Abstract
The biosynthesis of glycoconjugates such as N-glycoproteins and GPI-anchored proteins in eukaryotes and cell wall peptidoglycan and lipopolysaccharide in bacteria, requires lipid intermediates to be flipped rapidly across the endoplasmic reticulum or bacterial cytoplasmic membrane (so-called biogenic membranes). Rapid flipping is also required to normalize the number of glycerophospholipids in the two leaflets of the bilayer as the membrane expands in a growing cell. Although lipids diffuse rapidly in the plane of the membrane, the intrinsic rate at which they flip across membranes is very low. Biogenic membranes possess dedicated lipid transporters or flippases to increase flipping to a physiologically sufficient rate. The flippases are ‘ATP-independent’ and facilitate ‘downhill’ transport. Most predicted biogenic membrane flippases have not been identified at the molecular level, and the few flippases that have been identified by genetic approaches have not been biochemically validated. Here we summarize recent progress on this fundamental topic and speculate on the mechanism(s) by which biogenic membrane flippases facilitate transbilayer lipid movement.
Introduction
The processes of membrane differentiation, turnover and growth require the incorporation of building blocks - proteins and lipids - into pre-existing membranes (1). For the eukaryotic plasma membrane this is largely accomplished through the fusion of cargo-bearing secretory vesicles that originate in biosynthetic compartments, and by the non-vesicular delivery of lipids like cholesterol. In the case of biogenic or ‘self-synthesizing’ membranes, such as the endoplasmic reticulum (ER)1 or bacterial cytoplasmic membrane (bCM), newly synthesized components are directly integrated. The membrane insertion and integration of nascent polypeptide chains into the ER and bCM is relatively well understood - much of the requisite machinery has been structurally defined and plausible insertion mechanisms have been outlined (2, 3). The integration of membrane lipids on the other hand presents a topologically distinct problem since lipids occupy both leaflets of the bilayer. This necessitates reorientation - or flipping - of lipids from one side of the bilayer to the other.
While in-plane lateral and rotational movements of membrane lipids are rapid, the intrinsic rate at which lipids flip across membranes is very low because the polar headgroup of the lipid must transit through the hydrophobic interior of the membrane. Thermal fluctuations (~1 kcal/mol at ambient temperature) alone are not sufficient to scale the ~15–50 kcal/mol barrier that would be encountered by the common phospholipid phosphatidylcholine (PC) during reorientation from one side of a membrane bilayer to the other (4–6). Consistent with this, PC flips infrequently across artificial membranes (typical time constants range from hours to weeks). Yet PC and other phospholipids flip back and forth rapidly across biogenic membranes, on a timescale of tens of seconds or less.
Polar lipids must flip rapidly across biogenic membranes not only to sustain membrane growth, but also for a number of biosynthetic pathways, especially those involved in the synthesis of glycoconjugates. For example, assembly of bacterial peptidoglycan by the well-established penicillin- and lantibiotic-sensitive pathway requires flipping of a complex peptidoglycolipid across the bCM. Similarly, protein N-glycosylation and GPI-anchoring in eukaryotic cells require that glycolipid precursors are translocated rapidly across the ER. Since flipping is intrinsically slow, biogenic membranes must have a mechanism to increase its rate. While a number of different mechanisms have been discussed over the years, including the possibilities that non-bilayer arrangements of lipids, the mere presence of membrane proteins, or transient defects in bilayer structure could promote flipping, it is now clear that lipid flipping across biogenic membranes requires specific lipid transporters (flippases). These ‘biogenic membrane flippases’ facilitate ATP-independent, transbilayer diffusion of lipids; i.e., they catalyze ‘downhill’ transport of lipids. They are functionally distinct from the more familiar lipid transporters of the P-type ATPase or ABC (ATP-binding cassette) transporter families, typically found in the plasma membrane and late secretory organelles, that couple ATP hydrolysis to concentrative ‘uphill’ transport (7–9).
In this review we highlight the roles of lipid flipping in membrane biogenesis and glycolipid assembly in eukaryotes and prokaryotes. Our focus is on biogenic membranes, specifically the ER and bCM, where lipid flipping is characteristically ATP-independent. Most predicted biogenic membrane flippases have not been identified at the molecular level and the activities of flippases identified by genetic approaches have not been strictly biochemically validated by reconstitution of purified proteins. We describe these systems (thus answering the question posed in the title of this review) and conclude by speculating on the mechanism(s) by which these novel transport proteins might facilitate transbilayer lipid movement.
A note on nomenclature
We are guilty of ‘abusing’ the term flippase (10), since it has come to describe an ATP-driven transporter that unidirectionally transfers lipids from the exoplasmic to the cytoplasmic leaflet. The term floppase describes transporters that catalyze transport in the opposite direction, i.e., in (cyto-) to out (exo-). A biogenic membrane flippase is perhaps more properly referred to as a scramblase. To avoid confusion with the plasma membrane-localized Ca2+-dependent phospholipid scramblase activity (11), and for historical reasons, we continue to use the term ‘flippase’ for transporters that facilitate ATP-independent, bidirectional lipid diffusion across biogenic membranes.
Glycerophospholipid flip-flop across biogenic membranes
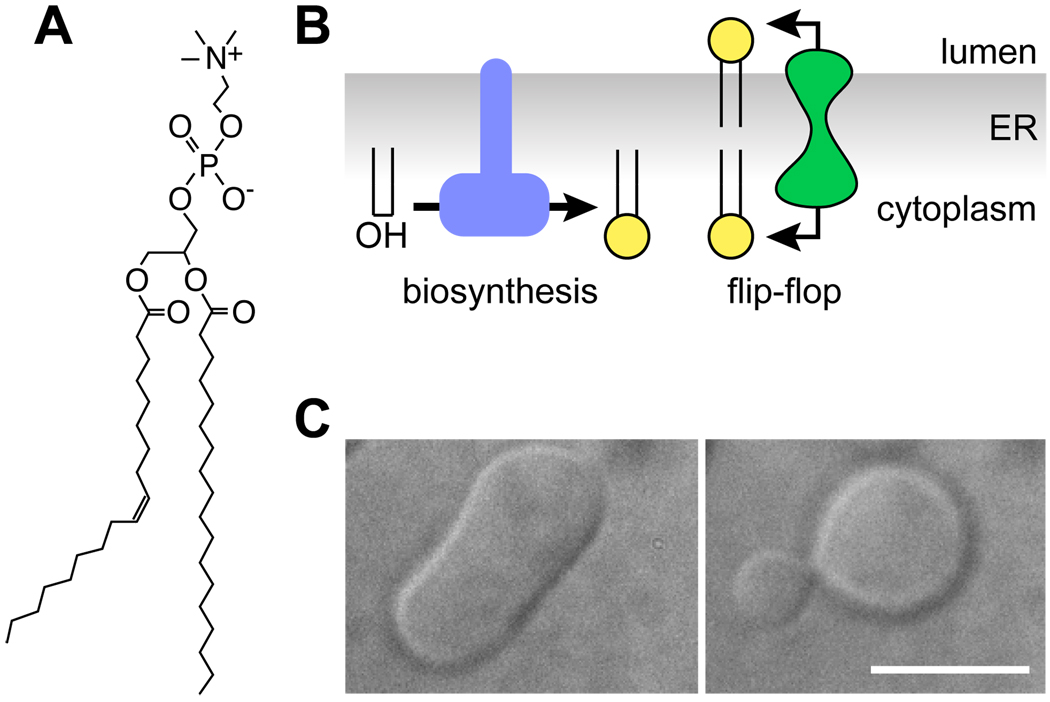
Glycerophospholipids (PLs; e.g., Fig. 1A) are synthesized on the cytoplasmic face of the ER and bCM (12–15). In order to grow the bilayer, newly synthesized PLs - or other PLs in the cytoplasmic leaflet - must flip across the membrane (Fig. 1B). Since the two leaflets of a membrane bilayer are coupled, dramatic membrane deformations can be anticipated if flipping does not occur and PLs accumulate in one leaflet at the expense of the other (10, 16). Fig. 1C shows an example of membrane bending induced by transbilayer lipid number asymmetry: when the outer leaflet of a prolate giant unilamellar vesicle (GUV) is enlarged by introducing a fraction of a mole percent of lyso-PC, the vesicle is deformed into a structure that appears to be on the verge of releasing a bud. Such deformations may have a physiological role in membrane budding and tubulation (17) but are clearly undesirable for bilayer propagation.
Figure 1. Glycerophospholipid synthesis and flip-flop at the ER.
(A) Phosphatidylcholine (PC), a major glycerophospholipid in eukaryotic cells. (B) A glycerophospholipid (PL) biosynthetic enzyme (left) converts diacylglycerol to PL on the cytoplasmic face of the ER. The PL flippase (right) facilitates bidirectional transbilayer translocation of PLs, allowing balanced growth of the bilayer. The ER PL flippase does not itself generate or maintain transbilayer lipid asymmetry. (C) Shape change induced in a prolate giant unilamellar vesicle (left) by the introduction of 0.1 mol percent lyso PC into the outer leaflet. Scale bar, 10 µm. Image courtesy of A. Papadopulos.
The rate of PL synthesis and flipping must match the demand for new membrane as the cell grows. In a bacterial cell that doubles every ~30 min, at least ~5000 PLs must flip across the bCM every second. For Gram-negative cells where PLs are also needed for outer membrane biogenesis, the demand for lipid synthesis and flipping is greater. Measurements of PL flipping across the bCM (14, 18, 19) indicate that it is rapid (t1/2 on the order of tens of seconds), occurring at a much greater rate than required for cell doubling. PL flipping is similarly rapid across the ER membrane (20–22). Biochemical reconstitution studies (23–25) indicate that both the ER and bCM have specific transport proteins (PL flippases) that facilitate flipping. Although their activity has been studied for over 30 years, the PL flippases have thus far eluded identification at the molecular level.
PL flip-flop across the ER membrane and bCM occurs bi-directionally and does not require a source of metabolic energy (26, 27). The PL flippase is nonselective with respect to glycerophospholipid structure (19–22, 26, 28). Not only are PLs with different headgroups flipped with similar rates, but PL stereoisomers containing sn-2,3-diacylglycerol (the isomer found in archaebacteria; PLs in eukaryotes and eubacteria contain sn-1,2-diacylglycerol) are also rapidly translocated (22).
