Abstract
Whereas drugs are intended to be selective, at least some bind to several physiologic targets, explaining both side effects and efficacy. As many drug-target combinations exist, it would be useful to explore possible interactions computationally. Here, we compared 3,665 FDA-approved and investigational drugs against hundreds of targets, defining each target by its ligands. Chemical similarities between drugs and ligand sets predicted thousands of unanticipated associations. Thirty were tested experimentally, including the antagonism of the β1 receptor by the transporter inhibitor Prozac, the inhibition of the 5-HT transporter by the ion channel drug Vadilex, and antagonism of the histamine H4 receptor by the enzyme inhibitor Rescriptor. Overall, 23 new drug-target associations were confirmed, five of which were potent (< 100 nM). The physiological relevance of one such, the drug DMT on serotonergic receptors, was confirmed in a knock-out mouse. The chemical similarity approach is systematic and comprehensive, and may suggest side-effects and new indications for many drugs.
The creation of target-specific “magic bullets” has been a therapeutic goal since Ehrlich1 and a pragmatic criterion in drug design for 30 years. Still, several lines of evidence suggest that drugs may have multiple physiologic targets.2-5 Psychiatric medications, for instance, notoriously act through multiple molecular targets and this “polypharmacology” is likely therapeutically essential.6 Recent kinase drugs, such as Gleevec and Sutent, though perhaps designed for specificity, modulate multiple targets and these “off-target” activities also may be essential for efficacy.7,8 Conversely, anti-Parkinsonian drugs such as Permax and Dostinex activate not only dopamine receptors but also 5-HT2B serotonin receptors, thereby causing valvular heart disease and severely restricting their use.9
Predicting drug polypharmacology
Drug polypharmacology has inspired efforts to predict and characterize drug-target associations.10-15 Several groups have used phenotypic and chemical similarities among molecules to identify those with multiple targets,16,17 and early drug candidates are screened against molecular target panels.18 To predict new targets for established drugs, Bork and colleagues looked for side-effects shared between two molecules,19 while Hopkins and colleagues linked targets by drugs that bind to more than one of them.20 Indeed, using easily accessible associations, one can map 332 targets by the 290 drugs that bind to at least two of them, resulting in a network with 972 connections (Figure 1a). It seemed interesting to calculate a related map that predicts new off-target effects.
Figure 1. Drug-target networks, before and after predicting off-targets.
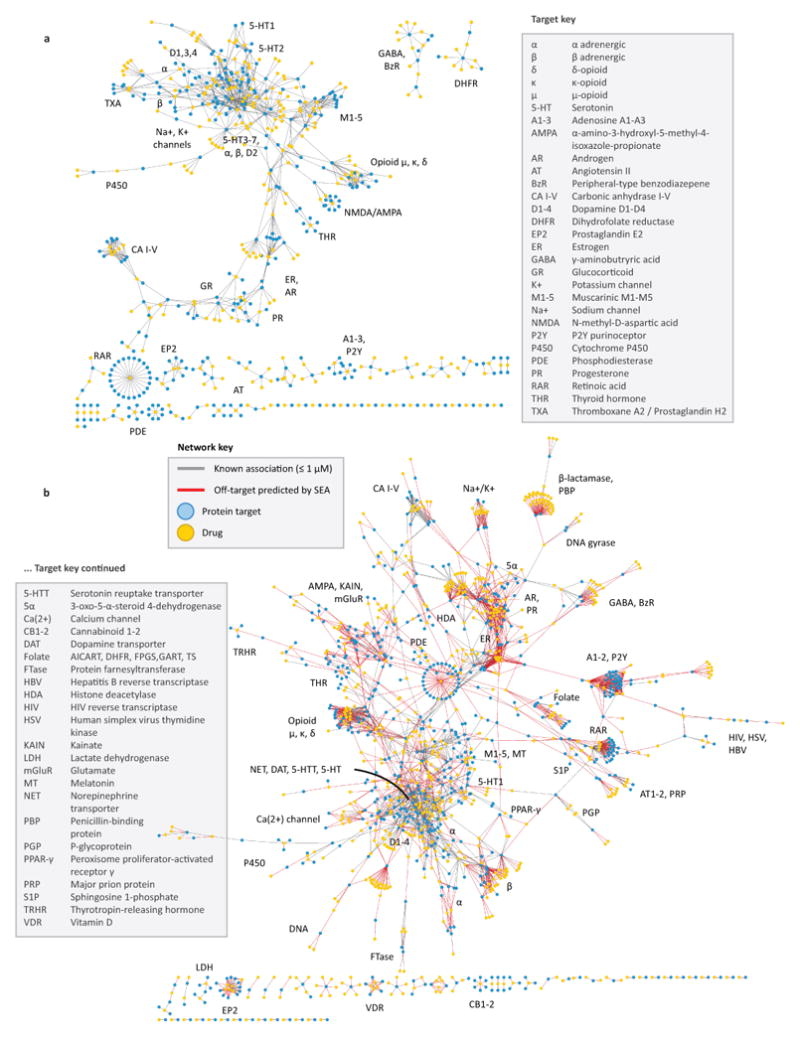
(A) Known drug-target network. Each drug (gold) is linked to its known protein targets (cyan) by a gray edge. Each edge denotes a Ki of 1 μM or better for that drug to its target. (B) Predicted drug-target network. Drugs and proteins are linked as per the known drug-target network in (A), but with the addition of red edges representing SEA off-target predictions with E-values ≤ 10-10.
