The SARS coronavirus protein nsp1 can suppress host gene expression at a post-transcriptional level, with previous work showing a reduction in mRNA abundance. Now a direct effect on protein synthesis is revealed, as nsp1 modifies transcripts and also inactivates the 40S ribosomal subunit.
Supplementary information
The online version of this article (doi:10.1038/nsmb.1680) contains supplementary material, which is available to authorized users.
Abstract
Severe acute respiratory syndrome coronavirus nsp1 protein suppresses host gene expression, including type I interferon production, by promoting host mRNA degradation and inhibiting host translation, in infected cells. We present evidence that nsp1 uses a novel, two-pronged strategy to inhibit host translation and gene expression. Nsp1 bound to the 40S ribosomal subunit and inactivated the translational activity of the 40S subunits. Furthermore, the nsp1–40S ribosome complex induced the modification of the 5′ region of capped mRNA template and rendered the template RNA translationally incompetent. Nsp1 also induced RNA cleavage in templates carrying the internal ribosome entry site (IRES) from encephalomyocarditis virus, but not in those carrying IRES elements from hepatitis C or cricket paralysis viruses, demonstrating that the nsp1-induced RNA modification was template-dependent. We speculate that the mRNAs that underwent the nsp1-mediated modification are marked for rapid turnover by the host RNA degradation machinery.
Supplementary information
The online version of this article (doi:10.1038/nsmb.1680) contains supplementary material, which is available to authorized users.
Main
Severe acute respiratory syndrome (SARS) coronavirus (SCoV), the causative agent of a newly emerged disease, SARS, is an enveloped virus that contains a single-stranded, positive-sense RNA genome of about 29.7 kb1. Upon infection, genome expression begins with the translation of gene 1, which constitutes the 5′-end two-thirds of the viral genome, to produce two large precursor polyproteins2,3 that are proteolytically processed by two virally encoded proteinases to generate 16 mature proteins, namely nsp1 to nsp16 (ref. 2). Although most of these gene-1 proteins are essential for viral RNA synthesis, some of them seem to have other biological functions4,5,6,7.
The most N-terminal gene-1 protein, nsp1, has interesting biological functions; expressed SCoV nsp1 induces host mRNA degradation and suppresses host translation8. The expressed nsp1 suppresses the host antiviral signaling pathways as well9. Furthermore, nsp1 suppresses host gene expression, including type I interferon (IFN) production, by promoting host mRNA degradation and host translation suppression in infected cells10. The nsp1 of a closely related mouse hepatitis virus also suppresses host gene expression and is a viral virulence factor11. These data suggest that the SCoV nsp1-mediated suppression of host genes is important in the pathogenesis of SARS. Accordingly, a delineation of the mechanisms of the nsp1-induced suppression of host gene expression is important for providing insight into SCoV pathogenesis at the molecular level. We designed the present study to uncover the mechanism of nsp1-induced suppression of host gene expression primarily by using an in vitro system. Our data revealed that nsp1 uses a novel, two-pronged strategy to inhibit host translation and gene expression.
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
Results
Nsp1 suppresses translation in vitro
We tested whether nsp1 protein could suppress translation in rabbit reticulocyte lysate (RRL). We expressed the full-length, wild-type nsp1 protein and its mutant form (nsp1-mt) carrying K164A and H165A mutations as glutathione S-transferase (GST)-tagged fusion proteins in Escherichia coli followed by the removal of the GST tag to generate recombinant nsp1 and nsp1-mt proteins, respectively; nsp1-mt neither suppresses host translation nor promotes host mRNA degradation in expressing cells and infected cells10. After incubation of the nsp1 protein with RRL at 4 °C for 30 min, we added different concentrations of capped and polyadenylated Renilla luciferase mRNA transcripts (rLuc RNA) to the mixture and incubated the samples in the presence of [35S]methionine for 30 min. In control samples in which rLuc RNA was incubated with GST and nsp1-mt proteins, rLuc activity and labeled rLuc protein abundance increased with rising mRNA concentrations. In contrast, the rLuc activity and labeled rLuc protein abundance in the nsp1-containing sample were substantially lower than in the control samples; the rLuc activity and the radiolabeled rLuc protein abundance in the presence of nsp1 were about 6%–8% of the levels observed with GST or nsp1-mt (Fig. 1a,b), which clearly demonstrates that nsp1 efficiently inhibited the rLuc protein synthesis from capped rLuc RNA in RRL.
Figure 1. Effects of nsp1 on cap-dependent translation and IRES-mediated translation in RRL.
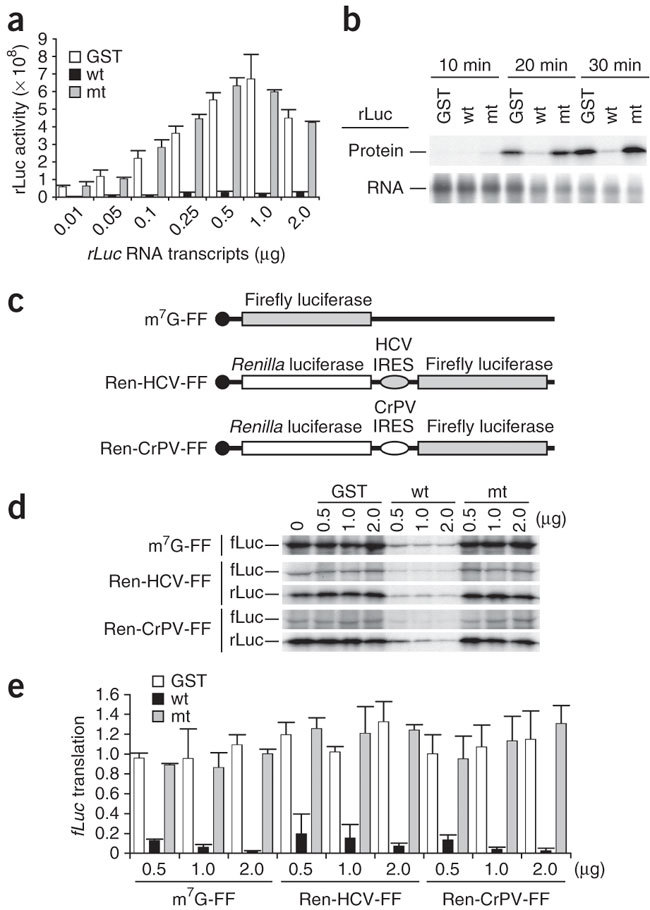
(a) Increasing amounts of rLuc RNA were incubated in RRL in the presence of 1 μg of nsp1 (wt), GST or nsp1-mt (mt) at 4 °C for 30 min; the molar ratios of rLuc RNA to nsp1 protein ranged from 1:10 to 1:2,000. Then the samples were incubated for 30 min at 30 °C, and the rLuc enzymatic activities were measured. y axis, light units. (b) A mixture of 0.25 μg of rLuc RNA and 1 μg of nsp1, GST or nsp1-mt was incubated in RRL in the presence of [35S]methionine at 30 °C for 10, 20 and 30 min. Translated products were analyzed on SDS-PAGE and detected by autoradiography. (c) Schematic diagram of various RNA transcripts used for d,e. (d) Increasing amounts of GST, nsp1, or nsp1-mt were incubated with RRL for 10 min at 4 °C. Then, the samples were incubated at 30 °C for 30 min with 0.5 μg of m7G-FF RNA, Ren-HCV-FF RNA, or Ren-CrPV-FF RNA in the presence of [35S]methionine. Translated products were analyzed on SDS-PAGE and detected by autoradiography. (e) For a given RNA group, the amount of fLuc protein, determined by densitometry, in each experimental group was normalized to the amount of the fLuc protein in a control group, to which the purified protein was not added. For a,d, averages of three independent experiments; error bars, s.d.
