Abstract
Metastasis is a complex multi-step process requiring the concerted action of many genes and is the primary cause of cancer deaths. Pathways that regulate metastasis enhancement and suppression both contribute to tumor dissemination process. In order to identify novel metastasis suppressors, we set up a forward genetic screen in a mouse model. We transduced a genome-wide RNAi library into the non-metastatic 168FARN breast cancer cell line, orthotopically transplanted the cells into mouse mammary fat pads, and then selected for cells that could metastasize to the lung and identified an RNAi for the KLF17 gene. Conversely, we demonstrate that ectopic expression of KLF17 in highly metastatic 4T1 breast cancer cell line inhibited their ability to metastasize from the mammary fat pad to the lung. We also show that suppression of KLF17 expression promotes breast cancer cell invasion and epithelial-mesenchymal transition (EMT) and that KLF17 functions by directly binding to the promoter of Id-1, a key metastasis regulator in breast cancer, to inhibit its transcription. Finally, we demonstrate that KLF17 expression is significantly down-regulated in primary human breast cancer samples and that the combined expression patterns of KLF17 and Id-1 can serve as a potential biomarker for lymph node metastasis in breast cancer.
Metastasis, the spread of cells from a primary tumor site followed by growth of secondary tumors in a new location, is responsible for most cancer deaths and remains one of the most poorly understood process in cancer biology1-8. Metastasis occurs via a multi-step process including invasion of local tissues, entrance of cancer cells into the blood stream, survival in the circulation, exit from blood vessels, initiation and maintenance of micrometastases at distant sites, and finally metastatic tumor development1-3, 8, 9. This process depends on balancing metastasis promotion and suppression programs in the tumor cells1-4. Tumor cells must both overcome the cellular suppression machinery and activate their promotion pathways to become metastatic4, 10, 11. Identification and characterization of the genes that suppress metastasis and of the mechanisms by which this suppression occurs will lead to the identification of new markers of metastasis and potential therapeutic targets for prevention and treatment.
Forward genetic screens provide an unbiased approach to the identification of genes associated with a phenotype of interest12-15. RNAi technology and the availability of genome sequences of model organisms have facilitated the construction of RNAi libraries and provided powerful tools for loss-of-function (RNAi mediated knockdowns) screens at the genome-wide level16-18. Because of the complexity of the metastatic process, application of forward genetic screens to animal models of metastasis should assist in the identification of genes that are critical to this process. Here we report the identification of KLF17 as a metastasis suppressor in human breast cancer using a genome-wide RNAi screen in an orthotopic mouse model.
Results
Identification of KLF17 as a metastasis suppressor using a forward genetic screen in an orthotopic mouse model
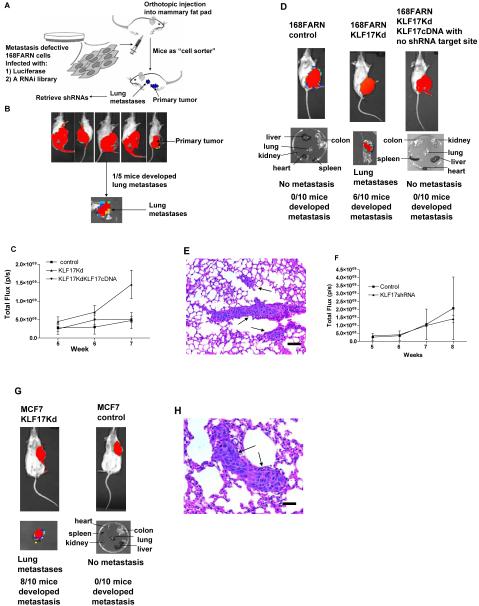
168FARN cells, originally isolated from a single mouse mammary tumor that arose spontaneously in a wild type Balb/cJ mouse19-22, were chosen for the initial screen as they are known to be deficient in the early steps of metastasis (i.e., tumor migration and invasion), but have full metastatic potential once they reach the blood stream and enter the lung23. Thus they are suitable for the isolation of genes that promote or suppress the early steps of metastasis where more effective approaches to controlling metastasis might be developed. We used a genome-wide lentiviral RNAi library (System Biosciences, CA) consisting of short hairpin RNAs (shRNAs) targeting 40,000 mouse genes for our screen, as it is possible to easily retrieve them from the positively selected cells by PCR23. We transplanted the transduced 168FARN tumor cells to the mouse mammary fat pad where they would normally remain. RNAi knock-down of a metastasis suppressor gene should allow the cells to metastasize to the lung. Lung metastases served as the selection system for these studies (Figure 1A). A proof-of-concept experiment was carried out using five Balbc/J mice, one of which developed a lung metastasis in seven weeks (Figure 1B). The one short hairpin RNA (shRNA) retrieved from the metastatic cells was found to correspond to the Krüppel-like transcription factor 17 (KLF17). We carried out following validation studies to support the identification of KLF17 as a negative regulator of metastasis.
Figure 1.
Identification of KLF17 as a metastasis-suppressing gene. (A) The scheme for the forward genetic screen in a mouse model. (B) Tumor cell growth in the primary site following injection of 168FARN-Luc cells containing a genome-wide RNAi library in mammary fat pad. One out of five mice developed lung metastasis. (C) Tumor growth in the primary sites following the transplantation of 168FARN cells expressing a control non-target shRNA; 168FARN expressing KLF17 shRNA; and 168FARN cells expressing both KLF17 shRNA and KLF17 cDNA in mammary fat pads. The growth rate is similar in all three groups before week 7. Tumor growth in 168FARN cells expressing KLF17 shRNA is higher than the other two groups at week 7 (p=0.001, t-test, n=10). (D) Transplantation of 168FARN cells stably expressing KLF17 shRNA in mammary fat pad leads to lung metastasis. Transplantation of 168FARN expressing a non-target shRNA does not lead to metastasis. No metastasis developed following the transplantation of 168FARN cells stably expressing both KLF17 shRNA and KLF17 cDNA which lacks shRNA binding site. (E) Histology analysis of metastasis in the lung following the transplantation of 168FARN cells expressing KLF17 shRNA. Arrows indicate micrometastases. Scale bar represents 100 microns. (F) Tumor growth in the primary site following the transplantation of MCF7 control cells and MCF7 expressing KLF17 shRNA in mammary fat pad. There is no significant difference between two groups (p=0.775, t-test, n=10). (G) Transplantation of human breast cancer MCF7 cells stably expressing KLF17 shRNA in mammary fat pad leads to lung metastasis whereas MCF7 cells expressing a non-target shRNA did not. (H) Histology analysis of metastasis in the lung following the transplantation of MCF7 cells expressing KLF17 shRNA. Arrow indicates micrometastases. Scale bar represents 50 microns.
KLF17 is a negative regulator of metastasis
KLF17 (ZNF393) is a member of Sp/KLF zinc finger protein family with diverse functions24-26. Krüppel-like factor (KLF) family members can function as either activators or repressors depending on the promoters they bind and the cellular proteins they interact with24, 25. They are critical regulators of various cellular processes including reprogramming of differentiated cells to stem cells27, erythropoiesis28, 29 and cell survival30. Moreover, some KLF family members have been implicated in tumor development31-33. The murine homologue of KLF17 is believed to play a role in gametogenesis and early embryogenesis based on its expression in early embryos, testes and ovary34, however, there is little functional information on either the human or mouse KLF17.
