Abstract
A positive-feedback loop is a simple motif that is ubiquitous to the modules and networks that comprise cellular signaling systems. Signaling behaviors that are synonymous with positive feedback include amplification and rapid switching, maintenance, and the coherence of outputs. Recent advances have been made towards understanding how positive-feedback loops function, as well as their mechanistic basis in controlling eukaryotic cell cycle progression. Some of these advances will be reviewed here, including: how cyclin controls passage through Start and maintains coherence of G1/S regulon expression in yeast; how Plk1 activation is driven by Bora and Aurora A, and its expression is stimulated by FoxM1 in mammalian cells; and how some of the various dynamic behaviors of spindle assembly and anaphase onset can be produced.
Keywords: positive feedback, cell cycle, spindle assembly, anaphase-promoting complex (APC), switching, coherence
Introduction
Eukaryotic cells are faced with many decisions. Division, differentiation, and death are a few major choices that must be made during the growth of the population, or development of the organism they comprise. It is not surprising that the pathways leading to these fates have evolved to culminate with a substantial degree of completeness—stereotypically known as “all-or-none” (digital) responses—although the inputs themselves that feed into these pathways can range from being switch-like to graded (analog) [1-3]. During growth and development, cells must survey many conditions and signals, both extrinsic and intrinsic. Are sufficient nutrients and growth factors available for a cell to commit to another cycle? Has it grown enough to initiate replicating its genome? Is the cell prepared to partition its duplicated genome and subcellular components and divide? Once conditions are met for a cell to initiate a fate—whether transitioning to a new cell cycle phase, or becoming functionally specified—it is implicit that the mechanism driving it should assure that the reaction moves forward decisively [4]. One example in which this applies is the transition from interphase to mitosis. It is imperative for mitotic initiation to be reasonably rapid in order to minimize the period of time during which interphase and mitotic conditions coexist. [5]. This means that in addition to the rapidity of the transition, the in vivo processes that drive these cellular changes often need to be coherent in both time and space [6]. Although the need for the decisiveness in specific cellular processes might be apparent, it is the hazards of indecisiveness that reinforce why it can be so critical. This is especially true in the case of proliferation and control of the cell cycle. Unless otherwise programmed, cells must replicate their DNA only once each cycle, they must successfully segregate their newly-replicated chromosomes, and their cytokinesis must occur precisely at the right time and place as mitosis is completed. Failure of any of these steps could be catastrophic to the cell – or worse, may lead to aneuploidy, or permit other conditions that increase the risk of malignancy [7-9]. With cells requiring rapidity and decisiveness in, as well as memory of particular responses, it is not surprising that positive feedback is a feature ubiquitous to their control systems.
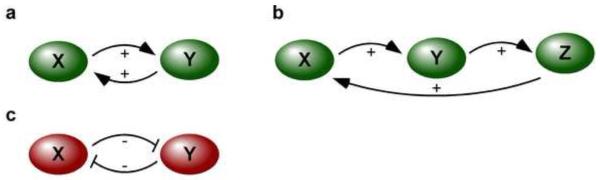
Recent studies of the transcriptional and signaling networks that govern cell cycle transitions have exemplified the importance of positive feedback in defining an array of systems-level behaviors [10-14]. Although it might not seem surprising in-and-of itself, the diversity of emergent properties that positive feedback can confer underscores why it is so common in the regulation of cellular phenomena. It can occur in simple and complex systems alike, and can be derived through two different elementary motifs (Fig. 1). First, one scenario could be a factor, X, activating an X-activating factor, Y – this topology is prevalent in schemes requiring signal amplification, as well as the increased sharpness of a switch. Additional stimulatory components can also be involved, resulting in a lengthened (and therefore, slower) positive-feedback loop: for instance, X activating Y which then activates the X-activating factor Z (Fig. 1b). Second, the basis for positive feedback could also arise from a double-negative feedback motif: a factor, X, inhibiting the X-inhibitor, Y (Fig. 1c). This type of feedback also produces a potent response like that caused by an activation-based positive-feedback loop, but there are other unique features to double-negative feedback. The first inhibitory step (X inhibiting Y) can serve to establish the threshold of a stimulus-response relationship: how much Y must accumulate until its activation? This initial inhibition can therefore also provide capacitance in the system prior to a response, resulting in an increase of the intensity of its output once the system is activated (i.e. Y is then stimulated to repress its inhibition by X). The layering of both positive- and double-negative-feedback loops together in biological systems—including in maturing Xenopus oocytes, and in the CDK1-driven mitosis activating system in Xenopus embryonic extracts—has been found to tune thresholds as well as to ensure the production of powerful and sustained responses [5,15]. In this review, an overview of some exciting recent experimental studies focusing on positive-feedback-driven behaviors such as signal amplification and rapid switching, response coherence, and output maintenance will be presented. Moreover, the biological contexts of these positive-feedback loops, the diverse functions that can be yielded, and the underlying molecular mechanisms behind them will be explored.
Fig. 1.
Topologies of some simple positive-feedback loops. (a) X activating an X-activating factor Y. (b) X activating Y activating an X-activating factor Z. (c) A double-negative (positive) feedback loop, where X inhibits an X-inhibiting factor Y.
The Switch into Start and the Coherence of G1/S Regulon Expression
As mentioned above, positive feedback has become synonymous with the establishment of mitotic onset and progression, and these topics will be discussed in more detail later in this review. But while division is the culmination of cellular growth and DNA replication, the process must start with cells first committing to initiating the cycle anew. The concept of “Start” control represents a commonly shared critical point for eukaryotic cells to ensure their preparedness to replicate their DNA, and then to live with increased ploidy until their division [16]. Although Start has been studied extensively in yeast, the basis of the dynamics that drive yeast cells to make this commitment is not well understood. It makes sense that Start initiation should be a switch-like and irreversible phenomenon, but past studies in yeast had not revealed any source of positive feedback that might promote these behaviors. The three G1 cyclins—Cln1, Cln2, and Cln3—are all able to activate expression of the G1/S regulon, but expression is first triggered by Cdc28/Cln3-phosphorylation of promoter-bound proteins and transcription factors, including SBF and MBF. Two of the genes in this regulon are CLN1 and CLN2, making it entirely possible for a positive-feedback loop to exist [17-19]. Evidence for a Cln1/Cln2-CLN2 transcriptional positive-feedback loop was found previously in a CLN3 null strain, but no difference in the kinetics of transcription from the CLN2 promoter was observed in populations of synchronized cells with or without CLN1 and CLN2 [17,18,20].