PL flippase activity has been demonstrated in proteoliposomes reconstituted from detergent-solubilized ER or bCM proteins (22–25, 29–34). Fractionation of the protein mixture prior to reconstitution indicates that the activity can be enriched, and also separated from other flippase activities including those required to flip isoprenoid-based lipids (see below). Two different proteins (or variants of the same protein) independently contribute to the PL flippase activity of the ER – one of these proteins is inactivated by diethylpyrocarbonate (DEPC), while the other is inactivated by the sulfhydryl alkylating reagent N-ethylmaleimide (NEM) (30). The two proteins appear broadly similar since they co-fractionate on a number of chromatographic supports such as ion exchange and hydroxyapatite; however, they may be separable by lectin affinity chromatography2. It is not known whether PL flippase activity in the bCM is similarly due to two proteins.
The E. coli inner membrane protein LplT (formerly ygeD), a non-essential polytopic membrane protein with 12 predicted transmembrane spans, has been proposed to flip 2-acyl-glycerophosphoethanolamine (2-acyl GPE) from the periplasmic leaflet to the cytoplasmic leaflet of the inner membrane (35). 2-acyl GPE is formed on the periplasmic face as a result of a transacylation reaction in which the 1-acyl chain of PE is transferred to the N-terminus of the major outer membrane lipoprotein. On its return to the cytoplasmic leaflet, 2-acyl GPE is acylated to regenerate PE. Reacylation is reduced by ~70% in cells that lack LplT; spheroplasted ΔlplT cells also have a reduced ability to take up a fluorescent 2-acyl GPE analog. These results suggest that LplT plays a major - but not unique - role in transporting 2-acyl GPE across the bCM.
LplT is a member of the major facilitator superfamily and likely acts by a uniport mechanism since its function does not require ATP or the proton motive force. Although it transports 2-acyl GPE across the bCM, LplT is unlikely to have a generalized PL flippase function. In some bacteria LplT is fused to the acyltransferase, creating a single protein which functions as a transport/acylation system to regenerate PE; this suggests a dedicated function. Also, since LplT is not found in Gram-positive bacteria there is clearly another protein with possibly wider species distribution that acts as a PL flippase. Even if LplT is not a general PL flippase, it appears to be the first example of a protein capable of facilitating the rapid transbilayer translocation of a glycerophospholipid. While this opens the door to analyses of the lipid transport mechanism of PL flippases, it will first be necessary to confirm LplT’s transport activity via biochemical reconstitution experiments.
Glycolipid flip-flop across the ER
Nascent polypeptide chains are N-glycosylated by oligosaccharyltransferase as they emerge from the protein translocon into the ER lumen (Fig. 2A); in some cases, they are subsequently modified with a glycosylphosphatidylinositol (GPI) lipid anchor (Fig. 2B). The N-glycan is derived from an isoprenoid-based glycolipid, oligosaccharide-diphosphate dolichol (typically Glc3Man9GlcNAc2-PP-dolichol; Fig. 2C), whereas GPI anchors (Fig. 2C) are transferred ‘as is’ to the protein. Oligosaccharide-diphosphate dolichol and GPI must be present in the lumenal leaflet of the ER to participate in these protein modification reactions, but their biosynthesis is initiated on the cytoplasmic face of the ER. Thus a biosynthetic lipid intermediate in each case must flip across the ER membrane in order for glycolipid assembly to be completed (36–39).
Figure 2. Protein N-glycosylation and GPI anchoring in the ER.
(A) Oligosaccharyltransferase (OST) transfers Glc3Man9GlcNAc2 from Glc3Man9GlcNAc2-PP-dolichol (left) to an asparagine residue (yellow) within a glycosylation sequon (Asn-X-Ser/Thr) in the nascent polypeptide (right) as it emerges into the ER lumen. (B) An ER-translocated protein with a C-terminal GPI-directing signal sequence (orange) is a substrate for GPI transamidase (GPIT). GPIT replaces the C-terminal signal sequence with a GPI anchor. The anchor is attached via an amide bond between an ethanolamine residue in the GPI and the α-carboxyl group of the C-terminal amino acid. Symbols are explained in Fig. 3. (C) Structure of Glc3Man9GlcNAc2-PP-dolichol (a 75-carbon yeast dolichol is depicted) and the mammalian GPI H8.
Biosynthesis of the N-glycan precursor involves the stepwise addition of components to dolichyl phosphate (Fig. 3A). The first seven steps convert dolichyl phosphate to Man5GlcNAc2-PP-dolichol (M5-DLO) on the cytoplasmic side of the ER. M5-DLO is then flipped across the bilayer into the ER lumen where the next seven sugars are added. These sugars are derived from the glycolipids mannose-phosphate-dolichol (MPD) and glucose-phosphate-dolichol (GPD), both of which are synthesized on the cytoplasmic face of the ER and must be flipped into the lumen to participate in glycosyltransfer reactions that convert M5-DLO to Glc3Man9GlcNAc2-PP-dolichol. After transfer of the glycan to asparagine residues within specific sequons in the nascent polypeptide, the dolichyl pyrophosphate byproduct is dephosphorylated to dolichyl phosphate in the ER lumen and recycled (40).
Figure 3. Lipid flipping in protein N-glycosylation and GPI anchoring in the ER.
(A) Assembly of Glc3Man9GlcNAc2-PP-dolichol. The first 7 steps, leading to the synthesis of M5-DLO from dolichyl phosphate occur on the cytoplasmic face of the ER. GDP-mannose and UDP-GlcNAc directly contribute the sugar moieties for these reactions. M5-DLO is then flipped into the ER lumen by M5-DLO flippase, and extended in a further 7 reactions to yield Glc3Man9GlcNAc2-PP-dolichol. The sugar donors for these lumenal reactions are MPD and GPD. MPD is synthesized from dolichyl phosphate and GDP-mannose on the cytoplasmic face of the ER, then flipped to the ER lumen by MPD flippase. In the lumen it is the mannosyl donor for the 4 reactions that convert M5-DLO to Man9GlcNAc2-PP-dolichol. GPD is synthesized from dolichyl phosphate and UDP-glucose (not shown) on the cytoplasmic face of the ER, then flipped to the ER lumen by GPD flippase. (B) GPI biosynthesis. GlcN-PI is synthesized in two steps from PI on the cytoplasmic face of the ER, then flipped into the ER lumen to be elaborated into a mature GPI anchor. Lumenal reactions include inositol acylation, and mannose and phosphoethanolamine addition. The inositol acyl group is derived from fatty acyl CoA (FA-CoA) which is transported into the ER by an unknown mechanism. Mannose and phosphoethanolamine residues are derived from MPD and PE, respectively; both these lipids have to be flipped into the ER lumen. (C) Structures of the lipids that are flipped across the ER membrane in the dolichol cycle and GPI biosynthesis pathways. Yeast dolichol (15 isoprene units) is depicted in the M5-DLO, MPD and GPD structures; mammalian dolichol is longer, typically 19 isoprene units.
GPI assembly (Fig. 3B) begins with the synthesis and de-N-acetylation of GlcNAc-PI on the cytoplasmic face of the ER. In mammalian cells GlcN-PI then flips across the ER membrane to the lumenal leaflet where it is inositol acylated, mannosylated and modified with phosphoethanolamine residues to generate a GPI anchor precursor that can be transferred to proteins (39). Variations of this basic biosynthetic scheme are found in almost all eukaryotes. Flipping of GlcN-PI was recently demonstrated in proteoliposomes reconstituted from detergent-solubilized rat liver ER membrane proteins (41). The results suggested that rather than being transported by a dedicated “GPI flippase”, GlcN-PI is likely translocated by the PL flippase in an ATP-independent manner. This conclusion is plausible given the unselective nature of the PL flippase, but remains to be verified. The inositol-linked fatty acid, mannose residues and phosphoethanolamine moieties that are added to GlcN-PI are derived from cytoplasmically synthesized fatty acyl CoA, MPD and PE, respectively, that must be transported to the ER lumen. Translocation of MPD and PE is facilitated by dedicated flippases. Nothing is known about how fatty acyl CoAs enter the ER lumen.
In summary, five different lipids (M5-DLO, MPD, GPD, GlcN-PI and PE; Fig. 3C) must flip across the ER membrane to support the biosynthesis of N-glycosylated and GPI-anchored proteins. In each case flipping is ATP-independent and bi-directional; however, since the flipped lipid is consumed in subsequent biosynthetic steps, flipping is effectively vectorial. A sixth lipid, dolichyl phosphate, generated in lumenal mannosyltransfer (including reactions leading to C- and O-mannosylation of proteins), glucosyltransfer and OST reactions must be returned to the cytoplasmic face of the ER to reenter the ‘dolichol cycle’. Even less is known about how this occurs except that it is likely to involve an ATP-independent mechanism. None of the flippases required for these translocation events has been identified. However, there has been considerable recent progress in characterizing the flippase activities responsible for translocating M5-DLO and MPD. Both activities have been biochemically reconstituted and separated from each other as well as from PL flippase activity. The next two sections summarize these new results.