In order to validate that KLF17 expression promotes metastasis, we generated a luciferase-tagged 168FARN cell line that stably expresses a KLF17 shRNA. The suppression of KLF17 expression was confirmed by immunoblot (Supplementary Figure 1A). The KLF17 deficient cells were then transplanted into mouse mammary fat pads using the same procedure as in the initial screen. The growth of primary tumors is similar in both KLF17 knockdown cells and control cells before week 6, but is higher in the knockdown cells after 6 weeks (Figure 1C). The KLF17 knockdown cells in the primary tumors could be seen to invade neighboring healthy muscle and fat tissues (Supplementary Figure 1B), demonstrating their invasive nature. Furthermore, lung metastases developed 7-8 weeks following mammary fat pad transplantation of the knockdown cells, demonstrating that down-regulation of KLF17 expression can promote tumor metastasis in vivo (Figure 1D, Supplementary figure 1C). Histological analysis confirmed the formation of micrometastases in the lungs (Figure 1E). KLF17 knockdown had little effect on the growth of metastases following transplantation via the tail vein (Supplementary figure 1D), further supporting the hypothesis that the KLF17 acts on the early steps of metastasis. To further confirm that the enhanced metastatic potential is specifically due to the inhibition of KLF17 expression, we introduced a KLF17 cDNA lacking the 3′UTR region which includes the target sequence for the KLF17 shRNA, into 168FARN cells stably expressing the KLF17 shRNA. The expression of KLF17 protein was confirmed by immunoblot (Supplementary figure 1E). These cells were then transplanted into the mouse mammary fat pad. The metastatic phenotype exhibited by the KLF17 shRNA knockdown cells was completely abolished by the introduction of the KLF17 cDNA lacking the target sequence demonstrating that the induction of the metastatic capabilities in 168FARN cells was indeed due to the suppression of KLF17 expression (Figure 1D).
To determine whether KLF17 played a similar role in metastasis of human breast cancers, we generated a luciferase-tagged human breast cancer MCF7 cell line stably expressing a KLF17 shRNA and confirmed loss of protein expression by immunoblot (Supplementary Figure 1F). The KLF17 knockdown MCF7 cells were transplanted into the mammary fat pads of SCID mice. There was no significant difference in primary tumor growth in KLF17 knockdown cells compared to the control cells (Figure 1F). Lung micrometastases developed 8-10 weeks following transplantation whereas MCF7 cells expressing a control non-target shRNA did not metastasize (Figure 1G, 1H, Supplementary Figure 1G), demonstrating that knock-down of KLF17 expression also promotes metastasis in human breast cancer cells.
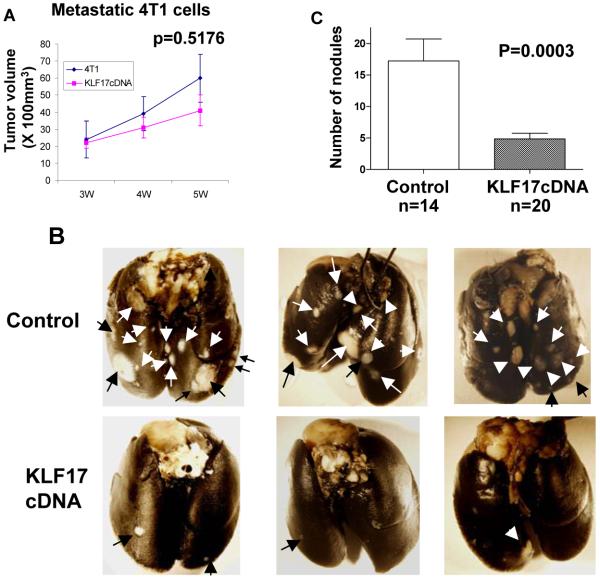
To further validate the metastasis suppression activity of KLF17, we did the converse experiment and generated a mouse 4T1 cell line that stably overexpressed a mouse KLF17 cDNA. 4T1 and 168FARN cells were originally isolated from the same mouse mammary tumor but differ dramatically from each other in their metastasis potentials19-22. Overexpression of KLF17 in 4T1 cells was confirmed by immunoblot (Supplementary Figure 1H). Although both cell lines are capable of forming primary mammary carcinomas, only 4T1 cells form metastases after mammary pad injection. 4T1 cells normally form visible metastatic nodules in the lung with high efficiency after transplantation to the mammary fat pad5, 19-23. The lung surface was examined for the presence of metastatic nodules 4-5 weeks following the transplantation of 4T1 cells stably overexpressing the KLF17 protein into mouse mammary fat pads. Although KLF17 overexpression was shown not to affect 4T1 growth in vivo (Figure 2A), the numbers of metastatic nodules were significantly less in 4T1 cells overexpressing KLF17 compared to the vector control, demonstrating that KLF17 functions to suppress metastasis in vivo (Figure 2B and 2C).
Figure 2.
(A) The volumes of primary tumors were measured following the transplantation of 4T1 cells stably overexpressing KLF17 or a vector control in mammary fat pad. There is no statistical difference in primary tumor growth between these two groups (t-test, n=14 for the control group, n=20 for the KLF17 group). (B and C) The metastastic nodules on the surface of the lungs (B) were counted following the transplantation of 4T1 cells stably overexpressing KLF17 or a vector control in mammary fat pad. Transplantation of 4T1 cells overexpressing KLF17 leads to significant less lung metastases compared to the control (t-test) (C).
Downregulation of KLF17 promotes EMT
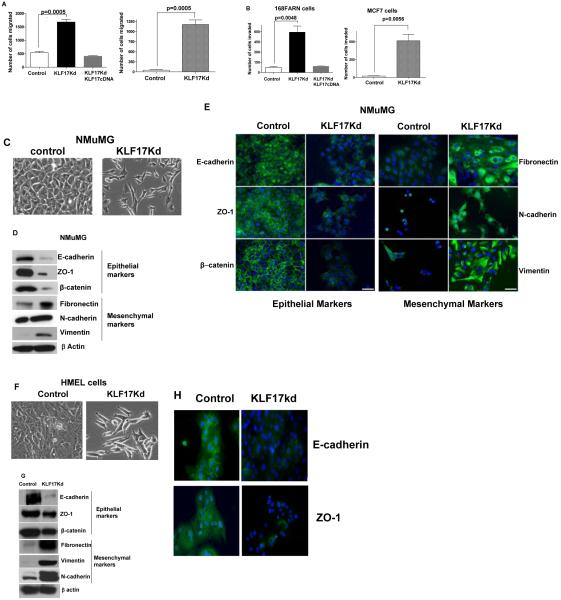
Because 168FARN cells are defective in the early steps of tumor metastasis19-23, we further examined whether KLF17 might play a role in the initial invasion of the tumor cells into neighboring tissues through processes such as migration, invasion and epithelial-mesenchymal transition (EMT). The effects of the KLF17 knockdown on these processes were evaluated in a transwell migration assay. Knockdown of KLF17 expression resulted in a clear and potent migratory phenotype in both mouse breast and human breast cancer cells as compared to their respective control cells (Figure 3A). Similar results were obtained in a Matrigel invasion assay (Figure 3B) indicating that knockdown of KLF17 promotes tumor cell migration and invasion.
Figure 3.
Suppression of KLF17 expression promotes tumor cell migration, invasion and EMT. (A and B) Breast cancer 168FARN and MCF7 cells stably expressing KLF17 shRNA alone or KLF17 shRNA and KLF17 cDNA were subjected to migration (A) and invasion (B) assays. The suppression of KLF17 expression in these cells leads to significant increase of cell migration and invasion. Data represent mean and s.d. (triplicates, t-test). The expression of KLF17 cDNA with no shRNA target site reverses the migratory and invasive phenotype induced by KLF17 knockdown. (C-H) The knockdown of KLF17 expression in NMuMG and HMEL cells causes epithelial-mesenchymal transition (EMT). (C, F) NMuMG and HMEL cells stably expressing KLF17 shRNA displayed spindle-like, fibroblastic morphology (20X magnification). (D, G) Immunoblots of epithelial and mesenchymal markers in NMuMG-KLF17Kd and HMEL-KLF17Kd cells show the inhibition of the expression of epithelial markers E-cadherin, ZO-1 and β-catenin and the increase of the expression of mesenchymal markers fibronectin, vimentin and N-cadherin; (E, H) Immunostaining of NMuMG-KLF17Kd and HMEL-KLF17Kd cells using antibodies shows the loss of epithelial markers in cell-cell contacts. Scale bar represents 50 microns.
