Abstract
Background and Objective
Photodynamic therapy (PDT) has been advocated as an alternative to antimicrobial agents to suppress subgingival species and treat periodontitis. Bacteria located within dense biofilms, such as those encountered in dental plaques, have been found to be relatively resistant to antimicrobial therapy. In the present study, we investigated the ability of PDT to affect bacteria resistant in biofilms by comparing the photodynamic effects of methylene blue (MB) on human dental plaque microorganisms in planktonic phase and in biofilms.
Material and Methods
Dental plaque samples were obtained from 10 subjects with chronic periodontitis. Suspensions of plaque microorganisms from 5 subjects were sensitized with MB (25 μg/ml) for 5 minutes followed by exposure to red light. Multi-species microbial biofilms developed from the same plaque samples were also exposed to MB (25 μg/ml) and the same light conditions as their planktonic counterparts. In a second set of experiments, biofilms were developed with plaque bacteria from 5 subjects and sensitized with 25 and 50 μg/ml MB followed by exposure to light as above. After PDT, survival fractions were calculated from colony-forming unit counts.
Results
In suspension, PDT produced approximately 63% killing of bacteria. In biofilms, the effect of PDT resulted in much lower reductions of microorganisms (32% maximal killing).
Conclusion
Oral bacteria in biofilms are less affected by PDT than bacteria in planktonic phase. The antibacterial effect of PDT is reduced in biofilm bacteria but not to the same degree as has been reported for treatment with antibiotics under similar conditions.
Keywords: dental plaque bacteria, planktonic phase, biofilms, photodynamic therapy
Introduction
Bacteria growing in biofilms adhere to a solid surface where they multiply and form microcolonies embedded in an extracellular polymeric matrix, which includes water and nutrient channels (1). Biofilms that colonize tooth surfaces and epithelial cells lining the periodontal pocket/gingival sulcus (subgingival dental plaques) are among the most varied and complex biofilms that exist in nature. These biofilms may include a subset of selected species from more than 700 bacterial species or phylotypes (2-4) and can lead to periodontal diseases. Mechanical removal of the periodontal biofilms is currently the most frequently used method of periodontal disease treatment. Antimicrobial agents are also used, but biofilm species exhibit several resistance mechanisms (5-7). In addition, disruption of the oral microflora and the difficulty of maintaining therapeutic concentrations of antimicrobials in the oral cavity are also problems associated with the use of these agents (8).
Photodynamic therapy (PDT) has been suggested as an alternative to chemical antimicrobial agents to eliminate subgingival species and treat periodontitis (9). PDT is based on the concept that non-toxic photosensitizers can be preferentially localized in certain tissues and subsequently activated by light of the appropriate wavelength to generate singlet oxygen and free radicals that are cytotoxic to cells of the target tissue (10). Several studies have shown that oral bacteria are susceptible to PDT in planktonic cultures (9, 11, 12) and plaque scrapings (9, 13, 14). Recent studies have reported that PDT-induced bacterial cell killing reduced bacterial numbers by more than 10-fold in Streptococcus mutans, Streptococcus sobrinus and Streptococcus sanguinis (15-18) biofilms using toluidine blue O or erythrosine as the photosensitizer. Data produced in our laboratory, however, have shown that eradication of oral bacteria is incomplete following biofilm sensitization with methylene blue (MB) and exposure to red light in Actinomyces naeslundii biofilms (19, 20) and multi-species biofilms produced from human saliva as inoculum (21).
In this study, we investigated effects on bacteria derived from human natural dental plaque exposed to PDT in vitro under planktonic or biofilm conditions. The goal of our research was to compare the susceptibility of dental plaque bacteria in suspension or biofilms to PDT after their sensitization with certain concentrations of MB and exposure to red light at 665 nm.
Materials and Methods
Subjects and plaque samples
Samples of dental plaque were taken from 10 subjects. Permission to collect dental plaque samples was authorized by Institutional Review Board-approved informant consent. All the subjects were diagnosed as having chronic periodontitis with probing depths greater than 5 mm. None of the subjects used antibiotics nor had undergone periodontal treatment during 3 months prior to sampling. Dental plaque samples were taken from supra- and subgingival mesiobuccal aspects of premolars or molars in each subject (4-8 samples/subject) with individual sterile Gracey curettes. After their removal, all of the samples from each subject were placed immediately into one vial containing of 4.5 ml pre-reduced, anaerobically sterilized (PRAS) Ringer's solution (Anaerobe System Morgan Hill, CA). Microorganisms were dispersed by sonication and repeated passage through Pasteur pipettes. Aliquots were measured in a spectrophotometer in 1 ml cuvettes (one optical density unit was considered as approximately 109 cells/ml at 600 nm). Then each sample from 5 subjects (subjects 1 to 5) was divided in two parts. The first part, in suspension, was exposed to PDT. The second part was used for the development of biofilms, which were also exposed to PDT a week later. In the first group, we compared the photodynamic effects of the same MB concentration (25 μg/ml) on both planktonic and biofilm bacteria. The samples from the other 5 subjects (subjects 6 to 10) were used only for the development of biofilms that were also exposed to PDT. In the second group, we compared the photodynamic effects of two different MB concentrations (25 and 50 μg/ml) on biofilm species.
