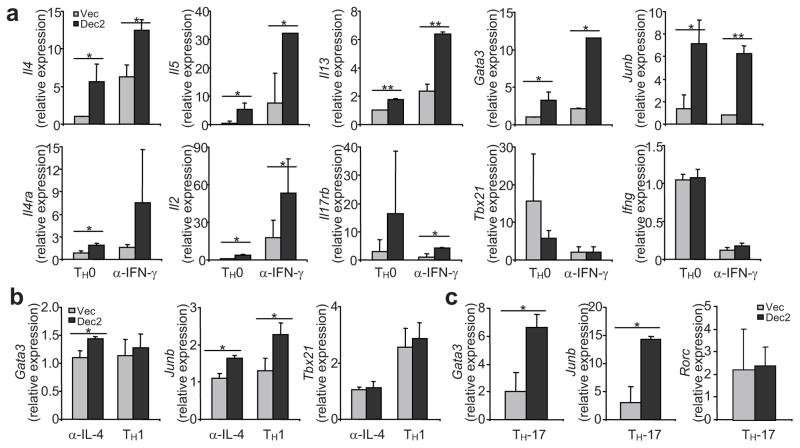
To understand if Dec2 is sufficient to initiate TH2 differentiation, we over-expressed Dec2 by retroviral transduction. Naïve CD4+ T cells from wild-type mice were activated with plate-bound anti-CD3 and anti-CD28 in the presence or absence of a blocking antibody specific for IFN-γ. Thirty-six hours later, the activated cells were infected with bicistronic retroviruses encoding GFP alone or with Dec2. Two days after infection, GFP+ cells were FACS-sorted and their gene expression profiles were assessed using real-time RT-PCR. In both the presence and the absence of anti-IFN-γ, Dec2 over-expression greatly enhanced Il2, Il4, Il5, Il13, Gata3, Junb, Il4ra and Il17rb mRNA expression, while Ifng and Tbx21 expression was not changed (Fig. 6a). Thus, Dec2 is sufficient to up-regulate TH2-specific gene expression, even under neutral conditions.
Fig. 6. Dec2 activates TH2 gene expression.
(a–c) Naïve CD4+ T cells from B6 mice were activated under (a) neutral conditions or with anti-IFN-γ, (b) with anti-IL-4 or under TH1 conditions, or (c) TH-17 conditions. Thirty-six hours later, cells were infected with bicistronic retroviruses encoding Dec2 and GFP or GFP alone. Two days after infection, GFP+ cells were sorted and mRNA expression was analyzed by real-time RT-PCR. Results were normalized to Actb and presented in arbitrary units. Values are means of 2–3 independent experiments and s.d. are indicated. Student t test, *, P < 0.05; **, P < 0.005.
We then asked whether Dec2 can up-regulate TH2-specific gene expression under non-favorable conditions. First, we over-expressed Dec2 in cells cultured in TH1 or TH-17-polarizing conditions or in the presence of a blocking antibody specific for IL-4. In the presence of anti-IL-4, low amounts of Il5 and Il13 mRNA expression were detected after over-expression of Dec2 (data not shown), which correlated with a very modest increase in Gata3 and Junb mRNA expression; no change in Tbx21 or Ifng expression was detected (Fig. 6b and data not shown). Under TH1-polarizing conditions, over-expression of Dec2 moderately elevated only JunB expression (Fig. 6b). Under TH-17-polarizing conditions, both Gata3 and Junb mRNA were significantly increased in cells infected with Dec2-encoding compared to control retrovirus, but no TH2 cytokine expression was detected (Fig. 6c and data not shown). In summary, Dec2 overexpression drives TH2 differentiation under neutral conditions but is not sufficient to promote TH2 differentiation in the TH1 or TH-17-polarizing conditions.