Accordingly, we used a statistics-based chemoinformatics approach to predict new off-targets for 878 purchasable FDA-approved small-molecule drugs and 2,787 pharmaceutical compounds. Unlike bioinformatics methods, which might use the sequence or structural similarity among targets, this Similarity Ensemble Approach (SEA)21 compares targets by the similarity of the ligands that bind to them, expressed as expectation values, adapting the BLAST algorithms21-23 (other methods such as naïve Bayesian classifiers23,24 may also be used, see Supplementary Table 1). The approach thus captures ligand-based similarities among what would otherwise be considered disparate proteins. The 3,665 drugs were compared against 65,241 ligands organized into 246 targets drawn from the MDL Drug Data Report (MDDR) database,25 yielding 901,590 drug-target comparisons.
Most drugs had no significant similarities to most ligand sets. However, 6,928 pairs of drugs and ligand sets were similar, with expectation values (E-values) better than 1×10-10. We analyzed these predictions retrospectively against known associations and prospectively for unreported drug polypharmacology.
Retrospective drug-target predictions
We first compared the predicted drug-target associations from the MDDR database against reported associations with affinities better than 1 μM in a second database, the World of Molecular Bioactivity (WOMBAT).26 For instance, the MDDR annotates Azopt (brinzolamide) only as an “antiglaucoma agent,” but WOMBAT reports that it binds carbonic anhydrase II at 3 nM. Correspondingly, when screened internally against all MDDR molecular targets, SEA associated this drug with “Carbonic anhydrase inhibitors” with an E-value of 8.32×10-139. For 184 of the 746 drugs in WOMBAT, the predicted MDDR target agreed with the annotated WOMBAT target with E-values of 1×10-10 or better, recapitulating 19% of the off-targets missing from the MDDR (Supplementary Table 2). Another 257 drug-target predictions were unannotated in either database, and may suggest new polypharmacology.
A second retrospective test predicted targets for the 3,665 drugs uncharacterized in either database but known in the literature. Of the 6,928 drug off-targets predicted, we discarded 430 as highly similar by structure to known target ligands, and another 2,666 as trivial. This left 3,832 predictions, of which we inspected 184 by literature search and by interrogating other databases. Of these, 42 turned out to be known associations (Supplementary Table 3). For instance, when we screened the drug Revanil (lisuride) against the MDDR ligand-target sets, its best E-value was as an α2 adrenergic antagonist, and when we screened the drug Permax (pergolide) it had an E-value of 8.70×10-29 as a 5-HT1D receptor agonist. Consistent with these predictions, Revanil has been reported to bind adrenergic α2 at 0.055 nM and Permax the 5-HT1D receptor at 13 nM (Supplementary Table 3), although neither activity was reported in the MDDR or WOMBAT databases.
New drug-target predictions
For many of these 184 predictions we found no literature precedent. We therefore tested 30 predictions that were experimentally accessible to us. In radioligand competition binding assays, 23 of these (77%) yielded binding constants (Ki's) less than 15 μM (lower Ki values indicate higher affinity) (Table 1, Table 2, Supplementary Figure 1). Fifteen of these 23 were to aminergic G-protein coupled receptors (GPCRs) (Table 1), and the remainder crossed major receptor classification boundaries (Table 2). For instance, the α1 antagonist Doralese was predicted and observed to bind to the dopamine D4 receptor—both α1 and D4 are aminergic GPCRs. Conversely, the HIV-1 reverse transcriptase (enzyme) inhibitor Rescriptor was predicted and observed to bind histamine H4; this prediction crosses major target boundaries. For several predictions, we tested multiple receptor subtypes because the MDDR left these unspecified; e.g., for a predicted “α1 adrenergic blocker,” we tested the drug at α1A, α1B, and α1D subtypes; we count these as a single target. In total, 14 drugs bound 23 previously unknown targets, with 13 having sub-micromolar and five having sub-100 nM affinities (Table 1, Table 2). In cases such as Doralese's, the affinity for the discovered off-target dopamine D4, to which it binds with a Ki of 18 nM, was better than that for its known therapeutic targets, α1A and α1B adrenergic receptors, for which its Ki values are 611 and 226 nM, respectively (Figure 2a).27
Table 1. Prediction and testing of new aminergic GPCR targets for drugs.
| Drug / Pharmacological Action | E-value | Predicted Target | Ki (nM)‡ | ||
|---|---|---|---|---|---|
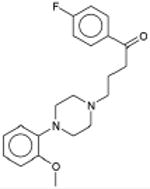 |
Sedalande Neuroleptic |
8.3×10-136 | α1 Adrenergic Blocker † | α1A | 1.2 |
| α1B | 14 | ||||
| α1D | 7.0 | ||||
| 1.7×10-14 | 5-HT1D Antagonist | 140 | |||
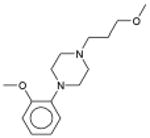 |
Dimetholizine Antihistamine; Antihypertensive |
1.6×10-129 | α1 Adrenergic Blocker † | α1A | 70 |
| α1B | 240 | ||||
| α1D | 170 | ||||
| 2.7×10-113 | 5-HT1A Antagonist | 110 | |||
| 7.4×10-56 | Dopamine D2 Antagonist | 180 | |||
 |
Kalgut Cardiotonic |
3.1×10-79 | β3 Adrenergic Agonist | 2.1×103 | |
 |
Fabahistin Antihistamine |
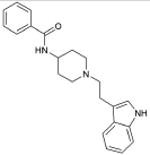
5.7×10-57 | 5-HT5A Antagonist | 130 | |
 |
Prantal Anticholinergic; Antispasmodic |
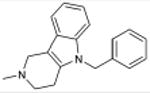
5.5×10-32 | δ Opioid Agonist | 1.4×104 | |
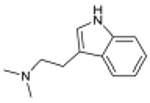 |
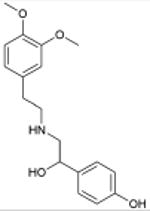
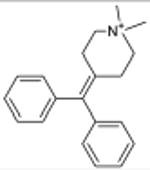
N,N-dimethyltryptamine Serotonergic Hallucinogen |
3.1×10-21 | 5-HT1B Agonist | 130 | |
| 1.2×10-13 | 5-HT2A Agonist § | 130 | |||
| 1.1×10-7 | 5-HT5A Antagonist | 2.1×103 | |||
| 5.0×10-6 | 5-HT7 Modulator | 210 | |||
 |
Doralese Adrenergic α1 Blocker; Antihypertensive; Antimigraine |
2.8×10-27 | Dopamine D4 Antagonist | 18 | |
 |
Prozac 5-HT Reuptake Inhibitor; Antidepressant |
3.9×10-15 | β Adrenergic Blocker † | β1 | 4.4×103 |
 |
Motilium Antiemetic; Peristaltic Stimulant |
4.8×10-11 | α1 Adrenergic Blocker † | α1A | 71 |
| α1B | 530 | ||||
| α1D | 710 | ||||
 |
Paxil 5-HT Reuptake Inhibitor; Antidepressive Agent |
1.3×10-7 | β Adrenergic Blocker † | β1 | 1.0×104 |
For the targets marked, the reference dataset did not specify the receptor subtype, requiring a separate assay for each one. For instance, the MDDR contains an “α1 adrenergic Blocker” set, for which it was necessary to test the α1A, α1B, and α1D subtypes.