EMT, a process by which tumor associated epithelial cells obtain mesenchymal features resulting in reduced cell-cell contact and increased motility, plays a critical role in metastasis35, 36. Several promoting factors of this process have been identified5, 37-40, however the roles of EMT negative regulators remain unclear. To assess the contribution of KLF17 to EMT regulation, we generated a mouse mammary epithelial cell line NMuMG stably expressing a KLF17 shRNA and confirmed knockdown by immunoblot (Supplementary Figure 2A). After KLF17 knockdown, NMuMG cells displayed spindle-like, fibroblastic morphology, one of the major characteristics of EMT (Figure 3C, Supplementary Figure 2B). The expression of molecular markers of EMT was confirmed by immunoblotting and immunocytochemistry to assess the presence of both epithelial and mesenchymal markers in these cells. The expression of epithelial markers, E-cadherin, zonula occludens-1 (ZO-1) and β-catenin, were significantly reduced in KLF17 knock-down cells (Figure 3D), and E-cadherin and ZO-1 was lost from cell-cell contacts (Figure 3E). In contrast, the expression of mesenchymal markers, fibronectin, vimentin and N-cadherin, whose expression positively correlate with EMT, were dramatically up-regulated in the KLF17 knock-down cells (Figure 3D, 3E). We further confirmed the EMT phenotype following KLF17 knockdown in an immortalized human mammary epithelial cell line (HMEL). Similar to NMuMG, expression of epithelial markers was reduced following KLF17 knockdown (Figure 3F, 3G, 3H) whereas the mesenchymal markers were up-regulated. Suppression of KLF17 expression in 168FARN cells also causes the downregulation of epithelial marker expression and upregulation of mesenchymal markers (Supplementary Figure 2C and 2D) demonstrating that knock-down of KLF17 promotes EMT.
Id1 is directly regulated by KLF17 and is required for KLF17 function
In order to identify the downstream molecules regulated by KLF17, gene expression studies were performed using the Mouse WG-6 Bead array from Illumina. Gene expression was analyzed in 168FARN cells that either stably expressed KLF17 shRNA, overexpressed KLF17 cDNA or which carried a control vector. We were specifically interested in genes whose expression were increased in KLF17 knockdown cells but decreased in KLF17 overexpressing cells or vice versa (Supplementary Table 1). Among the significant genes that met these criteria, Id1 expression was found to be up-regulated in KLF17 knock-down cells and down-regulated in KLF17 overexpressing cells. Id1 is a member of the Id protein family that serves as dominant negative regulators of the basic helix-loop-helix family of transcription factors41-43. The Id1 protein is a key regulator in development, cell cycle and tumorigenesis41-43. Recent findings suggest that Id1 is deregulated in various types of cancers and is important in the development of embryonic stem cell like phenotypes in cancer cells44. Id1 has also been shown to play a critical role in EMT, angiogenesis, invasion and metastasis45-47 and to cooperate with oncogenic Ras to induce mammary tumor metastasis48. Id1 up-regulation has been associated with poor clinical outcome in breast cancer49, 50 and suppression of Id1 by shRNAs or chemical inhibitors blocks invasion and metastasis in breast cancer51-53. Because both animal models and clinical studies indicated that Id1 is a key regulator of breast cancer development and metastasis, we further investigated the interactions of KLF-17 and Id-1.
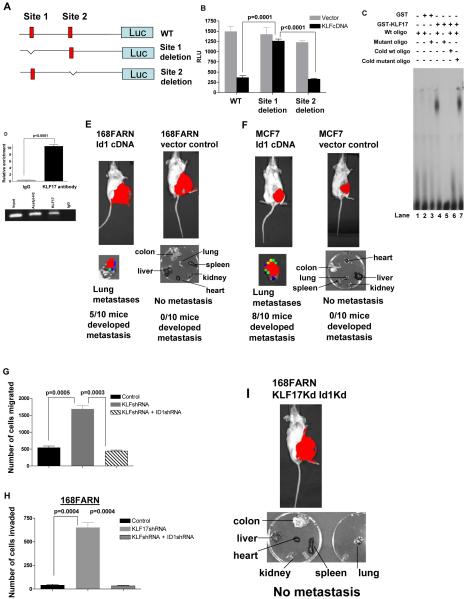
It has been shown that KLF17 binds to the DNA consensus sequence (5′-CACCC-3′) to regulate transcription 26. We scanned the promoter region of mouse Id1 for the KLF17 consensus sequence and found two potential binding sites, at -2127 to -2110 (site 1) and -1327 to -1316 (site 2) from the transcriptional initiation site (Figure 4A). We cloned the promoter region of mouse Id-1 upstream of a luciferase gene in a reporter plasmid (Figure 4A) and co-transfected the KLF17 cDNA construct and the Id1 reporter construct into 168FARN cells. KLF17 expression suppressed the luciferase signal from the reporter plasmid driven by the Id1 promoter region by 63% (Figure 4B) in a dose-dependent manner (Supplementary Figure 3A). Mutation of KLF17 potential binding site 1 relieved the suppression by KLF17 (Figure 4B) whereas mutation of binding site 2 did not (Figure 4B). Thus KLF17 can suppress Id1 transcription and requires binding site 1 but not 2.
Figure 4.
KLF17 directly binds to the promoter of Id1 and suppresses its expression. (A) Luciferase reporter constructs contain the Id-1 promoter with two potential KLF17 binding sites upstream of a luciferase gene, or Id1 promoter with the deletion of one potential binding site 1 or Id1 promoter with the deletion of potential binding site 2. Red indicates two potential binding sites (site 1: -2127 to -2110 and site 2: -1327 to -1316). (B) 168FARN cells were co-transfected with KLF17 cDNA, a control luciferase vector or luciferase reporter plasmids shown in (A) and a Renilla luciferase as a normalizing control. Luciferase activity was measured 48 hours following the transfection. The percentage of luciferase activity was determined by the activity of promoter reporter over the control luciferase vector. Data represent mean and s.d. (triplicates, t-test). (C) EMSA assay was performed using the recombinant GST-KLF17 protein and the isotope-labeled DNA probe comprising site 1. (D) Chromatin immunoprecipitation (ChIP) was performed using KLF17 antibody, acetyl-H3 antibody or control IgG. Id1 promoter region where KLF17 binds showed a significant enrichment following immunoprecipitation by KLF17 antibody. Data represent mean and s.d. (triplicates, t-test). PCR products following ChIP were run on an ethidium-stained gel. Anti-acetyl H3 antibody was used as a positive control. (E) Transplantation of 168FARN cells stably expressing mouse Id1 cDNA in mammary fat pad leads to lung metastasis. (F) Transplantation of human breast cancer MCF7 cells stably expressing human Id1 cDNA in the mammary fat pad of SCID mice leads to lung metastasis. (G-H) 168FARN cells stably expressing a KLF17 shRNA; or KLF17 and Id1 shRNAs; or a control non-target shRNA were subjected to migration (G), invasion (H) and metastasis (I) assays. Data represent mean and s.d. (triplicates, t-test). Knockdown of KLF17 significantly increase the migration, invasion and metastasis in 168FARN. Knockdown of both KLF17 and its target gene Id1 abrogates the migratory, invasive and metastasis phenotypes induced by KLF17 knockdown.
To further investigate whether KLF17 directly binds to the site 1 of Id-1 promoter, we carried out electrophoretic mobility shift assay (EMSA) using recombinant GST-KLF17 protein expressed in E. coli, and the isotope-labeled DNA probe comprising site 1. The recombinant GST-KLF17 binds specifically to the wildtype site 1 (Figure 4C, lane 4), but not to the mutant site 1 (Figure 4C, lane 5). Moreover, competition with an unlabeled wildtype site 1 oligo blocked the binding of GST-KLF17 to site 1 (Figure 4C, lane 6) whereas the addition of an unlabeled mutant site 1 oligo had no effect on the binding of GST-KLF17 to site 1 (Figure 4C, lane 7). These results demonstrate that KLF17 suppresses Id1 expression by directly binding to the Id1 promoter region.
To determine whether KLF17 binds to the Id1 promoter in vivo, we performed chromatin immunoprecipitation (ChIP) analysis using a KLF17 antibody. The Id1 promoter region where KLF17 binds in EMSA was significantly enriched as compared to a control IgG ChIP as assessed by quantitative PCR, further demonstrating that Id1 is a direct target of KLF17 in vivo (Figure 4D, Supplementary Figure 3B).