Dec2 binds to and activates Junb and Gata3 genes
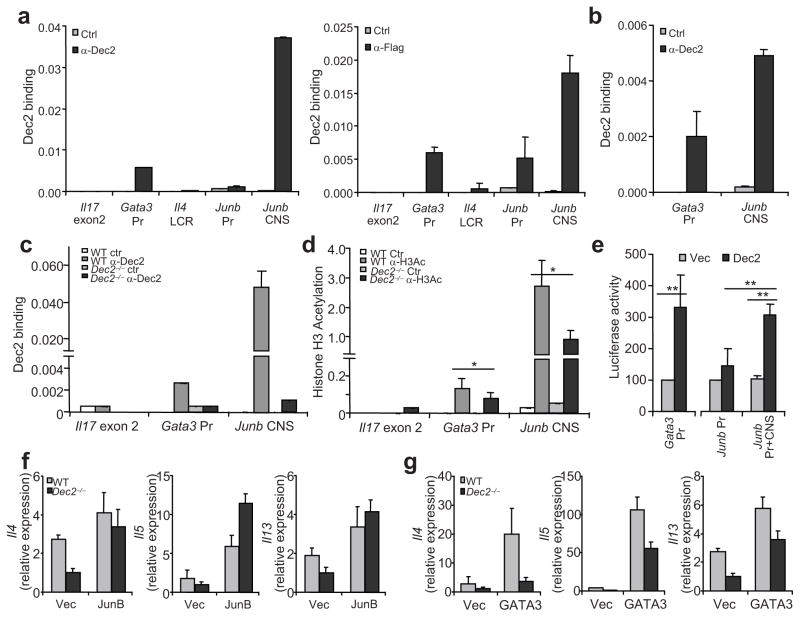
Dec2 can promote TH2 differentiation, which is IL-4 dependent. Junb and Gata3 expression was elevated by Dec2 overexpression. We thus reasoned that Dec2 may promote TH2 differentiation by directly regulating the expression of these factors. Using the VISTA program 27, 28, we searched for conserved non-coding sequences (CNSs) throughout the Gata3, Junb and Il5-Il13-Il4 gene loci by comparing mouse versus human sequences (data not shown). To predict Dec2 binding motifs in the promoters and CNS, we utilized the PROMO program 29, 30; we found a cluster of potential Dec2-binding sites in a Junb CNS approximately 6–7 kb upstream of the transcriptional start site (Supplementary Fig. 5) as well as some scattered putative Dec2 binding motifs in the Junb and Gata3 promoters and in the TH2 locus control region (data not shown). To determine if Dec2 directly binds these sites in vivo, we utilized a transgenic mouse that expresses a Flag-tagged Dec2 protein under a CD2 mini-locus 31 (Supplementary Fig. 6). Naïve CD4+ T cells from these mice were cultured in TH2-polarizing conditions in vitro. The cells were then crosslinked with paraformaldehyde, sonicated and immunoprecipitated with an antibody specific for the Flag tag, a polyclonal antibody specific for Dec2 with rat IgG or rabbit IgG control, respectively. The crosslinks were then reversed and the immunoprecipitated nucleic acid subjected to real-time PCR using primers specific for the predicted Dec2-binding regions. In Il17 exon 2, a negative control, there was no detectable binding of Dec2 (Fig. 7a). The Junb CNS and Gata3 promoter region were strongly bound by Dec2, but no Dec2 binding was detected in the Junb promoter region or in the TH2 locus control region (Fig. 7a). To substantiate the above results, we differentiated naïve CD4+ T cells from OT-II mice into TH2 cells in vitro using OT-II peptide and splenic APCs and then repeated the chromatin immunoprecipitation (ChIP) experiments using anti-Dec2 and control antibody. Consistent with the ChIP experiments from the Dec2 transgenic mice, we detected Dec2 binding to the Junb CNS and Gata3 promoter in these OT-II TH2 cells (Fig. 7b). This binding appears specific as no Dec2 binding to Junb CNS and Gata3 promoter was detected in Dec2-deficient T cells (Fig. 7c). To examine whether Dec2 regulates chromatin modifications in the Junb and Gata3 loci, naïve T cells from Dec2-deficient and wild-type mice were activated under TH2-polarizing conditions for 2 days and the abundance of acetylated histone H3 in the Junb CNS and Gata3 promoter were assessed by ChIP. Dec2-deficiency resulted in partially reduced histone H3 acetylation in the Gata3 promoter and the Junb CNS regions (Fig. 7d), suggesting a role for Dec2 in regulation of chromatin modifications in the Junb and Gata3 loci.
Fig. 7. Dec2 and regulates Junb and Gata3 transcription.
Naïve CD4+ T cells from CD2-flag-Dec2 transgenic mice (a–c), from OT-II mice (b) or from Dec2−/− and wild-type (WT) mice (c) were differentiated into TH2 cells and Dec2 binding to the indicated loci was assessed by ChIP. Pr, promoter. (d) Naïve CD4+ T cells from Dec2−/− and WT mice were cultured under TH2-polarizing conditions for 2 days and Histone H3 acetylation at the indicated loci was assessed by ChIP. Data represent 2 independent experiments with consistent results. *, Student t test, P < 0.05. Values were relative to the input abundance. (e) Dec2-expressing vector (Dec) or empty vector (Vec) was transfected into 293T cells with luciferase constructs containing the Junb promoter with or without the CNS element, or the Gata3 promoter. Renilla luciferase was used to normalize transfection efficiency and luciferase activity. Data shown are a combination of 2 (Gata3 Pr) or 3 (Junb Pr and Pr + CNS) independent tests. **, Student t test, P < 0.005. (f–g) Naïve CD4+ T cells from Dec2−/− and WT mice were activated in the presence of neutralizing antibody against IFN-γ and were infected with vectors encoding JunB (f) or GATA3 (g) and GFP, or GFP alone (Vec). Four days later, GFP+ cells were sorted and TH2 cytokine mRNA expression was assessed by real-time RT-PCR. Data shown represent 2 (f) or 3 (g) independent experiments with consistent results. Values are means and s.d.