Next, we examined the effect of nsp1 on translation mediated by the internal ribosome entry sites (IRES) using in vitro-synthesized, bicistronic mRNAs, Ren-HCV-FF RNA and Ren-CrPV-FF RNA, in which expression of the upstream rLuc open reading frame (ORF) was mediated by cap-dependent translation and the translation of downstream firefly luciferase (fLuc) ORF was driven by a hepatitis C virus (HCV) IRES and a cricket paralysis virus (CrPV) IRES, respectively (Fig. 1c). Nsp1 efficiently suppressed the translation of the 7-methylguanosine-capped fLuc RNA, m7G-FF, as well as translation driven by both IRES elements (Fig. 1d,e). According to current models of translation initiation, the CrPV-IRES recruits the 40S ribosomal subunit in the absence of any initiation factors12,13, which suggested to us that nsp1 could suppress the function of the 40S subunit or block translation subsequent to the initiation step.
Nsp1 protein interacts with the 40S ribosomal subunit
We tested whether nsp1 associates with the 40S ribosomal subunit and suppresses its function. We transfected human embryonic kidney (HEK) 293 cells with mRNAs encoding nsp1, nsp1-mt or chloramphenicol acetyltransferase (CAT) proteins; all of these encoded proteins carried a C-terminal myc-His tag. At 8 h after transfection, we prepared the cell extracts and performed polysome profile analysis (Fig. 2a–c). Expression of CAT and nsp1-mt yielded similar polysome profiles, whereas nsp1 expression resulted in the accumulation of 80S monosomes, along with a substantial reduction in the polysome abundance, in accordance with the observation that the expressed nsp1, but not nsp1-mt, suppresses host translation10. Western blot analysis of the resolved gradient fractions showed that the majority of nsp1, but not CAT and nsp1-mt, sedimented with the 40S ribosomal subunit (Fig. 2b). Likewise, only nsp1 sedimented with the 40S ribosomal sub-unit in RRL (Fig. 2d–f). To confirm the interaction of nsp1 with the 40S ribosomal subunit, we transfected 293 cells with in vitro-synthesized mRNAs encoding nsp1-myc-His fusion protein, CAT-myc-His protein or nsp1-mt-myc-His protein and immunoprecipitated the cell extracts with antibody to myc (anti-myc). The immune complexes were washed with 0.5 M KCl, a stringent, high-salt condition in which the 60S ribo-somal subunits and the translation initiation factors dissociate from the 40S subunits14. Consistent with our previous report10, the expression of nsp1 was substantially lower than that of nsp1-mt or CAT. Nevertheless, an efficient immunoprecipitation of 18S ribosomal RNA and S6 ribosomal protein, core components of the 40S subunit, with nsp1, but not with CAT and nsp1-mt (Fig. 2g–i), suggested the tight association of nsp1 with the 40S ribosomal subunit. Similarly, coimmunoprecipitation analysis showed that nsp1 also interacted with the 40S ribosomal subunit in RRL (Supplementary Fig. 1).
Figure 2. Binding of nsp1 to the 40S ribosomal subunits.
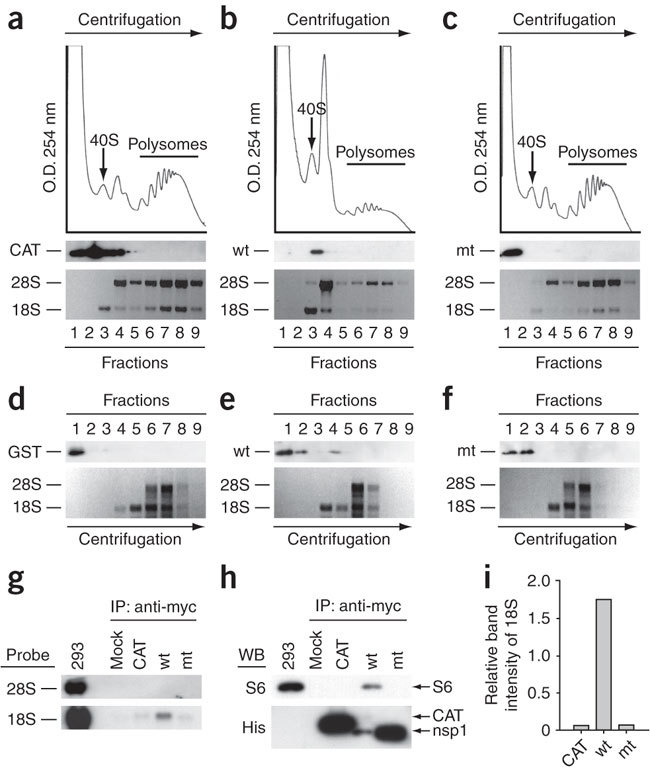
(a–c) CAT (a), nsp1 (b) or nsp1-mt (c) RNAs were transfected into 293 cells. Cell extracts were prepared 8 h after transfection and subjected to polysome profile analysis (top panels). CAT, nsp1 (wt) and nsp1-mt (mt) in each fraction were detected by western blot analysis using antibody to myc (middle panels). Bottom panels show rRNAs in each fraction. (d–f) After incubation of the mixture of 0.25 μg of rLuc RNA and 1 μg of nsp1, GST or nsp1-mt in RRL for 30 min at 30 °C, the samples were separated on 10%–50% sucrose gradient. GST, nsp1 (wt) and nsp1-mt (mt) in each fraction were detected by western blot analysis using anti-GST antibody and anti-nsp1 (ref. 8) antibody. The locations of GST (d), nsp1 (e) or nsp1-mt (f) in fractions are shown (all top panels). The bottom panels show rRNAs. (g–i) 293 cells were transfected with CAT RNA (CAT), nsp1 RNA (wt), or nsp1-mt RNA (mt). Cell extracts were prepared 8 h after transfection and subjected to immunoprecipitation using anti-myc antibody. Extracted RNAs from the immunoprecipitates were separated by agarose gel electrophoresis, and rRNAs were detected by northern blot analysis (g). The immunoprecipitated proteins were examined by western blot analysis using anti-S6 antibody (40S subunit specific) and anti-His antibody for CAT, nsp1 and nsp1-mt (h). Panel I represents the values, each of which was obtained by dividing the abundance of the immunoprecipitated 18S rRNA with the immunoprecipitated CAT protein, nsp1 protein or nsp1-mt protein in an arbitrary scale.