M5-DLO flippase: biochemical reconstitution studies
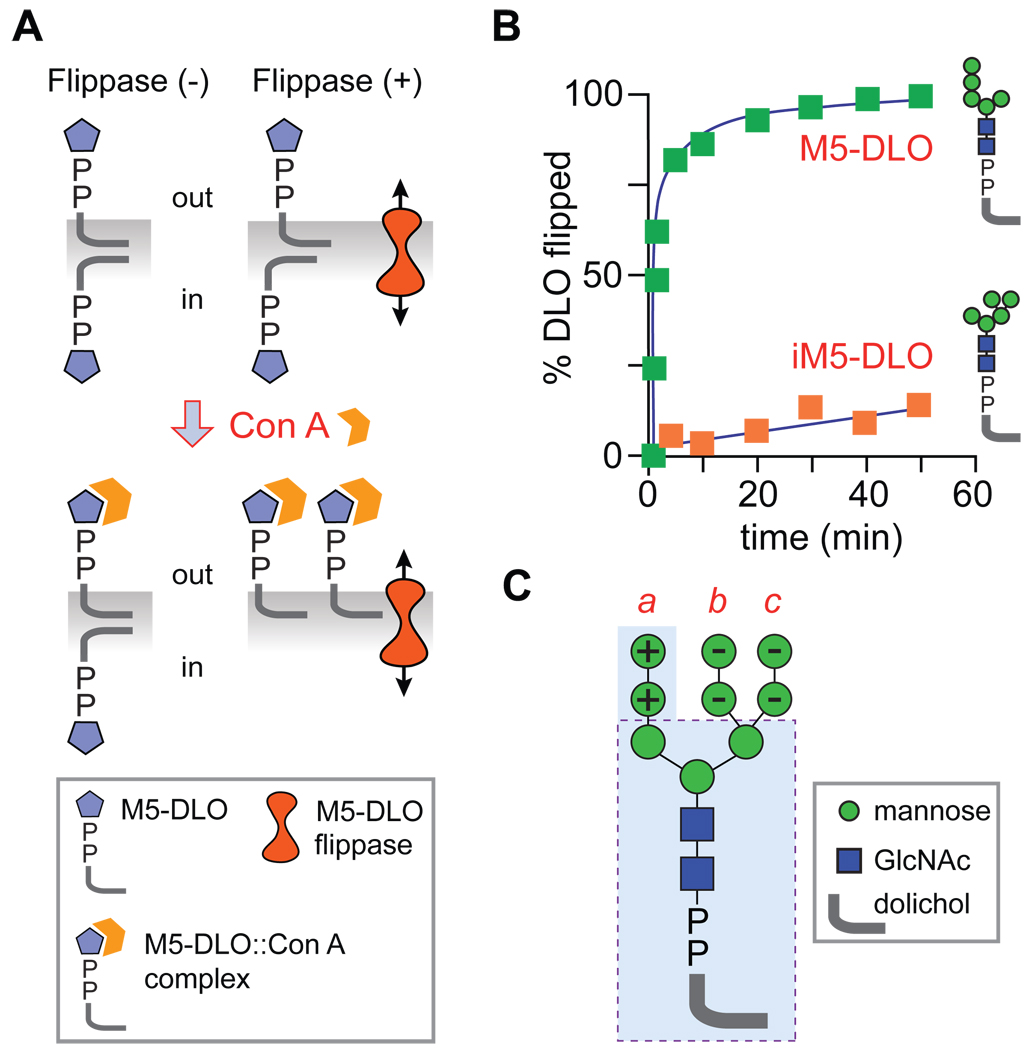
M5-DLO flipping was assayed in a reconstituted system (Fig. 4A) by exploiting the ability of the lectin Concanavalin A (Con A) to bind mannose-containing glycolipids, and in so doing render the lipids insoluble in organic solvents. To assay flipping, large unilamellar liposomes were prepared with trace amounts of [3H]M5-DLO distributed equally between the inner and outer leaflets; proteoliposomes were prepared similarly, except that Triton X-100-solubilized ER membrane proteins (source of M5-DLO flippase) were included during vesicle reconstitution. When protein-free liposomes are treated with Con A (a large, tetrameric protein that cannot cross the membrane), all the [3H]M5-DLO molecules in the outer leaflet are expected to bind the lectin whereas those in the inner leaflet are protected by the membrane barrier. Thus, 50% of the [3H] radioactivity (corresponding to the outer leaflet pool of [3H]M5-DLO) is expected to precipitate with Con A, while the remaining 50% (corresponding to the inner leaflet pool of [3H]M5-DLO) is expected to be organic solvent-extractable. When proteoliposomes containing M5-DLO flippase are assayed in the same way, 100% of the [3H]M5-DLO is expected to interact with Con A and precipitate. This is because in flippase-containing vesicles, the inner leaflet pool of [3H]M5-DLO can flip to the outer leaflet. The fraction of [3H]M5-DLO that can be precipitated in a given sample of vesicles is expected to range from 50–100%, depending on the proportion of vesicles that contain a functional M5-DLO flippase.
Figure 4. M5-DLO flipping in a reconstituted system.
(A) Principle of the M5-DLO flippase assay in reconstituted vesicles. Vesicles are reconstituted with trace amounts of [3H]M5-DLO. Protein-free liposomes or vesicles containing irrelevant membrane proteins are indicated as Flippase (−); proteoliposomes containing the M5-DLO flippase are indicated Flippase (+). On adding the lectin Con A to the vesicles, M5-DLO molecules in the outer leaflet bind Con A and are rendered insoluble in an organic solvent (a mixture of chloroform, methanol and water); M5-DLO molecules in the inner leaflet are protected. Thus, for Flippase (−) vesicles, 50% of the M5-DLO molecules are captured by Con A; for Flippase (+) vesicles, 100% are captured since those in the inner leaflet are translocated to the outer leaflet. (B) Kinetics of flipping (obtained from the rate of capture of M5-DLO by Con A in intact vesicles (42)) of M5-DLO and its structural isomer, iM5-DLO. Symbols are explained in panel C. (C) The structure of Man9GlcNAc2-PP-dolichol is depicted. The M5-DLO sub-structure is contained within the light blue shaded region. M3-DLO (structure enclosed within the dashed lines) is flipped almost as well as M5-DLO. DLO structures containing mannoses indicated “+” are flipped more rapidly whereas the presence of mannose residues indicated by “−” reduces the rate of flipping (42). These rules indicate that M5-DLO is the optimal substrate for the DLO flippase.
Using this assay, M5-DLO flipping was shown to be rapid (τ <1 min; this estimate is limited by the rate at which Con A binds M5-DLO) (Fig. 4B), protein-dependent and ATP-independent (31). M5-DLO flippase activity sedimented operationally at ~4S and could be enriched on a variety of chromatography media, including blue dye resin where it was completely resolved from PL flippase activity (31). Furthermore, the activity bound quantitatively to Con A-Sepharose indicating that M5-DLO flippase is either a glycoprotein or tightly associated with one2. Specificity tests indicated that a non-natural, triantennary isomer of M5-DLO was not flipped (τ ≫200 min)(Fig. 4B), while the rate at which biosynthetic intermediates (such as M6-DLO) were flipped depended on their structure (42). GlcNAc2-PP-citronellol, a water-soluble analog of GlcNAc2-PP-dolichol, was an effective competitor and could drastically reduce the rate of M5-DLO flipping2. The fine specificity revealed through these studies suggests that while M5-DLO is the optimal transport substrate, the M5-DLO flippase has a binding site that recognizes a core component, likely GlcNAc2-PP-dolichol or GlcNAc-PP-dolichol, within the full-length DLO structure (Fig. 4C). The specificity data also validate the reconstitution approach: it is extremely unlikely that residual detergent in the vesicle preparations or potential detergent-induced protein denaturation (43) provides a mechanism that can account for the substrate-specific flipping that is observed
A few years ago it was proposed that Rft1 is the M5-DLO flippase (44, 45). Rft1 is an essential, polytopic, ER membrane protein of yeast that is also found in all eukaryotes that synthesize mannosylated N-glycans. The conclusion that it is the M5-DLO flippase was based on genetic evidence. For example, yeast cells with reduced levels of Rft1 were shown to accumulate M5-DLO and hypoglycosylate proteins, consistent with the inability to transport M5-DLO to the lumenal leaflet. To determine whether Rft1 is indeed the M5-DLO flippase, flippase activity was assayed in proteoliposomes reconstituted with Triton X-100-solubilized yeast microsomal proteins. Activity was unaffected when the protein mixture used for reconstitution was derived from cells with reduced levels of Rft1, or if the mixture was treated with antibodies to quantitatively remove its content of Rft1 (46). Furthermore, M5-DLO flippase activity could be resolved from Rft1 by velocity sedimentation, ion exchange and blue dye resin chromatography of the Triton X-100-solubilized proteins (31, 46). These results indicate that Rft1 is not the M5-DLO flippase. In a more recent study, sealed microsomes from Rft1-depleted yeast were shown to be able to (i) elaborate in situ synthesized [3H]GlcNAc2-PP-dolichol to Man9GlcNAc2-PP-dolichol, a process that requires flipping of M5-DLO, and (ii) transport [3H]GlcNAc2-PP-dolichol15, a water-soluble analog of [3H]GlcNAc2-PP-dolichol, across the membrane into the vesicle interior. These data indicate that M5-DLO flippase activity is unaffected in Rft1-depleted microsomes (47), supporting the results of the biochemical reconstitution studies. The function of Rft1 in N-glycosylation remains to be established.
MPD flippase
MPD is synthesized on the cytoplasmic face of the ER (Fig. 3A) by Dpm1, an enzyme that acts alone in yeast, or as part of a ternary complex (Dpm1-3) in mammals (48). Yeast Dpm1 is a single-pass membrane protein whereas in mammals it owes its membrane association to the other subunits of the ternary complex, Dpm2 and Dpm3. It was proposed that Dpm1 itself functions as the MPD flippase. Initial findings with partially purified yeast Dpm1 and an elegant reconstitution-based approach supported this idea (49). However, it was subsequently shown that the essential function of Dpm1 was retained even in the absence of its transmembrane domain (50). This suggests that Dpm1 is not the MPD flippase.
MPD flipping across the ER has been studied with mannose-P-citronellol (Man-P-Cit), an analog of MPD (51). The analog contains the 10-carbon isoprenoid citronellol (instead of the ~75-carbon dolichol moiety of yeast MPD), making it water-soluble to ~50 mM. Since Man-P-Cit is a substrate for MPD-dependent mannosyltransferases that elongate M5-DLO to Man9GlcNAc2-PP-dolichol (Fig. 3A)(52), it was reasoned that it would be a suitable reporter of MPD flipping. Incubation of Man-P-Cit with sealed rat liver ER vesicles resulted in its transport into the intravesicular space. Transport occurred via a saturable, equilibrative, ATP-independent, trypsin- and DEPC-sensitive process. Manα-P-Cit (the non-natural isomer) was poorly transported, indicating that the MPD transport system is stereoselective (51). Interestingly, glucose-P-citronellol, a GPD analog that is itself transported into ER vesicles (53), did not compete with Man-P-Cit for uptake (51). The lack of competition between the two analogs suggests that MPD and GPD are translocated by different flippases.