Preparation of blood agar culture plates
An enriched agar medium was prepared containing 20 g/l trypticase soy agar (BBL, Cockeysville, MD), 26 g/l brain heart infusion (BHI) agar (Difco Laboratories, Detroit, MI), 10 g/l yeast extract (BBL) and 5 mg/l hemin (Sigma Chemical Co., St. Louis, MO). The medium was autoclaved and cooled to 50°C. Then 5% defribinated sheep blood (Northeast Labs, ME), 5 mg/ml menadione 6 (Sigma Chemical Co. St. Louis, MD) and 10 mg/ml N-acetylmuramic acid (Sigma) were aseptically added. Aliquots of 150 μl of the agar mixture was dispensed into wells of 96-well microtiter plates at volume of 150 μl per well respectively (NUNC, Rochester, NY) and allowed to dry.
Development of plaque-derived biofilms
The dental plaque samples collected from each subject were placed into one vial containing PRAS Ringer's solution. Under anaerobic conditions, the entire sample was dispersed and added to BHI broth (Beckton, Dickinson & Company, Sparks, MD). For biofilm development, the plaque/BHI broth inoculum contained approximately 107 cells/ml. One hundred and fifty μl of this inoculum (approximately 1.5 × 106 bacteria) was carefully pipetted to fill four blood agar wells in each 96-well plate. The plates were then incubated anaerobically (80% N2, 10% H2, and 10% CO2) at 35°C for 7 days. After initial incubation of 48 hours, the liquid medium was carefully aspirated from each well and the biofilms were replenished with fresh BHI broth. Then fresh BHI broth was daily added onto each well very slowly to avoid damage of the biofilm.
Biofilm characterization
a) Counts of biofilm microorganisms
On day 7 of their development, biofilms were gently scraped from blood agar in each well with a sterile bacteriological loop to remove the entire visible biomass. Then spectroscopy was performed to determine the total bacterial load.
b) Confocal scanning laser microscopy (CSLM)
A Leica SP2 confocal scanning fluorescence microscope (Leica Inc., Malvern, PA) with a 40x or 100x water-dipping objective lens was used to observe the distribution of deal/live microorganisms in biofilms. Biofilms were grown on agar in 24-well plates (to accommodate the confocal microscope objective) as described above. For optimum biofilm development, the plaque:BHI inoculum contained 109 cells/ml. Live and dead biofilm bacteria were simultaneously viewed using the reagents SYTO 9 stain and propidium iodide in the LIVE/DEAD BacLight Bacterial Viability Kit (Molecular Probes, Inc., Eugene, OR) according to the manufacturer's instructions. Biofilms were stained in the dark at room temperature for 15 min. An argon laser (476 nm) was used as the excitation source for the reagents and the fluorescence light emitted was collected by two separate emission filters at 500 nm (SYTO 9) and 635 nm (propidium iodide). Sections were collected at 20 μm intervals and these were then analyzed by image-processing techniques to assess the distribution of dead/live bacteria within the biofilm matrices.
c) Microbial analysis
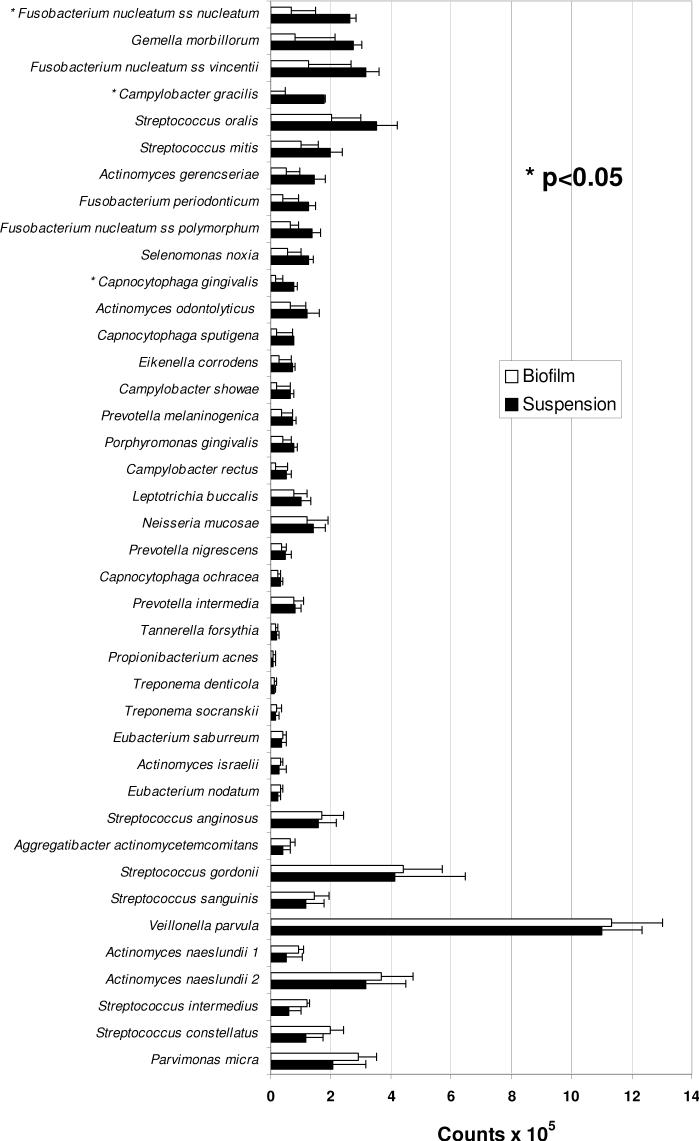
The microbial composition of biofilms was assayed using whole genomic probes to 40 oral microorganisms (see Fig. 2) as described below.
Figure 2.