To examine the effect of Id1 upregulation in tumor metastasis in vivo, we generated luciferase-tagged mouse 168FARN and human MCF7 cell lines stably expressing mouse or human Id1 cDNAs respectively. Id1 expression in the 168FARN and MCF7 cells was confirmed by immunoblot (Supplementary Figure 3C and 3D) and the over-expressing cells were transplanted into the mammary fat pads of Balb/c or SCID mice respectively. Lung metastases developed from Id1-upregulated cells following transplantation but not in controls, demonstrating that upregulation of Id1 expression promotes tumor metastasis in vivo (Figure 4E and 4F). Finally, we examined the importance of Id1 up-regulation in the process of KLF17 knockdown induced migration and invasion. We used a shRNA to knock-down Id1 expression in 168FARN KLF17 knockdown cells. Downregulation of Id1 in 168FARN KLF17 knockdown cells reverses the expression of epithelial and mesenchymal markers in these cells (Supplementary Figure 2C), indicating Id1 is a major mediator of KLF17-induced EMT. Figure 4G and 4H show that the knockdown of Id1 expression in 168FARN cells, in which KLF17 expression is suppressed, significantly reduces the number of migrating and invading cells. No metastasis occurred when the KLF17 and Id1 double knockdown 168FARN cells were transplanted into mammary fat pad (Figure 4I, Supplementary Figure 3E). Altogether, these results suggest that knock-down of KLF17 promotes tumor metastasis, at least in part, by increasing Id1 expression and that KLF17 is a negative regulator of Id1 expression.
Since it has also been shown that Id1 cooperates with oncogenic Ras to promote metastasis48, we examined whether the KLF17 knockdown also influences the function of oncogenic Ras in tumor metastasis. We found no primary tumor growth when NMuMG cells or NMuMG cells stably expressing KLF17 shRNA were transplanted into mouse mammary fat pads (data not shown). In contrast, NMuMG cells stably expressing both KLF17 shRNA and G12V oncogenic HRas or G12V oncogenic HRas alone developed both primary tumors and lung metastases (Supplementary Figure 4A). However, the number of lung metastases in the KLF17Kd + Ras group is 50% higher than the Ras-alone group (Supplementary Figure 4B). In addition, 50% of mice transplanted with NMuMG cells stably expressing both KLF17 shRNA and oncogenic Ras also developed metastases in liver, kidney and colon, whereas NMuMG cells expressing Ras alone did not produce metastases in any other organs except lung (Supplementary Figure 4A). These results indicate that knockdown of KLF17 enhances tumor metastasis promoted by oncogenic Ras. We further examined the role of Ras in cell migration and invasion in tumor cells when KLF17 expression is downregulated. We cloned and sequenced the coding region of HRas, NRas and KRas in 168FARN cells. 168FARN cells harbor a I55Y mutation in HRas and no mutations in either NRas or KRas. To our knowledge, this HRas mutation has not been reported and whether this mutation has any effect on HRas activity is not known. We then used the Ras farnesylation inhibitor FTI-277 to assess Ras functions in cell migration and invasion. Incubation of 168FARN cells stably expressing KLF17 shRNA with FTI-277 did not affect the expression of either epithelial markers or mesenchymal markers (data not shown). However, the activities of these migratory and invasive cells were significantly reduced in our in vitro migration and invasion assays (Supplementary Figure 4C), indicating that Ras plays an important role in tumor cell migration and invasion that is enhanced when KLF17 expression is downregulated.
KLF17 is down-regulated in human breast cancer tumor samples
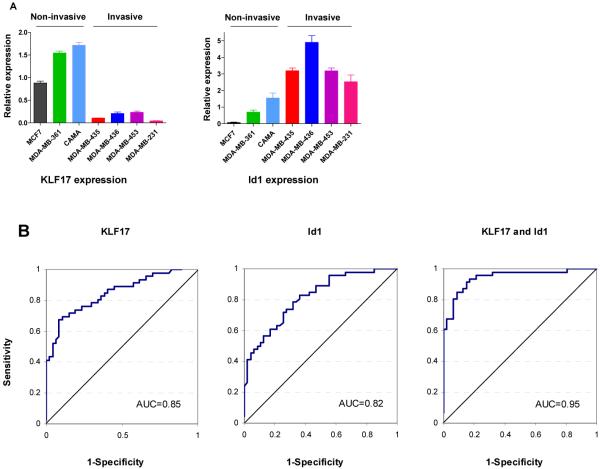
Finally we examined the expression levels of KLF17 and Id-1 in breast cancer cell lines and primary human breast cancer tumor samples. The invasive capability of seven breast cancer cell lines was determined in a Matrigel assay (data not shown). KLF17 expression was lower in breast cancer cell lines with an invasive phenotype and Id1 expression was higher in these cell lines (Figure 5A). Pearson correlation analysis indicated that the expression of KLF17 and Id-1 were inversely correlated in the cell lines (r=-0.75, p=0.05). Next, we examined the expression of these two genes in 93 human primary breast tumors with known lymph node metastasis status. 46 samples were lymph node positive and 47 samples were lymph node negative at the time of diagnosis. We found that the mean expression of KLF17 in the patients with histologically confirmed lymph node metastases was significantly lower than the mean expression in patients without metastases (Table 1, p<0.001, Supplementary Figure 5, Supplementary Table 2-4). Conversely, the mean expression of Id1 was higher in the patients with lymph node metastases than in those with no metastases (Table 1, p<0.001, Supplementary Figure 5, Supplementary Table 2-4). More importantly, based on the multivariate analysis, KLF17 and Id1 were independent predictors of lymph node metastasis with adjusted odds ratios for metastasis of 0.02 and 18.77, respectively (Table 2). To further explore KLF17 and Id1 as biomarkers for identifying lymph node metastasis, we computed the receiver operating curves (ROC) for these two biomarkers independently and in combination (Figure 5B). The area under the ROC curve (AUC) for KLF17 and Id1 individually were 0.85 and 0.82, respectively. The AUC increased to 0.95 using the predicted values from the logistic regression model that included both biomarkers. Cut-points were determined for each biomarker based on their ROC curves. In patients with tumors where the expression of KLF17 was high and the expression of Id1 was low, the percentage of patients with lymph node metastasis was very low (12.8%) (Table 3). In contrast, in tumors with low KLF17 expression and either high Id1 expression or low Id1 expression, percentages of patients with lymph node metastasis were very high ranging from 82.6% to 100% (Table 3). Taken together, these analyses indicated that specific combinations of KLF17 and Id1 had high sensitivity or specificity to detect those patients with or without lymph node metastasis, respectively, and together their expression can distinguish metastatic (lymph node positive) from non-metastatic (lymph node negative) breast cancer.
Figure 5.
KLF17 and Id1 expression in breast cancer cell lines and their predictive value of lymph node metastasis in human primary breast cancer samples. (A) KLF17 and Id1 expression in human breast cancer cells was determined by qRT-PCR. KLF17 expression is higher in non-invasive MCF7, MDA-MB-361 and CAMA cells, but lower in invasive MDA-MB-435, MDA-MB-436, MDA-MB-453 and MDA-MB-231 cells. Id1 expression is higher in invasive cells but lower in non-invasive cells. Pearson correlation analysis indicated that the expression of KLF17 and Id-1 was inversely correlated (r=-0.75, p=0.05). (B) KLF17 and Id1 expression are predictive markers of lymph node metastasis in human primary breast cancer samples. The receiver operating characteristic curves for KLF17 (left), Id-1 (middle) and the bivariate logistic regression model for KLF17 and Id1 (right).
Table 1. Mean expression of KLF17 and Id1 in primary breast cancer samples by qRT-PCR.
Mean Expression for Patients with and without Lymph Node Metastases
| Metastasis = No | Metastasis = Yes | Mean | ||||
|---|---|---|---|---|---|---|
| Mean | St.Dev | Mean | St.Dev | Difference | p-value* | |
| KLF17 | 2.02 | 0.22 | 0.75 | 0.09 | 1.27 | <0.001 |
| Id-1 | 1.38 | 0.11 | 3.09 | 0.35 | -1.71 | <0.001 |
t-test for two independent groups with Satterthwaite’s adjustment for unequal variances
Table 2. Association of KLF17 and Id1 expression with the presence of lymph node metastasis.