Ki values are accurate ± 20% at two significant figures.
5-HT2A is a known target of DMT, but is shown here with its retrospective SEA E-value for comparison purposes.
Table 2. Prediction and testing of new cross-boundary targets for drugs.
| Drug / Canonical Target | E-value | Predicted Target | Ki (nM)‡ | ||
|---|---|---|---|---|---|
 |
Xenazine VMAT2 (transporter) |
1.4×10-61 | α2 Adrenergic receptor † (GPCR) | α2A | 960 |
| α2C | 1.3×103 | ||||
 |
Rescriptor HIV-1 RT (enzyme) |
1.05×10-30 | Histamine H4 receptor (GPCR) | 5.3×103 | |
 |
Vadilex NMDAR (ion channel) |
5.14×10-13 | μ Opioid receptor (GPCR) | 1.4×103 | |
| 1.98×10-4 | 5-HTT; Serotonin transporter (transporter) | 77 | |||
 |
RO-25-6981 NMDAR (ion channel) |
1.53×10-8 | 5-HTT; Serotonin transporter (transporter) | 1.4×103 | |
| 1.94×10-6 | Dopamine D4 receptor (GPCR) | 120 | |||
| 3.61×10-6 | NET; Norepinephrine transporter (transporter) | 1.3×103 | |||
| 9.08×10-5 | κ Opioid receptor (GPCR) | 3.1×103 | |||
The MDDR database did not specify the α2 adrenergic receptor subtype, requiring a separate assay for each one (α2A, α2C).
Ki values are accurate ± 20% at two significant figures.
Figure 2. Testing new off-target activities.
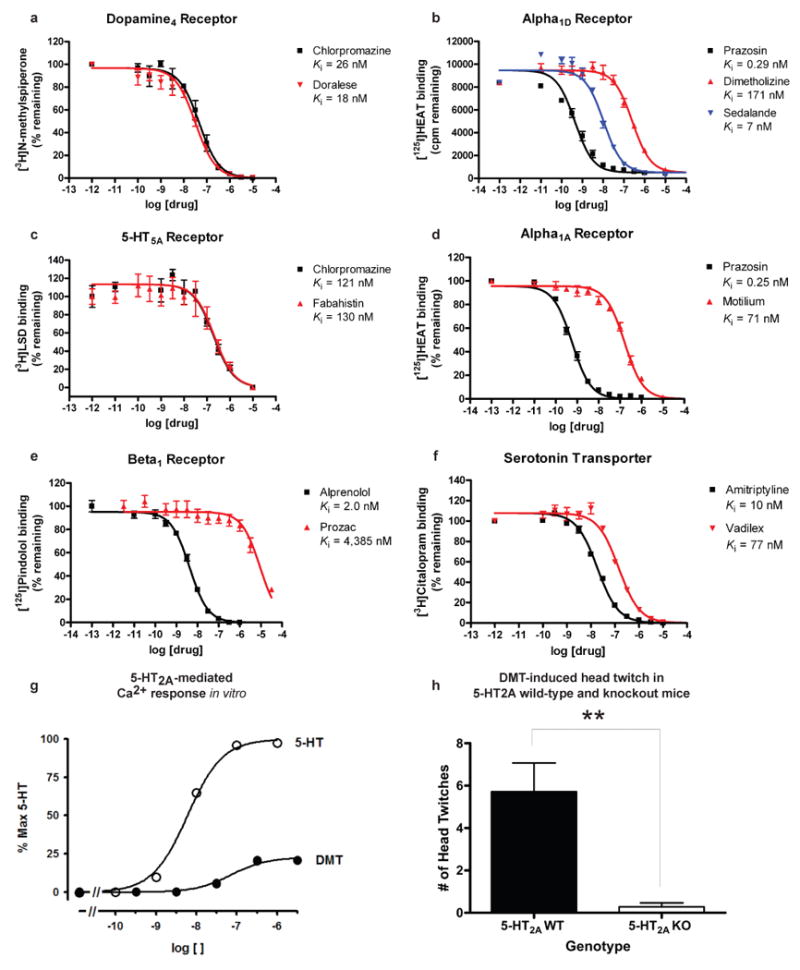
(A-F) Radioligand competition binding assays: (A) Doralese at D4, (B) Sedalande and Dimetholizine at α1D, (C) Fabahistin at 5-HT5A, (D) Motilium at α1A, (E) Prozac at β1, and (F) Vadilex at the serotonin transporter. (G-H) Investigating 5-HT2A as the target of DMT-induced hallucination: (G) 5-HT2A-mediated Ca2+ response was measured after treating HEK 293 cells stably expressing the human 5-HT2A receptor with DMT or 5-HT. DMT's EC50 was found to be 118±29 nM (vs. 5-HT's 6.6±0.4 nM baseline, n = 3), with an Emax of 23±0.4% (n = 3), confirming that DMT is a potent partial agonist at 5-HT2A receptors. (H) DMT elicited head twitch behavior only in 5-HT2A wild-type mice, confirming that it is a hallucinogenic 5-HT2A agonist. **, p < .01.