Profiles of mean DNA counts of 40 microorganisms in dental plaque samples (suspensions) and plaque-derived biofilms. Each bar is the mean (× 105) of values obtained from 5 subjects (6-10) with chronic periodontitis (data from each subject were representative of 3-4 independent suspensions or biofilms). Error bars denote the standard error of the mean. No significant differences in species levels were found in statistical comparisons between suspensions and biofilms after applying Bonferroni criteria (with overall alpha=0.10) to adjust for multiple comparisons.
Composition of pooled dental plaque and biofilms
A part of each dental plaque sample (4 × 108 bacteria) obtained from subjects 6 to 10 was split in 4 tubes with BHI broth (108 bacteria per ml). The bacterial solutions underwent serial dilutions and 100 μl aliquots were spread over the surfaces of blood agar plates, which were incubated anaerobically for 7 days. Then the microbial composition was assayed using whole genomic probe assay as described previously (22). In biofilms, which were developed from the same subjects and were not exposed to light and/or MB (L-MB-), the composition was assayed 7 days after PDT. Briefly, Tris-EDTA buffer (1.5 ml) was added to the plates and the bacterial colonies were scraped off the surface with sterile L-shaped glass rods. The suspensions were placed into individual Eppendorf tubes and sonicated for 10 sec to break up clumps. The optical density (OD) of each suspension was adjusted to a final OD of 1.0, which corresponded to approximately 109 cells. Ten μl of the suspension (107 cells) were removed and placed in another Eppendorf tube with 140 μl of TE buffer and 150 μl of 0.5M NaOH. The samples were lysed and the DNA was placed in lanes on positively charged nylon membrane using a Minislot device (Immunetics, Cambridge, MA, USA). After fixation of the DNA to the membrane, the membrane was placed in Miniblotter 45 (Immunetics) with the lanes of DNA at perpendicular to the lanes of the device. Digoxigenin-labeled whole genomic DNA probes to 40 bacteria species (Fig. 2) were hybridized in individual lanes of the Miniblotter. After hybridization, the membranes were washed at high stringency and the DNA probes were detected using antibody to digoxigenin conjugated with alkaline phosphatase for chemifluorescence detection. Signals were detected using AttoPhos substrate (Amersham Life Science, Arlington Heights, IL, USA) and were scanned using a Storm Fluorimager (Molecular Dynamics, Sunnyvale, CA, USA). Computer-generated images were analyzed to determine the fluorescence intensity associated with each sample and probe. Two lanes in each membrane contained DNA standards with 1 ng (105 bacteria) and 10 ng (106 bacteria) of each species. The sensitivity of the assay was adjusted to permit detection of 104 cells of a given species by adjusting the concentration of each DNA probe. The measured fluorescence intensities were converted to absolute counts by comparison with the standards on the same membrane. Failure to detect a signal was recorded as zero.
Photosensitizer
Methylene blue (Sigma, St Louis, MO) was dissolved in BHI broth to give solutions at concentrations of 25 and 50 μg/ml before use. The ultraviolet-visible absorption spectra of MB in BHI broth were recorded from 300 to 700 nm using quartz cuvetts with 1 cm path length on a diode array spectrophotometer (model 335907P-000, ThermoSpectronic, Rochester, NY), and were characterized by a long-wavelength maximum at 665 nm.
Light source
A diode laser (BWTEK Inc., Newark, DE) with an output power of 1 Watt and a central wavelength of 665 nm was used. The system was coupled to a 1 mm optical fiber that delivered light into a lens, which formed a uniform circular spot on the base of the 24- or 96-well plate, 2 cm in diameter. This spot of light was able to irradiate each time either one well in a 24-well plate or a group of 4 wells in a 96-well plate. The power density of incident radiation was measured with a power meter (Ophir Optronics, LTD, Danvers, MA). The distance between the lens and the illuminated plates was adjusted to create a 2 cm in diameter spot of light with a fixed power density of 100 mW/cm2.
Photodynamic treatment
The light parameters used in this study for bacterial suspensions and biofilms were 100 mW/cm2 (power density) and 30 J/cm2 (energy fluence). The MB concentration of 25 μg/ml was applied on both suspensions and biofilms that were derived from samples obtained from subjects 1 to 5 (Table 1). The MB concentrations of 25 and 50 μg/ml were applied on biofilms developed using plaque samples from subjects 6 to 10 (Table 2). The following groups were used: 1) L-MB- (No light, no MB), 2) L-MB+ (treated only with MB), 3) L+MB- (treated only with light), and 4) L+MB+ (treated with MB and light, PDT group). Groups 1 and 2 were kept in plates at room temperature covered with aluminum foil during irradiation.
TABLE 1.
Phototoxicity mediated by MB in planktonic dental plaque bacteria and plaque-derived biofilmsa
| Planktonic bacteria | Biofilm bacteria | |||||
|---|---|---|---|---|---|---|
| Subject no. | L+MB- | L-MB+ | L+MB+ | L+MB- | L-MB+ | L+MB+ |
| 1 | 91 | 90.6 | 40 | 112.8 | 135.5 | 85.1 |
| 2 | 112 | 49.8 | 28.9 | 111.6 | 94.5 | 38.3 |
| 3 | 52.8 | 103.8 | 23.7 | 81.5 | 79.6 | 79.2 |
| 4 | 75.2 | 59.2 | 34.7 | 91.9 | 75.3 | 75.3 |
| 5 | 88.5 | 61.9 | 58.6 | 127.6 | 66.9 | 66.9 |
| Mean survival fraction | 83.9 | 73.1 | 37.2 | 105.1 | 101.9 | 69 |
| SEM | 9.8 | 10.3 | 6 | 8.2 | 9.8 | 8.2 |
The percent survival of bacteria was assayed by the colony-forming assay following 5 min of treatment with 25 μg/ml MB and exposure to light (30 J/cm2) at 665 nm. Surviving bacteria were expressed as a percentage of dark controls (L-MB-). Each value represents the mean survival fraction from 3-4 independent experiments.