This led to Skotheim et al. hypothesizing that a population effect caused by the use of synchronized cells might have led to what had been considered the “linear” model of regulon activation [19]. The authors aimed to resolve this discrepancy by studying transcriptional activation on the single-cell level. They cleverly engineered a yeast strain to express unstable green fluorescent protein (GFP) under the control of the CLN2 promoter to monitor CLN2 expression, and tracked cell birth using GFP-tagged myosin (Myo1-GFP). The timing of CLN2 expression was then measured from the moment that the Myo1-GFP ring disappeared – marking cytokinesis completion. Indeed, this methodology revealed a striking difference in timing between WT yeast and the CLN mutants. CLN2pr-GFP expression occurred much sooner—and with less variability—in WT than the cln1Δcln2Δ mutant, and expression in this mutant depends on CLN3 since no reporter expression occurred in cln1Δcln2Δcln3Δ yeast. Both cln1ΔCLN2 and CLN1cln2 mutants activated the CLN2 promoter with similar timing, so not only is a positive-feedback loop intrinsic to the G1/S regulon, both CLN1 and CLN2 can drive it. Interestingly, on the single-cell level, the amplitude of GFP fluorescence was increased and signal prolonged in cln1Δcln2Δ yeast, possibly as a consequence of the delay between CLN2 and CLB2 expression in this mutant. This could cause the earliest individual expressers in an imperfectly synchronized population to obscure the average transcriptional timing of the whole. As shown in Fig. 2, differences between single-cell and population-level approaches could lead to an improper conclusion regarding timing of CLN2pr-GFP induction in the latter. Asynchrony in the mutant population and/or increased noise in CLN2 expression in the cln 1cln 2 mutant would contribute to this error. Wild-type yeast (Fig. 2a) expressed lower levels of GFP and turned it over more rapidly, relative to the clnΔ1clnΔ2 mutant strain (Fig. 2b) [19]. This mutant expressed higher levels of GFP and turned the protein over less effectively, and could indeed appear to have the same induction timing as WT yeast if the average levels of GFP in both populations was used as a metric (Fig. 2c; top). This was the case, as Skotheim et al. showed that summing the measurements of CLN2pr-GFP from individual cln1Δcln2Δ cells resulted in timing that was no earlier than WT cells [19]. This finding mirrored results found in prior studies that contributed to the earlier “linear” model of CLN2 transcription CLN [17,18,20]. Despite the differences in GFP expression and turnover between WT and mutant yeast, however, single-cell analysis of the appearance of GFP signal showed clearly that CLNpr-GFP expression was induced earlier and more sharply in the former (Fig. 2c; bottom). Once again, this demonstrated the importance of performing single-cell measurements to avoid population-level errors.
Fig. 2.
Differences in population-level versus single-cell analysis of CLN2pr-GFP expression in yeast. (a) Production of GFP in ten WT yeast daughters throughout a time course. (b) Production of GFP in ten cln1Δcln2Δ yeast daughters throughout a time course. (c) Schematized plots of mean GFP fluorescence in all ten cells combined (top) and the fraction of individual cells out of ten in which GFP expression was induced (bottom), with numerical labels and vertical hash lines representing the three time points represented in (a) and (b).”Birth” is denoted as the disappearance of GFP-tagged myosin (represented by the green oval at the cell-to-cell junction) shown in the prior step. For simplicity, all ten daughters shown in (a) and (b) are not depicted as budding, although budding would normally be observed at steps 2 and 3. Plots in (c) are adapted from [19].
Although positive feedback was found to enable rapid switching of expression from the CLN2 promoter, the G1/S regulon confers the expression of a wide array of genes at Start – including those activated by SBF and MBF. So, how is post-Start production of the machinery responsible for cell cycle progression triggered in unison? Using CLN2pr-GFP, as well as mCherry fusions to the promoters of two other regulon members—RAD27 and RAF1—Skoltheim et al. compared their timing of expression. In most of the cln1Δcln2Δ cells analyzed, expression from both RAD27pr and RAF1pr was delayed behind CLN2pr-GFP. Loss of G1 cyclins was not a cause of this, as overexpression of Cln3 in those cells did not reduce the incoherence of expression from RAD27 and CLN2 promoters. Therefore, the Cln1/Cln2-driven positive feedback is not only necessary for switching on CLN2 expression, but is likely to simultaneously induce expression from an ensemble of SBF- and MBF-controlled genes [19,21]. This concept manifested physiologically in these experiments, with an appreciable number of cln1Δcln2Δ cells arresting in an unbudded state.
With CLN1 and CLN2 deletion causing cell cycle arrest, the authors hypothesized that incoherent expression from some regulon members might promote improper expression of the SBF inhibitor, cyclin Clb2, and lead to arrest. Removal of CLB2 or overexpression of a nonphosphorylatable Cdh1—a mitotic cyclin-targeting co-factor of APC—decreased the population of unbudded arrested cln1Δcln2Δ cells. The rapid exit of the remaining pool of transcriptional inhibitor Whi5 from the nucleus was also found to depend on the positive feedback generated by Cln1/Cln2. This rate increase occurs in tandem with a slow nuclear export of Whi5 that is driven by Cln3, and loss of Cln1/Cln2 makes this process less switch-like. Could there be a direct regulatory connection between Cln and Whi5? To answer this question, expression of a GFP-fused WHI5 allele missing half of its twelve Cln-dependent phosphorylation sites (WHI56A) was tested for its effect on nuclear shuttling and regulon expression. Export of WHI56A-GFP from the nucleus was slowed and incomplete, with expression from CLN2pr and RAD27pr being less coherent than in WT cells, but more coherent than cln1Δcln2Δ cells. This supported the idea that Cln-dependent phosphorylation on Whi5 and its switch-like movement contributes to the positive-feedback loop and regulon coherence [19,21]. However, additional WHI5-independent feedback may exist. Cells deleted for WHI5 alone activated CLN2pr-directed transcription earlier than cln1Δcln2Δwhi5Δ cells, suggesting that one or more factors in addition to Whi5—possibly Cln3—may work in conjunction with Cln1/Cln2 to contribute the positive feedback that controls the coherence of G1/S regulon expression.
The phenotypes observed in yeast with reduced positive feedback in their Start network epitomized its importance in the control of genetic and physiological outputs related to distinct aspects of G1 regulation. First, the expression of several genes from the G1/S regulon lagged. Second, the synchronicity of their expression was lost. Lastly, the somewhat premature activation of the mitotic cyclin Clb2—possibly due to the incoherence in regulon expression—coincided with the arrest of unbudded cells [19]. Altogether, Skoltheim et al. demonstrated that positive feedback is necessary for ensuring cell cycle entry at Start in yeast is sharp and switch-like. Due to this feedback, and the exclusion of Whi5 from the nucleus, it appears that the passage through Start itself also involves the promotion of a driving force for S-phase initiation by way of Cln-dependent events. The Cln1/Cln2-derived positive feedback may very well be a necessary attribute that permits cells to both establish and progress past Start, and through the remainder of the yeast cell cycle.