Univariate and Multivariate Logistic Regression for Metastasis at Diagnosis
| Unadjusted | Adjusted | |||
|---|---|---|---|---|
| Odds Ratio | p-value | Odds Ratio | p-value | |
| KLF17 | 0.15 | 0.0001 | 0.02 | 0.0003 |
| Id1 | 3.27 | 0.0001 | 18.77 | 0.0002 |
| HER2 | 0.99 | 0.2336 | 0.97 | 0.0526 |
| ER | 1.00 | 0.9170 | 0.99 | 0.4914 |
Table 3. KLF17 and Id1 expression as predictive markers of lymph node metastasis.
Metastasis Rates by Groups Defined by Biomarkers
| Biomarker | Values | Metastasis | ||||
|---|---|---|---|---|---|---|
| No | Yes | All | Met Rate | |||
| hKLF17 > 0.772 | Id1 ≤ 2.75 | 41 | 6 | 47 | 12.8 | Good |
| hKLF17 > 0.772 | Id1 > 2.75 | 2 | 9 | 11 | 81.8 | Poor |
| hKLF17 ≤ 0.772 | Id1 ≤ 2.75 | 4 | 19 | 23 | 82.6 | Poor |
| hKLF17 ≤ 0.772 | Id1 > 2.75 | 0 | 12 | 12 | 100.0 | Poor |
| Total | 47 | 46 | 93 | 49.5 | ||
Discussion
We have identified a negative regulator of metastasis using a loss-of-function screen with 168FARN cells. The 168FARN cell line is only defective in the early steps of metastasis such as migration and invasion23. Thus only the metastasis suppressors that inhibit these early steps could be identified and likely accounts for lack of isolation of other metastasis-suppressor genes in the screen. Screens using cell lines defective in other steps of metastasis may identify additional suppressors as this proof-of-concept screen demonstrates the feasibility of this loss-of-function in vivo selection approach. A saturated or comprehensive screen of metastatic regulators that function at different steps in the metastasis process would require larger numbers of animals.
We have also demonstrated that the loss of KLF17 promotes breast cancer metastasis at least in part through the direct regulation of Id1. Other targets of KLF17 that may play a role in tumor migration, invasion and EMT remain to be investigated. Our results also indicate that the downregulation of KLF17 expression cooperates with Ras to promote metastasis. Studies to understand the regulation of KLF17 and the mechanisms that suppress KLF17 expression and promote cancer metastasis are under way. Our results with human breast cancer samples also demonstrate that low KLF17 and high Id1 expression levels can potentially be used to predict the metastatic state of primary breast cancer samples and indicate that KLF17/Id-1 reciprocal expression is a critical pathway in the development of breast cancer metastasis. Suppression of this pathway may serve as a basis for the development of therapies that target early metastatic events.
Supplementary Material
Acknowledgements
We would like to thank Dr. Fred Miller for providing 168FARN and 4T1 cells; Dr. Robert Weinberg for providing HMEL cell line; Dr. Janet Price for providing MDA-MB-435 cell line. We would like to thank Drs. Celia Chang and Wenhwai Horng for assistance in microarray analysis, James Hayden and Frederick Keeney for assistance in microscopy. Q.H. is supported by Breast Cancer Alliance, Pardee Foundation, V Foundation. Q.H. and L.C.S. are supported by Commonwealth Universal Research Enhancement Program, Pennsylvania Department of Health, P30 CA10815 and (LCS) PA DOH grant SAP 4100020718. G.C. and L.Z. are supported by NCI ovarian SPORE P50-CA83638, Ovarian Cancer Research Fund and Mary Kay Ash Charitable Foundation.
Appendix
Material and Methods
Functional screen in an orthotopic animal model and validation of metastasis-suppressing activity
To establish the animal model for the forward genetic screen, 1 × 106 168FARN-Luc cells containing a genome-wide mouse RNAi library covering 40,000 genes (System Biosciences, CA) were orthotopically transplanted into the mammary fat pads of 10-week-old BALB/c mice (National Cancer Institute, MD). 5 mice were used in the initial screen. Mice bearing luciferase positive tumors were imaged 7-8 weeks after transplantation with the aid of IVIS 200 Imaging system (Xenogen Corporation, Hopkinton, MA). Lung metastasis nodules were isolated and genomic DNA of lung metastatic cells was isolated with the Genomic DNA Purification kit according to supplier’s instruction (Qiagen, Valencia, CA). PCRs were performed according to the manufacturers protocol (SBI, system Biosciences). PCR products were cloned with a TA cloning kit (Invitrogen) and sequenced. To validate the hits from the screen, 168FARN-Luc cells were transduced with lentiviruses containing the KLF17shRNA or KLF17shRNA and KLF17 cDNA which lacks shRNA binding site; 1 × 106 cells were transplanted into the mammary fat pads of mice and imaged 7-8 weeks after transplantation. MCF-7 human breast cancer cells (7 × 106 cells/mouse) stably expressing KLF17shRNA were transplanted into the mammary fat pad of the female SCID mice (6-8 weeks old). A slow-release pellet of 17β-estradiol (1.7 mg, 90-day release; Innovative Research of America, Sarasota, FL) was implanted subcutaneously in the dorsal interscapular region before the transplantation of MCF7 cells. Mice bearing luciferase positive tumors were imaged 6-12 weeks after transplantation with the aid of IVIS 200 Imaging system (Xenogen Corporation, Hopkinton, MA).
To analyze the effect of overexpression of KLF17 on 4T1 cells, 4T1 cells were transduced with lentivirus containing mouse KLF17cDNA and transplanted as described above. 168FARN-Luc cells containing a non-target shRNA as a control were also developed as above. To determine the number of metastasis nodules on the surface of the lungs, mouse lungs were harvested and fixed with Fekete’s solution (60% ethanol, 3% formaldehyde, and 4% glacial acetic acid) after intratracheal injection of a 15% india ink solution as described54. The total number of unstained nodules on the lung surface was counted 4-5 weeks following transplantation.
To analyze the effect of overexpression of Id1 on 168FARN and MCF7 cells, 168FARN and MCF7 cells were transduced with lentivirus containing mouse or human Id1 cDNA respectively and transplanted into female Balb/c or SCID mice respectively as described above. Mice bearing luciferase positive tumors were imaged with the aid of IVIS 200 Imaging system (Xenogen Corporation, Hopkinton, MA) as described above.
To study the effect of downregulation of KLF17 expression and Ras in tumor metastasis, NmuMG cells stably expressing a control vector, KLF17 shRNA, oncogenic HRas or coexpressing KLF17 shRNA and oncogenic HRas were transplanted into SCID mice as described above. Mice bearing luciferase positive tumors were imaged with the aid of Xenogen IVIS 200 Imaging system as described above.
Lentiviral shRNA transduction
Cell lines stably expressing KLF17 shRNA (KLF17Kd), Id1 shRNA (Id1Kd) or control non-target shRNA were established using vector based shRNA technique. All shRNAs were purchased from Sigma. Mouse KLF17 shRNA targets gcctggaaagttctggagtta; human KLF17 shRNA targets cgacagtaccttctgacgaaac; mouse Id1 shRNA targets cctactagtcaccagagactt. Briefly, Lentiviral KLF17 shRNA, Id1 shRNA and control non-target shRNA were purchased from Sigma and lentiviruses were produced by co-transfecting subconfluent human embryonic kidney (HEK) 293T cells with the KLF17shRNA, Id1 shRNA or control non-target shRNA expression plasmid and packaging plasmids (pMDLg/pRRE and RSV-Rev) using Fugene6 as a transfection reagent. Infectious lentiviruses were collected 48 h after transfection, centrifuged to remove cell debris and filtered through 0.45 μm filters (Millipore). 168FARN, MCF7, NMuMG and HMEL cells were transduced with the lentivirus containing KLF17 shRNA or Id1 shRNA or non-target shRNA. The KLF17 or Id1 knockdown efficiency was determined by quantitative real time PCR. To generate cells stably overexpressing KLF17, Id1 or oncogenic HRas, full-length mouse KLF17 cDNA, Id1 cDNA or oncogenic HRas was cloned into lentiviral vector and produced virus as described above and transduced into cells respectively.