Nsp1 inhibits 80S formation
The finding that nsp1 inhibited CrPV-IRES-driven translation suggested that nsp1 could affect an event or events downstream of the 43S complex formation that involve the binding of eIF2–GTP–Met-tRNA ternary complex and other initiation factors, such as eIF3, to the 40S subunit. To test the effect of nsp1 on 48S complex formation, in which the 43S complex binds to the mRNA, and 80S monosome assembly, we examined, by sucrose gradient fractionation, the accumulation of these complexes on a radiolabeled mRNA template in the presence of nsp1. To analyze the effect of nsp1 on 80S formation, we incubated 32P-labeled rLuc RNA template with nsp1, GST or nsp1-mt in RRL in the presence of cycloheximide (CHX). CHX treatment inhibits the elongation step, but does not affect 80S monosome assembly15. We used hippuristanol, which blocks the function of initiation factor eIF4A and inhibits 48S complex formation, as a control16. CHX treatment induced 80S complex accumulation, and hippuristanol inhibited it (Fig. 3a). Nsp1, but not GST and nsp1-mt, suppressed 80S complex formation (Fig. 3b). We tested the effect of nsp1 on 48S complex formation by incubating the samples with GMP-PNP, a non-hydrolyzable analog of GTP, and subsequently subjecting them to sucrose gradient centrifugation; GMP-PNP does not affect 48S complex formation, whereas it blocks GTP hydrolysis by eIF2 in eIF2-GTP-Met-tRNA complexes and inhibits the release of eIF2 and subsequent joining of the 60S ribosomal subunit17. The 48S complex accumulated in the presence of GMP-PNP, but not in the presence of hippuristanol (Fig. 3c). Nsp1, nsp1-mt and GST did not suppress the 48S complex formation (Fig. 3d). These data show that nsp1 did not inhibit 48S complex formation, but it suppressed 60S subunit joining.
Figure 3. Ribosome binding assays of radiolabeled rLuc RNA.
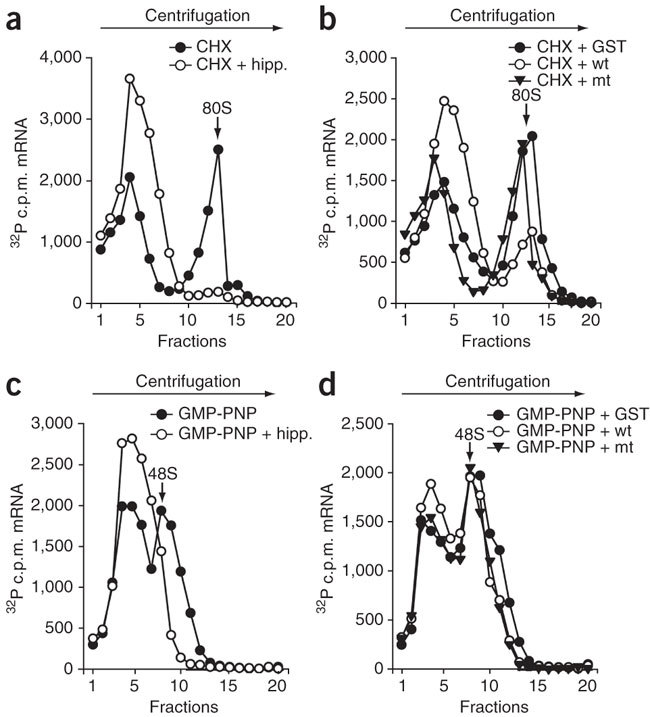
Representative data from three independent experiments. (a) rLuc RNA was incubated in RRL in the presence of CHX only or a mixture of CHX and hippuristanol (hipp). The percentages of the rLuc RNA bound in 80S complexes were, for CHX, 17.4%; CHX + hipp., 1.0%. (b) rLuc RNA was incubated in RRL with GST, nsp1 (wt) or nsp1-mt (mt) in the presence of CHX. The percentages of the rLuc RNA bound in 80S complexes were, for GST, 13.3%; nsp1, 5.4%; nsp1-mt, 14.8%. (c) rLuc RNA was incubated in RRL in the presence of GMP-PNP or a mixture of GMP-PNP and hipp. The percentages of the rLuc RNA bound in 48S complexes were, for GMP-PNP, 11.7%; GMP-PNP + hipp., 2.9%. (d) rLuc RNA was incubated in RRL with GST, nsp1 (wt) or nsp1-mt (mt), in the presence of GMP-PNP. The percentages of the rLuc RNA bound in 48S complexes were, for GST, 13.1%; nsp1, 12.1%; nsp1-mt, 12.0%.
Characterization of the 48S complex by toeprinting analysis
Next, we performed a toeprinting analysis, which defines the positions of stalled ribosomes on the mRNA chain18. After initial incubation of RRL with nsp1, GST or nsp1-mt in the presence of CHX or a mixture of CHX and GMP-PNP, we added rLuc RNA that had been preannealed with a 5′-end-labeled primer to the samples. Subsequently, we subjected the samples to primer extension without extracting the RNAs. In control groups, using GST and nsp1-mt, we detected a correctly positioned toeprint (TP-AUG), a premature primer extension termination signal that was induced by the presence of the stalled 40S subunit. The toeprint was ∼15–17 nt downstream from the translation initiation codon in the presence of GMP-PNP and CHX, which allow 48S complex formation but not 60S joining (Fig. 4a). In the presence of CHX, the control groups showed an extra band approximately 3 nt downstream from TP-AUG, which was most probably generated by the joining of the 60S subunits to the 40S complex positioned at the AUG codon (Fig. 4a). Consistent with the observation that nsp1 suppressed the 80S complex formation (Fig. 3b), nsp1 caused a clear reduction in the amount of the toeprint for the 80S complex (Fig. 4a). Compared to the observations in the control groups, the intensity of the TP-AUG band was also lower in the sample containing nsp1 (Fig. 4a), which suggests that nsp1 impaired start-codon recognition. Unexpectedly, we detected several more bands, both upstream and downstream of the TP-AUG, in the sample incubated with nsp1, but not in controls; a longer exposure of the CHX-treated sample clearly showed the same signals (data not shown). These data suggest that nsp1 induced the modification of rLuc RNA at several discrete sites, which blocked cDNA elongation on these modified RNAs.
Figure 4. Toeprinting and primer extension analyses.