These findings in yeast raise the following question: are there similar positive-feedback loops that function within mammalian restriction point and G1-S control? One well-known loop is the expression of cyclin E under the control of the transcription factor, E2F. Positive feedback arises from Cdk2/cyclin E phosphorylating and releasing the bound retinoblastoma protein (Rb) from E2F, which stimulates further E2F-driven cyclin E transcription [22,23]. Loss of the three mammalian E2F factors in mouse embryonic fibroblasts—E2F1, 2, and 3—increases the expression of p21 and reduces S-phase cyclin-dependent kinase (CDK) activity and Rb phosphorylation [24]. This underscores the importance of transcriptional and post-translational positive-feedback loops in the mammalian G1-S transition. More recently, proteolysis has also been shown to be fundamental to mammalian restriction point control. Skp2 (of the SCFSkp2 E3 ubiquitin ligase complex) gene expression is induced by E2F1 [25]. SCFSkp2 targets the CDK2/cyclin E inhibitor p27 for proteolysis, and CDK2/cyclin E phosphorylation of Rb releases E2F1 to further induce transcription of its target genes, as described above [26]. Indeed, positive-feedback loops pervade the transcriptional and signaling systems that govern the restriction point and the G1-S transitions in yeast and mammalian cells, and provide mechanisms that yield switch-like and coherent responses within them.
Switching on the Activation and Synthesis of the M-Phase Machinery
Once a eukaryotic cell has traversed past Start and through S phase, another critical decision must be made. Are the requirements of cellular growth and health (e.g. no DNA damage) satisfied for mitotic initiation to proceed? If this is the case then the transition from G2 phase to mitosis can ensue, and this also involves multiple positive-feedback loops. One loop that is synonymous with the G2-M transition involves the mitosis-stimulating kinase, cyclin-dependent kinase 1 (CDK1), and its activating phosphatase, Cdc25 [27,28]. After binding cyclin, CDK1 is first phosphorylated by the inhibitory kinases Wee1 and Myt1 [29-32] – this is the first step of the double-negative-feedback loop. CDK1 activation is then somehow triggered, and it inhibits the repressors Wee1 and Myt1 – this is the second leg of the double-negative-feedback loop. CDK1 is then further stimulated through the increased activation of Cdc25, which dephosphorylates and activates the remaining inactive CDK1/cyclin complexes. CDK1 activation at M-phase onset is a classic example of signal amplification and a sharpened switch-like response initiating a complex cellular transition [33]. The importance of these positive-feedback loops has been demonstrated through both computational modeling [14,34] and experimentation [5,35,36]. What contributes to this triggering mechanism at mitotic onset remains unclear, but recent progress has further delineated the signaling architecture that underlies M-phase initiation. What comprises the inner workings of this positive-feedback-enhanced switch? Polo-like kinase (Plk1) has long been implicated as a stimulator of the mitotic transition, and this is likely since it is involved in several cellular functions during M-phase, including centrosome maturation, spindle establishment, and mitotic progression [37]. Its biochemical connection with mitotic entry is evidenced by its activation of Cdc25B and Cdc25C [38-41], and inactivation of Wee1 via proteolysis [42,43]. The steps that lead to its activation, however, are not well understood. Extensive studies performed by Seki et al. revealed that as Bora protein is produced in G2 phase, it interacts with the C-terminal Polo-box domain (PBD) of Plk1 [44]. This interaction exposes the activating phosphorylation site of Plk1, which is then subsequently phosphorylated by Aurora A kinase (AurA) and leads to Plk1 activation at the G2/M transition. RNAi knockdown of Bora in HeLa cells did not affect transitions between G1, S, and G2 phases, but it delayed mitotic entrance, supporting its M-phase-stimulatory role. The authors suggest that during the binding of Bora to Plk1 the Plk1-Cdc25-Cdk1 positive-feedback loop is initiated, but that Plk1 activation is not complete. At this stage, Bora binding precluding the interaction of Plk1 with phosphorylated substrates may not permit the complete activation of Plk1 [45]. Only after Bora is phosphorylated by Plk1 and degraded through the ubiquitin-proteasome pathway can Plk1 become maximally activated [44]. This event coincides with continued M-phase progression and fully active Plk1 targeting its mitotic substrates.
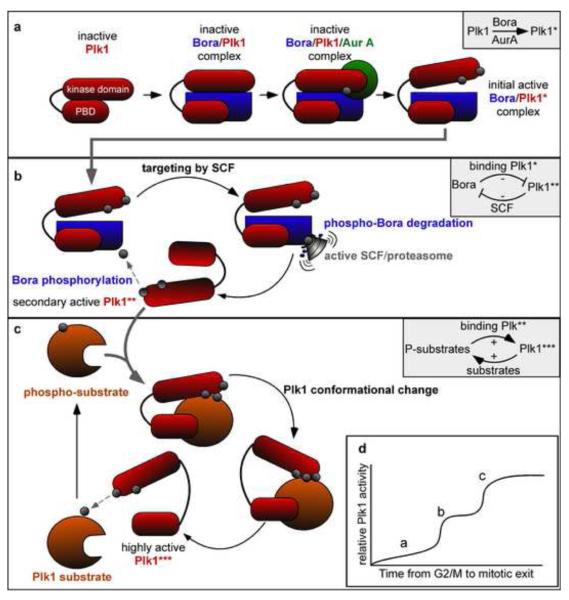
While there have been no single-cell resolution studies revealing what dynamic changes might occur in Plk1 activity during M-phase progression, its regulation by Bora could conceivably help ensure that the proper amount of Plk1 activity is delivered at the right time. The regulation of Plk1 by Bora could indeed have some interesting consequences on the dynamics of Plk1 activation. This relationship suggests that one, and possibly even more positive-feedback loops could exist in addition to the relationship between Plk, Bora, and AurA that was identified by Seki et al. (Fig. 3). Plk1 becoming activated initially by AurA phosphorylation on T210 with the assistance of Bora, followed by a Bora-derived inhibition of additional Plk1 activation might first serve to limit the amount of initial Plk1 activity (Fig. 3a; Plk1*). But with Bora first acting as activator, and then activity limiter, Plk1 phosphorylation of its β-TrCP-targeting degron could then eventually initiate a positive-feedback loop. β-TrCP-driven proteolysis of Plk1-phosphorylated Bora would rapidly remove this inhibitor as more Plk1 is activated (Fig. 3b; Plk**). This double-negative-feedback loop could potentially serve to initially build an increasing amount of Bora-inhibited Plk1. Once enough Bora is phosphorylated by Plk1 to spark its destruction by SCF, a second rapid activation of Plk1 could occur. Lastly, as this more highly activated, non-Bora-bound Plk1 phosphorylates and binds substrates, a third positive feedback is generated and could conceivably activate Plk1 even further (Fig. 3c; Plk***). What purpose might these dynamics serve? They could plausibly ensure the switch-like nature of mitotic progression during mid- to late mitosis, and the potential dual-stage activation of Plk1 could be important for delivering different amplitudes of activity. First, a slow increase of Plk1 activity could serve to initiate the Cdc25-CDK1 positive-feedback loop (Fig. 3d; see “a”). Once the spindle checkpoint is relieved, progression from metaphase to anaphase might necessitate a rapidly increased Plk1 activity. Destruction of Bora by SCF driving a (double-negative) positive-feedback loop could provide this signal amplification (Fig. 3d; see “b”). Lastly, interaction of Plk1 with phosphorylated substrates could conceivably create a second positive-feedback loop, where an increased number of phosphorylated substrates binding to Plk1 further increase Plk1 activity (Fig. 3d; see “c”). With Bora itself acting as a stimulatory interface for Plk1 activation by AurA, and then as a limiter of Plk1 activity, it is plausible that Bora chaperones two of three different phases of Plk1 activation that is essential for proper mitotic progression. And indeed, Seki et al. showed that non-degradable Bora permits mitotic onset but delays metaphase exit and anaphase onset [46]. This suggests that Bora destruction—while not essential for mitosis—may be critical to define the kinetics underlying mitotic entrance, as well as spindle structure and function during mitotic progression. It seems that Bora adds another level of regulation and complexity to the positive feedback that underlies M-phase control. It will be important to learn if and how other members of the CDK1 regulatory network interface with this protein during entry and passage through mitosis. These findings also reinforce the need for further quantitative studies on Plk1 activation and function, particularly on the single-cell level.