TABLE 2.
Phototoxicity mediated by MB in plaque-derived biofilmsa
| Subject no. | L+MB- | L-MB+ (25 μg/ml) | L-MB+ (50 μg/ml) | L+MB+ (25 μg/ml) | L+MB+ (50 μg/ml) |
|---|---|---|---|---|---|
| 6 | 98.6 | 96.2 | 76.5 | 96.3 | 45.5 |
| 7 | 91.2 | 98.3 | 101.8 | 92.1 | 83.6 |
| 8 | 95 | 83.4 | 69.7 | 84.7 | 64.4 |
| 9 | 115.4 | 95.9 | 95.5 | 104.8 | 88.1 |
| 10 | 106 | 106 | 97.5 | 80.5 | 58.3 |
| Mean survival fraction | 103.2 | 95.9 | 88.2 | 91.6 | 67.9 |
| SEM | 5.2 | 3.6 | 6.3 | 4.2 | 7.9 |
The percent survival of bacteria was assayed by the colony-forming assay after incubation with 25 or 50 μg/ml MB for 5 min followed by exposure to light (30 J/cm2) at 665 nm. Surviving bacteria were expressed as a percentage of dark controls (L-MB-). Each value represents the mean survival fraction from 3-4 independent experiments.
a) Plaque samples
Suspensions of bacteria (108/ml) were incubated with MB (25 μg/ml) for 5 minutes in the dark at room temperature in tetraplicate. Following incubation, bacterial suspensions were placed in the wells of 24-well plates and exposed to light of 665 nm from above for 5 minutes in the dark at room temperature. Two neighboring wells with bacterial suspensions were separated by at least two empty wells to avoid any overlapping exposure of wells. During PDT, 24-well plates remained covered with a lid, and special care was taken not to disturb the plates. After illumination of the appropriate wells, bacterial suspensions underwent serial dilutions in BHI broth and 100 μl aliquots were plated on blood agar plates for anaerobic incubation for 7 days.
b) Biofilms
Four wells of 96-well plates containing the biofilms were exposed to MB (25 or 50 μg/ml) for 5 min. These wells were then irradiated with red light simultaneously. There was only one group of 4 wells with biofilms in each 96-well plate thereby avoiding any adjacent well exposure. During PDT 96-well plates remained covered with a lid and were not disturbed. After illumination, adherent bacteria were gently scraped from blood agar in each well with a sterile bacteriological loop to remove the biofilm and dispersed in BHI broth. The same experienced researcher removed all of the biofilms to assure that the scrapings collected the entire biofilm and did not add variability to the results. Aliquots were measured in a spectrophotometer in 1 ml cuvettes. Then serial dilutions were prepared and 100 μl aliquots were spread over the surfaces of blood agar plates. The plates were incubated anaerobically at 35°C for 7 days.
Data analysis
The multiple comparisons of 40 individual species in suspensions and biofilms were evaluated against a Bonferroni adjusted P-value (with overall alpha=0.10). Survival fractions in each group (L+MB+, L-MB+, L+MB-) were calculated by dividing the mean number of colony-forming units (CFU) by CFU from dark controls (L-MB-), planktonic or biofilm as appropriate, from the same subject. Survival fractions in Tables 1 and 2 were evaluated by repeated measures analysis of variance to compare treatment groups while controlling variation across subjects. Pair-wise comparisons were done by Least Significant Difference (LSD) tests.
Results
Characterization of biofilms
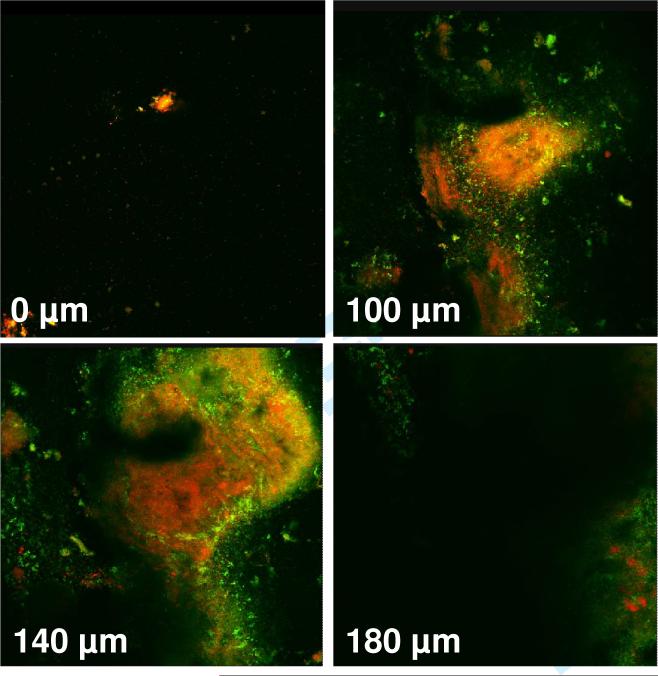
The average number of microorganisms obtained from 50 independent biofilms was approximately 109. Confocal images (X-Y) that were obtained from dental plaque-derived biofilms on day 7 of their growth showed a mixture of dead and live microorganisms extending to a depth of 180-200 μm (Fig. 1). No fluorescent signal was observed below 200 μm.