As Dec2 was originally identified as a transcriptional repressor, we tested the activity of Dec2 on Gata3 promoter and Junb CNS using a dual luciferase reporter system. Dec2 overexpression significantly increased Gata3 promoter activity (Fig. 7e). Moreover, Dec2 enhanced the activity of the Junb promoter but only in the presence of the Junb CNS (Fig. 7e). Therefore, Dec2 may directly bind to the Junb CNS and the Gata3 promoter to activate their transcription.
Our above results suggest Gata3 and Junb are direct and biologically meaningful downstream targets of Dec2. To confirm this hypothesis, we overexpressed GATA3 and JunB in Dec2-deficient T cells. Naïve T cells from Dec2-deficient and wild-type mice were activated in the presence of a blocking antibody specific for IFN-γ and were infected with GATA3- or JunB-encoding retroviruses or a control retrovirus. Two days after infection, GFP+ cells were FACS-sorted and expression of Il4, Il5 and Il13 mRNA was analyzed by real-time RT-PCR. Overexpression of JunB fully restored TH2 cytokine mRNA expression in Dec2-deficient cells (Fig. 7f). However, overexpression of GATA3 only partially restored enhanced TH2 cytokine mRNA expression in Dec2-deficient cells (Fig. 7g). Taken together, Dec2 directly binds to Gata3 and Junb loci and activates their transcription; reciprocally, GATA3 induces Dec2 expression. Our results thus suggest a feed-forward regulatory circuit during TH2 differentiation (Supplementary Fig. 7).
Discussion
Effector T cell differentiation is regulated by environmental cytokine milieu which establishes T cell-intrinsic lineage-specific transcriptional programs. TH2 differentiation is driven by autocrine IL-4 production. However, the initiation and maintenance of IL-4 expression is not well studied. In the current study, we showed that the transcription factor Dec2 is selectively expressed by TH2 cells, that Dec2 promotes expression of JunB and GATA3, and that Dec2 facilitates autocrine IL-4 and IL-2 expression and TH2 differentiation. Our data support a crucial role for Dec2 in TH2 cell differentiation in vitro and in vivo.
In response to anti-CD3 and anti-CD28 stimulation, Dec2-deficient naïve CD4+ T cells also exhibited impaired proliferation compared with wild-type cells; this defect is consistent with reduced IL-2 production by Dec2-deficient T cells. Notably, under neutral conditions, Dec2-deficient T cell populations also contained greatly reduced numbers of IFN-γ+ cells. Reduced IL-2 and IFN-γ expression also likely accounted for the enhanced IL-17 expression by Dec2-deficient T cells cultured with TGF-β and IL-6. However, Dec2-deficient mice did not exhibit any significant defect in TH-17 differentiation in vivo, and Dec2-deficient mice showed reduced TH1 responses in the S. mansoni egg challenge but not in the OVA + alum or OVA + CFA challenge models. In contrast, both in vitro and in vivo, Dec2-deficiency led to defective production of TH2 cytokines. Dec2-deficient mice showed impaired TH2 cytokine expression in response to S. mansoni egg challenge. In an allergic asthma model, Dec2-deficient mice had greatly reduced numbers of cells including eosinophils in BAL; this reduction was associated with a defect in TH2 cytokine production but not in IL-5-mediated eosinophilia.
Our analysis of Dec2 expression provides an interesting insight into the mechanisms underpinning TH2 differentiation. Keeping with the importance of costimulatory signals, especially those emanating from ICOS, in TH2 cell polarization, Dec2 expression was boosted by simultaneous stimulation of ICOS and the TCR. In addition, IL-25, a cytokine established as able to promote TH2 differentiation 7, also enhanced Dec2 expression.