How interesting and biologically relevant are these new off-targets? This can be evaluated by the following criteria: when the new targets contribute to the primary activity of the drug, when they may mediate drug side effects, or when they are unrelated by sequence, structure and function to the canonical targets. Whereas not all of the newly predicted off-targets fall into these three categories, several fall into each.
New targets as primary sites of action
The new targets can improve our understanding of drug action. N,N-dimethyltryptamine (DMT) is an endogenous metabolite and a notorious hallucinogen. Recently the molecule was characterized as a σ1-receptor regulator at micromolar concentrations, an association implicated in its hallucinogenic properties.28,29 This surprised us because many drugs, including non-hallucinogens, bind promiscuously to the σ1 receptor with higher affinity than DMT.30 Also, DMT's hallucinogenic characteristics are consistent with other hallucinogens thought to act through serotonergic receptors, some of which the molecule is known to bind.31-33 We therefore screened DMT against the 1,133 WOMBAT targets. SEA predicted it to be similar against multiple serotonergic (5-HT) ligand sets, with expectation values ranging from 9.2×10-81 to 7.4×10-6. Upon testing, we find DMT binds 5-HT1A, 5-HT1B, 5-HT1D, 5-HT2A, 5-HT2B, 5-HT2C, 5-HT5A, 5-HT6, and 5-HT7 receptors with affinities from 39 nM to 2.1 μM (Supplementary Table 4, Supplementary Figure 2). Of these, three were previously unknown (Table 1), and all had substantially greater affinities for DMT than that represented by its 14.75 μM Kd for σ1.28 To further investigate the role of serotonin receptors in DMT-induced hallucination, we turned to a cell-based assay and an animal model that are predictive of hallucinatory actions.34 Consistent with SEA prediction, we find that DMT not only is a potent partial agonist at 5-HT2A (Figure 2g) as has been reported,31 but also that it induces head twitch response in wild type but not 5-HT2A knockout mice (Figure 2h), which is new to this study. The EC50 of DMT at 5-HT2A is 100-fold lower (better) than that observed for σ1.28 These observations support 5-HT2A as the primary target for DMT's hallucinogenic effects.
Similarly, the new off-targets for Sedalande, a neuroleptic and anxiolytic derived from haloperidol, may illuminate this drug's therapeutic effects. Although used in psychiatric clinical trials as far back as the early 1960s,35 neither its mechanism of action in the central nervous system (CNS), nor that of the related Dimetholizine, is well understood. In addition to new activities against α1 adrenergic receptors (1.2 nM – 240 nM, Figure 2b, Table 1), Dimetholizine was found to bind the D2 and 5-HT1A receptors and Sedalande to bind the 5-HT1D receptor (Table 1, Supplementary Figure 1). This likely contributes to the CNS activity of both drugs, given the association of the former with anxiety and aggression modulation, and the activity of many antipsychotics against the D2 receptor. We also found analogs of Sedalande that were active against 5-HT1D, often at affinities comparable to or greater than those of Sedalande itself (Supplementary Table 5, Supplementary Figure 3). This supports the possibility of optimizing these drugs for new indications.
An example of a drug now being investigated for a new indication is Fabahistin. Used since the 1950s as a symptomatic antihistamine, Fabahistin is now being investigated for Alzheimer's disease. When screened against 1,133 WOMBAT targets, SEA found an extraordinary similarity to 5-HT5A ligands, with an expectation value of 2.0×10-58. When we measured its binding to the 5-HT5A receptor, Fabahistin had a Ki of 130 nM (Figure 2c, Table 1). This is another example of a drug whose new, “off-target” affinity is much better than that for its canonical H1 receptor target.36 Its activity against 5-HT5A and related serotonergic receptors37 may have implications for Fabahistin's role as an Alzheimer's disease therapeutic.
Off-targets as side-effect mediators
Some of the new off-targets may contribute to a drug's adverse reactions. Motilium is an antiemetic and dopamine D1/2 antagonist that achieves peak plasma concentrations of 2.8 μM38 on intravenous administration. This formulation was withdrawn due to adverse cardiovascular effects, with the US FDA citing cardiac arrest, arrhythmias, and sudden death.39 While Motilium binds the hERG channel with an IC50 of 5 μM,40 the 71 - 710 nM affinities observed here against α1A, α1B, and α1D may also contribute to these cardiovascular effects (Figure 2d, Table 1, Supplementary Figure 1).