Man-P-Cit uptake was also demonstrated in sealed proteoliposomes formed from an octylglucoside-solubilized mixture of egg phospholipids and rat liver ER proteins (54). Uptake in the reconstituted system recapitulated characteristics of Man-P-Cit uptake by ER vesicles: it was ATP-independent, stereoselective and sensitive to DEPC. Fractionation of the octylglucoside-solubilized ER protein mixture prior to reconstitution revealed that Man-P-Cit transport activity could be enriched by anion exchange chromatography. Using a membrane-impermeant oxidant as a topological probe, we recently developed an assay for MPD flipping in a reconstituted system using the radiolabeled natural lipid ([3H]MPD), rather than Man-P-Cit2. With this assay we showed that MPD flippase is not a glycoprotein, and can be resolved from M5-DLO flippase by lectin affinity and ion exchange chromatography. Competition experiments with water-soluble isoprenoid phosphates revealed a surprising specificity: [3H]MPD flipping was inhibited by citronellyl phosphate, but not with neryl phosphate. Since nerol differs from citronellol solely by a cis-unsaturated α-isoprene, this result suggests that MPD flippase recognizes the α-isoprene unit of the carrier lipid, in addition to the mannose residue as described above. The observation that Cit-P, a dolichyl phosphate analog, is recognized by MPD flippase raises the possibility that MPD flippase - and perhaps also GPD flippase - may play a role in recycling dolichyl phosphate.
Lec35/MPDU1 mutants of Chinese hamster ovary cells synthesize MPD normally but are unable to mannosylate M5-DLO (55, 56). The mutant cells are also unable to C-mannosylate proteins and cannot add mannose to GlcN-acylPI in GPI biosynthesis. These results suggest that Lec35, a polytopic ER membrane protein, is somehow involved in making MPD available for lumenal mannosyltransfer reactions. But is Lec35 the MPD flippase? The ER lumenal mannosylation defects displayed by the Lec35 mutant are not recapitulated in sealed microsomes prepared from the mutant cells, but can be reproduced if gentler methods are used to prepare an ‘in vitro’ sample. Thus, Lec35 cells permeabilized with the pore-forming toxin Streptolysin O cannot elaborate M5-DLO to Man9GlcNAc2-PP-dolichol when supplemented with GDP-mannose. However, when the permeabilized cells are supplemented with Man-P-Cit, Man9GlcNAc2-PP-dolichol synthesis is restored. These data suggest that Lec35 is not the MPD flippase, but may play a role in regulating access of MPD to the flippase, or to the various lumenal mannosyltransferases (56), a requirement that can be bypassed by Man-P-Cit. The observation that Lec35 cells also display a GPD utilization defect suggests that Lec35 has a more general role in utilization of dolichyl-P-monosaccharides (56).
Bacterial cell wall assembly: flipping of Lipid II across the bCM
The bacterial cell wall envelopes the cell and protects it against osmotic stress. It lies outside the bCM in the periplasmic/exoplasmic space and consists primarily of peptidoglycan, a polymer of alternating, β1,4 glycosidically-linked N-acetylmuramic acid (MurNAc, a derivative of GlcNAc) and GlcNAc residues. The polymer chains are crosslinked to each other via short peptides that are attached to MurNAc. Biosynthesis of peptidoglycan (Fig. 5A) is a multi-step process that starts on the cytoplasmic face of the bCM and results in delivery of MurNAc(pentapeptide)-β1,4GlcNAc building blocks to the periplasmic/exoplasmic space (Fig. 5A)(57–60). The building blocks are assembled on a carrier lipid (undecaprenol), and transferred across the membrane as a result of lipid flipping.
Figure 5. Flipping of glycolipids across the bCM.
(A) Peptidoglycan assembly. MurNAc-pentapeptide is transferred from UDP-MurNAc-pentapeptide to undecaprenyl phosphate to form Lipid I. Addition of GlcNAc from UDP-GlcNAc to Lipid I generates Lipid II on the cytoplasmic face of the bCM. Lipid II is translocated by a flippase to the exoplasmic/periplasmic surface. Here, its GlcNAc-MurNAc-peptide headgroup is transferred to a growing peptidoglycan polymer by transglycosylation, following which undecaprenyl diphosphate is recycled. Transpeptidation reactions link polymer chains to form the cell wall (orange bricks). Penicillin binding proteins (PBPs), many of which are bi-functional and contain both transglycosylase and transpeptidase activities, catalyze these later steps. (B) O-antigen and lipopolysaccharide assembly in Gram-negative bacteria. Hexa-acylated Lipid A, synthesized on the cytoplasmic surface of the bCM, is modified with 3-deoxy-d-manno-octulosonic Acid (Kdo) residues and a core oligosaccharide before being translocated to the periplasmic face by the ABC transporter MsbA. In parallel, the undecaprenyl diphosphate-linked repeat unit of the O-antigen is flipped across the bCM; the flippase is presumed to be a member of the Wzx family of proteins. The O-antigen units are polymerized as shown, then ligated to Lipid A-Kdo2-core. (C) Structure of Lipid II (shown with lysine in its pentapeptide side-chain; diaminopimelic acid is also commonly found instead of lysine), GlcNAc-PP-undecaprenol and aminoarabinose-P-undecaprenol and Lipid A-Kdo2-core oligosaccharide (decorated with O-antigen and aminoarabinose (in blue); these modifications to Lipid A-Kdo2-core oligosaccharide occur on the periplasmic face of the bCM). GlcNAc-PP-undecaprenol is recognized by the O-antigen assembly system, resulting in transfer of GlcNAc to lipid A-core (72). An analog of GlcNAc-PP-undecaprenol was used to test the role of WzxE as a flippase (71).
In the first step of biosynthesis, catalyzed by the polytopic membrane protein MraY, MurNAc(pentapeptide)-phosphate is transferred from UDP-MurNAc-pentapeptide to undecaprenyl phosphate to form ‘Lipid I’. Addition of GlcNAc from UDP-GlcNAc to Lipid I, catalyzed by the peripheral membrane protein MurG, forms Lipid II on the cytoplasmic face of the bCM. Lipid II is flipped across the bCM to serve as a substrate for the polymerization reactions that lead to peptidoglycan production in the exoplasmic space. Each polymerization step generates undecaprenyl pyrophosphate as a byproduct. Like dolichyl pyrophosphate released during protein N-glycosylation in the ER, undecaprenyl pyrophosphate is dephosphorylated to the monophosphate form and recycled.
Lipid II must be flipped across the bCM at a rate of ~5000 molecules.s−1 to match the rate at which the peptidoglycan polymer is assembled in rapidly growing E. coli cells (61). Flip-flop of Lipid II was recently studied (62) using NBD-Lipid II, a fluorescently tagged version in which the lysine residue in the pentapeptide side-chain (Fig. 5C) was modified with a fluorescent NBD group. To assay transport of NBD-Lipid II across E. coli inner membrane vesicles, the lipid was synthesized in situ by introducing the precursors UDP-MurNAc-(NBD)pentapeptide and UDP-GlcNAc into the vesicle interior by a freeze-thaw-resealing process. Antibodies were used to detect and trap NBD-Lipid II molecules that flipped from the inner leaflet of the sealed vesicles to the vesicle exterior. Using this approach it was shown that NBD-Lipid II flipped relatively rapidly across E. coli inner membrane vesicles with t1/2 ~5 min at 14°C (62). Transport did not require ATP or the proton motive force, but clearly required the participation of one or more bCM proteins since spontaneous flipping of NBD-Lipid II across liposomal membranes could not be detected over a 3 hr period.
Two groups proposed that the polytopic membrane protein MviN is the putative Lipid II flippase (63, 64). MviN is essential in E. coli. When MviN function is reduced by regulating the level of expression of the protein or by replacing the endogenous protein with a temperature-sensitive variant and growing the cells at the non-permissive temperature, a specific growth defect is observed. The cells lyse in normal media and form spheroplasts in the presence of an osmotic stabilizer such as sucrose. They also accumulate nucleotide and lipid precursors of peptidoglycan, including Lipid II. Since Lipid II accumulates and peptidoglycan synthesis appears to be compromised, these data suggest that MviN is the Lipid II flippase. However, recent results argue against this conclusion (65).
Peptidoglycan synthesis is essential in Bacillus subtilis as it is in E. coli, and so it is reasonable to expect that the protein responsible for flipping Lipid II should be essential and conserved in this organism. B. subtilis has four homologs of MviN; strains harboring mutations in each of these homologs, or all four of them simultaneously, display normal growth and morphology (65) indicating that the MviN homologs are not essential in B. subtilis. This suggests that MviN and its homologs are not directly responsible for flipping Lipid II. It may be that there are additional (redundant) proteins that flip Lipid II in B. subtilis, or that MviN has an accessory role in Lipid II flipping that is essential in E. coli but not essential in B. subtilis. Resolution of this point and definitive identification of the lipid II flippase await biochemical tests.
The cell envelope of Gram-negative bacteria: flipping of undecaprenol-based glycolipids
Lipopolysaccharide (LPS), a major component of the cell surface of Gram-negative bacteria, is composed of lipid A, a core oligosaccharide (including inner Kdo residues) and in some cases, O-antigen (Fig. 5C). Lipid A (endotoxin) is a multiply acylated glucosamine-based phospholipid that serves as the hydrophobic anchor of LPS. O-antigen is a repeat unit polysaccharide that accounts for O-serotype specificity. There are two mechanisms for the biosynthesis of O-antigens, only one of which (the wzy polymerase-dependent pathway) concerns us here. In this pathway (Fig. 5B), O-antigen oligosaccharide units (comprising 2–6 sugar residues) are assembled on a lipid carrier. Biosynthesis of the core-lipid A structure and undecaprenyl-PP-linked O-antigen units occurs on the cytoplasmic face of the inner membrane. These lipids are flipped to the periplasmic face where the O-antigen units are polymerized by Wzy, then attached to core-lipid A by the ligase WaaL. The resulting glycoconjugate is a giant hexa-acylated lipid that must be transported to the outer face of the outer membrane.
Here we discuss the transbilayer translocation of undecaprenyl-PP-linked O-antigen units and also that of undecaprenyl-P-aminoarabinose, a lipid required for the modification of the 4’-phosphate of Lipid A on the periplasmic face of the inner membrane. Lipid A itself is translocated across the bCM via an ATP-dependent mechanism, catalyzed by the ABC transporter MsbA (66, 67). After being modified at the periplasmic face of the bCM, it is transported to the outer face of the outer membrane by a route that requires the Lpt proteins. The subject of Lipid A transport across the bCM and to the outer membrane is beyond the scope of this review; more information may be found in reference (68).