During TH2 cell differentiation, Dec2 shared a similar expression pattern and induction kinetics to GATA3, indicating that they are co-regulated. In fact, overexpression GATA3 greatly enhanced Dec2 mRNA expression, independently of IL-4 signaling. However, after deleting Gata3 in established TH2 cells, Dec2 expression remained stable for 1 week, although it diminished thereafter. In addition, deletion of Gata3 during the early TH2 differentiation did not significantly alter Dec2 expression. Thus, GATA3 may be dispensable for the induction and maintenance of Dec2 expression. When T cells were activated under the neutral conditions or in the presence of anti-IFN-γ, overexpression of Dec2 markedly elevated expression of GATA3 as well as IL-4, JunB, IL-4Rα and IL-17RB. However, Dec2 could not upregulate GATA3 expression when T cells were activated under TH1-polarizing conditions. Thus, Dec2 may require other factors, such as STAT6, to activate GATA3 expression. Nonetheless, our results altogether suggest a reciprocal regulation model in which Dec2 and GATA3 participate in regulating TH2 cell development.
High doses of exogenous IL-4 partially restored TH2 cytokine expression in Dec2-deficient cells in vitro, suggesting that Dec2 may be involved in the regulation of early autocrine IL-4 production. This hypothesis was confirmed using Dec2-deficient T cells. NFATc1 and JunB are important in regulating early IL-4 production 32. As NFATc1 expression was not altered in Dec2-deficient cells, we focused on GATA3 and JunB, whose expression was greatly reduced by Dec2 deficiency. Under neutral and TH-17 but not TH1-polarizing conditions, forced expression of Dec2 greatly enhanced Junb and Gata3 mRNA expression. As shown in a ChIP assay, a CNS upstream the Junb gene and the Gata3 promoter were directly bound by Dec2. Dec2-deficiency led to reduced histone H3 acetylation at the JunB CNS and the Gata3 promoter, implicating a role of Dec2 in regulation of chromatin modification at these two loci. Although Dec2 is considered as a transcriptional repressor, overexpression of Dec2 greatly enhanced Gata3 promoter and Junb CNS activity. Our data thus indicated that GATA3 and JunB are direct targets of Dec2. In fact, overexpression of JunB fully restored TH2 cytokine expression in Dec2-deficient cells. Overexpression of GATA3 partially restored TH2 cytokine expression. Our results thus indicate that Dec2 activates JunB as well as GATA3 expression, that GATA3 further regulates Dec2 expression to reinforce an autoregulatory loop and that in turn JunB and GATA3 together with NFATc1 regulate early IL-4 and IL-2 production, which aid in establishing the genetic program of TH2 differentiation.
Methods
Mice
A targeting vector was generated by introduction of 2 LoxP sites into the Dec2 locus with Neor as positive and Diphtheria toxin A as negative selection markers (Supplementary Fig. 1). Targeted ES cell clones were selected and injected into C57BL/6 (B6) blastocysts to generate chimeras. High percentage chimeras were bred with B6 mice for germline transmission. Targeted mice were crossed with CMV-Cre mice (Jackson Laboratory), and the excision of the promoter and exons 1–3 resulted in the generation of Dec2+/− mice. Dec2+/− mice were backcrossed with B6 for 6–8 generations and were then interbred to generate Dec2−/− mice. Dec2−/− mice on the B6 background were used with age and sex matched B6 wild-type controls except those in Fig. 2b were on a mixed B6.129 background. The genotyping primers were F, 5′-tgctgcaaaacaagccctgtcg, R1, 5′-cccaaatgcacgcgcactggagc and R2, 5′-gctgctcagttaaggctgttagc. The primers F and R1 amplify a 233-bp wild-type band and/or a 439-bp LoxP band, while the primers F and R2 give a 331-bp KO band. For the generation of Flag-Dec2 transgenic mice, Flag-Dec2 cDNA was inserted into phCD2 containing a human CD2 mini-locus 31 and a XhoI and XbaI fragment was isolated and microinjected into B6 mice at the Genetic Engineering Mouse Facility at M. D. Anderson Cancer Center. Transgenic founders were maintained by breeding with B6 mice. Gata3fl/fl mice were previous described 24. 4-get and Il4−/− mice were purchased from the Jackson laboratory. The animal experiments were performed at the age of 6–10 weeks using protocols approved by Institutional Animal Care and Use Committee.
Dec2-specific antibody
A Dec2-specific rabbit polyclonal antibody was raised against bacterially produced recombinant mouse Dec2 protein fragments encompassing amino acids 245–410. The specific antibody was purified using Affinity Chromatography.