Similarly, the micromolar activity against the β-adrenergic receptors of the widely used selective serotonin reuptake inhibitor (SSRI) antidepressants Prozac and Paxil (Figure 2e, Table 1, Supplementary Figure 1) may explain several of their adverse effects. Abrupt withdrawal of Paxil raises standing heart rate, a symptom of the SSRI discontinuation syndrome.41 This is counterintuitive, as relieving blockade of serotonin reuptake should reduce synaptic serotonin, inconsistent with the cardiovascular syndrome.42 β-blockade by these SSRIs may partially explain this effect since β-blockers induce a similar rebound tachycardia upon abrupt withdrawal, due to β receptor up-regulation and sensitization. Despite its higher affinity for β receptors, Prozac has a longer half-life than Paxil, and its withdrawal does not induce SSRI discontinuation syndrome. Also, both SSRIs and many β-blockers can induce sexual dysfunction.43 Since both serotonergic and adrenergic signaling are involved in sexual response, the binding of Paxil and Prozac to the β1-receptor may explain why they induce greater dysfunction than other SSRIs.
Drug binding across major protein boundaries
Whereas many of the predicted off-targets occur among aminergic GPCRs, a target class for which cross-activity is well-known (see below),44 four of the drugs bound to targets unrelated by sequence or structure to their canonical targets (Table 2). For instance, the reverse transcriptase (enzyme) inhibitor Rescriptor was predicted and shown to bind to the histamine H4 receptor, a GPCR. These two targets share no evolutionary history, functional role, or structural similarity whatsoever. Intriguingly, while Rescriptor's Ki for the H4 receptor is high at 5.3 μM (Table 2, Supplementary Figure 1), this is within its steady-state plasma concentration (Cmin averages 15 μM) and is consistent with the painful rashes associated with Rescriptor use;45 likewise, H4 dysregulation has been associated with atopic dermatitis.46 Similarly, the vesicular monoamine transporter (VMAT) inhibitor47 Xenazine binds two different GPCRs at sub-micromolar concentrations (Table 2, Supplementary Figure 1). Despite its use over the last 50 years, Xenazine has not been reported to bind any GPCR. Finally, the selective ion channel inhibitors Vadilex and RO-25-6981 were predicted and found to bind to GPCRs and to transporters to which they were previously unknown to bind (Figure 2f, Table 2, Supplementary Figure 1). Whereas these ion channel drugs have known polypharmacology (Figure 3), a key point is that the new targets for these four drugs are unrelated to their main therapeutic targets except in the similarity of the ligands that modulate their activities.
Figure 3. Discovered off-targets network.
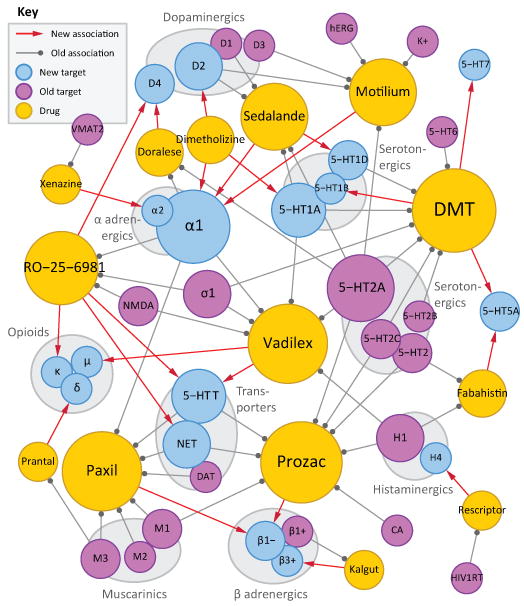
Bipartite network where drugs (gold) are linked by gray edges to their known targets (violet) and by red arrows to their discovered off-targets (cyan). Gray edges denote binding at 1 μM or better, where these affinities are known. Node sizes increase with number of incident edges. Target abbreviations: 5-HTx, serotonin receptor type x; 5-HTT, serotonin transporter; β1+, β1 adrenergic agonist; β1-, β1 adrenergic antagonist; β3+, β3 adrenergic agonist; σ1, σ1-receptor; CA, carbonic anhydrase; DAT, dopamine transporter; HIV1RT, HIV-1 reverse transcriptase; hERG, human Ether-a-go-go Related Gene channel; K+, Potassium channel; NET, norepinephrine transporter; NMDA, N-methyl-D-aspartate receptor; VMAT2, vesicular monoamine transporter 2.
More broadly, the protein target with highest sequence similarity to any of a drug's known targets is rarely predicted by the SEA approach. Rather, the target predicted by ligand similarity is typically well down in the sequence similarity ranking. Thus for Xenazine, the off-target α2 adrenergic receptor is 78th most similar to the known target VMAT2 and in fact has no significant similarity at all, with a PSI-BLAST E-value of 125 (Supplementary Table 6), while for Rescriptor, H4 is the 167th most similar receptor to HIV-1 RT, and even for Prantal, the aminergic δ-opioid receptor is only 26th most similar to its known muscarinic M3 target.
Certain caveats merit mention. Not all of the new off-targets predicted here would surprise specialists. For instance, Dimetholizine has antihypertensive activity and so its affinity for adrenergic receptors is not wholly unanticipated. Similarly, Kalgut is classified as a “selective β1 agonist,” thought to have little activity on other adrenergic receptors.48 Whereas the observation that it does bind to the β3 receptor goes against this classification, structurally this seems easy to credit (Table 1, Supplementary Figure 1). Indeed, ten of the fourteen drugs reported here are active against aminergic GPCRs (Figure 3), and so their cross-activities against other aminergic GPCRs has some precedent.44 Finally, whereas most of the drugs were active at their predicted off-targets, a third were not; these are examples of the false-positives to which this method is susceptible (Supplementary Table 7). Thus, the anxiolytics Valium and Centrax scored well against Cholecystokinin B ligands, the antipsychotic Emilace was predicted to bind 5-HT4, the anaesthetic Duocaine the κ-opioid receptor, the antihypertensive Doralese neurokinin receptors, and the narcotic Dromoran and the bradycardic Zatebradine scored well against the D2 and D1 receptors. None of these bound their predicted off-targets with affinities better than 10 μM. SEA ignores pharmacophores in its predictions, comparing drugs to ligand sets based on all shared chemical patterns. This is at once a strength, in that it is model-free, and a weakness, in that it may predict activity for drugs that share many features with the ligands of a target, and yet miss a critical chemotype.