Flipping of lipid-linked O-antigen units
Undecaprenyl-PP-linked O-antigen units are synthesized from undecaprenyl phosphate and nucleotide sugars. The first reaction transfers a monosaccharide phosphate (frequently hexose-P or hexosamine-P), accounting for the diphosphate bridge in the lipid. The Wzx family of proteins has been proposed to flip undecaprenyl-PP-linked O-units across the inner membrane (69). These proteins are polytopic membrane proteins, typically with twelve predicted transmembrane spans (70). Sequence analysis provides no clues as to their mechanism of action. In a wzx mutant strain, undecaprenyl-PP-O units are synthesized and accumulate in the bCM, but O units are not ligated to core-lipid A (69). This is consistent with a defect in undecaprenyl-PP-O unit translocation due to lack of Wzx. However, there is no definitive evidence that the undecaprenyl-PP-O units accumulate on the cytoplasmic face of the bCM as would be expected in the absence of flippase activity.
The flippase function of a Wzx family member was recently analyzed in a study of WzxE. WzxE is involved in the synthesis of enterobacterial common antigen (ECA), a cell surface polysaccharide found in all enteric bacteria. Like O-antigen, it is made up of oligosaccharide repeat units. The ECA unit, a trisaccharide of amino sugars with GlcNAc at the reducing end, is synthesized as the undecaprenyl diphosphate-linked intermediate. This lipid is flipped across the bCM where the ECA units are polymerized and transferred primarily to phosphoglycerides. It has been proposed that WzxE is responsible for flipping undecaprenyl-PP-linked ECA units. A radiolabeled, water-soluble analog of GlcNAc-PP-undecaprenol was used as a reporter to test flippase activity of sealed, everted cytoplasmic membrane vesicles from WzxE-competent and wzxE null E. coli strains. The analog, GlcNAc-PP-nerol, was transported into the vesicles by a WzxE-dependent, ATP-independent and bidirectional process (71). While this result would appear to provide clear support for the proposal that WzxE can flip GlcNAc-PP-undecaprenol and is likely the flippase for undecaprenyl diphosphate-linked ECA units, there are complications. The vesicles used for these assays have the O-antigen Wzx protein, WzxO16, which is expected to recognize GlcNAc-PP-undecaprenol based on in vivo studies (72–74). Despite this apparent functional redundancy, deletion of wzxO16 had no effect on GlcNAc-PP-nerol transport, indicating that uptake of the analog is solely linked to WzxE. It is possible that WzxO16 is expressed at much lower levels than WzxE in the E. coli strain from which the vesicles were prepared, or that it cannot transport the short chain analog efficiently. Although the discrepancy between the in vivo and in vitro results remains to be clarified, the analog experiments provide the first evidence suggesting that a Wzx family member can function as a lipid transporter.
Modification of Lipid A
Lipid A has phosphate groups at the 1 and 4’ positions, one on each of the glucosamine residues that make up its structural backbone. These phosphates can be modified under certain growth conditions or in some mutants. For example, In Salmonella, the modification of lipid A phosphates occurs upon activation of the PmrA-PmrB regulon (75) which governs resistance to the antimicrobial peptide polymyxin. Here, 4-amino-4-deoxy-l-Arabinose (l-Ara4N) is added to the 4' phosphate of lipid A, reducing its net negative charge and lowering electrostatic interactions between the cell surface and cationic antimicrobial peptides. l-Ara4N is transferred to undecaprenyl phosphate by ArnC at the cytoplasmic surface of the inner membrane; the resulting undecaprenyl-P-aminoarabinose lipid is flipped to the periplasmic face where it supplies l-Ara4N for modification of lipid A. A heterodimeric complex of PmrL and PmrM (renamed ArnE and ArnF) is implicated in undecaprenyl-P-aminoarabinose flipping (76). Mutants of arnE-arnF generated in a polymyxin resistant parental strain regain polymyxin sensitivity, and despite high levels of undecaprenyl phosphate-l-Ara4N, lipid A modification is abolished. Chemical modification of undecaprenyl phosphate-l-Ara4N with an inner membrane impermeable reagent showed that the lipid was less accessible in the mutant strain, relative to the parent, indicating that it could not be translocated across the inner membrane. ArnE and ArnF are predicted to have four transmembrane spans each; the ArnE-ArnF heterodimeric complex resembles those of small multidrug resistance transporters that use the energy of electrochemical gradients to transport substrates. While it appears likely that the ArnE-ArnF complex is the undecaprenyl-P-aminoarabinose flippase, its mechanism of action - specifically its metabolic energy requirement - remains to be addressed through biochemical reconstitution of its activity.
How does a biogenic membrane flippase work?
There is no simple answer to this question since many of the transporters have yet to be identified and, for proteins such as LplT and Wzx, flipping activity remains to be verified by biochemical reconstitution. However, it is clear that there are at least two types of ATP-independent flipping mechanism that must be considered, one used by the relatively unspecific PL flippases and another used by the more specific isoprenoid-lipid flippases. Here we ignore the spirit of the Racker aphorism ‘don’t waste clean thoughts on dirty enzymes’ (77) by speculating on possible mechanisms.
General considerations
The lipid headgroup may be regarded as a solute, albeit an unusual solute because of its membrane-anchoring hydrocarbon chain(s). There are a number of paradigms for the facilitated transport of solutes by dedicated transport proteins and it is interesting to consider whether these can be adapted to understand lipid translocation. In the ‘alternating access’ model (78), the transport protein switches between two conformations in which a central cavity is open to one or other side of the membrane. The conformational cycle may be linked to solute binding and dissociation. A solute entering the cavity from one side can exit on the other side of the bilayer when the corresponding open conformation is available. In the case of ion channels, ions move through a water-filled pore within the channel as they diffuse from one side of the membrane to the other (79). Selectivity and gating ensure that the pore transmits only particular ions in a regulated fashion. A flippase could operate by alternating access, or via a pore mechanism, provided that it has an amphipathic architecture that allows the hydrocarbon portion of the lipid to remain in the hydrophobic interior of the bilayer even as the transiting lipid headgroup is accommodated within an aqueous cavity or pore. Another possibility would be for the transport machinery to encapsulate the entire lipid during its transmembrane passage; structures capable of binding an entire lipid have been described for numerous cytoplasmic lipid transport proteins, e.g., Sec14 (80).
Pore models
Atomic scale molecular dynamics simulations suggest that transbilayer movement of PLs in protein-free bilayers occurs rapidly in the presence of membrane-spanning water pores (81, 82). The simulations show that once the lipid encounters the pore, diffusive translocation begins – the headgroup moves through the bilayer along the pore, while the acyl chains desorb from the membrane leaflet and enter the opposing leaflet. Midway, the lipid becomes oriented parallel to the plane of the bilayer. Spontaneous pore formation is infrequent, hence rate-limiting, accounting for the low rate at which lipids flip across artificial membranes.
The simulation data suggest that a stable, proteinaceous pore of suitable geometry could act as flippase in the manner described above for a naked water pore. This is seen in the case of certain pore-forming peptides such as the antibiotic magainin, an amphipathic α-helical peptide from frog skin that forms toroidal pores in membranes and causes fluorescent PL analogs to flip in a few minutes (83)(see ref (84) for a discussion of phospholipid flip-flop induced by other pore-forming peptides). The topology of the pore, which is sustained by a cluster of at least four magainin peptides, is such that the two leaflets of the bilayer form a continuum. Since the pore lumen is lined by both peptides and PL headgroups, PLs can diffuse laterally from one leaflet of the bilayer to the other. Presumably the headgroup of the lipid moves through the aqueous pore while its hydrocarbon chains, intercalating between the magainin peptides, remain in the hydrophobic interior of the membrane. The ability of a lipid to exchange between leaflets through such a pore may depend on the size of its headgroup and the flexibility of its hydrocarbon chain(s). It would be interesting to test whether isoprenoid-based lipids could exchange between membrane leaflets by such a mechanism. As suggested below, the unusual conformation of the isoprenoid chain may preclude transport of these lipids through a pore.
It has been hypothesized that the protein translocon, a conserved component of biogenic membranes, could act as a PL flippase by virtue of its pore geometry (25): it not only has an aqueous channel for translocating proteins but also a hydrophobic lateral avenue for integration of transmembrane helices into the lipid bilayer. However reconstitution experiments showed that the protein translocon does not contribute to PL flippase activity in the ER and bCM (32, 34). While there is no evidence one way or another to support the idea that biogenic membrane flippases have a toroidal pore geometry, a pore model nevertheless provides a useful way to think about the mechanism of flipping. Gating the pore, or twisting its peptide scaffold to narrow the pore diameter would restrict solute permeation while retaining lipid exchange capability. A variation of the overt pore model is shown in Fig. 6A. Here, the flippase is a polytopic membrane protein that has a number of water molecules in the interior of its transmembrane region as reported for Class A G-protein coupled receptors (85). These molecules would comprise the ‘pore’ with which the PL headgroup could interact while translocating across the bilayer.
Figure 6. Flippase mechanisms.
(A) A pore model for the PL flippase provides a central hydrophilic path for the transiting headgroup while leaving the hydrophobic chains of the lipid in the bilayer. The lipid is shown (in a number of snapshots) intercalating between transmembrane spans of the flippase. In this model the lipid is not specifically recognized, but conformationally constrained hydrophobic entities such as dolichol would prevent MPD or M5-DLO from intercalating into and being transported by the PL-flippase. (B) Slip-pop model for phospholipid transport. The dynamic behavior of hydrophobic single-membrane-spanning proteins causes transient defects in the lipid-helix interface that allow phospholipids to flip-flop across the bilayer. (C) Possible mechanism for the M5-DLO flippase (flippases for MPD (Fig. 3C) and other isoprenoid-based lipids such as Lipid II (Fig. 5C), may use a similar mechanism). Snapshots of a transiting M5-DLO molecule are shown. M5-DLO specifically binds to the flippase at one or other membrane interface, then exchanges with a symmetrically located binding site from which it is released into the bilayer. A single, centrally located binding site can also be envisaged, possibly located in a ‘thin’ portion of the membrane.