Fig. 3.
A model for positive feedback involvement during the multiple phases of Plk1 activation during M phase. (a) Stepwise activation of Plk1 by AurA phosphorylation of Plk/Bora complex. This would generate the first level of Bora-bound active Plk1 (Plk*; see inset box). (b) A possible double-negative/positive-feedback loop caused by Plk1 phosphorylation of Bora, followed by phospho-Bora destruction by SCF-β-TrCP/proteasome activity, leading to further activation of Plk1 (Plk**; see inset box). (c) Binding of phospho-substrate to secondary active Plk1 causes its conformational change and produces more highly active Plk1 (Plk***). This may also yield a positive-feedback loop (see inset box) with highly active Plk1 producing additional phospho-substrates. (d) Schematized increases of Plk1 activity resulting from the reactions described in (a), (b), and (c), with proposed positive-feedback loops yielding rapid increases in activity in the latter two.
While positive-feedback loops arising from posttranslational modifications are ubiquitous to the initiation of mitotic entry, Plk1 itself is embedded in a feedback loop that influences its own expression during that time. The mammalian transcription factor Forkhead Box M1 (FoxM1) controls the expression of multiple M-phase regulatory genes at the G2/M transition [47,48], and Fu et al. recently identified FoxM1 as a binding partner of Plk1 in a yeast-2-hybrid screen [49]. Plk1 was also found to phosphorylate CDK1-phosphorylated FoxM1 at its Ser715 and Ser724 residues. A luciferase-encoding plasmid under the control of a 6xFoxM1-TATA promoter was used to test the function of either wild-type FoxM1 or mutants where both serines were replaced with Ala (the non-phosphorylatable form) or Glu (a phospho-mimetic form). FoxM1(EE) induced expression of the luciferase reporter, whereas FoxM1(AA) failed, supporting the notion that these phosphorylation sites are indeed functional targets of Plk1. Fu et al. then validated the function of the Plk1/FoxM1 relationship in vivo, by knocking down FoxM1 and attempting rescue with either a wild-type or Plk1-non-phosphorylatable version. Knockdown of the endogenous protein yielded cell cycle abnormalities familiar to loss of FoxM1 function. These included prolonged G2, and for cells that entered M-phase, prolonged mitosis, failed cytokinesis, and accumulation of aneuploid and polyploid cells [47,48]. These defects were mirrored in FoxM1 knockdown cells that stably-expressed a Plk1-non-phosphorylatable mutant that was resistant to RNAi, but were relieved by expression of the wild-type protein. Using this same RNAi/RNAi-resistant-rescue combination, FoxM1(Ser715A/Ser724A) down-regulated the transcription of cyclin B1, Aurora B, and Plk1 itself. Expression of the Plk1-phosphorylatable version, however, prevented this reduction. Lastly, mitotic defects caused by pharmacological inhibition of Plk1, including prolonged early mitotic phases and prometaphase arrest, were shown to be partially rescued by stable expression of Plk1-phosphorylation-mimicking FoxM1(Ser715E/Ser724E). This provided further evidence that phosphorylation of FoxM1 by Plk1 serves a critical functional role, in vivo. The discovery of Plk1 being embedded within its own transcriptional positive-feedback loop adds even further to its functional complexity and importance – as a hub to both posttranslational and transcriptional control of mitosis.
Taken together, these recent studies by the Fang and Chen groups demonstrate the crucial roles that Plk1-driven positive feedback plays to ensure that mitosis is initiated in a timely fashion. They also provide some understanding towards how Plk1 activity could be tuned to provide a succession of activities required for M-phase initiation and progression. This might occur in two ways. First, the Bora protein serves as a catalytic interface between Plk1 and AurA, and Plk1-phosphorylation of its β-TrCP degron and ubiquitylation by SCF could yield a double-negative-feedback loop that further activates Plk1 [44]. This is followed by another loop caused by the interaction of Plk1 with phosphorylated substrates of Plk1, which further increases its activity [45]. Second, a transcriptional positive-feedback loop where active Plk1 stimulates its own expression and expression of other mitotic genes by phosphorylating the transcription factor FoxM1 [49]. Due to the critical nature of these highly regulated steps during mitotic progression, it will be important to gain a fundamental understanding of any other connections between additional mitotic regulators and Bora, AurA, and Plk1.
Mitotic Maintenance and Progression
Mitotic entrance, anaphase onset, and mitotic exit each having a switch-like character and occurring as a highly organized progression makes sense, considering that a mitotic cell has only one opportunity to properly partition its genetic complement to its daughters. In addition to the abruptness of these transitions, there is now convincing evidence from studies in mammalian cells that positive feedback is important to sustain the delay that must occur during spindle formation, prior to APC activation and the onset of anaphase. Cells face the dilemma that CDK1 activation drives mitotic initiation while APC activity also must be stifled for a period of time—to stabilize cyclin B—so CDK1/cyclin B can help direct proper assembly of the mitotic spindle. Although this sounds like the task reserved for the spindle checkpoint, APC activity must be inhibited before the spindle is fully formed, and prior to the kinetochores being capable of checkpoint signaling [50,51].