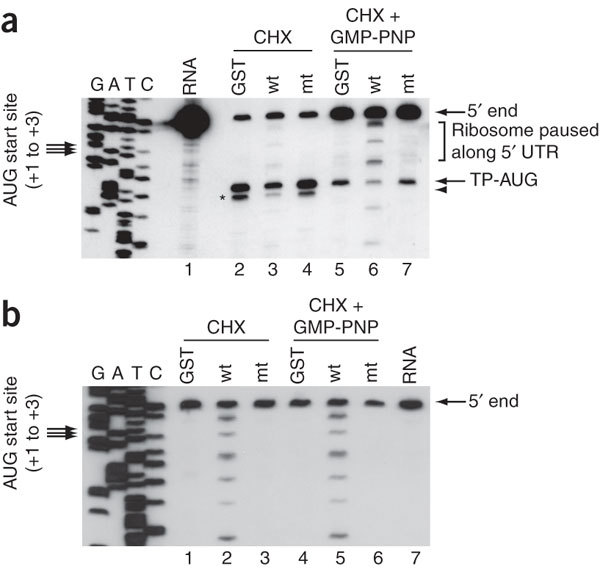
(a) Toeprinting. Samples shown in lanes 2–4 and 5–7 were incubated with CHX and a mixture of CHX and GMP-PNP, respectively, and toeprinting analysis was performed in the presence of GST (lanes 2 and 5), nsp1 (lanes 3 and 6) or nsp1-mt (lanes 4 and 7). Lane 1, the primer extension products using the rLuc RNA that was hybridized with a 5′-end labeled primer; 5′ end, the primer extension product that was extended to the 5′ end of rLuc RNA; UTR, untranslated region; TP-AUG, a correctly positioned toeprint. Arrowhead and asterisk represent a signal generated by the joining of the 60S subunits to the 40S complex positioned at the AUG codon. A dideoxynucleotide sequence of the rLuc gene generated with the same primer was run in parallel (left four rows).Translation initiation codon is shown at the left side of the gel. (b) Primer extension analysis. RRL was incubated with GST (lanes 1 and 4), nsp1 (lanes 2 and 5), or nsp1-mt (lanes 3 and 6) in the presence of CHX (lanes 1–3) or a mixture of CHX and GMP-PNP (lanes 4–6) for 5 min at 30 °C. Then rLuc RNA was added to each sample, and the samples were incubated for another 10 min at 30 °C. The RNAs were extracted and subjected to primer extension analysis. Lane 7, the primer extension products using the rLuc RNA that was hybridized with a 5′-end labeled primer.
Nsp1 renders the capped mRNA translationally inactive
We performed primer extension analyses to determine whether nsp1 indeed induced template RNA modification. After incubation of rLuc RNA with GST, nsp1 or nsp1-mt in RRL, as described above, we extracted the RNA and performed primer extension analysis using the same 5′-end labeled primer. In addition to the full-length primer extension product, only samples incubated with nsp1 showed several premature primer-extension termination products, all of which corresponded to those detected in the toeprinting analysis performed in the presence of nsp1 (Fig. 4b). Northern blot analysis of rLuc RNA, extracted after incubation with nsp1 in RRL under the same conditions, did not show significantly less of the full-length rLuc transcript than the control groups (Supplementary Fig. 2). These data demonstrate that nsp1 induced the modification of capped rLuc RNA at several sites in the 5′ region without inducing extensive template RNA degradation.
To further characterize the nsp1-induced template RNA modification, we incubated cap-labeled rLuc RNA and 3′-end-labeled rLuc RNA, independently, with nsp1 in RRL in the presence of CHX and GMP-PNP; GST and nsp1-mt served as controls. After incubation, we analyzed the extracted RNAs on a 5% sequencing gel. The amount of intact cap-labeled rLuc RNA remaining after incubation with nsp1 was clearly lower than that in the control incubations (Fig. 5a,b), demonstrating that the 5′-end region of the capped rLuc RNA was removed in the presence of nsp1. Incubation of the 3′-end-labeled rLuc RNA with nsp1 resulted in the appearance of an extra band that migrated slightly faster than the intact RNA (Fig. 5a). The total radioactivity of the intact 3′-end labeled rLuc RNA and the fast-migrating RNA band in nsp1-treated samples was similar to that of the intact 3′-end labeled rLuc RNA in the controls (Fig. 5b). To determine the approximate size of the fast-migrating band, we incubated 3′-end-labeled, non-polyadenylated rLuc RNA with nsp1 in RRL; we used non-polyadenylated rLuc RNA because some of the polyadenylated rLuc preparations had mRNA species with shorter poly(A) tails, which interfered with the positive identification of the truncated RNA products in the gel. We observed two main fast-migrating bands, and their sizes were approximately 20 to 40 nt shorter than the full-length RNA template (Supplementary Fig. 3). These data demonstrate that nsp1 induced the removal of the 5′ end, but not the 3′ end, from the rLuc RNA.
Figure 5. Nsp1-induced modification of rLuc RNA in RRL.
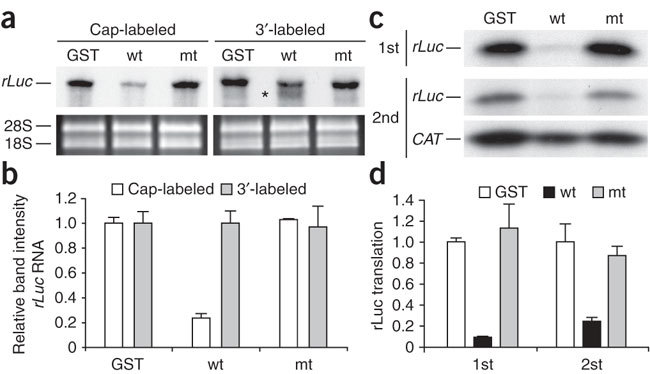
(a) Cap-labeled rLuc RNA and the 3′-end-labeled rLuc RNA were incubated with GST, nsp1 (wt) or nsp1-mt (mt) in RRL for 15 min at 30 °C in the presence of CHX and GMP-PNP. RNAs were extracted, resolved on a sequencing gel and visualized by autoradiography. Asterisk, a signal that migrated slightly faster than the full-length rLuc in the sample incubated with nsp1. The 28S and 18S rRNAs, detected by ethidium bromide staining, are shown as loading controls. (b) Densitometric determination of rLuc RNAband intensities in a, normalized to the value of the GST-incubated sample. For the 3′-labeled rLuc RNA in the nsp1 sample, graph shows a sum of the signal intensities of the full-length rLuc RNA band and the band marked by the asterisk in a. (c) RRL was incubated with GST, nsp1 (wt), or nsp1-mt (mt) for 10 min at 4 °C. Then rLuc RNA was added and samples incubated at 30 °C for 20 min in the presence of [35S]methionine (1st translation). Aliquots were analyzed by SDS-PAGE. After addition of the same amount of CAT RNA to each sample, RNAs were extracted and translated using fresh RRL in the presence of [35S]methionine (2nd translation), and the samples were analyzed on a 12.5% SDS-PAGE gel. (d) Quantitation of rLuc protein abundances in the 1st and 2nd translation reactions in experiment in c. For b,d, averages of three independent experiments; error bars, s.d.
Next, we tested the translational competence of the rLuc RNA that had undergone the nsp1-induced modification. After incubation of the rLuc RNA in RRL in the presence of nsp1, GST or nsp1-mt, we added the same amount of CAT RNA to each sample as an internal control RNA, extracted the RNAs and performed a second in vitro translation reaction without adding nsp1, GST or nsp1-mt. We performed this analysis to examine the translational competence of RNAs that were extracted from the first in vitro translation reaction. As expected, nsp1 suppressed the translation of rLuc RNA in the initial translation (Fig. 5c,d). The rLuc RNA that had undergone the nsp1-mediated modification was not efficiently translated into rLuc protein (Fig. 5c,d), which demonstrated that the modified rLuc RNA was translationally inactive.