Flipping facilitated by transmembrane peptides
In an interesting series of experiments, Kol et al. (86, 87) showed that certain fluorescent PL analogs flip rapidly in vesicles reconstituted with α-helical peptides that do not form pores. The peptides used in these studies were designed to mimic the transmembrane segments of membrane proteins. NBD-PG (a fluorescent analog of phosphatidylglycerol) was flipped with a t1/2~10 min when the lysine-flanked peptide KALP23 (acetyl-GKKL(AL)8KKA-amide) was incorporated into vesicles at a concentration of ~1000 peptides per vesicle,. Flipping was lipid specific since NBD-PE was flipped much more slowly (t1/2~1000 min). Neither lipid was flipped in the absence of peptide, as expected, and fluorescent analogs of PC and PS were not flipped (a recent independent study confirmed that KALP23 did not flip natural PC (6)). Further studies revealed that the rate of NBD-PG flipping was (i) sensitive to the flanking residue in the peptide and the lipid composition of the vesicle, and (ii) a linear function of peptide concentration. The latter result suggests that flipping is mediated by peptide monomers. Kol et al. extended their studies to membrane proteins (88) and showed that the rate of NBD-PG flipping in vesicles containing bacterial leader peptidase or the tetrameric potassium channel (~200 copies of protein per vesicle) was increased relative to protein-free vesicles or vesicles containing other membrane proteins such as MsbA. However, the rate of NBD-PG flipping under these conditions was slower than in vesicles containing the same mole percent of KALP23 peptide.
Based on these results, it was hypothesized that the mere presence of membrane proteins may endow biogenic membranes with the ability to flip PLs, and that high cholesterol levels would prevent this from occurring at the eukaryotic plasma membrane where it is important to preserve transbilayer lipid asymmetry. Indeed, the presence of cholesterol was found to depress the rate of peptide-induced NBD-PG flipping (87). Since it is possible to reconstitute particular mixtures of ER or bCM membrane proteins into proteoliposomes without conferring PL flippase activity to the vesicles (24, 25, 31, 32), this proposal cannot be correct as stated. Nevertheless, the idea that flipping occurs via peptide-induced local defects is an interesting one that could be adapted to understand how a specific flippase might work. Thus, in the “slip-pop” model of Kol et al. (89), local perturbations induced by dynamic movements of a transmembrane helix would cause a phospholipid to “slip” into the membrane interior into a transition state and “pop” out either on the same or on the opposite surface of the membrane (Fig. 6B). Related to this proposal is the idea that PLs in the vicinity of a membrane protein can become organized in a ‘hairpin bend’ with their headgroups aligned against the protein surface (27). This would effectively connect the two leaflets of the membrane, allowing lipid exchange via rapid lateral diffusion.
Flipping of isoprenoid-based lipids
We have discussed how the relatively unspecific flipping of PLs might occur. What about isoprenoid-based lipids such as M5-DLO, MPD and Lipid II? There are two points to consider. First, polyisoprenoids such as yeast and mammalian dolichols (C75 and C95, respectively) and bacterial undecaprenol (C55) are considerable hydrocarbon chains that are predicted to adopt a compact, tripartite L-shaped structure (60, 90), quite different from that of the diacylglycerol moiety of common PLs. It is easy to appreciate their unusual structure by noting that when fully extended they are longer than the thickness of a membrane bilayer. The potential conformational constraints imposed by the compacted structure of these long chain hydrocarbons within the membrane may prevent isoprenoid-based lipids from accessing the ‘unspecific’ modes of flipping discussed above. The second point is that the flipping of isoprenoid-based lipids has been experimentally determined to be quite specific. For example, M5-DLO flippase does not flip the isomeric structure iM5-DLO (Fig. 4B), and MPD flippase distinguishes between MPD and GPD and also recognizes the isoprene unit proximate to the phosphate group. These and other data indicate that the isoprenoid lipid flippases have a binding site that recognizes elements of the lipid headgroup in conjunction with a portion of the isoprenoid support. The binding site would likely be positioned at the membrane-water interface. Since the flippases are bidirectional, they must have two binding sites, one at each side of the membrane; alternatively, the binding site could be centrally located in a thinner region of the protein. Flipping would proceed by specific binding of a lipid headgroup to one side of the flippase, followed by translocation to the other side with the hydrophobic portion of the lipid remaining in the interior of the membrane (Fig. 6C).
Concluding remarks
Flipping of polar lipids, an intrinsically unfavorable process, has to occur rapidly for membrane growth, as well as for the topologically split pathways through which essential extracellular glycoconjugates are synthesized in eukaryotes and prokaryotes. Whereas the biosynthetic reactions of glycoconjugate biosynthesis have been largely defined, with genes/proteins assigned to each step, the crucial lipid flipping events remain a mystery. With the possible exception of the Wzx protein family - and even here the evidence is far from complete - the molecular identity of the ATP-independent flippases is unknown. Traditional impediments to the biochemical analysis of membrane transport processes appear to have been overcome, at least in some instances, with recently developed reconstitution-based assays through which flipping of PLs, M5-DLO and MPD has been characterized and the corresponding activities separated. The highly specific flipping of isoprenoid-based lipids suggests that it may be possible to label or inhibit the flippases with specific reagents, providing a clear opportunity to bring chemical approaches to bear on this problem. Conflation of these methods with genetic and genomic approaches should allow the flippases to be identified and the study of their novel transport mechanism(s) to begin.
Acknowledgments
This work was supported by NIH grant GM71041 and the Cornell-Rockefeller-Sloan-Kettering Tri-Institutional Training Program in Chemical Biology. We acknowledge http://www.bobdylan.com/#/songs/who-killed-davey-moore for a portion of the title, Charles Rock, Jonathan Dworkin, Matthijs Kol, Stanley Lo, Arun Radhakrishnan and Jeff Rush for comments on different sections of the manuscript, Andreas Papadopulos for Figure 1C, and Jeremy Dittman for help with figure preparation.
Footnotes
Abbreviations: ABC, ATP-binding cassette; 2-acyl GPE, 2-acyl-glycerophosphoethanolamine; bCM, bacterial cytoplasmic membrane; ECA, enterobacterial common antigen; ER, endoplasmic reticulum; Glc, glucose; GlcNAc, N-acetylglucosamine; GPI, glycosylphosphatidylinositol; LPS, lipopolysaccharide; M5-DLO, Man5GlcNAc2-PP-dolichol; Man, mannose; Man-P-Cit, mannose-P-citronellol; MPD, mannosylphosphoryl dolichol; MurNAc, N-acetylmuramic acid; NBD, nitrobenz-2-oxa-1,3-diazole; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PL, glycerophospholipid
SS, AKM; unpublished results
References
- 1.Palade GE. Membrane biogenesis: an overview. Methods Enzymol. 1983;96:xxix–lv. doi: 10.1016/s0076-6879(83)96004-4. [DOI] [PubMed] [Google Scholar]
- 2.Blobel G. Protein targeting (Nobel lecture) Chembiochem. 2000;1:86–102. doi: 10.1002/1439-7633(20000818)1:2<86::AID-CBIC86>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 3.White SH, von Heijne G. How translocons select transmembrane helices. Annu Rev Biophys. 2008;37:23–42. doi: 10.1146/annurev.biophys.37.032807.125904. [DOI] [PubMed] [Google Scholar]
- 4.Kornberg RD, McConnell HM. Inside-outside transitions of phospholipids in vesicle membranes. Biochemistry. 1971;10:1111–1120. doi: 10.1021/bi00783a003. [DOI] [PubMed] [Google Scholar]
- 5.Liu J, Conboy JC. 1,2-diacyl-phosphatidylcholine flip-flop measured directly by sum-frequency vibrational spectroscopy. Biophys J. 2005;89:2522–2532. doi: 10.1529/biophysj.105.065672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakano M, Fukuda M, Kudo T, Matsuzaki N, Azuma T, Sekine K, Endo H, Handa T. Flip-flop of phospholipids in vesicles: kinetic analysis with timeresolved small-angle neutron scattering. J Phys Chem B. 2009;113:6745–6748. doi: 10.1021/jp900913w. [DOI] [PubMed] [Google Scholar]
- 7.Lenoir G, Williamson P, Holthuis JC. On the origin of lipid asymmetry: the flip side of ion transport. Curr Opin Chem Biol. 2007;11:654–661. doi: 10.1016/j.cbpa.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 8.Graham TR. Flippases and vesicle-mediated protein transport. Trends Cell Biol. 2004;14:670–677. doi: 10.1016/j.tcb.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 9.Pomorski T, Menon AK. Lipid flippases and their biological functions. Cell Mol Life Sci. 2006;63:2908–2921. doi: 10.1007/s00018-006-6167-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Devaux PF, Herrmann A, Ohlwein N, Kozlov MM. How lipid flippases can modulate membrane structure. Biochim Biophys Acta. 2008;1778:1591–1600. doi: 10.1016/j.bbamem.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 11.Daleke DL. Phospholipid flippases. J Biol Chem. 2007;282:821–825. doi: 10.1074/jbc.R600035200. [DOI] [PubMed] [Google Scholar]
- 12.Bell RM, Ballas LM, Coleman RA. Lipid topogenesis. J Lipid Res. 1981;22:391–403. [PubMed] [Google Scholar]