T cell differentiation
Naïve CD4+CD25−CD62LhiCD44lo T cells were FACS sorted as described 33, 34 and were activated with plate-bound 1 μg/ml anti-CD3 and 2 μg/ml anti-CD28 and 50 unit/ml rhIL-2 in the presence of polarizing cytokines or/and blocking antibodies. For TH1 polarization we used 10 μg/ml anti-IL-4 (11B11) and 5 ng/ml IL-12 (Peprotech). For TH2 polarization, we used10 μg/ml anti-IFN-γ (XMG1.2) and 5 ng/ml (or indicated doses of) IL-4 (Peprotech). For TH-17 polarization we used 20 ng/ml IL-6 (Peprotech) and 2.5 ng/ml TGF-β (Peprotech) with or without anti-IFN-γ and anti-IL-4. For iTreg differentiation we used 5 ng/ml TGF-β.
OVA immunization
Mice (3–5 per group) were immunized with OVA (0.5 mg/ml) emulsified in CFA (0.5 mg/ml) at the tail base (100 μl each mouse) or with OVA (0.5 mg/ml) mixed in aluminium hydroxide (alum) intraperitoneally. Five to seven days later, splenocytes from the immunized mice were cultured in vitro in the presence of OVA protein for 2–3 days and cytokine expression was analyzed by ELISA. For adoptive transfers, CD4+ T cells from wild-type or Dec2−/− mice were enriched by MACS using anti-CD4 beads (Miltenyi Biotec.) and mixed with B cells from wild-type mice at 1:1 ratio before injection i.v. into Rag1−/− mice (10 × 106 CD4+ T cells per mouse). The recipient mice were immunized with OVA in alum i.p. on the next day. Five days after immunization, splenocytes were restimulated with OVA protein for 3 days. Cytokine expression was determined by ELISA.
Schistosoma mansoni egg challenge
Mice were i.p. injected with S. mansoni eggs (gift of Y. Belkaid, NIAID, NIH) that were inactivated by repeated freeze-thaw cycles (3000 eggs per mouse). Eight days later, splenocytes from the challenged mice were restimulated with SEA for 3 days and cytokine expression was analyzed by ELISA. SEA was prepared as described 35 with modification. In brief, 5000 eggs were homogenized in 1 ml ice-cold PBS and centrifuged at 10000×g for 20 min. The supernatant was collected and sterilized by Spin-X columns (Corning). SEA protein concentration was determined by a Bradford method (Bio-Rad).
Asthma induction
Allergic asthma was induced and analyzed as described 26.
Retroviral transduction
pGFP-RV containing IRES-regulated GFP, and pGFP-RV-GATA3 were gifts from K. Murphy (Washington University, St. Louis, MO) 36. The genes encoding JunB (GenBank Acc. # NM_008416.1) and Flag-tagged Dec2 (GenBank Acc. # NM_024469) were ampilified (the latter using a primer attached to a Flag-tag sequence) and cloned into pGFP-RV. Naïve CD4+CD25−CD62LhiCD44lo T cells from B6 mice were FACS sorted and transduced as described 37.
Quantitative real-time PCR
Gene expression was examined with a Bio-Rad iCycler Optical System using an iQ™ SYBR green real-time PCR kit (Bio-Rad Laboratories, Inc.). The data were normalized to an Actb or Cd4 reference gene. The primers were, Dec2, forward, 5′-aacatggacgaaggaatccctc, and reverse, 5′-taaggctgttagcgctttcaag; Irf4, forward, 5′-tcctctggatggctccagatgg, and reverse, 5′-caccaaagcacagagtcacctg; Gata3, forward, 5′-agggacatcctgcgcgaactgt, and reverse, 5′-catcttccggtttcgggtctgg; Il17rb, forward, 5′-ccatccctccagatgacaac, and reverse, 5′-tgctccttccttgcctccaagtta; Il2, forward, 5′-cactcctcacagtgacctcaag, and reverse, 5′-gggcaagtaaaatttgaaggtg. Cd4 primters were purchased from ABI (Cat# Mm00442754_m1). Junb 38, Nfatc1p1 39, Actb, Rorc(γt), Tbx21, Ifng, Il4, Il5, and Il13 7, 37 were amplified as described before.
Chromatin immunoprecipitation
ChIP assays were carried out as described 37, 40. The antibodies used were a polyclonal antibody to Dec2, anti-Flag (M2, Sigma), anti-acetyl-Histone H3 (Milipore) or rat IgG. The resulting DNA was analyzed by real-time PCR. The primers were, Junb CNS, forward, 5′-tttggcagagcctatcgtggca, and reverse, 5′-gtaggtgttttctgctgggcac; Junb promoter, forward, 5′-tggcgctcaacctggcggatcc, and reverse, 5′-gatcaagcgctccagttccgtg; Gata3 promoter, forward, 5′-ctggctgtcggaggtgtgctgc, and reverse, 5′-tgggtgctgaccgttgaggacc; Il5-Il13-Il4 LCR, forward, 5′-agctcgcttaggagcactgcca, and reverse, 5′-cagtgtgctttactctgagacg, and a negative control, Il17 exon 2, forward, 5′-tcaaccgttccacgtcaccctggac, and reverse, 5′-tcagcattcaacttgagctctcatgc.