Transwell migration and invasion assay
In vitro cell migration assays were performed as described previously55 using Trans-well chambers (8μM pore size; Costar). Cells were allowed to grow to subconfluency (~75-80%) and were serum-starved for 24 h. After detachment with trypsin, cells were washed with PBS, resuspended in serum-free medium and 250 μl cell suspensions (2 × 105 cells ml-1) was added to the upper chamber. Complete medium was added to the bottom wells of the chambers. The cells that had not migrated were removed from the upper face of the filters using cotton swabs, and the cells that had migrated to the lower face of the filters were fixed with 5% glutaraldehyde solution and stained with 0.5% solution of Toluidine Blue in 2% sodium carbonate. Images of three random ×10 fields were captured from each membrane and the number of migratory cells was counted. The mean of triplicate assays for each experimental condition was used. Similar inserts coated with Matrigel were used to determine invasive potential in the invasion assay.
Immunoblotting
Standard methods were used for western blotting. Cells were lysed in lysis buffer and total protein contents were determined by the Bradford method. 30 μg of proteins were separated by SDS-PAGE under reducing conditions and blotted onto a polyvinylidene difluoride membrane (Millipore). Membranes were probed with specific antibodies Blots were washed and probed with respective secondary peroxidase-conjugated antibodies, and the bands visualized by chemoluminescence (Amersham Biosciences). The following antibodies were used: Rabbit polyclonal E-cadherin, mouse monoclonal N-cadherin and ß catenin (Cell Signaling Technology), rabbit polyclonal ZO-1 (Zymed Laboratories) mouse monoclonal Fibronectin and ß-actin (Sigma -Aldrich), mouse monoclonal vimentin (Calbiochem) and KLF17 monoclonal antibody. 1:1000 dilution of the primary antibodies and 1:5000 dilution of the secondary antibodies were used for immunoblotting.
Immunocytochemistry
Cells were grown on glass cover slips in a six well plate and washed 3 times with PBS then fixed in 4% formaldehyde solution and permeabilized with 0.1% Triton X-100 in PBS for 5 min. Cells were blocked with 1% BSA in PBS for 30min at Room temperature. Cover slips were incubated with respective primary antibodies at 1:100 dilutions for 1 h and then washed with PBS and incubated for 1h with fluorescein-conjugated secondary antibodies at 1:200 dilutions (Molecular Probes). Cells were further washed in PBS and mounted with Vectashield® mounting medium containing 4′,6-diamidino-2-phenylindole (DAPI), (Vector Laboratories, Burlingame, CA) and analyzed using fluorescence microscopy.
Luciferase Reporter Assay
The DNA fragments of -2442 to +1 bp (upstream 2442bp from transcription initiation site; containing two KLF17 consensus binding sites), site 1 deletion (same as wildtype except binding site 1 is deleted) and site 2 deletion (same as wildtype except binding site 2 is deleted) of mouse Id-1 promoter region were cloned into the pGL3-Enhancer vector (Promega, Madison, WI). 168FARN cells were co-transfected with various amounts of mouse KLF17 cDNA, 0.5μg of pGL3 or pGL3 reporter and 0.2ng of Renilla luciferase (pRL-TK) as normalizing control. Luciferase activity was determined using Dual-Luciferase Reporter Assay (Promega, Madison, WI) according to the manufacturer’s instructions 48 hours following transfection. The activity was normalized to the Renilla luciferase activity. Data presented are means ± SD of three independent experiments and are given as the percentage of promoter reporter luciferase activity over vector control pGL3-Enhancer luciferase activity.
Recombinant GST-KLF17 fusion protein
The GST fusion protein of KLF17 was cloned into pGEX4T-1 vector (Amersham Biosciences, Piscataway, NJ) and expressed in Escherichia coli. The KLF17 fusion protein with N-terminal GST tag was purified from the IPTG induced bacterial pellet by sonicating in a buffer (HEPES, pH 8.0, 100 mM NaCl, 5% glycerol, 0.25 mM ZnCl2, 0.1 mM EDTA, 1% Triton X-100, 0.01% Nonidet P-40, 1 mM dithiothreitol and phenymethylsulfonyl fluoride at 1 mM). The supernatant was applied to a glutathione-sepharose 4B column (Amersham/Pharmacia, Piscataway, NJ). After washing, the protein was eluted with 5 mM glutathione in the same buffer, and then dialyzed against 10 mM Tris-HCl buffer, pH 7.9, containing 0.1 M NaCl and 10% glycerol. Control GST protein was purified as described above.
Electrophoretic mobility shift assay (EMSA)
Electrophoretic mobility shift assay (EMSA) was performed using recombinant GST-KLF17 or GST proteins and DNA probes. Double-stranded oligonucleotides corresponding to nucleotides to -2134 to -2097 (5′ CCCCCTTCACCCCACCCCACACCCAATTAGAATAAAC 3′) of the mouse Id-1 promoter with KLF17 consensus binding site or the mutant site 1 (5′ CCCCCTTAGCTAAGCCCCAAGTACAATTAGAATAAAC 3′) were annealed and end-labeled with [γ-32P] ATP and T4 polynucleotide kinase (New England Biolabs). 0.3ng of the labeled probe was incubated with 100ng of recombinant proteins in a binding buffer containing 20 mM HEPES, pH 7.4, 50 mM NaCl, 5 mM dithiothreitol, 1 mM EDTA, 5% glycerol, 1 μg of poly (dI:dC) in a total volume of 20 μl, at room temperature for 15 min. In the competition assays, corresponding double stranded DNA oligonucleotides were end-filled with Klenow using cold dCTP and added to the binding mixture. DNA-protein complexes were resolved on a 6% polyacrylamide gel with 0.5x TBE (25 mM Tris base, 25 mM boric acid and 0.5 mM EDTA) buffer at 150 V for 2 h at room temperature. The gels were dried and the labeled complexes were detected by autoradiography.
Chromatin Immunoprecipitation (ChIP) Assay
168 FARN cells overexpressing KLF17 were fixed in 1% formaldehyde at 37 °C for 10 min. Cells were washed twice with ice-cold phosphate-buffered saline containing protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 1 μg/ml aprotinin, and 1 μg/ml pepstatin A), scraped, and centrifuged at 4 °C. Cell pellets were resuspended in lysis buffer and sonicated to shear DNA. After sonication, the lysate was centrifuged and the supernatant was diluted 10 fold with ChIP dilution buffer (0.01% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.0, 150 mM NaCl, and protease inhibitors). Anti-mouse KLF17, or anti-acetyle H3, or normal mouse IgG were added to the supernatant and incubated overnight at 4 °C with rotation. The immunocomplex was precipitated with protein A/G-agarose and washed sequentially with low salt buffer, high salt buffer, and lithium chloride wash buffer and eluted with elution buffer (1% SDS, 0.1 M NaHCO3, and 200 mM NaCl). Reversal of cross-linking was done by heating at 65 °C overnight in the presence of NaCl. DNA was purified using PureLink PCR Purification kit (Invitrogen Carlsbad, CA). The amount of immunoprecipitated DNA was analyzed in triplicates using mouse ID1 promoter primer sequence by real-time PCR on an ABI 7500 detection system (Applied Biosystems, Foster City, CA) with SYBR green PCR kit. Data were analyzed using the 2-ΔCt method and normalized with input samples as described56.
RNA Isolation, reverse transcription and real-time PCR analysis
Total RNA was extracted from breast cancer cell lines, frozen primary and metastasis tissues using Trizol total RNA isolation reagent (Invitrogen), according to the manufacturer’s specifications and treated with DNase. cDNA was synthesized from total RNA using random hexamers with TaqMan cDNA Reverse Transcription Kit (Applied Biosystems). To determine the levels of KLF17 and Id1 expression, gene primers were designed using Primer Express v3.0 Software and real- time PCR was performed using SYBR Green Jumpstart Taq ReadyMix (Sigma) with Applied Biosystems 7500 Fast Real Time PCR system. The relative amount of expression was calculated from a relative standard curve obtained by using log dilutions of cDNA containing the gene of interest. The average of three independent analyses for each gene and sample was calculated and was normalized to the endogenous reference control gene GAPDH.