- 13.Vance DE, Choy PC, Farren SB, Lim PH, Schneider WJ. Asymmetry of phospholipid biosynthesis. Nature. 1977;270:268–269. doi: 10.1038/270268a0. [DOI] [PubMed] [Google Scholar]
- 14.Huijbregts RP, de Kroon AI, de Kruijff B. Topology and transport of membrane lipids in bacteria. Biochim Biophys Acta. 2000;1469:43–61. doi: 10.1016/s0304-4157(99)00014-3. [DOI] [PubMed] [Google Scholar]
- 15.Cronan JE. Bacterial membrane lipids: where do we stand? Annu Rev Microbiol. 2003;57:203–224. doi: 10.1146/annurev.micro.57.030502.090851. [DOI] [PubMed] [Google Scholar]
- 16.Sheetz MP, Singer SJ. Biological membranes as bilayer couples. A molecular mechanism of drug-erythrocyte interactions. Proc Natl Acad Sci U S A. 1974;71:4457–4461. doi: 10.1073/pnas.71.11.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burger KN. Greasing membrane fusion and fission machineries. Traffic. 2000;1:605–613. doi: 10.1034/j.1600-0854.2000.010804.x. [DOI] [PubMed] [Google Scholar]
- 18.Rothman JE, Kennedy EP. Rapid transmembrane movement of newly synthesized phospholipids during membrane assembly. Proc Natl Acad Sci U S A. 1977;74:1821–1825. doi: 10.1073/pnas.74.5.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hrafnsdóttir S, Nichols JW, Menon AK. Transbilayer movement of fluorescent phospholipids in Bacillus megaterium membrane vesicles. Biochemistry. 1997;36:4969–4978. doi: 10.1021/bi962513h. [DOI] [PubMed] [Google Scholar]
- 20.Buton X, Morrot G, Fellmann P, Seigneuret M. Ultrafast glycerophospholipid-selective transbilayer motion mediated by a protein in the endoplasmic reticulum membrane. J Biol Chem. 1996;271:6651–6657. doi: 10.1074/jbc.271.12.6651. [DOI] [PubMed] [Google Scholar]
- 21.Marx U, Lassmann G, Holzhutter HG, Wustner D, Muller P, Hohlig A, Kubelt J, Herrmann A. Rapid flip-flop of phospholipids in endoplasmic reticulum membranes studied by a stopped-flow approach. Biophys J. 2000;78:2628–2640. doi: 10.1016/S0006-3495(00)76807-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vishwakarma RA, Vehring S, Mehta A, Sinha A, Pomorski T, Herrmann A, Menon AK. New fluorescent probes reveal that flippase-mediated flip-flop of phosphatidylinositol across the endoplasmic reticulum membrane does not depend on the stereochemistry of the lipid. Org Biomol Chem. 2005;3:1275–1283. doi: 10.1039/b500300h. [DOI] [PubMed] [Google Scholar]
- 23.Backer JM, Dawidowicz EA. Reconstitution of a phospholipid flippase from rat liver microsomes. Nature. 1987;327:341–343. doi: 10.1038/327341a0. [DOI] [PubMed] [Google Scholar]
- 24.Hrafnsdóttir S, Menon AK. Reconstitution and partial characterization of phospholipid flippase activity from detergent extracts of the Bacillus subtilis cell membrane. J Bacteriol. 2000;182:4198–4206. doi: 10.1128/jb.182.15.4198-4206.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Menon AK, Watkins WE, Hrafnsdóttir S. Specific proteins are required to translocate phosphatidylcholine bidirectionally across the endoplasmic reticulum. Curr Biol. 2000;10:241–252. doi: 10.1016/s0960-9822(00)00356-0. [DOI] [PubMed] [Google Scholar]
- 26.Bishop WR, Bell RM. Assembly of the endoplasmic reticulum phospholipid bilayer: the phosphatidylcholine transporter. Cell. 1985;42:51–60. doi: 10.1016/s0092-8674(85)80100-8. [DOI] [PubMed] [Google Scholar]
- 27.Langley KE, Kennedy EP. Energetics of rapid transmembrane movement and of compositional asymmetry of phosphatidylethanolamine in membranes of Bacillus megaterium. Proc Natl Acad Sci U S A. 1979;76:6245–6249. doi: 10.1073/pnas.76.12.6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herrmann A, Zachowski A, Devaux PF. Protein-mediated phospholipid translocation in the endoplasmic reticulum with a low lipid specificity. Biochemistry. 1990;29:2023–2027. doi: 10.1021/bi00460a010. [DOI] [PubMed] [Google Scholar]
- 29.Gummadi SN, Menon AK. Transbilayer movement of dipalmitoylphosphatidylcholine in proteoliposomes reconstituted from detergent extracts of endoplasmic reticulum. Kinetics of transbilayer transport mediated by a single flippase and identification of protein fractions enriched in flippase activity. J Biol Chem. 2002;277:25337–25343. doi: 10.1074/jbc.M203809200. [DOI] [PubMed] [Google Scholar]
- 30.Chang QL, Gummadi SN, Menon AK. Chemical modification identifies two populations of glycerophospholipid flippase in rat liver ER. Biochemistry. 2004;43:10710–10718. doi: 10.1021/bi049063a. [DOI] [PubMed] [Google Scholar]
- 31.Sanyal S, Frank CG, Menon AK. Distinct flippases translocate glycerophospholipids and oligosaccharide diphosphate dolichols across the endoplasmic reticulum. Biochemistry. 2008;47:7937–7946. doi: 10.1021/bi800723n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vehring S, Pakkiri L, Schroer A, Alder-Baerens N, Herrmann A, Menon AK, Pomorski T. Flip-flop of fluorescently labeled phospholipids in proteoliposomes reconstituted with Saccharomyces cerevisiae microsomal proteins. Eukaryot Cell. 2007;6:1625–1634. doi: 10.1128/EC.00198-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kubelt J, Menon AK, Muller P, Herrmann A. Transbilayer movement of fluorescent phospholipid analogues in the cytoplasmic membrane of Escherichia coli. Biochemistry. 2002;41:5605–5612. doi: 10.1021/bi0118714. [DOI] [PubMed] [Google Scholar]
- 34.Watkins WE, Menon AK. Reconstitution of phospholipid flippase activity from E. coli inner membrane: a test of the protein translocon as a candidate flippase. Biol Chem. 2002;383:1435–1440. doi: 10.1515/BC.2002.162. [DOI] [PubMed] [Google Scholar]
- 35.Harvat EM, Zhang YM, Tran CV, Zhang Z, Frank MW, Rock CO, Saier MH., Jr Lysophospholipid flipping across the Escherichia coli inner membrane catalyzed by a transporter (LplT) belonging to the major facilitator superfamily. J Biol Chem. 2005;280:12028–12034. doi: 10.1074/jbc.M414368200. [DOI] [PubMed] [Google Scholar]
- 36.Schenk B, Fernandez F, Waechter CJ. The ins(ide) and out(side) of dolichyl phosphate biosynthesis and recycling in the endoplasmic reticulum. Glycobiology. 2001;11:61R–70R. doi: 10.1093/glycob/11.5.61r. [DOI] [PubMed] [Google Scholar]
- 37.Hirschberg CB, Snider MD. Topography of glycosylation in the rough endoplasmic reticulum and Golgi apparatus. Annu Rev Biochem. 1987;56:63–87. doi: 10.1146/annurev.bi.56.070187.000431. [DOI] [PubMed] [Google Scholar]
- 38.Lennarz WJ. Protein glycosylation in the endoplasmic reticulum: current topological issues. Biochemistry. 1987;26:7205–7210. doi: 10.1021/bi00397a001. [DOI] [PubMed] [Google Scholar]
- 39.Orlean P, Menon AK. Thematic review series: lipid posttranslational modifications. GPI anchoring of protein in yeast and mammalian cells, or: how we learned to stop worrying and love glycophospholipids. J Lipid Res. 2007;48:993–1011. doi: 10.1194/jlr.R700002-JLR200. [DOI] [PubMed] [Google Scholar]
- 40.Rush JS, Gao N, Lehrman MA, Waechter CJ. Recycling of dolichyl monophosphate to the cytoplasmic leaflet of the endoplasmic reticulum after the cleavage of dolichyl pyrophosphate on the lumenal monolayer. J Biol Chem. 2008;283:4087–4093. doi: 10.1074/jbc.M707067200. [DOI] [PubMed] [Google Scholar]
- 41.Vishwakarma RA, Menon AK. Flip-flop of glycosylphosphatidylinositols (GPI's) across the ER. Chem Commun (Camb) 2005:453–455. doi: 10.1039/b413196g. [DOI] [PubMed] [Google Scholar]
- 42.Sanyal S, Menon AK. Specific transbilayer translocation of dolichol-linked oligosaccharides by an endoplasmic reticulum flippase. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0810225106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Helenius J, Ng DTW, Marolda CL, Walter P, Valvano M, Aebi M. Does Rft1 flip an N-glycan lipid precursor? Helenius et al. reply. Nature. 2008;454:E4–E5. doi: 10.1038/nature07165. [DOI] [PubMed] [Google Scholar]
- 44.Ng DT, Spear ED, Walter P. The unfolded protein response regulates multiple aspects of secretory and membrane protein biogenesis and endoplasmic reticulum quality control. J Cell Biol. 2000;150:77–88. doi: 10.1083/jcb.150.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Helenius J, Ng DT, Marolda CL, Walter P, Valvano MA, Aebi M. Translocation of lipid-linked oligosaccharides across the ER membrane requires Rft1 protein. Nature. 2002;415:447–450. doi: 10.1038/415447a. [DOI] [PubMed] [Google Scholar]
- 46.Frank CG, Sanyal S, Rush JS, Waechter CJ, Menon AK. Does Rft1 flip an N-glycan lipid precursor? Nature. 2008;454:E3–E4. doi: 10.1038/nature07165. discussion E4–5. [DOI] [PubMed] [Google Scholar]
- 47.Rush JS, Gao N, Lehrman MA, Matveev S, Waechter CJ. Suppression of Rft1 expression does not impair the transbilayer movement of Man5GlcNAc2-P-P-dolichol in sealed microsomes from yeast. J Biol Chem. 2009 doi: 10.1074/jbc.M109.000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maeda Y, Kinoshita T. Dolichol-phosphate mannose synthase: structure, function and regulation. Biochim Biophys Acta. 2008;1780:861–868. doi: 10.1016/j.bbagen.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 49.Haselbeck A, Tanner W. Dolichyl phosphate-mediated mannosyl transfer through liposomal membranes. Proc Natl Acad Sci U S A. 1982;79:1520–1524. doi: 10.1073/pnas.79.5.1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zimmerman JW, Robbins PW. The hydrophobic domain of dolichyl-phosphate-mannose synthase is not essential for enzyme activity or growth in Saccharomyces cerevisiae. J Biol Chem. 1993;268:16746–16753. [PubMed] [Google Scholar]
- 51.Rush JS, Waechter CJ. Transmembrane movement of a water-soluble analogue of mannosylphosphoryldolichol is mediated by an endoplasmic reticulum protein. J Cell Biol. 1995;130:529–536. doi: 10.1083/jcb.130.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rush JS, Shelling JG, Zingg NS, Ray PH, Waechter CJ. Mannosylphosphoryldolichol-mediated reactions in oligosaccharide-P-P-dolichol biosynthesis. Recognition of the saturated alpha-isoprene unit of the mannosyl donor by pig brain mannosyltransferases. J Biol Chem. 1993;268:13110–13117. [PubMed] [Google Scholar]