Luciferase reporter assay
Dec2-encoding or empty vector was transfected into 293T cells with luciferase constructs containing the Junb promoter (−876 to +590 relative to the translation start site, GenBank Acc. # NM_008416.1) with or without CNS element (504 bp, the cloning primers were 5′-ttggcagagcctatcgtggca and 5′-agggactagcccaacaggttcc), or the Gata3 promoter (−830 to +398 relative to translation start site, GenBank Acc. # NM_008091). Firefly and Renilla luciferase activity were measured by using a dual-luciferase reporter system (Promega) and Renilla luciferase was used to normalize transfection efficiency and luciferase activity.
Statistical analysis
Results were expressed as mean ± standard deviation (s.d.). Differences between groups were calculated for statistical significance using the unpaired Student’s t test. P ≤ 0.05 was considered as significant.
Supplementary Material
Acknowledgments
We thank S. Rivera and J. Parker-Thornburg at MD Anderson Genetic Engineered Mouse Facility for their help in generation of knockout and transgenic animals, K. Murphy (Washington University, St. Louis, MO) for GFP-RV and GFP-RV-GATA3 vectors, Y. Belkaid (National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD) for S. mansoni eggs and suggestion, and the Dong lab members for their help and discussion. The work is supported by research grants from NIH, and MD Anderson Center for Targeted Therapy (to CD and RN), and American Heart Association (to XOY) and by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health (to JZ). RN receives a Scientist Development Award from American Heart Association, JP receives a fellowship from Ministry of Education of China, and CD is a Trust Fellow of the MD Anderson Cancer Center, and a Leukemia and Lymphoma Society Scholar.
Footnotes
Author Contributions
C.D., X.O.Y. and B.S. designed the research, analyzed and interpreted the results. X.O.Y., P.A., J.Z., J.P., Z.L., R.N., X.L., Y.C. and S.H.C. performed the experiments, and X.O.Y., B.S. and C.D. prepared the manuscript.
References
- 1.Dong C. Diversification of T-helper-cell lineages: finding the family root of IL-17-producing cells. Nat Rev Immunol. 2006;6:329–333. doi: 10.1038/nri1807. [DOI] [PubMed] [Google Scholar]
- 2.Glimcher LH, Murphy KM. Lineage commitment in the immune system: the T helper lymphocyte grows up. Genes Dev. 2000;14:1693–1711. [PubMed] [Google Scholar]
- 3.Martinez GJ, Nurieva RI, Yang XO, Dong C. Regulation and function of proinflammatory TH17 cells. Ann N Y Acad Sci. 2008;1143:188–211. doi: 10.1196/annals.1443.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dong C. Regulation and pro-inflammatory function of interleukin-17 family cytokines. Immunol Rev. 2008;226:80–86. doi: 10.1111/j.1600-065X.2008.00709.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dong C, Flavell RA. Cell fate decision: T-helper 1 and 2 subsets in immune responses. Arthritis Res. 2000;2:179–188. doi: 10.1186/ar85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lederer JA, Perez VL, DesRoches L, Kim SM, Abbas AK, Lichtman AH. Cytokine transcriptional events during helper T cell subset differentiation. J Exp Med. 1996;184:397–406. doi: 10.1084/jem.184.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Angkasekwinai P, Park H, Wang YH, Chang SH, Corry DB, Liu YJ, Zhu Z, Dong C. Interleukin 25 promotes the initiation of proallergic type 2 responses. J Exp Med. 2007;204:1509–1517. doi: 10.1084/jem.20061675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nurieva RI, Chuvpilo S, Wieder ED, Elkon KB, Locksley R, Serfling E, Dong C. A costimulation-initiated signaling pathway regulates NFATc1 transcription in T lymphocytes. J Immunol. 2007;179:1096–1103. doi: 10.4049/jimmunol.179.2.1096. [DOI] [PubMed] [Google Scholar]