Nsp1 induces RNA cleavage in an IRES-driven mRNA template
In addition to cap-dependent translation, nsp1 also suppressed translation mediated by IRES elements derived from HCV and CrPV (Fig. 1). To address the question whether the nsp1-induced template RNA modification is specific for cap-dependent RNA template, we tested whether nsp1 induced the modification of IRES in RNA templates carrying HCV, CrPV and encephalomyocarditis virus (EMCV)-derived IRES elements. We incubated the RNA transcripts Ren-HCV-FF, Ren-CrPV-FF and Ren-EMCV-FF—a bicistronic RNA carrying EMCV IRES between the upstream rLuc ORF and the downstream fLuc ORF (Fig. 6a)—in RRL in the presence of nsp1, GST or nsp1-mt. After incubation, we extracted the RNAs and analyzed them by northern blot (Fig. 6). Nsp1 did not induce any cleavage of Ren-HCV-FF and Ren-CrPV-FF (Fig. 6b and Fig. 6c, respectively). In contrast, nsp1 did induce cleavage of Ren-EMCV-FF (Fig. 6d). A cleaved RNA fragment, detected by a probe that binds to the rLuc ORF, migrated similarly to control RNA1, carrying the rLuc ORF and EMCV IRES (Fig. 6d), and the size of the other RNA fragment, detected by a probe that binds to the fLuc ORF, was similar to that of the fLuc ORF in Ren-EMCV-FF. These data suggest that nsp1 induced the cleavage at or near the 3′ region of the EMCV IRES in Ren-EMCV-FF (Fig. 6d). These results demonstrate that nsp1 induced a template-dependent RNA cleavage in IRES-driven RNAs and that the nsp1-induced RNA modification was not specific for capped RNA template.
Figure 6. Nsp1 induces cleavage at the IRES in Ren-EMCV-FF RNA.
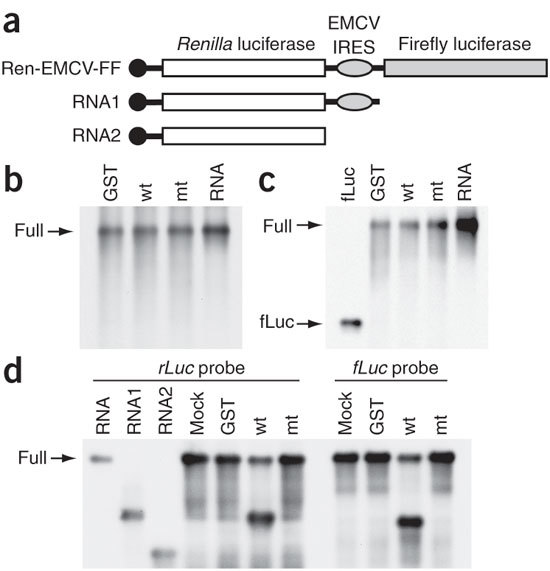
(a) Schematic diagram of Ren-EMCV-FF RNA and control RNAs RNA1 and RNA2. (b,c) After incubation of RRL with GST, nsp1 (wt), or nsp1-mt (mt) for 10 min at 4 °C, Ren-HCV-FF RNA (b) or Ren-CrPV-FF RNA (c) was added to each sample and incubated at 30 °C for 25 min. Then RNAs were extracted and analyzed by northern blot using a digoxigenin-labeled probe that binds to the fLuc ORF. RNA lane in b, the untreated Ren-HCV-FF RNA; fLuc lane in c, the RNA encoding fLuc ORF; RNA, the untreated Ren-CrPV-FF RNA. (d) RRL was incubated with GST, nsp1 (wt), or nsp1-mt (mt) for 10 min at 4 °C. Ren-EMCV-FF RNA was added to each sample and incubated at 30 °C for 10 min. RNAs were extracted and analyzed by northern blot using a probe that binds to the rLuc ORF (rLuc probe) or fLuc ORF (fLuc probe). Samples in the first three lanes, including untreated Ren-EMCV-FF (RNA), RNA1 and RNA2, served as size markers.
The nsp1-40S subunit complex exerts template RNA modification
To assess the biological significance of nsp1 binding to 40S sub-units, we examined whether nsp1 could modify the rLuc RNA without ribosomes. After pelleting the ribosomes in RRL by centrifugation, we incubated the ribosome-free supernatant with the cap-labeled rLuc RNA and nsp1 in the presence of CHX and GMP-PNP; GST and nsp1-mt served as controls. Gel electrophoresis analysis of the extracted RNAs showed that the amounts of the cap-labeled rLuc RNA were similar among the three samples (Fig. 7a,b). To determine whether the rLuc RNA that had been incubated with nsp1 in the absence of the ribosomes was translationally competent, we incubated the capped rLuc RNA with nsp1, GST or nsp1-mt in the ribosome-free RRL. As expected, rLuc protein synthesis did not occur in the ribosome-free lysate (Fig. 7c, top panel). After incubation, we added the same amount of CAT RNA to the samples, extracted the RNAs and subjected them to in vitro translation without adding nsp1, GST or nsp1-mt. The rLuc RNA that had been incubated with nsp1 in the ribosome-free RRL was translationally competent in the second in vitro translation assay (Fig. 7c, middle panel). These data suggest that the 40S subunit–bound nsp1 modified and inactivated the rLuc RNA.
Figure 7. Importance of ribosomes for the function of nsp1.
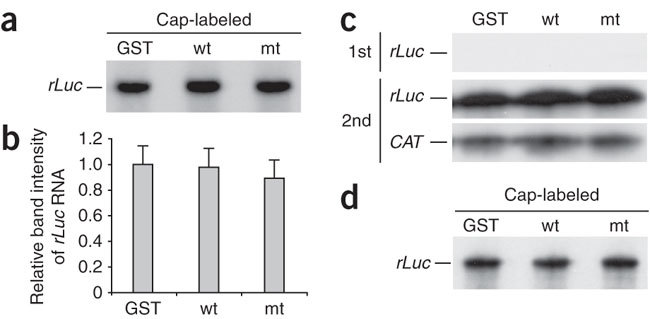
(a) Cap-labeled rLuc RNA was incubated with GST, nsp1 (wt), or nsp1-mt (mt) in ribosome-free RRL for 15 min at 30 °C in the presence of CHX and GMP-PNP. RNAs were extracted, resolved on a sequencing gel and visualized by autoradiography. (b) The band intensity of rLuc RNA shown in a was determined by densitometric analysis. Mean of three independent experiments. (c) Experiments performed as described in Figure 5c, except that ribosome-free RRL was used for the initial translation reaction. (d) Cap-labeled rLuc RNA was incubated with GST, nsp1 (wt), or nsp1-mt (mt) in RRL in the presence of hippuristanol. RNAs were extracted, resolved on a sequencing gel and visualized by autoradiography.
We further tested whether the association of rLuc RNA with the 43S complex was required for nsp1-mediated template RNA modification by incubating the cap-labeled rLuc RNA with nsp1, GST or nsp1-mt in RRL in the presence of hippuristanol. Nsp1 did not affect the amount of the labeled RNA in the presence of hippuristanol (Fig. 7d), leading us to suggest that the modification of the capped RNA requires the association of template RNA with the nsp1–40S complex.