- 53.Rush JS, van Leyen K, Ouerfelli O, Wolucka B, Waechter CJ. Transbilayer movement of Glc-P-dolichol and its function as a glucosyl donor: protein-mediated transport of a water-soluble analog into sealed ER vesicles from pig brain. Glycobiology. 1998;8:1195–1205. doi: 10.1093/glycob/8.12.1195. [DOI] [PubMed] [Google Scholar]
- 54.Rush JS, Waechter CJ. Functional reconstitution into proteoliposomes and partial purification of a rat liver ER transport system for a water-soluble analogue of mannosylphosphoryldolichol. Biochemistry. 2004;43:7643–7652. doi: 10.1021/bi036083o. [DOI] [PubMed] [Google Scholar]
- 55.Lehrman MA, Zeng Y. Pleiotropic resistance to glycoprotein processing inhibitors in Chinese hamster ovary cells. The role of a novel mutation in the asparagine-linked glycosylation pathway. J Biol Chem. 1989;264:1584–1593. [PubMed] [Google Scholar]
- 56.Anand M, Rush JS, Ray S, Doucey MA, Weik J, Ware FE, Hofsteenge J, Waechter CJ, Lehrman MA. Requirement of the Lec35 gene for all known classes of monosaccharide-P-dolichol-dependent glycosyltransferase reactions in mammals. Mol Biol Cell. 2001;12:487–501. doi: 10.1091/mbc.12.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Breukink E, de Kruijff B. Lipid II as a target for antibiotics. Nat Rev Drug Discov. 2006;5:321–332. doi: 10.1038/nrd2004. [DOI] [PubMed] [Google Scholar]
- 58.van Heijenoort J. Lipid intermediates in the biosynthesis of bacterial peptidoglycan. Microbiol Mol Biol Rev. 2007;71:620–635. doi: 10.1128/MMBR.00016-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bouhss A, Trunkfield AE, Bugg TD, Mengin-Lecreulx D. The biosynthesis of peptidoglycan lipid-linked intermediates. FEMS Microbiol Rev. 2008;32:208–233. doi: 10.1111/j.1574-6976.2007.00089.x. [DOI] [PubMed] [Google Scholar]
- 60.de Kruijff B, van Dam V, Breukink E. Lipid II: a central component in bacterial cell wall synthesis and a target for antibiotics. Prostaglandins Leukot Essent Fatty Acids. 2008;79:117–121. doi: 10.1016/j.plefa.2008.09.020. [DOI] [PubMed] [Google Scholar]
- 61.Mengin-Lecreulx D, van Heijenoort J. Effect of growth conditions on peptidoglycan content and cytoplasmic steps of its biosynthesis in Escherichia coli. J Bacteriol. 1985;163:208–212. doi: 10.1128/jb.163.1.208-212.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van Dam V, Sijbrandi R, Kol M, Swiezewska E, de Kruijff B, Breukink E. Transmembrane transport of peptidoglycan precursors across model and bacterial membranes. Mol Microbiol. 2007;64:1105–1114. doi: 10.1111/j.1365-2958.2007.05722.x. [DOI] [PubMed] [Google Scholar]
- 63.Ruiz N. Bioinformatics identification of MurJ (MviN) as the peptidoglycan lipid II flippase in Escherichia coli. Proc Natl Acad Sci U S A. 2008;105:15553–15557. doi: 10.1073/pnas.0808352105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Inoue A, Murata Y, Takahashi H, Tsuji N, Fujisaki S, Kato J. Involvement of an essential gene, mviN, in murein synthesis in Escherichia coli. J Bacteriol. 2008;190:7298–7301. doi: 10.1128/JB.00551-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fay A, Dworkin J. B. subtilis homologs of MviN (MurJ), the putative E. coli Lipid II flippase, are not essential. J Bacteriol. 2009 doi: 10.1128/JB.00605-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou Z, White KA, Polissi A, Georgopoulos C, Raetz CR. Function of Escherichia coli MsbA, an essential ABC family transporter, in lipid A and phospholipid biosynthesis. J Biol Chem. 1998;273:12466–12475. doi: 10.1074/jbc.273.20.12466. [DOI] [PubMed] [Google Scholar]
- 67.Doerrler WT. Lipid trafficking to the outer membrane of Gram-negative bacteria. Mol Microbiol. 2006;60:542–552. doi: 10.1111/j.1365-2958.2006.05130.x. [DOI] [PubMed] [Google Scholar]
- 68.Ruiz N, Kahne D, Silhavy TJ. Transport of lipopolysaccharide across the cell envelope: the long road of discovery. Nat Rev Microbiol. 2009;7:677–683. doi: 10.1038/nrmicro2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu D, Cole RA, Reeves PR. An O-antigen processing function for Wzx (RfbX): a promising candidate for O-unit flippase. J Bacteriol. 1996;178:2102–2107. doi: 10.1128/jb.178.7.2102-2107.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cunneen MM, Reeves PR. Membrane topology of the Salmonella enterica serovar Typhimurium Group B O-antigen translocase Wzx. FEMS Microbiol Lett. 2008;287:76–84. doi: 10.1111/j.1574-6968.2008.01295.x. [DOI] [PubMed] [Google Scholar]
- 71.Rick PD, Barr K, Sankaran K, Kajimura J, Rush JS, Waechter CJ. Evidence that the wzxE gene of Escherichia coli K-12 encodes a protein involved in the transbilayer movement of a trisaccharide-lipid intermediate in the assembly of enterobacterial common antigen. J Biol Chem. 2003;278:16534–16542. doi: 10.1074/jbc.M301750200. [DOI] [PubMed] [Google Scholar]
- 72.Feldman MF, Marolda CL, Monteiro MA, Perry MB, Parodi AJ, Valvano MA. The activity of a putative polyisoprenol-linked sugar translocase (Wzx) involved in Escherichia coli O antigen assembly is independent of the chemical structure of the O repeat. J Biol Chem. 1999;274:35129–35138. doi: 10.1074/jbc.274.49.35129. [DOI] [PubMed] [Google Scholar]
- 73.Marolda CL, Vicarioli J, Valvano MA. Wzx proteins involved in biosynthesis of O antigen function in association with the first sugar of the O-specific lipopolysaccharide subunit. Microbiology. 2004;150:4095–4105. doi: 10.1099/mic.0.27456-0. [DOI] [PubMed] [Google Scholar]
- 74.Marolda CL, Tatar LD, Alaimo C, Aebi M, Valvano MA. Interplay of the Wzx translocase and the corresponding polymerase and chain length regulator proteins in the translocation and periplasmic assembly of lipopolysaccharide o antigen. J Bacteriol. 2006;188:5124–5135. doi: 10.1128/JB.00461-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gunn JS, Lim KB, Krueger J, Kim K, Guo L, Hackett M, Miller SI. PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol Microbiol. 1998;27:1171–1182. doi: 10.1046/j.1365-2958.1998.00757.x. [DOI] [PubMed] [Google Scholar]
- 76.Yan A, Guan Z, Raetz CR. An undecaprenyl phosphate-aminoarabinose flippase required for polymyxin resistance in Escherichia coli. J Biol Chem. 2007;282:36077–36089. doi: 10.1074/jbc.M706172200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Miller C. Efraim Racker 1913–1991. J Membr Biol. 1992;125:97–98. doi: 10.1007/BF00233349. [DOI] [PubMed] [Google Scholar]
- 78.Lemieux MJ, Huang Y, Wang DN. The structural basis of substrate translocation by the Escherichia coli glycerol-3-phosphate transporter: a member of the major facilitator superfamily. Curr Opin Struct Biol. 2004;14:405–412. doi: 10.1016/j.sbi.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 79.Gouaux E, Mackinnon R. Principles of selective ion transport in channels and pumps. Science. 2005;310:1461–1465. doi: 10.1126/science.1113666. [DOI] [PubMed] [Google Scholar]
- 80.Schaaf G, Ortlund EA, Tyeryar KR, Mousley CJ, Ile KE, Garrett TA, Ren J, Woolls MJ, Raetz CR, Redinbo MR, Bankaitis VA. Functional anatomy of phospholipid binding and regulation of phosphoinositide homeostasis by proteins of the sec14 superfamily. Mol Cell. 2008;29:191–206. doi: 10.1016/j.molcel.2007.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tieleman DP, Marrink SJ. Lipids out of equilibrium: energetics of desorption and pore mediated flip-flop. J Am Chem Soc. 2006;128:12462–12467. doi: 10.1021/ja0624321. [DOI] [PubMed] [Google Scholar]
- 82.Gurtovenko AA, Vattulainen I. Molecular mechanism for lipid flip-flops. J Phys Chem B. 2007;111:13554–13559. doi: 10.1021/jp077094k. [DOI] [PubMed] [Google Scholar]
- 83.Matsuzaki K. Magainins as paradigm for the mode of action of pore forming polypeptides. Biochim Biophys Acta. 1998;1376:391–400. doi: 10.1016/s0304-4157(98)00014-8. [DOI] [PubMed] [Google Scholar]
- 84.Fattal E, Nir S, Parente RA, Szoka FC., Jr Pore-forming peptides induce rapid phospholipid flip-flop in membranes. Biochemistry. 1994;33:6721–6731. doi: 10.1021/bi00187a044. [DOI] [PubMed] [Google Scholar]
- 85.Angel TE, Chance MR, Palczewski K. Conserved waters mediate structural and functional activation of family A (rhodopsin-like) G protein-coupled receptors. Proc Natl Acad Sci U S A. 2009;106:8555–8560. doi: 10.1073/pnas.0903545106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kol MA, de Kroon AI, Rijkers DT, Killian JA, de Kruijff B. Membrane-spanning peptides induce phospholipid flop: a model for phospholipid translocation across the inner membrane of E. coli. Biochemistry. 2001;40:10500–10506. doi: 10.1021/bi010627+. [DOI] [PubMed] [Google Scholar]
- 87.Kol MA, van Laak AN, Rijkers DT, Killian JA, de Kroon AI, de Kruijff B. Phospholipid flop induced by transmembrane peptides in model membranes is modulated by lipid composition. Biochemistry. 2003;42:231–237. doi: 10.1021/bi0268403. [DOI] [PubMed] [Google Scholar]
- 88.Kol MA, van Dalen A, de Kroon AI, de Kruijff B. Translocation of phospholipids is facilitated by a subset of membrane-spanning proteins of the bacterial cytoplasmic membrane. J Biol Chem. 2003;278:24586–24593. doi: 10.1074/jbc.M301875200. [DOI] [PubMed] [Google Scholar]
- 89.Kol MA, de Kroon AI, Killian JA, de Kruijff B. Transbilayer movement of phospholipids in biogenic membranes. Biochemistry. 2004;43:2673–2681. doi: 10.1021/bi036200f. [DOI] [PubMed] [Google Scholar]
- 90.Zhou GP, Troy FA., 2nd NMR studies on how the binding complex of polyisoprenol recognition sequence peptides and polyisoprenols can modulate membrane structure. Curr Protein Pept Sci. 2005;6:399–411. doi: 10.2174/138920305774329377. [DOI] [PubMed] [Google Scholar]