- 9.Dong C, Nurieva RI, Prasad DV. Immune regulation by novel costimulatory molecules. Immunol Res. 2003;28:39–48. doi: 10.1385/IR:28:1:39. [DOI] [PubMed] [Google Scholar]
- 10.Cote-Sierra J, Foucras G, Guo L, Chiodetti L, Young HA, Hu-Li J, Zhu J, Paul WE. Interleukin 2 plays a central role in Th2 differentiation. Proc Natl Acad Sci U S A. 2004;101:3880–3885. doi: 10.1073/pnas.0400339101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamane H, Zhu J, Paul WE. Independent roles for IL-2 and GATA-3 in stimulating naive CD4+ T cells to generate a Th2-inducing cytokine environment. J Exp Med. 2005;202:793–804. doi: 10.1084/jem.20051304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liao W, Schones DE, Oh J, Cui Y, Cui K, Roh TY, Zhao K, Leonard WJ. Priming for T helper type 2 differentiation by interleukin 2-mediated induction of interleukin 4 receptor alpha-chain expression. Nat Immunol. 2008;9:1288–1296. doi: 10.1038/ni.1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Azmi S, Taneja R. Embryonic expression of mSharp-1/mDEC2, which encodes a basic helix-loop-helix transcription factor. Mech Dev. 2002;114:181–185. doi: 10.1016/s0925-4773(02)00049-7. [DOI] [PubMed] [Google Scholar]
- 14.Fujimoto K, Shen M, Noshiro M, Matsubara K, Shingu S, Honda K, Yoshida E, Suardita K, Matsuda Y, Kato Y. Molecular cloning and characterization of DEC2, a new member of basic helix-loop-helix proteins. Biochem Biophys Res Commun. 2001;280:164–171. doi: 10.1006/bbrc.2000.4133. [DOI] [PubMed] [Google Scholar]
- 15.Honma S, Kawamoto T, Takagi Y, Fujimoto K, Sato F, Noshiro M, Kato Y, Honma K. Dec1 and Dec2 are regulators of the mammalian molecular clock. Nature. 2002;419:841–844. doi: 10.1038/nature01123. [DOI] [PubMed] [Google Scholar]
- 16.Azmi S, Ozog A, Taneja R. Sharp-1/DEC2 inhibits skeletal muscle differentiation through repression of myogenic transcription factors. J Biol Chem. 2004;279:52643–52652. doi: 10.1074/jbc.M409188200. [DOI] [PubMed] [Google Scholar]
- 17.Sato F, Bhawal UK, Kawamoto T, Fujimoto K, Imaizumi T, Imanaka T, Kondo J, Koyanagi S, Noshiro M, Yoshida H, Kusumi T, Kato Y, Kijima H. Basic-helix-loop-helix (bHLH) transcription factor DEC2 negatively regulates vascular endothelial growth factor expression. Genes Cells. 2008;13:131–144. doi: 10.1111/j.1365-2443.2007.01153.x. [DOI] [PubMed] [Google Scholar]
- 18.Li B, Tournier C, Davis RJ, Flavell RA. Regulation of IL-4 expression by the transcription factor JunB during T helper cell differentiation. Embo J. 1999;18:420–432. doi: 10.1093/emboj/18.2.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hartenstein B, Teurich S, Hess J, Schenkel J, Schorpp-Kistner M, Angel P. Th2 cell-specific cytokine expression and allergen-induced airway inflammation depend on JunB. Embo J. 2002;21:6321–6329. doi: 10.1093/emboj/cdf648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bunting K, Wang J, Shannon MF. Control of interleukin-2 gene transcription: a paradigm for inducible, tissue-specific gene expression. Vitam Horm. 2006;74:105–145. doi: 10.1016/S0083-6729(06)74005-5. [DOI] [PubMed] [Google Scholar]
- 21.Jain J, Loh C, Rao A. Transcriptional regulation of the IL-2 gene. Curr Opin Immunol. 1995;7:333–342. doi: 10.1016/0952-7915(95)80107-3. [DOI] [PubMed] [Google Scholar]
- 22.Jain J, Valge-Archer VE, Rao A. Analysis of the AP-1 sites in the IL-2 promoter. J Immunol. 1992;148:1240–1250. [PubMed] [Google Scholar]
- 23.Mohrs M, Shinkai K, Mohrs K, Locksley RM. Analysis of type 2 immunity in vivo with a bicistronic IL-4 reporter. Immunity. 2001;15:303–311. doi: 10.1016/s1074-7613(01)00186-8. [DOI] [PubMed] [Google Scholar]
- 24.Zhu J, Min B, Hu-Li J, Watson CJ, Grinberg A, Wang Q, Killeen N, Urban JF, Jr, Guo L, Paul WE. Conditional deletion of Gata3 shows its essential function in T(H)1-T(H)2 responses. Nat Immunol. 2004;5:1157–1165. doi: 10.1038/ni1128. [DOI] [PubMed] [Google Scholar]