Predicting polypharmacology on a large scale
Notwithstanding these caveats, it is the model-free nature of these predictions that allows a comprehensive exploration of drug-target interactions, most of which remain unexplored. We have focused on a thin slice of pharmacological targets, one dominated by aminergic drugs (Figure 3). Stepping back to view the larger space, 364 additional off-targets for 158 drugs are predicted with E-values better than 1×10-50, while 1,853 new off-targets are predicted with E-values better than 1×10-10 (Figure 1b). This compares to the only 972 off-target activities already annotated in the databases (Figure 1a). The Similarity Ensemble Approach and related chemoinformatics methods16-20 provide tools to explore these associations systematically, both to understand drug effects and explore new opportunities for therapeutic intervention.
Methods Summary
Prediction of off-targets
A collection of 3,665 FDA-approved and investigational drug structures was computationally screened against a panel of over 1,400 protein targets. The drug collection was extracted from the MDL Comprehensive Medicinal Chemistry database. Each target was represented solely by its set of known ligands, which were extracted from three sources of annotated molecules: the MDL Drug Data Report, the World of Molecular Bioactivity (WOMBAT),26 and the StARlite databases. The 2D structural similarity of each drug to each target's ligand set was quantified as an expectation value (E-value) using the Similarity Ensemble Approach (SEA).21
Experimental testing
Predicted “off-targets” with strong SEA E-values were evaluated for novelty against orthogonal databases and the literature. Those off-targets without precedent were subjected to radioligand competition binding assays using standard techniques49 at the NIMH Psychoactive Drug Screening Program. The role of 5-HT2A agonism in DMT-induced hallucination was examined in cell-based and in knock-out mouse models.34 Derivatives of Sedalande were identified in the ZINC50 database by substructure search, and their affinities for 5-HT1D tested using standard techniques.49
Drug-target networks and out-group analysis
Comprehensive networks of known drug-target associations (by WOMBAT) and predicted off-targets (by SEA) were constructed. Additionally, SEA off-target predictions were compared to those derived from naïve Bayesian classifiers and from PSI-BLAST21-23 comparisons of a drug's known protein target(s) against the panel of potential protein targets.
Supplementary Material
Acknowledgments
Supported by grants from the NIH supporting chemoinformatics (to B.K.S. and J.J.I.) and NIH grants and contracts supporting drug discovery and receptor pharmacology (to B.L.R). M.J.K., J.H., and C.L. were supported by fellowships from the National Science Foundation, the 6th FP of the European Commission, and the Max Kade Foundation, respectively. B.L.R. was also supported by a Distinguished Investigator Award from the NARSAD and the Michael Hooker Chair. We thank T. Oprea of Sunset Molecular for WOMBAT, Elsevier MDL for the MDDR, Scitegic for PipelinePilot, J. Overington of the European Bioinformatics Institute (EMBL-EBI) for StARlite, Daylight Chemical Information Systems Inc. for the Daylight toolkit, and J. Gingrich for 5-HT2A KO mice.
Footnotes
Supplementary Information accompanies this paper on www.nature.com/nature.
Author contributions: B.K.S., J.J.I., and M.J.K. developed the ideas for SEA. M.J.K. wrote the SEA algorithms, undertook the calculations, and identified the off-targets reported here, typically vetted with J.J.I. and B.K.S., unless otherwise noted below. M.J.K. wrote the Naïve Bayesian classifier algorithms with assistance from J.H. With assistance from B.K.S. and J.J.I., C.L. identified off-targets for Fabahistin, K.L.H.T. identified off-targets for Prozac and Paxil, and D.D.E. identified the off-target for Rescriptor. V.S. and B.L.R. designed empirical tests of the predictions, analyzed and interpreted data, and performed experiments. T.B.T., R.W., R.C.M., A.A., N.H.J., and M.B.K. performed empirical testing of the predictions. S.J.H. and R.A.G. generated materials for the experiments. M.J.K., B.K.S., and B.L.R., the senior author, wrote the manuscript with contributions and review from B.L.R. and V.S. All authors discussed the results and commented on the manuscript.
The authors declare competing financial interests: details accompany the full-text HTML version of the paper at www.nature.com/nature.