The nsp1-40S subunit complex is translationally inactive
Nsp1 suppressed translation mediated by the CrPV IRES (Fig. 1) but did not induce cleavage of the Ren-CrPV-FF RNA (Fig. 6b). These data suggest that, in addition to modifying template RNA, the association of nsp1 with 40S ribosomal subunit also inactivated the translational function of 40S ribosomes. To further examine this possibility, we subjected Ren-CrPV-FF RNA to in vitro translation in RRL in the presence of nsp1, GST or nsp1-mt. After incubation, we analyzed aliquots of the samples by SDS-PAGE. As expected, nsp1, but not GST or nsp1-mt, suppressed both cap-dependent and IRES-dependent translation (Fig. 8a). We added the same amount of the CAT RNA to the remaining samples, extracted RNAs and performed a second in vitro translation analysis without adding nsp1, GST or nsp1-mt (Fig. 8a,b). The cap-dependent rLuc gene translation, from Ren-CrPV-FF RNA that was exposed to nsp1 in the first in vitro translation reaction, was inhibited, but the IRES-mediated fLuc gene translation was not inhibited. Thus, nsp1 suppressed CrPV IRES-mediated translation in the first in vitro translation reaction but did not induce the modification that inactivated the translational activity of CrPV IRES in the second in vitro translation assay. As with cap-labeled rLuc RNA (Fig. 5), incubation of cap-labeled Ren-CrPV-FF RNA with nsp1 resulted in the removal of the 5′ region from this RNA (Fig. 8c). We interpreted these results to mean that binding of nsp1 to 40S ribosomal subunits inactivated the translational functions of 40S ribosomes, leading to the inhibition of CrPV IRES–driven translation of Ren-CrPV-FF. Collectively, these data show that nsp1 suppressed translation by inactivating the translational functions of 40S ribosomal subunits and modifying the template RNA.
Figure 8. Nsp1 inactivates the translational function of 40S ribosomal subunit.
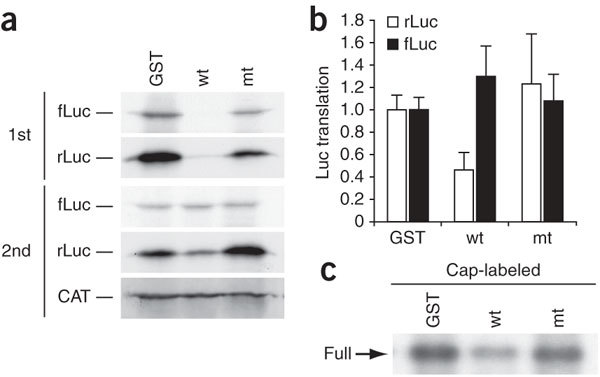
RRL was incubated with GST, nsp1 (wt), or nsp1-mt (mt) for 10 min at 4 °C. Ren-CrPV-FF RNA was added to each sample and incubated at 30 °C for 25 min in the presence of [35S]methionine. Aliquots of the samples were used for SDS-PAGE (a, 1st). RNAs were extracted from the remaining samples after addition of the same amount of CAT RNA to each sample. Experiments were performed as described in Figure 5c, except that Ren-CrPV-FF was used as the template RNA. rLuc and fLuc are expressed by cap-dependent and CrPV IRES-mediated mechanisms, respectively (a, 2nd). (b) Experiments described in a were repeated three times, and the amounts of fLuc and rLuc proteins in the 2nd translation reaction are shown. Error bars, s.d. (c) Cap-labeled Ren-CrPV-FF RNA was incubated with GST, nsp1, or nsp1-mt in RRL for 15 min at 30 °C in the presence of CHX and GMP-PNP. RNAs were extracted, resolved on a sequencing gel and visualized by autoradiography.
Discussion
Host translational suppression, often detected in virus-infected cells, is an effective viral defensive strategy for suppressing host innate immune responses and freeing host translational machinery for viral-specific gene expression. Although many viral proteins that suppress host translation have been identified19,20,21,22, SCoV nsp1 (ref. 8) and several herpesvirus proteins23,24,25,26, including herpes simplex virus-1 vhs protein27, are known to promote destabilization of host mRNAs and suppress host translation. Vhs protein, which has a sequence similarity to known RNases, is indeed an RNase28, induces RNA cleavage in RRL29 and degrades both host and viral mRNAs30. Unlike vhs, SCoV nsp1 has similarity in sequence and structure with no known proteins. The biological activities of SCoV nsp1 also substantially differed from those of vhs, as nsp1 inactivated the translational functions of the 40S ribosomal subunit and modified RNA transcripts only in the context of its association with the 40S subunit. The strategy that SCoV nsp1 has evolved to bind to 40S subunits to exert its function seems to be an excellent one for blocking translation of both capped mRNAs and IRES-containing mRNAs, and for efficiently accessing host mRNAs, including those involved in host innate immune functions, and inactivating them.
By binding to the 40S ribosomal subunits and inactivating them, SCoV nsp1 efficiently suppressed cap-dependent and IRES-mediated translation, including translation mediated by CrPV IRES. In contrast to SCoV nsp1, CrPV IRES-mediated translation is relatively insensitive to Drosophila Reaper, a potent apoptosis inducer, which also inhibits translation by binding directly to the 40S subunit31.
Further studies are needed to determine how the binding of nsp1 to 40S ribosomal subunits inactivates their translational function. Using rLuc RNA as a template, we showed that the formation of the 80S complex, but not the 48S complex, was inhibited in the presence of nsp1. Because the nsp1–40S complex also modified the 5′ region of the rLuc RNA, it is possible that the nsp1-mediated RNA modification affected the formation of 48S and/or 80S complexes on these modified RNA templates (Fig. 3). We obtained somewhat conflicting results, which showed that nsp1 did not suppress the 48S assembly on rLuc RNA (Fig. 3d) but that the 48S TP-AUG signal was reduced in the presence of nsp1 (Fig. 4a). These data may imply that some of the multiple toeprints upstream of the TP-AUG, observed in nsp1-treated sample, represent arrested 48S translation intermediates as a result of nsp1 inactivating the function of 40S ribosomal subunit, leading to inefficient start site recognition; these arrested 48S translation intermediates could contribute to the signal observed for 48S complex formation (Fig. 3d). Furthermore, the nsp1-induced RNA modification alone cannot fully account for the substantially reduced luciferase protein synthesis from capped rLuc RNA (Fig. 1), highlighting the contribution of the translation inhibition function of nsp1 to the suppression of reporter protein expression in RRL. To clarify the exact step in the translation of capped mRNAs that nsp1 inhibits, it would be useful to isolate an nsp1 mutant that retains the 40S subunit inactivating function but lacks the RNA modification function and test the effect of this mutant on formation of 48S and 80S complexes.