How do cells ensure that CDK1/cyclin B remains active and the APC inactive early in mitosis, while the spindle is still being constructed? Performing biochemical analyses in Xenopus egg extracts, as well as live-cell studies using HeLa and HCT116 cells, Ban et al. uncovered a positive-feedback-driven mechanism that helps cells cope with this predicament in M-phase control [50]. Early mitotic inhibitor 1 (Emi1) represses APC activity throughout S and G2 phases to permit the accumulation of cyclins A and B. Once mitosis is initiated, a majority of Emi1 is degraded through SCF-βTrCP-directed proteolysis [52,53]. Immediately following prophase and up until anaphase, however, the remaining pool of Emi1 and the core APC subunit, Cdc27, co-localizes to the spindle poles [54]. Destabilization of microtubules by nocodazole abolishes this localization, whereas nocodazole washout, as well as the formation of microtubule asters in the presence of taxol permits their interaction. Because localization of Emi1 to the spindle poles was found to not overlap with γ-tubulin/centrosomes, Ban et al. tested if disrupting the dynein-dynactin motor complex—responsible for minus-end-directed movement—would displace Emi1. Indeed Emi1 and the APC/C were both displaced under this condition, whereas another cytoskeletal-binding protein, Eg5, remained associated. Immunoprecipitation of Emi showed that it was not only in a complex with APC/C, but that the nuclear matrix and spindle assembly protein NuMA (nuclear mitotic apparatus) was also present. In fact, each component could be precipitated individually while containing the entire complex—including dynein/dynactin and the APC/C subunit, Cdc23—and the association of these proteins coincided with hypophosphorylation of Cdc27. This implied that the entire complex associates with a spindle-bound APC/C [55]. To determine if Emi1 plays a role in bridging NuMA and APC/C, cell extracts were depleted of either Emi1 or NuMA. This removal of Emi1 disrupted the Cdc27 and NuMA interaction, and while immunoprecipitated NuMA completely depleted the pool of Emi1, depletion of the Emi1 did not eliminate the NuMA from the extract. Further experiments demonstrated that disrupting the NuMA interaction with the spindle causes delocalization of Emi1, and that NuMA both recruits to, and protects at the spindle poles this small remaining pool of Emi1. Hence, Emi1 is a limiting factor facilitating the interaction between APC/C and NuMA during mitosis, while NuMA is essential to deliver Emi1 to the poles and prevent its APC/C-directed proteolysis at the spindle during M-phase.
It is apparent that a stable Emi1-NuMA-APC/C aggregate forms during mitosis, but is this the complex that prevents premature cyclin B destruction on the spindle? RNAi knockdown of Emi1, or injection of Emi1-inactivating antibodies both caused defects in spindle formation and chromosome congression [50]. Phenotypes ranged from highly elongated and misshaped spindles with chromosome scattering, to poles with a barely formed spindle and disorganized chromosomes. Emi1 knockdown not only hindered spindle formation, but it also prohibited the movement of dynactin subunits p50 and p150 to the poles, whereas HSET and Kif2 motors localized properly. In vivo depletion of Emi1 also prevented Cdc27 from co-localizing to the mitotic spindle, suggesting that Emi1 and dynein/dynactin work in tandem to properly organize APC/C on the mitotic spindle. Since CDK1/cyclin B activity and inactivity coincide with spindle organization and disassembly, respectively, the authors proposed that cyclin B was a strong candidate to benefit from the inhibition of APC/C by Emi1at the spindle poles. Indeed, RNAi knockdown of Emi1 showed a relative loss of localization of cyclin B at the spindle and spindle poles. Depletion of Emi1 from human mitotic cell extracts conferred a premature destruction of cyclin B and reduced levels of aster formation. The requirement for Emi1 to block cyclin B proteolysis was demonstrated in vitro by adding non-degradable cyclin B to Emi1-depleted extracts, and this treatment recovered aster formation.
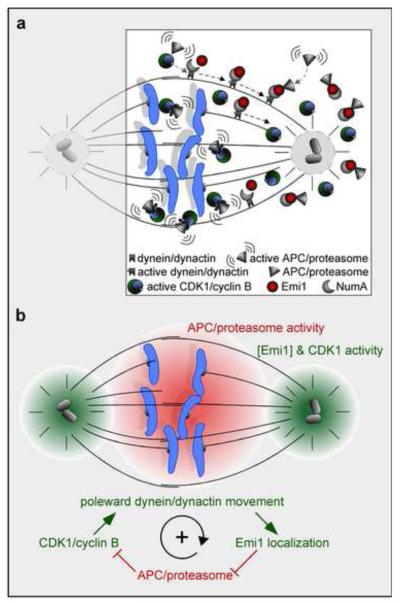
This substantial body of work by Ban et al. demonstrated the function of the Emi1-NuMA complex, in vivo, and invited the following question: how can the signal to not degrade cyclin B be reinforced during this period of CDK1 activity? What was uncovered is a positive-feedback loop that can sustain the localized inhibition of APC/C at the spindle poles (Fig. 4). The activation of the minus-end directed motor dynein-dynactin by CDK1 phosphorylation serves to deliver the matrix and spindle assembly protein NuMA to the spindle poles (Fig. 4a). It is at this location that Emi1 is maintained by its interaction with, and delivery by NuMA/dynein-dynactin. This CDK1-stimulated movement of dynein-dynactin results in a population of Emi1-inhibited APC/C at the spindle poles (Fig. 4b). By stimulating the inhibition (through Emi1) of its inhibitor (APC/C), CDK1 can maintain its own activity through what the authors' deemed the END (Emi1/NuMA/dynein-dynactin) network [50]. It is through this END-network-derived positive-feedback loop that CDK1 can facilitate the proper formation and maintenance of the spindle during early mitosis in mammalian cells. This network prevents premature inactivation of CDK1/cyclin B by APC/C before the spindle checkpoint becomes functional.
Fig. 4.
Positive feedback of the END network. (a) Schematic of the Emi1/NuMA/dyneindynactin network (depicted only at the right spindle pole) functioning in a pro-metaphase cell, where active CDK1/cyclin B phosphorylates dynein-dynactin and stimulates the poleward movement of its Emi1/NuMA cargo. This inhibits APC/proteasome activity at the spindle poles. (b) CDK1/cyclin B-driven transport of Emi1 to the spindle poles increases cyclin B1 stability (due to APC/proteasome inhibition), resulting in a positive-feedback loop that sustains CDK1 activity in a gradient that is highest at the centrosomes (green). The gradients of [Emi1] and CDK1 activity diminishes towards the central spindle, where the gradient of APC/proteasome activity increases due to decreasing concentrations of Emi1 (red).