Figure 1.
Confocal images (horizontal X-Y sections) obtained by dental-plaque derived biofilms grown on agar in 24-well plates. Live bacteria with intact membranes were stained fluorescent green by SYTO 9 stain, while dead bacteria with damaged membranes were stained fluorescent orange by propidium iodide. The fluorescent signals were obtained to a depth of 180 μm.
Microbial analysis
DNA probe analysis of plaque samples and biofilms demonstrated that the composition of each were similar. Although there were, on average, slightly more bacteria in the suspension (5.9 × 106/sample) than in the biofilms (4.6 × 106/sample), these differences were not statistically significant. No significant differences in species levels were found in statistical comparisons between suspensions and biofilms after applying Bonferroni criteria (with overall alpha=0.10) to adjust for multiple comparisons. Although counts were consistently somewhat lower in biofilms, species profiles were generally similar for biofilms and suspensions.
Photodynamic treatment of planktonic bacteria versus biofilms
The effects of light with/without MB (25 μg/ml) were evaluated on dental plaque bacteria from 5 subjects in planktonic versus biofilm phase (Table 1). Pair-wise comparisons by LSD tests indicated significant differences (P<0.05) between L+MB+ relative to MB alone (L-MB+) and light alone (L+MB-) in both planktonic and biofilm states. The synergism of light and MB did not fully kill plaque microorganisms. The survival fractions for the PDT groups were approximately 37% and 69% in planktonic and biofilm cultures, respectively, compared with dark controls (L-MB-). Samples for all 5 subjects had higher survival fractions for L+MB+ in the biofilms relative to corresponding planktonic values.
Photodynamic treatment of biofilm bacteria
The effects of light with/without MB (either 25μg/ml or 50μg/ml) were evaluated on biofilms from subgingival plaque samples of 5 additional subjects (Table 2). LSD tests indicated that 50 μg/ml MB + light was significantly lower in bacterial number (P<0.05) than all other groups. Differences among mean survival fractions for the other treatment groups were quite modest.
Discussion
Several studies have reported that oral microorganisms in planktonic cultures (11-13), plaque scrapings (14) and biofilms (17, 18, 23) are susceptible to PDT. Recently, it was reported that PDT induced bacterial cell killing greater than 1 log10 in oral mono-species biofilms using erythrosine (15, 16), which is currently used clinically as a dental plaque-disclosing agent. However, other studies have demonstrated incomplete destruction of oral pathogens in plaque scrapings (20, 24), mono-species biofilms (19, 20) and multi-species biofilms derived from human saliva (21). In the present study, we investigated the photodynamic effects of MB on human dental plaque microorganisms in planktonic phase versus biofilm phase. Methylene blue, whose intravenous administration is FDA approved for methemoglobinemia, has been tested as a promising candidate for PDT of cancer (25) and has also been used in PDT for targeting various gram-positive and gram-negative oral bacteria (26). The hydrophilicity of MB (27), its low molecular weight, and the positive charge allow passage across the porin-protein channels in the outer membrane of gram-negative bacteria. MB predominantly interacts with the anionic macromolecule lipopolysaccharide resulting in the generation of MB dimmers (28), which participate in the photosensitization process (28, 29)
In our study PDT produced approximately 63% killing of bacteria in planktonic phase (Table 1), whereas in biofilms derived from the same plaque samples the effect of light resulted in much lower reductions of microorganisms (31% killing) (Table 1). Although PDT was less effective in treatment of bacteria within dense biofilms formed by dental bacteria than in planktonic culture, the difference was only 2-fold, whereas antibiotics have been reported to be approximately 250-fold less effective under these conditions (30). In comparing biofilm with planktonic effects, a degree of reduced efficacy would be expected of any penetrant molecule species. Incomplete bacterial killing by PDT is not limited to MB. In a previous study (20), a conjugate between the photosensitizer chlorine6 and a poly-L-lysine failed to completely eradicate microorganisms in dental plaque scrapings. Recently, incomplete elimination of microorganisms in subgingival scrapings was reported after their sensitization with toluidine blue, a phenothiazinium-based photosensitizer such as MB, and their subsequent exposure to red light at 635 nm (24). There are several explanations for the lowered PDT effect in dental plaque microorganisms. First, the reduced susceptibility to PDT may be related to the distinct and protected phenotypes expressed by them once they attach to the tooth (31). These phenotypic changes, which are critical for the development of dental biofilm resistance (32), are still carried by dental plaque bacteria in suspension. Second, the photodynamic effects of MB on dental plaque bacteria were probably affected by the presence of serum proteins in BHI broth (20, 33, 34). In the present study, MB was dissolved in BHI broth since proteins from both saliva and gingival crevicular fluid would also reduce the effect of MB in the hypothetical case of its in vivo application (34). Third, it has been shown that phenothiazinium-based photosensitizers, including MB, toluidine blue O and 1,9-dimethylmethylene blue, are substrates of multi-drug resistance pumps in bacteria (35).
The microcosm biofilm model that was employed in this study originates directly from the whole-mixed natural dental plaque, is technically simple to prepare and maintain, and, possibly, reflects the complexity of dental plaque. Microbial analysis (Fig. 2) showed the establishment of a mixed microflora, whereas CSLM (Fig. 1) showed a biofilm structure, which resembled that of natural dental plaque. The growth of microorganisms from pooled human dental plaque on blood agar has been demonstrated by other investigators (36, 37). Plaque microcosms are functional models for studying drug delivery and targeting (38). The characterization of the biofilm model used in the present study has been reported previously (39), whereas its validity has been demonstrated using novel drug delivery and therapeutic procedures (21).