Microarray Studies
The Illumina MouseWG-6_v2 bead chip was used for the analysis. 400 ng of total RNA was amplified according to Illumina and 1.2 ug of aRNA hybridized to the arrays. Three biological replicates were analyzed for each condition. Data is processed by Bead Studio and the expression levels for signal and control probes are exported. A set of negative control probes is used to calculate average background level and to determine signal detection threshold. The probe expression data is normalized using quantile normalization. The data is checked for outliers by calculating an outlier score for each of the samples. First, Spearman correlation coefficients are calculated for every sample pair. Correlation between arrays was >99%.
In order to reduce the experimental noise, the data is filtered by removing non-informative probes, i.e. probes that are not detected in majority of samples (more than 95%) or probes that do not change at least 1.2 fold between at least two samples. Analysis was carried out using ANOVA and p<0.05 genes selected for further analysis. GEO accession number of this study is GSE17081.
Clinical Specimens
The breast cancer specimens used in this study were collected at the time of surgery from previously untreated patients. Samples were snap-frozen immediately and stored at -80°C. Total RNA was isolated from frozen tissue with Trizol reagent (Invitrogen). Specimens were collected under local Institutional review board approval and processed under procedures approved by the HIPAA act. The clinical outcome was the presence of lymph node metastases at the time of diagnosis.
References
- 1.Gupta GP, Massague J. Cancer Metastasis: Building a framework. Cell. 2006;127:679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 2.Steeg PS. Tumor metastasis: mechanistic insights and clinical challenges. Nature Rev. Med. 2006;12:895–904. doi: 10.1038/nm1469. [DOI] [PubMed] [Google Scholar]
- 3.Fidler IJ. The pathogenesis of cancer metastasis: the “seed and soil” hypothesis revisited. Nature Rev. Cancer. 2003;3:1–6. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 4.Steeg PS. Metastasis suppressors alter the signal transduction of cancer cells. Nature Rev. Cancer. 2003;3:55–63. doi: 10.1038/nrc967. [DOI] [PubMed] [Google Scholar]
- 5.Yang J, Mani SA, Donaher JL, Ramaswarmy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 6.Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordon-Cardo C, Guise TA, Massague J. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–549. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- 7.Muller A, Homey B, Soto H, Ge N, Catron D, Buchanan M, McClanahan T, Murphy E, Yuan W, Wagner SN, Barrera JL, Mohar A, Verastegu E, Zlotnik A. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410:50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 8.Eccles SA, Welch DR. Metastasis: recent discoveries and novel treatment strategies. Lancet. 2007;369:1742–1757. doi: 10.1016/S0140-6736(07)60781-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nature Rev. Cancer. 2002;2:563–572. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]
- 10.Stafford LJ, Vaidya KS, Welch DR. Metastasis suppressors genes in cancer. Inter. J. Biochem. Cell Biol. 2008;40:874–891. doi: 10.1016/j.biocel.2007.12.016. [DOI] [PubMed] [Google Scholar]
- 11.Yoshida BA, Sokoloff MM, Welch DR, Rinker-Schaeffer CW. Metastasis-suppressor genes: a review and perspective on an emerging field. J. Natl. Cancer Inst. 2000;92:1717–1730. doi: 10.1093/jnci/92.21.1717. [DOI] [PubMed] [Google Scholar]
- 12.Grimm S. The art and design of genetic screens: mammalian culture cells. Nature Rev. Genetics. 2004;5:179–189. doi: 10.1038/nrg1291. [DOI] [PubMed] [Google Scholar]
- 13.Schlabach M, Luo J, Solimini NL, Hu G, Xu Q, Li MZ, Zhao Z, Smogorzewska A, Sowa ME, Ang XL, Westbrook TF, Liang AC, Chang K, Hackett JA, Harper W, Hannon GJ, Elledge SJ. Cancer proliferation gene discovery through functional genomics. Science. 2008;319:620–624. doi: 10.1126/science.1149200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dasgupta R, Kaykas A, Moon RT, Perrimon N. Functional genomic analysis of the Wnt-Wingless signaling pathway. Science. 2005;308:826–832. doi: 10.1126/science.1109374. [DOI] [PubMed] [Google Scholar]
- 15.Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J. Functional genomic analysis of C. elegans chromosome I by systemic RNA interference. Nature. 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- 16.Paddison PJ, Silva JM, Conklin DS, Schlabach M, Li M, Aruleba S, Vivekanand B, O’Shaughnessy A, Gnoj L, Scobie K, Chang K, Westbrook T, Cleary M, Schidanandam R, McCombie R, Elledge SJ, Hannon GJ. A resource for large-scale RNA-interference-based screens in mammals. Nature. 2004;428:427–431. doi: 10.1038/nature02370. [DOI] [PubMed] [Google Scholar]
- 17.Berns K, Hijmans EM, Mullenders J, Brummelkamp TR, Velds A, Heimerikx M, Kerkhoven RM, Madiredjo M, Nijkamp W, Weigelt B, Agami R, Ge W, Cavet G, Linsley PS, Beijersbergen RL, Bernards R. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature. 2004;428:431–7. doi: 10.1038/nature02371. [DOI] [PubMed] [Google Scholar]
- 18.Kolfschoten IG, van Leeuwen B, Berns K, Mullenders J, Beijersbergen RL, Bernards R, Voorhoeve PM, Agami RA. A genetic screen identifies PITX1 as a suppressor of RAS activity and tumorigenicity. Cell. 2005;121:849–58. doi: 10.1016/j.cell.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 19.Aslakson CJ, Miller FR. Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopulations of a mouse mammary tumor. Cancer Research. 1992;52:1399–1405. [PubMed] [Google Scholar]
- 20.Aslakson CJ, Rak JW, Miller BE, Miller FR. Differential influence of organ site on three subpopulations of a single mouse mammary tumor at two distinct steps in metastasis. Int. J. Cancer. 1991;47:466–472. doi: 10.1002/ijc.2910470327. [DOI] [PubMed] [Google Scholar]
- 21.Miller F, Jones RF, Jacob J, Kong YC, Wei YZ. From breast cancer immunology to her-2 DNA vaccine and autoimmune sequelae. Breast Disease. 2004;20:43–51. doi: 10.3233/bd-2004-20106. [DOI] [PubMed] [Google Scholar]
- 22.Dexter DL, Kowalski HM, Blazer BA, Fligiel Z, Vogel R, Heppner GH. Heterogeneity of tumor cells from a single mouse mammary tumor. Cancer Res. 1978;38:3174–3181. [PubMed] [Google Scholar]
- 23.Gumireddy K, Sun F, Klein-Szanto AJ, Gibbins GM, Gimotty PA, Saunders AJ, Schultz PG, Huang Q. An in vivo selection for metastasis promoting genes in the mouse. Proc. NAtl. Acad. Sci. USA. 2007;104:6696–6701. doi: 10.1073/pnas.0701145104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lomberk G, Urrutia R. The family feud: turning off Sp1 by Sp1-like KLF proteins. Biochem. J. 2005;392:1–11. doi: 10.1042/BJ20051234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaczynski J, Cook T, Urrutia R. Sp1- and Krüppel-like transcription factors. Genome Biol. 2003;4:206. doi: 10.1186/gb-2003-4-2-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vliet JV, Crofts LA, Quinlan KGR, Czolij R, Perkins AC, Crossley M. Human KLF17 is a new member of the Sp/KLF family of transcription factors. Genomics. 2005;87:474–482. doi: 10.1016/j.ygeno.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 27.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 28.Nuez B, Michalovich D, Bygrave A, Ploemacher R, Grosveld F. Defective haematopoiesis in fetal liver resulting from inactivation of the EKLF gene. Nature. 1995;375:316–318. doi: 10.1038/375316a0. [DOI] [PubMed] [Google Scholar]