While other mitotic processes may be more clearly switch-like, the dissolution of cohesion between sister chromatids once the metaphase checkpoint is relieved is also quite sudden, and irreversible. This ensures that disjunction of the chromosome pairs is well synchronized. Even with such switch-like character, there had been no basis for positive feedback uncovered in the metaphase-to-anaphase regulatory system. This “anaphase switch” is under the control of securin, the separase inhibitor whose degradation permits proteolysis of cohesin, and subsequent separation of the daughter chromosomes [56-58]. Recent studies of securin and its regulator by Holt et al. demonstrated that positive-feedback loops do indeed exist within the securin/separase system of yeast [59]. Mass spectrometry revealed CDK1 phosphorylation of securin occurred at previously uncharacterized N-terminal sites—T27 and S71—near a destruction box motif. N-terminally-phosphorylated securin (with mutated C-terminal CDK1 sites) was found to be poorly ubiquitylated in vitro, in contrast to phospho-securin that had been first dephosphorylated by treatment with Cdc14 phosphatase. The fact that separase stimulates Cdc14 activity completes this positive-feedback loop: initiation of securin degradation activates separase, which then activates Cdc14, leading to securin dephosphorylation and further subsequent securin ubiquitylation by APC-Cdc20 and proteolysis. A second positive-feedback loop could also exist in the form of a double-negative loop, postulated as APC activity on securin being first blocked by Cdk1/Clb5, followed by the continued destruction of securin by APC as Clb5 proteolysis continues [59]. In other words, as APC becomes active and targets Clb5 for destruction, this effectively reduces the rate of CDK1/Clb5 phosphorylation onto securin and increases the activity of APC onto that substrate. This double-negative feedback loop could help contribute to producing a high enough CDK1 activity prior to securin destruction and anaphase onset, and might also contribute to the sharpening of securin degradation as CDK1 is then inactivated by cyclin proteolysis.
Using gene replacement to swap out the endogenous securin, Holt et al. performed some elegant studies dissecting the function of this feedback loop, in vivo [59]. Insensitive to CDK1 phosphorylation at its N-terminus, securin-2A (securin T27A/S71A) bypasses the Cdc14-securin-separase positive-feedback loop. In live-cell imaging experiments, a dsRed-tagged Spc42 spindle-pole body protein and GFP-labeled loci integrated on both chromosomes IV (a LAC operator array) and V (a TET operator array) were tracked to quantitate the rates and timings of spindle elongation and chromosome separation, respectively. With the two GFPs integrated close enough in proximity to their sister chromosomes' respective centromeres, fluorescence appeared as a single focused spot until cohesin was degraded by separase and anaphase was initiated. This allowed precise measurements of the timing of chromosome segregation in strains defective in securin regulation. Chromosome V was found to segregate invariably after chromosome IV, and in securin-2A-expressing cells the lag time of this segregation was nearly double that of its wild-type counterpart. While loss of Cdc14 (in a Cdc14-1 mutant) or stabilizing Clb5 (by deleting its destruction box) undoubtedly caused some pleiotropic effects, both of these reductions of positive feedback in the securin destruction pathway mirrored the behavior of the securin-2A strain. The synchrony of anaphase was greatly reduced in both instances, and the lag time of chromosome IV and V segregation in securinΔ cells was more than tripled. These experiments demonstrated that securin itself is a major source of abruptness in the metaphase-to-anaphase transition, and validated the existence of a positive-feedback loop between Cdc14, securin, and separase in yeast. This feedback appears to be crucial to provide further sharpness of anaphase onset, as well as synchrony of chromosome segregation. Additional experiments using these strains scrutinized the relationship between securin regulation and its role in controlling anaphase spindle dynamics. Indeed, the phosphoregulation of securin—with its brief stabilization relative to cyclin at the anaphase transition—contributes to switch-like activation of separase and Cdc14. CDK1 repression of securin ubiquitylation was found to be critical to coordinating chromosome segregation with spindle dynamics. A loss of positive feedback in securin destruction correlated with defects in spindle elongation and maintenance, and increased the frequency of chromosome missegregation. From this, Holt et al. speculated that separase activity itself might have to exceed particular thresholds to dictate the segregation of particular chromosomes during mitosis [21,59]. This is a tantalizing proposition, and could very well explain how the ordering of sister chromatid separation is directed and maintained so precisely from cycle to cycle in eukaryotic cells.
In summary, through extensive biochemical and cellular studies, the Jackson and Morgan groups have uncovered how positive feedback contributes to two critical facets of mitotic progression control. With the END network blocking APC activation at the spindle poles during early mitosis, it affords time for the spindle checkpoint and its own inhibition of APC activity to be established. In yeast, once all kinetochores are attached by microtubules at metaphase, the Cdc14-securin-separase positive-feedback loop provides a sharp anaphase switch, and it may even contribute to the ordered movement of sister chromatids. Even with the differences in some of the molecular players in various eukaryotic systems, it is intriguing to think that the positive-feedback loops of the END network and the anaphase switch might in principle function in tandem to safely guide cells through mitosis. It is also enticing to think that studies of these different systems may eventually reveal how the molecular basis for generating these types of positive-feedback loops overlap, and may possibly recur in a similar functional context that spans plant and animal kingdoms.
Conclusion
The array of functions that a positive-feedback loop can confer to a biochemical system—from amplification and switching of an activity, to maintenance of a response, to providing coherence between multiple responses—is indispensable for directing the physiological outcomes required for cell proliferation, growth, and survival. This is particularly true for eukaryotic cell cycle control Cells must not merely make the decision to continue proliferating, but must ensure that gene expression and signaling pathways properly drive growth, genome replication, and the treacherous passage through mitosis. Although it is apparent that the recent and rapid emergence of positive feedback in cellular control systems exemplifies it as one of the most critical motifs in signaling, experimental and computational studies must continue for us to gain a better understanding of where, when, and how these loops function. If positive feedback serves as such a critical biological linchpin for so many signaling pathways in normal cells, might it also one day be an ideal place to target therapies to treat malignancies and other hyper-proliferative diseases? It may turn out that the challenges of trying to understand diseases and their abnormal molecular pathologies could take us full circle back to focus on the simpler relationships that predominate within the normal cellular context. Indeed, positive feedback may have been a key to better understand how to end these maladies all along, right from the very beginning.
Acknowledgements
The author would like to thank members of the Pomerening lab for critical reading of the manuscript, reviewers for their constructive criticisms and helpful comments, and sincerely apologizes to any authors not cited in this review due to space limitations. J.R. Pomerening is supported by a Pew Biomedical Scholar Award and a grant from the National Institutes of Health (NIGMS).