Biofilm bacteria showed resistance to PDT with killing not exceeding 32% compared with dark controls (Table 2). Although differences in the photodynamic sensitivity of biofilms at 25 μg/ml MB as illustrated in Tables 1 and 2 appear substantial (91.6% versus 69% reduction of CFU, P=0.05 by t-test), these differences would not be considered significant if corrected for multiple testing. Biofilms were developed using dental plaque obtained from different donors, and therefore biofilm variability may reflect differences in responses to PDT. Recently, Müller et al. (40) reported less than 1 log10 destruction of bacteria in six-species oral biofilms developed on bovine-enamel discs after their sensitization with MB followed by irradiation with red light at 665 nm. Incomplete destruction of bacteria was reported previously after their sensitization with MB and exposure to red light in Actinomyces naeslundii biofilms (19, 20) as well as in microcosm laboratory biofilms developed on agar in the wells of 24-well plates using human saliva as inoculum (21). In these studies, the reduced susceptibility of biofilms to PDT was attributed to reduced penetration of MB as revealed by confocal scanning laser microscopy, an explanation that has been introduced previously (41). Similar findings were obtained by O'Neill et al. (42). In their study, confocal scanning laser microscopy images of saliva-derived biofilms revealed that photodestruction occurred predominantly in the outer layers of biofilm clusters after exposure to toluidine blue O and light. It has been suggested that water channels can carry solutes into or out of the depths of a biofilm, but they do not guarantee access to the interior of the cell clusters (43), whose diameter may range from 20 to 600 μm (44). The mechanism responsible for the reduced susceptibility of biofilms to PDT may also be related to inactivation of MB (45), the existence of biofilm bacteria in a slow growing or starved state (46), and to distinct and protected phenotypes expressed by biofilm species when they attached to agar surface (32). Although the optimal PDT parameters for eradication of microorganisms in oral microcosm biofilms remain to be determined, preliminary results obtained in our laboratory using 50 μg/ml MB and light with energy fluence of 60 J/cm2 (two-fold greater fluence than that used in this study) produced incomplete (40%) killing of bacteria in biofilms developed using human dental plaque as inoculum (47). Despite the reduced efficacy of PDT, however, the effect is much greater than seen with antibiotic therapy and is amenable to modifications that could increase efficacy. In addition, in PDT one is able to use smaller, more permeant molecular species, more capable of negotiating the water channels of established biofilm structure. A recent in vivo study showed that scaling and root planing combined with PDT using MB led to significant improvements of the investigated clinical parameters over the use of scaling and root planing alone (48). The role of PDT in the clinical treatment of periodontal disease, either in combination with traditional methods of periodontal care or by itself, warrants further investigation. Novel delivery and targeting approaches may need to be developed to overcome the reduced susceptibility of complex dental biofilms to antimicrobial therapy.
ACKNOWLEDGEMENTS
Dr. Nikos Soukos is indebted to Dr. J. Max Goodson for his many years of encouragement, support, and inspiration. Carla R. Fontana was a fellow of CAPES Foundation, Brazilian government (process BEX 0547/05-0). This work was supported by NIDCR grants RO1-DE-14360 and RO1-DE-16922.
REFERENCES
- 1.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: A common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 2.Kroes I, Lepp P, Relman DA. Bacterial diversity within the human subgingival crevice. Proc Natl Acad Sci USA. 1999;96:14547–14552. doi: 10.1073/pnas.96.25.14547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kumar PS, Griffen AL, Moeschberger ML, Leys EJ. Identification of candidate periodontal pathogens and beneficial species by quantitative 16S clonal analysis. J Clin Microbiol. 2005;43:3944–3955. doi: 10.1128/JCM.43.8.3944-3955.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sakamoto M, Umeda M, Benno Y. Molecular analysis of human oral microbiota. J Periodontal Res. 2005;40:277–285. doi: 10.1111/j.1600-0765.2005.00793.x. [DOI] [PubMed] [Google Scholar]
- 5.Anderson GG, O'Toole GA. Innate and induced resistance mechanisms of bacterial biofilms. Curr Top Microbiol Immunol. 2008;322:85–105. doi: 10.1007/978-3-540-75418-3_5. [DOI] [PubMed] [Google Scholar]
- 6.del Pozo JL, Patel R. The challenge of treating biofilm-associated bacterialinfections. Clin Pharmacol Ther. 2007;82:204–209. doi: 10.1038/sj.clpt.6100247. [DOI] [PubMed] [Google Scholar]