In addition to the modification of the 5′ region of capped rLuc RNA (Fig. 5), Nsp1 also inactivated the translational competence of capped CAT or fLuc RNAs (data not shown), suggesting that nsp1 could modify capped RNAs in general. Nsp1 induced a substantial reduction in the amount of full-length cap-labeled rLuc RNA (Fig. 5a), which suggested that the very 5′ end of the rLuc RNA, including the 7-methylguanosine cap and the radiolabeled phosphate group, may have been removed. Primer extension analysis of the rLuc RNA that had been incubated with nsp1 (Fig. 4b) revealed that nsp1 caused the modification of several different regions at the 5′ end of rLuc RNA. An RNA signal that migrated slightly faster than the full-length rLuc RNA was detected after incubation of the 3′-end labeled rLuc in the presence of nsp1 (Fig. 5a). Analysis of the nsp1-treated non-polyadenylated rLuc RNA further showed that the fast-migrating species of rLuc RNA shown in Fig. 5a consisted of at least two bands that were roughly 20 to 40 nt shorter than the full-length rLuc RNA template (Supplementary Fig. 3). These data point to a possibility that the several premature primer extension termination products, which were obtained using nsp1-treated rLuc RNA (Fig. 4b), were derived from rLuc RNA species lacking different regions from their 5′ ends. Because nsp1 induced an internal RNA cleavage in Ren-EMCV-FF (Fig. 6), we suspect that nsp1 also triggered a similar endonucleolytic RNA cleavage in the 5′ region of rLuc RNA. We also noted that the amounts of the full-length primer extension product obtained from rLuc RNA that had been incubated with nsp1 were not substantially different from the corresponding signal obtained from the control group (Fig. 4b). These data imply that nsp1 induced the removal of the entire or a portion of the cap structure without affecting the first 5′-end nucleotide from the full-length rLuc RNA. Probably the putative nsp1-induced endonucleolytic cleavage and decapping activities both contributed to the translational inactivation of nsp1-treated rLuc RNA.
Nsp1 induced RNA cleavage in the EMCV IRES of Ren-EMCV-FF, but it did not induce the RNA cleavage in Ren-CrPV-FF and Ren-HCV-FF RNAs, revealing the template-dependent nature of nsp1-induced RNA cleavage. Furthermore, these data demonstrated that nsp1-induced RNA modification was not limited to cap-dependent RNA templates. Because the translation initiation factors eIF4G and eIF4A are required for cap-dependent and EMCV IRES-mediated translation but not for translation driven by the IRES elements of HCV and CrPV32, these factors may be involved in the induction of nsp1-mediated endonucleolytic cleavage of template RNAs. Alternatively, nsp1 may recognize putative unique RNA element(s), present in EMCV IRES and absent in HCV and CrPV IRES elements, to induce the RNA cleavage in the EMCV IRES. Further studies using various RNAs carrying different types of IRES elements will be necessary to distinguish between these two possibilities.
Two different mechanisms are conceivable for how the nsp1-40S ribosome complex induces the RNA modification at the 5′-end region of capped rLuc RNA and at the EMCV IRES of Ren-EMCV-FF RNA. One is that the activated nsp1, induced by the binding of nsp1 to the 40S subunits, modifies these RNAs. Another possibility is that binding of nsp1 to the 40S subunits induces the activation or recruitment of a host factor(s) that modifies the template RNAs. It is also possible that some ribosome-associated factors are involved in nsp1-induced RNA modification. Indeed, host endonucleases can destabilize host mRNAs during translational elongation in polysomes33,34. Because nsp1 modified the rLuc RNA during translation initiation (Figs. 3,4,5), the nsp1-mediated modification of the 5′ region of the capped RNA presents a novel strategy for modifying capped RNAs.
The expression of nsp1 in mammalian cells caused both host mRNA degradation and host translation inhibition8. Because host mRNAs that are not involved in active translation are delivered to host mRNA degradation pathways35, we hypothesize that host capped mRNAs that underwent the nsp1-mediated translation inhibition and modification in expressing cells and SCoV-infected cells are delivered to the host mRNA degradation machinery36 and efficiently degraded.
Efficient SCoV-specific gene expression occurs in infected cells, in which nsp1 suppresses host gene expression10, which implies that SCoV gene expression is somehow immune to the nsp1-induced suppression of gene expression in infected cells. Like many host mRNAs, coronavirus mRNAs are capped and polyadenylated37. Several mechanisms are conceivable for an apparent selective immunity of SCoV gene expression to the nsp1-induced translational suppression. One is that the SCoV-specific translation occurs in selected subcellular compartments, where the localization and the access of nsp1 is limited. Another possibility is that viral or host-derived protein(s) selectively bind to SCoV mRNAs and facilitate SCoV-specific translation in the presence of nsp1. Alternatively, the efficient accumulation of SCoV mRNAs outcompetes the nsp1-induced translational suppression.
Methods
Plasmid construction.
We cloned pST-fLuc by inserting the cDNA encoding the entire fLuc ORF region from the pGL3 control vector (Promega) into pSPT18 (Roche). We cloned the PCR-amplified full-length nsp1 and nsp1-mt genes into the pGEX vector (GE Healthcare), yielding pGEX-wt and pGEX-mt, respectively. We constructed pRL-EMCV-FL by replacing the fragment containing HCV IRES, fLuc gene and the 3′ noncoding region of pRC22F (ref. 38) with that containing EMCV IRES, fLuc gene and the 3'-noncoding region of pT7-IRES-fLuc (ref. 39).
In vitro RNA synthesis and RNA transfection.
We generated capped and polyadenylated CAT RNA, nsp1 RNA, nsp1-mt RNA, m7G-FF RNA, rLuc RNA, Ren-HCV-FF RNA and Ren-CrPV-FF RNA by linearizing and transcribing the plasmids pcD-CAT, pcD-Nsp1-wt, pcD-Nsp1-mt (ref. 10), pST-fluc, pRL-SV40 (Promega), pRC22F (ref. 38) and CrPV1.1, respectively, using an mMESSAGE mMACHINE T7 Ultra kit (Ambion). We synthesized capped and non-polyadenylated Ren-EMCV-FF RNA from pRL-EMCV-FL after linearization by ClaI digestion 288 nt upstream of the 3′ end of the fLuc gene. We used TransIT mRNA (Mirus) for transfecting in vitro RNA transcripts into subconfluent 293 cells.
Expression and purification of recombinant nsp1 protein.
We purified GST-fused proteins expressed in E. coli BL21-CodonPlus DE3 cells (Stratagene) using glutathione Sepharose 4B affinity beads (GE Healthcare) followed by cleavage of GST from the GST-fused protein using PreScission protease (GE Healthcare).
In vitro translation assay.
We performed the translation reactions using the Retic Lysate IVT kit (Ambion). After preincubating RRL with nsp1, GST or nsp1-mt for 10 min at 4 °C, we added RNA and incubated the samples at 30 °C in the presence of [35S]methionine (1,000 Ci mmol−1; MP Biomedicals). We resolved the translation products by 12% SDS-PAGE and visualized by autoradiography. We performed densitometric analysis using ImageJ software to quantify the bands.
Immunoprecipitation of rRNA.
At 8 h after transfection of 293 cells with RNAs, we prepared the cell lysates using a buffer containing 50 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 100 mM KCl and 1% (v/v) Triton-X-100 and incubated the lysates with an anti-myc antibody (Upstate Biotech.) at 4 °C. We collected the immune complexes after incubation with Protein G Plus-Agarose (Santa Cruz Biotech.) at 4 °C followed by centrifugation. After washing the pellets three times in a buffer containing 10 mM HEPES (pH 7.4), 500 mM KCl, 2.5 mM MgCl2 and 1 mM DTT, we extracted RNA and performed northern blot analysis using digoxigenin-labeled oligonucleotide probes40 to detect 18S and 28S rRNAs. We resolved the immunoprecipitated proteins in the pellets by SDS-PAGE and western blot analysis.