Abbreviations
- CDK1
cyclin-dependent kinase 1
- APC
anaphase-promoting complex
- Plk1
Polo-like kinase 1
- FoxM1
Forkhead Box M1
- Emi1
early mitotic inhibitor 1
- NuMA
nuclear mitotic apparatus
- END
Emi1/NuMA/dynein-dynactin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ferrell JE., Jr. Self-perpetuating states in signal transduction: positive feedback, double-negative feedback and bistability. Curr Opin Cell Biol. 2002;14:140–8. doi: 10.1016/s0955-0674(02)00314-9. [DOI] [PubMed] [Google Scholar]
- 2.Pomerening JR. Uncovering mechanisms of bistability in biological systems. Curr Opin Biotechnol. 2008;19:381–8. doi: 10.1016/j.copbio.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 3.Tyson JJ, Chen KC, Novak B. Sniffers, buzzers, toggles and blinkers: dynamics of regulatory and signaling pathways in the cell. Current Opinion in Cell Biology. 2003;15:221–31. doi: 10.1016/s0955-0674(03)00017-6. [DOI] [PubMed] [Google Scholar]
- 4.O'Farrell PH. Triggering the all-or-nothing switch into mitosis. Trends Cell Biol. 2001;11:512–9. doi: 10.1016/s0962-8924(01)02142-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pomerening JR, Kim SY, Ferrell JE., Jr. Systems-level dissection of the cell-cycle oscillator: bypassing positive feedback produces damped oscillations. Cell. 2005;122:565–78. doi: 10.1016/j.cell.2005.06.016. [DOI] [PubMed] [Google Scholar]
- 6.Kholodenko BN. Cell-signalling dynamics in time and space. Nat Rev Mol Cell Biol. 2006;7:165–76. doi: 10.1038/nrm1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ganem NJ, Storchova Z, Pellman D. Tetraploidy, aneuploidy and cancer. Curr Opin Genet Dev. 2007;17:157–62. doi: 10.1016/j.gde.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 8.King RW. When 2+2=5: the origins and fates of aneuploid and tetraploid cells. Biochim Biophys Acta. 2008;1786:4–14. doi: 10.1016/j.bbcan.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takaki T, Trenz K, Costanzo V, Petronczki M. Polo-like kinase 1 reaches beyond mitosis--cytokinesis, DNA damage response, and development. Curr Opin Cell Biol. 2008;20:650–60. doi: 10.1016/j.ceb.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 10.Brandman O, Ferrell JE, Jr., Li R, Meyer T. Interlinked fast and slow positive feedback loops drive reliable cell decisions. Science. 2005;310:496–8. doi: 10.1126/science.1113834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brandman O, Meyer T. Feedback loops shape cellular signals in space and time. Science. 2008;322:390–5. doi: 10.1126/science.1160617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Novak B, Tyson JJ. Design principles of biochemical oscillators. Nat Rev Mol Cell Biol. 2008;9:981–91. doi: 10.1038/nrm2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shaw AS, Filbert EL. Scaffold proteins and immune-cell signalling. Nat Rev Immunol. 2009;9:47–56. doi: 10.1038/nri2473. [DOI] [PubMed] [Google Scholar]
- 14.Tsai TY, Choi YS, Ma W, Pomerening JR, Tang C, Ferrell JE., Jr. Robust, tunable biological oscillations from interlinked positive and negative feedback loops. Science. 2008;321:126–9. doi: 10.1126/science.1156951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Justman QA, Serber Z, Ferrell JE, Jr., El-Samad H, Shokat KM. Tuning the activation threshold of a kinase network by nested feedback loops. Science. 2009;324:509–12. doi: 10.1126/science.1169498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Toone WM, Aerne BL, Morgan BA, Johnston LH. Getting started: regulating the initiation of DNA replication in yeast. Annu Rev Microbiol. 1997;51:125–49. doi: 10.1146/annurev.micro.51.1.125. [DOI] [PubMed] [Google Scholar]
- 17.Cross FR, Tinkelenberg AH. A potential positive feedback loop controlling CLN1 and CLN2 gene expression at the start of the yeast cell cycle. Cell. 1991;65:875–83. doi: 10.1016/0092-8674(91)90394-e. [DOI] [PubMed] [Google Scholar]
- 18.Dirick L, Nasmyth K. Positive feedback in the activation of G1 cyclins in yeast. Nature. 1991;351:754–7. doi: 10.1038/351754a0. [DOI] [PubMed] [Google Scholar]
- 19.Skotheim JM, Di Talia S, Siggia ED, Cross FR. Positive feedback of G1 cyclins ensures coherent cell cycle entry. Nature. 2008;454:291–6. doi: 10.1038/nature07118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bean JM, Siggia ED, Cross FR. Coherence and timing of cell cycle start examined at single-cell resolution. Mol Cell. 2006;21:3–14. doi: 10.1016/j.molcel.2005.10.035. [DOI] [PubMed] [Google Scholar]
- 21.Santos SD, Ferrell JE. Systems biology: On the cell cycle and its switches. Nature. 2008;454:288–9. doi: 10.1038/454288a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohtani K, DeGregori J, Nevins JR. Regulation of the cyclin E gene by transcription factor E2F1. Proc Natl Acad Sci U S A. 1995;92:12146–50. doi: 10.1073/pnas.92.26.12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weinberg RA. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–30. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 24.Wu L, et al. The E2F1-3 transcription factors are essential for cellular proliferation. Nature. 2001;414:457–62. doi: 10.1038/35106593. [DOI] [PubMed] [Google Scholar]
- 25.Vernell R, Helin K, Muller H. Identification of target genes of the p16INK4A-pRB-E2F pathway. J Biol Chem. 2003;278:46124–37. doi: 10.1074/jbc.M304930200. [DOI] [PubMed] [Google Scholar]
- 26.Yung Y, Walker JL, Roberts JM, Assoian RK. A Skp2 autoinduction loop and restriction point control. J Cell Biol. 2007;178:741–7. doi: 10.1083/jcb.200703034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumagai A, Dunphy WG. Regulation of the cdc25 protein during the cell cycle in Xenopus extracts. Cell. 1992;70:139–51. doi: 10.1016/0092-8674(92)90540-s. [DOI] [PubMed] [Google Scholar]
- 28.Hoffmann I, Clarke P, Marcote M, Karsenti E, Draetta G. Phosphorylation and activation of human cdc25-C by cdc2-cyclin B and its involvement in the self-amplification of MPF at mitosis. The EMBO Journal. 1993;12:53–63. doi: 10.1002/j.1460-2075.1993.tb05631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McGowan C, Russell P. Cell cycle regulation of human WEE1. The EMBO Journal. 1995;14:2166–75. doi: 10.1002/j.1460-2075.1995.tb07210.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mueller PR, Coleman TR, Dunphy WG. Cell cycle regulation of a Xenopus Wee1-like kinase. Molecular Biology of the Cell. 1995;6:119–34. doi: 10.1091/mbc.6.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mueller PR, Coleman TR, Kumagai A, Dunphy WG. Myt1: a membrane-associated inhibitory kinase that phosphorylates Cdc2 on both threonine-14 and tyrosine-15. Science. 1995;270:86–90. doi: 10.1126/science.270.5233.86. [DOI] [PubMed] [Google Scholar]