- 7.Fux CA, Costerton JW, Stewart PS, Stoodley P. Survival strategies of infectious biofilms. Trends Microbiol. 2005;13:34–40. doi: 10.1016/j.tim.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 8.Wilson M. Lethal photosensitization of oral bacteria and its potential application in the photodynamic therapy of oral infections. Photochem Photobiol Sci. 2004;3:412–418. doi: 10.1039/b211266c. [DOI] [PubMed] [Google Scholar]
- 9.Wilson M. Photolysis of oral bacteria and its potential use in the treatment of caries and periodontal disease: a review. J App Bacteriol. 1993;75:299–306. doi: 10.1111/j.1365-2672.1993.tb02780.x. [DOI] [PubMed] [Google Scholar]
- 10.Dougherty TJ, Gomer CJ, Henderson BW, Jori G, Kessel D, Korbelik M, Moan J, Peng Q. Photodynamic therapy. J Nat Cancer Inst. 1998;90:889–905. doi: 10.1093/jnci/90.12.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilson M, Dobson J, Sarkar S. Sensitisation of periodontopathogenic bacteria to killing by light from a low-power laser. Oral Microbiol Immunol. 1993;8:182–187. doi: 10.1111/j.1399-302x.1993.tb00663.x. [DOI] [PubMed] [Google Scholar]
- 12.Soukos NS, Ximenez-Fyvie LA, Hamblin MR, Socransky SS, Hasan T. Targeted antimicrobial photochemotherapy. Antimicrob Agents Chemother. 1998;42:2595–2601. doi: 10.1128/aac.42.10.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams JA, Pearson GJ, Colles MJ, Wilson M. The effect of variable energy input from a novel light source on the photoactivated bactericidal action of toluidine blue O on Streptococcus mutans. Caries Res. 2003;37:190–193. doi: 10.1159/000070443. [DOI] [PubMed] [Google Scholar]
- 14.Sarkar S, Wilson M. Lethal photosensitization of bacteria in subgingival plaque samples from patients with chronic periodontitis. J Periodontal Res. 1993;28:204–210. doi: 10.1111/j.1600-0765.1993.tb01070.x. [DOI] [PubMed] [Google Scholar]
- 15.Metcalf D, Robinson C, Devine D, Wood S. Enhancement of erythrosine-mediated photodynamic therapy of Streptococcus mutans biofilms by light fractionation. J Antimicrob Chemother. 2006;58:190–192. doi: 10.1093/jac/dkl205. [DOI] [PubMed] [Google Scholar]
- 16.Wood S, Metcalf D, Devine D, Robinson C. Erythrosine is a potential photosensitizer for the photodynamic therapy of oral plaque biofilms. J Antimicrob Chemother. 2006;57:680–684. doi: 10.1093/jac/dkl021. [DOI] [PubMed] [Google Scholar]
- 17.Zanin IC, Lobo MM, Rodrigues LK, Pimenta LA, Hofling JF, Goncalves RB. Photosensitization of in vitro biofilms by toluidine blue O combined with a light-emitting diode. Eur J Oral Sci. 2006;114:64–69. doi: 10.1111/j.1600-0722.2006.00263.x. [DOI] [PubMed] [Google Scholar]
- 18.Zanin IC, Goncalves RB, Junior AB, Hope CK, Pratten J. Susceptibility of Streptococcus mutans biofilms to photodynamic therapy: an in vitro study. J. Antimicrob Chemother. 2005;56:324–330. doi: 10.1093/jac/dki232. [DOI] [PubMed] [Google Scholar]
- 19.Soukos NS, Socransky SS, Mulholland SE, Lee S, Doukas AG. Photomechanical drug delivery into bacterial biofilms. Pharm Res. 2000;17:405–409. doi: 10.1023/a:1007568702118. [DOI] [PubMed] [Google Scholar]
- 20.Soukos NS, Mulholland SE, Socransky SS, Doukas AG. Photodestruction of human dental plaque bacteria: enhancement of the photodynamic effect by photomechanical waves in an oral biofilm model. Lasers Surg Med. 2003;33:161–168. doi: 10.1002/lsm.10208. [DOI] [PubMed] [Google Scholar]
- 21.Ogura M, Abernethy AD, Blissett RD, Ruggiero K, Som S, Goodson JM, Kent R, Doukas AG, Soukos NS. Photomechanical wave-assisted molecular delivery in oral biofilms. World J Microbiol Biotechnol. 2007;23:1637–1646. [Google Scholar]
- 22.Socransky SS, Smith C, Martin L, Paster BJ, Dewhirst FE, Levin AE. “Checkerboard” DNA-DNA hybridization. Biotechniques. 1994;17:788–92. [PubMed] [Google Scholar]
- 23.Wood S, Nattress B, Kirkham J, Shore R, Brookes S, Griffiths J, Robinson C. An in vitro study of the use of photodynamic therapy for the treatment of natural oral plaque biofilms formed in vivo. J Photochem Photobiol B. 1999;50:1–7. doi: 10.1016/S1011-1344(99)00056-1. [DOI] [PubMed] [Google Scholar]
- 24.Qin Y, Luan X, Bi L, He G, Bai X, Zhou C, Zhang Z. Toluidine blue-mediated photoinactivation of periodontal pathogens from supragingival plaques. Lasers Med Sci. 2008;23:49–54. doi: 10.1007/s10103-007-0454-x. [DOI] [PubMed] [Google Scholar]
- 25.De Rosa MC, Crutchley RJ. Photosensitized singlet oxygen and its applications. Coordination Chemistry Reviews. 2002;233:351–371. [Google Scholar]
- 26.Harris F, Chatfield LK, Phoenix DA. Phenothiazinium based photosensitizers -Photodynamic agents with a multiplicity of cellular targets and clinical applications. Curr. Drug Targets. 2005;6:615–627. doi: 10.2174/1389450054545962. [DOI] [PubMed] [Google Scholar]