- 25.Pearce EJ, CMK, Sun J, JJT, McKee AS, Cervi L. Th2 response polarization during infection with the helminth parasite Schistosoma mansoni. Immunol Rev. 2004;201:117–126. doi: 10.1111/j.0105-2896.2004.00187.x. [DOI] [PubMed] [Google Scholar]
- 26.Yang XO, Chang SH, Park H, Nurieva R, Shah B, Acero L, Wang YH, Schluns KS, Broaddus RR, Zhu Z, Dong C. Regulation of inflammatory responses by IL-17F. J Exp Med. 2008;205:1063–1075. doi: 10.1084/jem.20071978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mayor C, Brudno M, Schwartz JR, Poliakov A, Rubin EM, Frazer KA, Pachter LS, Dubchak I. VISTA : visualizing global DNA sequence alignments of arbitrary length. Bioinformatics. 2000;16:1046–1047. doi: 10.1093/bioinformatics/16.11.1046. [DOI] [PubMed] [Google Scholar]
- 29.Farre D, Roset R, Huerta M, Adsuara JE, Rosello L, Alba MM, Messeguer X. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 2003;31:3651–3653. doi: 10.1093/nar/gkg605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Messeguer X, Escudero R, Farre D, Nunez O, Martinez J, Alba MM. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics. 2002;18:333–334. doi: 10.1093/bioinformatics/18.2.333. [DOI] [PubMed] [Google Scholar]
- 31.Zhumabekov T, Corbella P, Tolaini M, Kioussis D. Improved version of a human CD2 minigene based vector for T cell-specific expression in transgenic mice. J Immunol Methods. 1995;185:133–140. doi: 10.1016/0022-1759(95)00124-s. [DOI] [PubMed] [Google Scholar]
- 32.Dong C, Flavell RA. Control of T helper cell differentiation--in search of master genes. Sci STKE 2000. 2000:PE1. doi: 10.1126/stke.2000.49.pe1. [DOI] [PubMed] [Google Scholar]
References in Methods
- 33.Nurieva R, Yang XO, Martinez G, Zhang Y, Panopoulos AD, Ma L, Schluns K, Tian Q, Watowich SS, Jetten AM, Dong C. Essential autocrine regulation by IL-21 in the generation of inflammatory T cells. Nature. 2007;448:480–483. doi: 10.1038/nature05969. [DOI] [PubMed] [Google Scholar]
- 34.Yang XO, Panopoulos AD, Nurieva R, Chang SH, Wang D, Watowich SS, Dong C. STAT3 regulates cytokine-mediated generation of inflammatory helper T cells. J Biol Chem. 2007;282:9358–9363. doi: 10.1074/jbc.C600321200. [DOI] [PubMed] [Google Scholar]
- 35.Boros DL, Warren KS. Delayed hypersensitivity-type granuloma formation and dermal reaction induced and elicited by a soluble factor isolated from Schistosoma mansoni eggs. J Exp Med. 1970;132:488–507. doi: 10.1084/jem.132.3.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ouyang W, Ranganath SH, Weindel K, Bhattacharya D, Murphy TL, Sha WC, Murphy KM. Inhibition of Th1 development mediated by GATA-3 through an IL-4-independent mechanism. Immunity. 1998;9:745–755. doi: 10.1016/s1074-7613(00)80671-8. [DOI] [PubMed] [Google Scholar]
- 37.Yang XO, Pappu BP, Nurieva R, Akimzhanov A, Kang HS, Chung Y, Ma L, Shah B, Panopoulos AD, Schluns KS, Watowich SS, Tian Q, Jetten AM, Dong C. T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR alpha and ROR gamma. Immunity. 2008;28:29–39. doi: 10.1016/j.immuni.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Voice J, Donnelly S, Dorsam G, Dolganov G, Paul S, Goetzl EJ. c-Maf and JunB mediation of Th2 differentiation induced by the type 2 G protein-coupled receptor (VPAC2) for vasoactive intestinal peptide. J Immunol. 2004;172:7289–7296. doi: 10.4049/jimmunol.172.12.7289. [DOI] [PubMed] [Google Scholar]
- 39.Evans KE, Fox SW. Interleukin-10 inhibits osteoclastogenesis by reducing NFATc1 expression and preventing its translocation to the nucleus. BMC Cell Biol. 2007;8:4. doi: 10.1186/1471-2121-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Akimzhanov AM, Yang XO, Dong C. Chromatin remodeling of interleukin-17 (IL-17)-IL-17F cytokine gene locus during inflammatory helper T cell differentiation. J Biol Chem. 2007;282:5969–5972. doi: 10.1074/jbc.C600322200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.