- 29.Perkins AC, Sharpe AH, Orkin SH. Lethal beta-thalassaemia in mice lacking the erythroid CACCC-transcription factor EKLF. Nature. 1995;375:318–322. doi: 10.1038/375318a0. [DOI] [PubMed] [Google Scholar]
- 30.Kuo CT, Veselits ML, Leiden JM. LKLF: a transcriptional regulator of single-positive T cell quiescence and survival. Science. 1997;277:1986–1990. doi: 10.1126/science.277.5334.1986. [DOI] [PubMed] [Google Scholar]
- 31.Foster KW, Frost AR, McKie-Bell P, Lin CY, Engler JA, Grizzle WE, Ruppert JM. Increase of GKLF messenger RNA and protein expression during progression of breast cancer. Cancer Res. 2000;60:6488–6495. [PubMed] [Google Scholar]
- 32.Narla G, Heath KE, Reeves HL, Li D, Giono LE, Kimmelman AC, Glucksman MJ, Narla J, Eng FJ, Chan AM, Ferrari AC, Martignetti JA, Friedman SL. KLF6, a candidate tumor suppressor gene mutated in prostate cancer. Science. 2001;294:2563–2566. doi: 10.1126/science.1066326. [DOI] [PubMed] [Google Scholar]
- 33.Ghaleb AM, Nandan MO, Chanchevalap S, Dalton WB, Hisamuddin IM, Yang VW. Krüppel-like factors 4 and 5: the yin and yang regulators of cellular proliferation. Cell Res. 2005;15:92–96. doi: 10.1038/sj.cr.7290271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yan W, Burns KH, Ma L, Matzuk MM. Identification of Zfp393, a germ-cell specific gene encoding a novel zinc finger protein. Mech. of Dev. 2002;118:233–239. doi: 10.1016/s0925-4773(02)00258-7. [DOI] [PubMed] [Google Scholar]
- 35.Hugo H, Ackland ML, Blick T, Lawrence MG, Clements JA, Williams ED, Thompson EW. Epithelial-mesenchymal and mesenchymal-epithelial transition in carcinoma progression. J. Cell. Physiol. 2007;213:374–383. doi: 10.1002/jcp.21223. [DOI] [PubMed] [Google Scholar]
- 36.Huber MA, Kraut N, Beug H. Molecular requirements for epithelial-mesenchymal transition during tumor progression. Curr. Opin. Cell Biol. 2005;17:548–558. doi: 10.1016/j.ceb.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 37.Zavadil J, Cermak L, Soto-Nieves N, Bottinger EP. Integration of TGF-β/Smad and Jagged1/Notch signaling in endothelial to mesenchymal transition. EMBO J. 2004;23:1155–1165. doi: 10.1038/sj.emboj.7600069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liebner S, Cattelino A, Gallini R, Rudini N, Iurlaro M, Piccolo S, Dejana E. β-catinin is required for endothelial-mesenchymal transformation furing heart cushion development in the mouse. J. Cell. Biol. 2004;166:359–367. doi: 10.1083/jcb.200403050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moody SE, Perez D, Pan T, Sarkisian CJ, Portocarrero CP, Sterner CJ, Notorfrancesco KL, Cardiff RD, Chodosh LA. The transcriptional repressor Snail promotes mammary tumor recurrence. Cancer Cell. 2005;8:197–209. doi: 10.1016/j.ccr.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 40.Hartwell KA, Muir B, Reinhardt F, Carpenter AE, Sgroi D, Winberg RA. The spemann organizer gene, goosecoid, promotes tumor metastasis. Proc. NAtl. Acad. Sci. USA. 2006;103:18969–18974. doi: 10.1073/pnas.0608636103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruzinova MB, Benezra R. Id proteins in development, cell cycle and cancer. Trends in Cell Biol. 2003;13:410–418. doi: 10.1016/s0962-8924(03)00147-8. [DOI] [PubMed] [Google Scholar]
- 42.Sikder HA, Devlin MK, Dunlap S, Ryu B, Alani R. Id proteins in cell growth and tumorigenesis. Cancer Cell. 2003;3:525–530. doi: 10.1016/s1535-6108(03)00141-7. [DOI] [PubMed] [Google Scholar]
- 43.Ivarone A, Lasorella A. ID proteins as targets in cancer and tools in neurobiology. Trends in Mol. Med. 2006;12:588–594. doi: 10.1016/j.molmed.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 44.Ying Q, Nichols J, Chambers I, Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115:281–292. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- 45.Li Y, Yang J, Luo J, Dedhar S, Liu Y. Tubular epithelial cell dedifferentiation is driven by the Helix-loop-helix transcriptional inhibitor Id1. J. Am. Soc. Nephrol. 2007;18:449–460. doi: 10.1681/ASN.2006030236. [DOI] [PubMed] [Google Scholar]
- 46.Lyden D, Young AZ, Zagzag D, Yan W, Gerald W, O’Reilly R, Bader BL, Hynes RO, Zhuang Y, Manova K, Benezra R. Id1 and Id3 are required for neurogenesis, angiogenesis and vascularization of tumor xenograft. Nature. 1999;401:670–677. doi: 10.1038/44334. [DOI] [PubMed] [Google Scholar]
- 47.Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, Viale A, Olshen AB, Gerald WL, Massagué J. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swarbrick A, Roy E, Allen T, Bishop JM. Id1 cooperates with oncogenic Ras to induce metastastic mammary carcinoma by subversion of the cellular senescence response. Proc. NAtl. Acad. Sci. USA. 2008;105:5402–5407. doi: 10.1073/pnas.0801505105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schoppmann SF, Schindl M, Bayer G, Aumayr K, Dienes J, Horvat R, Rudas M, Gnant M, Jakesz R, Birner P. Overexpression of Id-1 is associated with poor clinical outcome in node negative breast cancer. Int. J. Cancer. 2003;104:677–682. doi: 10.1002/ijc.11009. [DOI] [PubMed] [Google Scholar]
- 50.Lin CQ, Singh J, Murata K, Itahana Y, Parrinello S, Liang SH, Gillett CE, Campisi J, Desprez PY. A role for Id-1 in the aggressive phenotype and steroid hormone response of human breast cancer cells. Cancer Res. 2000;60:1332–1340. [PubMed] [Google Scholar]
- 51.Gupta GP, Perk J, Acharyya S, de Candia P, Mittal V, Todorova-Manova K, Gerald WL, Brogi E, Benezra R, Massagué J. ID genes mediate tumor reinitiation during breast cancer lung metastasis. Proc. NAtl. Acad. Sci. USA. 2007;104:19506–19511. doi: 10.1073/pnas.0709185104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Henke E, Perk J, Vider J, de Candia P, Chin Y, Solit DB, Ponomarev V, Cartegni L, Manova K, Rosen N, Benezra R. Peptide-conjugated antise oligonucleotides for targeted inhibition of a transcription regulator in vivo. Nat. Biotechnol. 2008;26:91–100. doi: 10.1038/nbt1366. [DOI] [PubMed] [Google Scholar]
- 53.McAllister s. D., Christian RT, Horowitz MP, Garcia A, Desprez P. Cannabidiol as a novel inhibitor of Id-1 gene expression in aggressive breast cancer cells. Mol. Cancer Ther. 2007;6:2921–2927. doi: 10.1158/1535-7163.MCT-07-0371. [DOI] [PubMed] [Google Scholar]
- 54.Kataoka M, Schumacher G, Cristiano RJ, Atkinson EN, Roth JA, Mukhopadhyay T. An agent that increases tumor suppressor transgene product coupled with systemic transgene delivery inhibits growth of metastatic lung cancer in vivo. Cancer Res. 1998;58:4761–4765. [PubMed] [Google Scholar]
- 55.Huang Q, Gumireddy K, Schrier M, le Sage C, Nagel R, Nair S, Egan DA, Li A, Huang G, Klein-Szanto AJ, Gimotty PA, Katsaros D, Coukos G, Zhang L, Puré E, Agami R. The microRNAs miR-373 and miR-520c promote tumor invasion and metastasis. Nature Cell Biology. 2008;10:202–210. doi: 10.1038/ncb1681. [DOI] [PubMed] [Google Scholar]
- 56.Mukhopadhyay A, Deplancke B, Walhout AJM, Tissenbaum HA. Chromatin immunoprecipitation (ChIP) coupled to detection by quantitative real-time PCR to study transcription factors binding to DNA in Caenorhabditis elegans. Nature Protocols. 2008;3:698–709. doi: 10.1038/nprot.2008.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.