References
- 1.Ehrlich P. The Theory and Practice of Chemotherapy. Folia Serologica. 1911;7:697–714. [Google Scholar]
- 2.Peterson RT. Chemical biology and the limits of reductionism. Nat Chem Biol. 2008;4:635–638. doi: 10.1038/nchembio1108-635. [DOI] [PubMed] [Google Scholar]
- 3.Nobeli I, Favia AD, Thornton JM. Protein promiscuity and its implications for biotechnology. Nat Biotechnol. 2009;27:157–167. doi: 10.1038/nbt1519. [DOI] [PubMed] [Google Scholar]
- 4.Marona-Lewicka D, Nichols DE. Further evidence that the delayed temporal dopaminergic effects of LSD are mediated by a mechanism different than the first temporal phase of action. Pharmacol Biochem Behav. 2007;87:453–461. doi: 10.1016/j.pbb.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 5.Marona-Lewicka D, Nichols DE. WAY 100635 produces discriminative stimulus effects in rats mediated by dopamine D(4) receptor activation. Behav Pharmacol. 2009;20:114–118. doi: 10.1097/FBP.0b013e3283242f1a. [DOI] [PubMed] [Google Scholar]
- 6.Roth BL, Sheffler DJ, Kroeze WK. Magic shotguns versus magic bullets: selectively non-selective drugs for mood disorders and schizophrenia. Nat Rev Drug Discov. 2004;3:353–359. doi: 10.1038/nrd1346. [DOI] [PubMed] [Google Scholar]
- 7.Rix U, et al. Chemical proteomic profiles of the BCR-ABL inhibitors imatinib, nilotinib, and dasatinib reveal novel kinase and nonkinase targets. Blood. 2007;110:4055–4063. doi: 10.1182/blood-2007-07-102061. [DOI] [PubMed] [Google Scholar]
- 8.Hopkins AL. Network pharmacology. Nat Biotechnol. 2007;25:1110–1111. doi: 10.1038/nbt1007-1110. [DOI] [PubMed] [Google Scholar]
- 9.Roth BL. Drugs and valvular heart disease. The New England journal of medicine. 2007;356:6–9. doi: 10.1056/NEJMp068265. [DOI] [PubMed] [Google Scholar]
- 10.Bajorath J. Computational analysis of ligand relationships within target families. Curr Opin Chem Biol. 2008;12:352–358. doi: 10.1016/j.cbpa.2008.01.044. [DOI] [PubMed] [Google Scholar]
- 11.Oprea TI, Tropsha A, Faulon JL, Rintoul MD. Systems chemical biology. Nat Chem Biol. 2007;3:447–450. doi: 10.1038/nchembio0807-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Newman DJ. Natural products as leads to potential drugs: an old process or the new hope for drug discovery? J Med Chem. 2008;51:2589–2599. doi: 10.1021/jm0704090. [DOI] [PubMed] [Google Scholar]
- 13.Siegel MG, Vieth M. Drugs in other drugs: a new look at drugs as fragments. Drug Discov Today. 2007;12:71–79. doi: 10.1016/j.drudis.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 14.Miller JR, et al. A class of selective antibacterials derived from a protein kinase inhibitor pharmacophore. Proc Natl Acad Sci U S A. 2009;106:1737–1742. doi: 10.1073/pnas.0811275106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walsh CT, Fischbach MA. Repurposing libraries of eukaryotic protein kinase inhibitors for antibiotic discovery. Proc Natl Acad Sci U S A. 2009;106:1689–1690. doi: 10.1073/pnas.0813405106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Young DW, et al. Integrating high-content screening and ligand-target prediction to identify mechanism of action. Nat Chem Biol. 2008;4:59–68. doi: 10.1038/nchembio.2007.53. [DOI] [PubMed] [Google Scholar]
- 17.Wagner BK, et al. Large-scale chemical dissection of mitochondrial function. Nat Biotechnol. 2008;26:343–351. doi: 10.1038/nbt1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krejsa CM, et al. Predicting ADME properties and side effects: the BioPrint approach. Curr Opin Drug Discov Devel. 2003;6:470–480. [PubMed] [Google Scholar]
- 19.Campillos M, Kuhn M, Gavin AC, Jensen LJ, Bork P. Drug target identification using side-effect similarity. Science. 2008;321:263–266. doi: 10.1126/science.1158140. [DOI] [PubMed] [Google Scholar]
- 20.Paolini GV, Shapland RHB, Hoorn WPv, Mason JS, Hopkins AL. Global mapping of pharmacological space. Nat Biotechnol. 2006;24:805–815. doi: 10.1038/nbt1228. [DOI] [PubMed] [Google Scholar]
- 21.Keiser MJ, et al. Relating protein pharmacology by ligand chemistry. Nat Biotechnol. 2007;25:197–206. doi: 10.1038/nbt1284. [DOI] [PubMed] [Google Scholar]
- 22.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 23.Hert J, Keiser MJ, Irwin JJ, Oprea TI, Shoichet BK. Quantifying the relationships among drug classes. J Chem Inf Model. 2008;48:755–765. doi: 10.1021/ci8000259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nigsch F, Bender A, Jenkins JL, Mitchell JB. Ligand-target prediction using Winnow and naive Bayesian algorithms and the implications of overall performance statistics. J Chem Inf Model. 2008;48:2313–2325. doi: 10.1021/ci800079x. [DOI] [PubMed] [Google Scholar]
- 25.Schuffenhauer A, et al. An ontology for pharmaceutical ligands and its application for in silico screening and library design. J Chem Inf Comput Sci. 2002;42:947–955. doi: 10.1021/ci010385k. [DOI] [PubMed] [Google Scholar]
- 26.Oprea TI. Chemoinformatics in drug discovery. Wiley-VCH; 2005. [Google Scholar]
- 27.Lomasney JW, et al. Molecular cloning and expression of the cDNA for the alpha 1A-adrenergic receptor. The gene for which is located on human chromosome 5. J Biol Chem. 1991;266:6365–6369. [PubMed] [Google Scholar]