- 27.Wainwright M, Phoenix DA, Gaskell M, Marshall B. Photobactericidal activity of methylene blue derivatives against vancomycin-resistant Enterococcus spp. J Antimicrobial Chemother. 1999;44:823–825. doi: 10.1093/jac/44.6.823. [DOI] [PubMed] [Google Scholar]
- 28.Usacheva MN, Teichert MC, Biel MA. The interaction of lipopolysaccharides with phenothiazine dyes. Lasers Surg Med. 2003;33:311–319. doi: 10.1002/lsm.10226. [DOI] [PubMed] [Google Scholar]
- 29.Bartlett JA, Indig GL. Effect of self-association and protein binding on the photochemical reactivity of triarylmethans. Implication of noncovalent interactions on the competition between photosensitization mechanisms Type I and Type II. Photochem Photobiol. 1999;70:490–498. [PubMed] [Google Scholar]
- 30.Sedlacek MJ, Walker C. Antibiotic resistance in an in vitro subgingival biofilm model. Oral Microbiol Immunol. 2007;22:333–339. doi: 10.1111/j.1399-302X.2007.00366.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davies DG, Parsek MR, Pearson JP, Iglewski BH, Costerton JW, Greenberg EP. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- 32.Whiteley M, Gita Bangera M, Bumgarner RE, Parsek MR, Teitzel GM, Lory S, Greenberg EP. Gene expression in Pseudomonas aeruginosa biofilms. Nature. 2001;413:860–864. doi: 10.1038/35101627. [DOI] [PubMed] [Google Scholar]
- 33.Bhatti M, MacRobert A, Meghji S, Henderson B, Wilson M. Effect of dosimetric and physiological factors on the lethal photosensitization of Porphyromonas gingivalis in vitro. Photochem Photobiol. 1997;65:1026–1031. doi: 10.1111/j.1751-1097.1997.tb07964.x. [DOI] [PubMed] [Google Scholar]
- 34.Kömerik N, Wilson M. Factors influencing the susceptibility of gram-negative bacteria to toluidine blue-mediated lethal photosensitisation. J Appl Microbiol. 2002;92:618–623. doi: 10.1046/j.1365-2672.2002.01567.x. [DOI] [PubMed] [Google Scholar]
- 35.Tegos GP, Hamblin MR. Phenothiazinium antimicrobial photosensitizers are substrates of bacterial multidrug resistance pumps. Antimicrob Agents Chemother. 2006;50:196–203. doi: 10.1128/AAC.50.1.196-203.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Jong MH, Van der Hoeven JS, Van Os JH. Growth of micro-organisms from supragingival dental plaque on saliva agar. J Dent Res. 1986;65:85–88. doi: 10.1177/00220345860650021601. [DOI] [PubMed] [Google Scholar]
- 37.Feres M, Haffajee AD, Allard K, Som S, Goodson JM, Socransky SS. Antibiotic resistance of subgingival species during and after antibiotic therapy. J Clin Periodontol. 2002;29:724–735. doi: 10.1034/j.1600-051x.2002.290809.x. [DOI] [PubMed] [Google Scholar]
- 38.Sissons CH. Artificial dental plaque biofilm model systems. Adv Dent Res. 1997;11:110–126. doi: 10.1177/08959374970110010201. [DOI] [PubMed] [Google Scholar]
- 39.Som S, Goodson JM, Abernethy A, Ruggiero K, Dunham J, Skobe Z, Rogers RA, Tegos G, Hamblin MR, Doukas AG, Soukos N. Characterization and validation of a dental plaque microcosm laboratory biofilm. J Dent Res. 2004;83 (abstract) [Google Scholar]
- 40.Müller P, Guggenheim B, Schmidlin PR. Efficacy of gasiform ozone and photodynamic therapy on a multispecies oral biofilm in vitro. Eur J Oral Sci. 2007;115:77–80. doi: 10.1111/j.1600-0722.2007.00418.x. [DOI] [PubMed] [Google Scholar]
- 41.Stewart PS, Grab L, Diemer JA. Analysis of biocide transport limitation in an artificial biofilm system. J Appl Microbiol. 1998;85:495–500. doi: 10.1046/j.1365-2672.1998.853529.x. [DOI] [PubMed] [Google Scholar]
- 42.O'Neill JF, Hope CK, Wilson M. Oral bacteria in multi-species biofilms can be killed by red light in the presence of toluidine blue. Lasers Surg Med. 2002;31:86–90. doi: 10.1002/lsm.10087. [DOI] [PubMed] [Google Scholar]
- 43.Stewart PS. Diffusion in biofilms. J Bacteriol. 2003;185:1485–1491. doi: 10.1128/JB.185.5.1485-1491.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rani SA, Pitts B, Stewart PS. Rapid diffusion of fluorescent tracers into Staphylococcus epidermidis biofilms visualized by time lapse microscopy. Antimicrob Agents Chemother. 2005;9:728–732. doi: 10.1128/AAC.49.2.728-732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Foley I, Gilbert P. Antibiotic resistance of biofilms. Biofouling. 1996;10:331–346. doi: 10.1080/08927019609386290. [DOI] [PubMed] [Google Scholar]
- 46.Brown SM, Allison DG, Gilbert P. Resistance of bacterial biofilms to antibiotics: A growth-rate related effect? J Antimicrol Chemother. 1988;22:777–783. doi: 10.1093/jac/22.6.777. [DOI] [PubMed] [Google Scholar]
- 47.Fontana CR, Ruggiero K, Doucette S, Doukas AG, Boussios C, Marcantonio RA, Goodson JM, Soukos NS. NIDCR. New Orleans: 2007. Phototargeting of dental plaque bacteria in planktonic phase versus biofilms. [Google Scholar]
- 48.Andersen R, Loebel N, Hammond D, Wilson M. Treatment of periodontal disease by photodisinfection compared to scaling and root planning. J Clin Dent. 2007;18:34–38. [PubMed] [Google Scholar]