- 32.Tang Z, Coleman T, Dunphy W. Two distinct mechanisms for negative regulation of the Wee1 protein kinase. The EMBO Journal. 1993;12:3427–36. doi: 10.1002/j.1460-2075.1993.tb06017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Solomon MJ, Glotzer M, Lee TH, Philippe M, Kirschner MW. Cyclin activation of p34cdc2. Cell. 1990;63:1013–24. doi: 10.1016/0092-8674(90)90504-8. [DOI] [PubMed] [Google Scholar]
- 34.Novak B, Tyson J. Numerical analysis of a comprehensive model of M-phase control in Xenopus oocyte extracts and intact embryos. Journal of Cell Science. 1993;106:1153–68. doi: 10.1242/jcs.106.4.1153. [DOI] [PubMed] [Google Scholar]
- 35.Pomerening J, Sontag E, Ferrell J., Jr. Building a cell cycle oscillator: hysteresis and bistability in the activation of Cdc2. Nature Cell Biology. 2003;5:346–51. doi: 10.1038/ncb954. [DOI] [PubMed] [Google Scholar]
- 36.Sha W, Moore J, Chen K, Lassaletta A, Yi C, Tyson J, Sible J. Hysteresis drives cell-cycle transitions in Xenopus laevis egg extracts. Proceedings of the National Academy of Sciences USA. 2003;100:975–80. doi: 10.1073/pnas.0235349100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Archambault V, Glover DM. Polo-like kinases: conservation and divergence in their functions and regulation. Nat Rev Mol Cell Biol. 2009;10:265–75. doi: 10.1038/nrm2653. [DOI] [PubMed] [Google Scholar]
- 38.Bonnet J, Mayonove P, Morris MC. Differential phosphorylation of Cdc25C phosphatase in mitosis. Biochem Biophys Res Commun. 2008;370:483–8. doi: 10.1016/j.bbrc.2008.03.117. [DOI] [PubMed] [Google Scholar]
- 39.Lobjois V, Jullien D, Bouche JP, Ducommun B. The polo-like kinase 1 regulates CDC25B-dependent mitosis entry. Biochim Biophys Acta. 2009;1793:462–8. doi: 10.1016/j.bbamcr.2008.12.015. [DOI] [PubMed] [Google Scholar]
- 40.Qian YW, Erikson E, Taieb FE, Maller JL. The polo-like kinase Plx1 is required for activation of the phosphatase Cdc25C and cyclin B-Cdc2 in Xenopus oocytes. Mol Biol Cell. 2001;12:1791–9. doi: 10.1091/mbc.12.6.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Toyoshima-Morimoto F, Taniguchi E, Nishida E. Plk1 promotes nuclear translocation of human Cdc25C during prophase. EMBO Rep. 2002;3:341–8. doi: 10.1093/embo-reports/kvf069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Vugt MA, Medema RH. Checkpoint adaptation and recovery: back with Polo after the break. Cell Cycle. 2004;3:1383–6. doi: 10.4161/cc.3.11.1248. [DOI] [PubMed] [Google Scholar]
- 43.Watanabe N, Arai H, Nishihara Y, Taniguchi M, Watanabe N, Hunter T, Osada H. M-phase kinases induce phospho-dependent ubiquitination of somatic Wee1 by SCFbeta-TrCP. Proc Natl Acad Sci U S A. 2004;101:4419–24. doi: 10.1073/pnas.0307700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seki A, Coppinger JA, Jang CY, Yates JR, Fang G. Bora and the kinase Aurora A cooperatively activate the kinase Plk1 and control mitotic entry. Science. 2008;320:1655–8. doi: 10.1126/science.1157425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Elia AE, et al. The molecular basis for phosphodependent substrate targeting and regulation of Plks by the Polo-box domain. Cell. 2003;115:83–95. doi: 10.1016/s0092-8674(03)00725-6. [DOI] [PubMed] [Google Scholar]
- 46.Seki A, Coppinger JA, Du H, Jang CY, Yates JR, 3rd, Fang G. Plk1-and beta-TrCP-dependent degradation of Bora controls mitotic progression. J Cell Biol. 2008;181:65–78. doi: 10.1083/jcb.200712027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Laoukili J, Kooistra MR, Bras A, Kauw J, Kerkhoven RM, Morrison A, Clevers H, Medema RH. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat Cell Biol. 2005;7:126–36. doi: 10.1038/ncb1217. [DOI] [PubMed] [Google Scholar]
- 48.Wang IC, et al. Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) ubiquitin ligase. Mol Cell Biol. 2005;25:10875–94. doi: 10.1128/MCB.25.24.10875-10894.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fu Z, Malureanu L, Huang J, Wang W, Li H, van Deursen JM, Tindal DJ, Chen J. Plk1-dependent phosphorylation of FoxM1 regulates a transcriptional programme required for mitotic progression. Nat Cell Biol. 2008;10:1076–82. doi: 10.1038/ncb1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ban KH, Torres JZ, Miller JJ, Mikhailov A, Nachury MV, Tung JJ, Rieder CL, Jackson PK. The END network couples spindle pole assembly to inhibition of the anaphase-promoting complex/cyclosome in early mitosis. Dev Cell. 2007;13:29–42. doi: 10.1016/j.devcel.2007.04.017. [DOI] [PubMed] [Google Scholar]
- 51.Meraldi P, Draviam VM, Sorger PK. Timing and checkpoints in the regulation of mitotic progression. Dev Cell. 2004;7:45–60. doi: 10.1016/j.devcel.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 52.Guardavaccaro D, et al. Control of meiotic and mitotic progression by the F box protein beta-Trcp1 in vivo. Dev Cell. 2003;4:799–812. doi: 10.1016/s1534-5807(03)00154-0. [DOI] [PubMed] [Google Scholar]
- 53.Margottin-Goguet F, Hsu JY, Loktev A, Hsieh HM, Reimann JD, Jackson PK. Prophase destruction of Emi1 by the SCF(betaTrCP/Slimb) ubiquitin ligase activates the anaphase promoting complex to allow progression beyond prometaphase. Dev Cell. 2003;4:813–26. doi: 10.1016/s1534-5807(03)00153-9. [DOI] [PubMed] [Google Scholar]
- 54.Hansen DV, Loktev AV, Ban KH, Jackson PK. Plk1 regulates activation of the anaphase promoting complex by phosphorylating and triggering SCFbetaTrCP-dependent destruction of the APC Inhibitor Emi1. Mol Biol Cell. 2004;15:5623–34. doi: 10.1091/mbc.E04-07-0598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kraft C, Herzog F, Gieffers C, Mechtler K, Hagting A, Pines J, Peters JM. Mitotic regulation of the human anaphase-promoting complex by phosphorylation. Embo J. 2003;22:6598–609. doi: 10.1093/emboj/cdg627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cohen-Fix O. The making and breaking of sister chromatid cohesion. Cell. 2001;106:137–40. doi: 10.1016/s0092-8674(01)00439-1. [DOI] [PubMed] [Google Scholar]
- 57.Cohen-Fix O, Peters JM, Kirschner MW, Koshland D. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 1996;10:3081–93. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- 58.Zachariae W, Nasmyth K. Whose end is destruction: cell division and the anaphase-promoting complex. Genes Dev. 1999;13:2039–58. doi: 10.1101/gad.13.16.2039. [DOI] [PubMed] [Google Scholar]
- 59.Holt LJ, Krutchinsky AN, Morgan DO. Positive feedback sharpens the anaphase switch. Nature. 2008;454:353–7. doi: 10.1038/nature07050. [DOI] [PMC free article] [PubMed] [Google Scholar]