- 28.Fontanilla D, et al. The hallucinogen N,N-dimethyltryptamine (DMT) is an endogenous sigma-1 receptor regulator. Science. 2009;323:934–937. doi: 10.1126/science.1166127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Su TP, Hayashi T, Vaupel DB. When the endogenous hallucinogenic trace amine N,N-dimethyltryptamine meets the sigma-1 receptor. Sci Signal. 2009;2:pe12. doi: 10.1126/scisignal.261pe12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roth BL, Kroeze WK, Patel S, Lopez E. The Multiplicity of Serotonin Receptors: Uselessly diverse molecules or an embarrasment of riches? The Neuroscientist. 2000;6:252–262. [Google Scholar]
- 31.Smith RL, Canton H, Barrett RJ, Sanders-Bush E. Agonist properties of N,N-dimethyltryptamine at serotonin 5-HT2A and 5-HT2C receptors. Pharmacol Biochem Behav. 1998;61:323–330. doi: 10.1016/s0091-3057(98)00110-5. [DOI] [PubMed] [Google Scholar]
- 32.Kohen R, et al. Cloning, characterization, and chromosomal localization of a human 5-HT6 serotonin receptor. J Neurochem. 1996;66:47–56. doi: 10.1046/j.1471-4159.1996.66010047.x. [DOI] [PubMed] [Google Scholar]
- 33.Pierce PA, Peroutka SJ. Hallucinogenic drug interactions with neurotransmitter receptor binding sites in human cortex. Psychopharmacology (Berl) 1989;97:118–122. doi: 10.1007/BF00443425. [DOI] [PubMed] [Google Scholar]
- 34.Abbas AI, et al. PSD-95 is essential for hallucinogen and atypical antipsychotic drug actions at serotonin receptors. J Neurosci. 2009;29:7124–7136. doi: 10.1523/JNEUROSCI.1090-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kurland AA, Mc CK, Michaux WW. Clinical trial of haloanisone (R-2028) with hospitalized psychiatric patients. J New Drugs. 1962;2:352–360. doi: 10.1177/009127006200200605. [DOI] [PubMed] [Google Scholar]
- 36.Gankina EM, et al. Effect of some antihistamine preparations on binding of3H-mepyramine and3H-cimetidine to histamine receptors in rat brain. Khimiko-farmatsevticheskii Zhurnal. 1992;26:9–11. [Google Scholar]
- 37.Gankina EM, et al. The effect of antihistaminic preparations on the binding of labelled mepyramine, ketanserin and quinuclidinyl benzilate in the rat brain. Eksp Klin Farmakol. 1993;56:22–24. [PubMed] [Google Scholar]
- 38.Heykants J, et al. On the pharmacokinetics of domperidone in animals and man. IV. The pharmacokinetics of intravenous domperidone and its bioavailability in man following intramuscular, oral and rectal administration. Eur J Drug Metab Pharmacokinet. 1981;6:61–70. doi: 10.1007/BF03189516. [DOI] [PubMed] [Google Scholar]
- 39.FDA Warns Against Women Using Unapproved Drug, Domperidone, to Increase Milk Production. US Food and Drug Administration Talk Paper. 2004 [Google Scholar]
- 40.Stork D, et al. State dependent dissociation of HERG channel inhibitors. Br J Pharmacol. 2007;151:1368–1376. doi: 10.1038/sj.bjp.0707356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Michelson D, et al. Interruption of selective serotonin reuptake inhibitor treatment. Double-blind, placebo-controlled trial. Br J Psychiatry. 2000;176:363–368. doi: 10.1192/bjp.176.4.363. [DOI] [PubMed] [Google Scholar]
- 42.Berger M, Gray JA, Roth BL. The Extended Pharmacology of Serotonin. Annual Reviews in Medicine. 2009;60:355–366. doi: 10.1146/annurev.med.60.042307.110802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Waldinger MD, Hengeveld MW, Zwinderman AH, Olivier B. Effect of SSRI antidepressants on ejaculation: a double-blind, randomized, placebo-controlled study with fluoxetine, fluvoxamine, paroxetine, and sertraline. J Clin Psychopharmacol. 1998;18:274–281. doi: 10.1097/00004714-199808000-00004. [DOI] [PubMed] [Google Scholar]
- 44.Peters JU, Schnider P, Mattei P, Kansy M. Pharmacological promiscuity: dependence on compound properties and target specificity in a set of recent Roche compounds. ChemMedChem. 2009;4:680–686. doi: 10.1002/cmdc.200800411. [DOI] [PubMed] [Google Scholar]
- 45.Scott LJ, Perry CM. Delavirdine: a review of its use in HIV infection. Drugs. 2000;60:1411–1444. doi: 10.2165/00003495-200060060-00013. [DOI] [PubMed] [Google Scholar]
- 46.Dijkstra D, et al. Human inflammatory dendritic epidermal cells express a functional histamine H4 receptor. J Invest Dermatol. 2008;128:1696–1703. doi: 10.1038/sj.jid.5701250. [DOI] [PubMed] [Google Scholar]
- 47.Mehvar R, Jamali F, Watson MW, Skelton D. Pharmacokinetics of tetrabenazine and its major metabolite in man and rat. Bioavailability and dose dependency studies. Drug Metab Dispos. 1987;15:250–255. [PubMed] [Google Scholar]
- 48.Inamasu M, Totsuka T, Ikeo T, Nagao T, Takeyama S. Beta 1-adrenergic selectivity of the new cardiotonic agent denopamine in its stimulating effects on adenylate cyclase. Biochem Pharmacol. 1987;36:1947–1954. doi: 10.1016/0006-2952(87)90493-x. [DOI] [PubMed] [Google Scholar]
- 49.Jensen NH, et al. N-desalkylquetiapine, a potent norepinephrine reuptake inhibitor and partial 5-HT1A agonist, as a putative mediator of quetiapine's antidepressant activity. Neuropsychopharmacology. 2008;33:2303–2312. doi: 10.1038/sj.npp.1301646. [DOI] [PubMed] [Google Scholar]
- 50.Irwin JJ, Shoichet BK. ZINC--a free database of commercially available compounds for virtual screening. J Chem Inf Model. 2005;45:177–182. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