Polysome analysis.
At 8 h after transfection of 293 cells with RNAs, we prepared the cell lysates in a buffer containing 50 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 100 mM KCl, 1% (v/v) Triton-X-100, 2 mM DTT, 100 μg ml−1 CHX and 0.5 mg ml−1 heparin, applied them onto a 10% to 50% sucrose gradient containing the same buffer and centrifuged at 39,000 r.p.m. in a Beckman SW41 rotor at 4 °C for 2 h. After fractionating the gradient fractions using a gradient fractionator, we monitored the absorbance at 254 nm using an ISCO UA-6 spectrophotometer. We precipitated the proteins from each fraction using trichloroacetic acid and detected them by western blot analysis. We extracted rRNAs from the fractions and visualized them on a gel by staining with ethidium bromide.
Ribosome binding assay.
We performed the ribosome binding assay as described previously16. After preincubating RRL with GST, nsp-1 or nsp1-mt for 10 min at 4 °C, we incubated the samples with [α-32P]UTP-labeled, in vitro transcribed rLuc RNA in the presence of 0.6 mM CHX or 1 mM GMP-PNP for 10 min at 30 °C. After centrifugation through 10% to 40% sucrose gradients (38,000 r.p.m. for 3.5 h in a Beckman SW41 rotor), we collected fractions from each gradient and determined the radioactivity in each fraction in a scintillation counter.
Toe printing analysis.
We performed toe printing analysis as described previously18,41. After pre-annealing the 5′ 32P-labeled primer DNA (5′-TTTTTCTGAATCATAATAATTAA-3′) (40,000 c.p.m.) to rLuc RNA by heating for 1 min at 65 °C, followed by incubation at 37 °C for 10 min, we preincubated RRL with 1 μg of GST, nsp1, or nsp1-mt for 10 min at 4 °C and then incubated the samples with 0.6 mM CHX or 0.6 mM CHX and 1 mM GMP-PNP for 5 min at 30 °C. After incubation, we added the primer–mRNA complexes to the samples and incubated for an additional 10 min at 30 °C. We diluted the reactions 20-fold with buffer containing 0.5 mM CHX, 7 mM magnesium acetate, 100 mM potassium acetate, 0.5 mM dATP, dCTP, dGTP, and dTTP, 20 mM Tris-HCl, pH 7.4, 2 mM DTT and 1 unit μl−1 SuperScript III (Invitrogen), and incubated them at 30 °C for 10 min. We resolved the extracted primer extension products on an 8% sequencing gel.
Primer extension analysis.
After preincubating RRL with GST, nsp-1, or nsp-1-mt for 10 min at 4 °C, we incubated the samples with 0.6 mM CHX or 0.6 mM CHX and 1 mM GMP-PNP for 5 min at 30 °C. Then, we added rLuc RNA to the samples and incubated them for 10 min at 30 °C. After extracting total RNAs, we incubated the RNAs with the 5′-end labeled primer as described above and performed primer extension using a Primer Extension System (Promega). We resolved the extracted primer extension products on 8% sequencing gel.
Assay for modification of radiolabeled in vitro-synthesized RNAs in RRL.
We generated the cap-labeled rLuc RNA by incubating uncapped rLuc RNA with vaccinia virus guanylyltransferase (Ambion) in the presence of [α-32P]GTP (3,000 Ci mmol−1; MP Biomedicals) at 37 °C for 1 h. We generated the 3′-end labeled rLuc RNA by incubating rLuc RNA with T4 RNA ligase (Ambion) in the presence of [32P]pCp (cytidine-3′,5′-bisphosphate; 3,000 Ci mmol−1, MP Biomedicals) and ATP at 4 °C for 16 h. We purified all labeled RNA transcripts with the RNeasy kit (Qiagen). We incubated the 32P-labeled RNA transcripts with nsp1, GST or nsp1-mt in RRL. After 15 min incubation at 30 °C in the presence of CHX and GMP-PNP, we extracted the RNAs and resolved them on 5% sequencing gel.
Supplementary information
Supplementary Figures 1–3 (PDF 84 kb)
Acknowledgements
We thank P. Sarnow (Stanford University) for the CrPV1.1 plasmid, S. Lemon (University of Texas Medical Branch at Galveston) for the pRC22F plasmid and J. Pelletier (McGill University) and J. Tanaka (University of the Ryukyus) for hippuristanol. This work was supported by US National Institutes of Health Public Health Service grant AI72493. W.K. and C.H. were supported by the James W. McLaughlin Fellowship Fund.
Author Contributions
W.K., C.H. and K.G.L. generated the reagents. W.K. and C.H. performed the experiments. W.K., C.H., K.N. and S.M. designed the study. W.K., K.N. and S.M. wrote the paper.
Footnotes
Wataru Kamitani and Cheng Huang: These authors contributed equally to this work.
References
- 1.Poon LL, Guan Y, Nicholls JM, Yuen KY, Peiris JS. The aetiology, origins, and diagnosis of severe acute respiratory syndrome. Lancet Infect. Dis. 2004;4:663–671. doi: 10.1016/S1473-3099(04)01172-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thiel V, et al. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 2003;84:2305–2315. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- 3.Prentice E, McAuliffe J, Lu X, Subbarao K, Denison MR. Identification and characterization of severe acute respiratory syndrome coronavirus replicase proteins. J. Virol. 2004;78:9977–9986. doi: 10.1128/JVI.78.18.9977-9986.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barretto N, et al. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 2005;79:15189–15198. doi: 10.1128/JVI.79.24.15189-15198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Graham RL, Sims AC, Brockway SM, Baric RS, Denison MR. The nsp2 replicase proteins of murine hepatitis virus and severe acute respiratory syndrome coronavirus are dispensable for viral replication. J. Virol. 2005;79:13399–13411. doi: 10.1128/JVI.79.21.13399-13411.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lindner HA, et al. The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J. Virol. 2005;79:15199–15208. doi: 10.1128/JVI.79.24.15199-15208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sulea T, Lindner HA, Purisima EO, Menard R. Deubiquitination, a new function of the severe acute respiratory syndrome coronavirus papain-like protease? J. Virol. 2005;79:4550–4551. doi: 10.1128/JVI.79.7.4550-4551.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kamitani W, et al. Severe acute respiratory syndrome coronavirus nsp1 protein suppresses host gene expression by promoting host mRNA degradation. Proc. Natl. Acad. Sci. USA. 2006;103:12885–12890. doi: 10.1073/pnas.0603144103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wathelet MG, Orr M, Frieman MB, Baric RS. Severe acute respiratory syndrome coronavirus evades antiviral signaling: role of nsp1 and rational design of an attenuated strain. J. Virol. 2007;81:11620–11633. doi: 10.1128/JVI.00702-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Narayanan K, et al. Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J. Virol. 2008;82:4471–4479. doi: 10.1128/JVI.02472-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zust R, et al. Coronavirus non-structural protein 1 is a major pathogenicity factor: implications for the rational design of coronavirus vaccines. PLoS Pathog. 2007;3:e109. doi: 10.1371/journal.ppat.0030109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jan E, Sarnow P. Factorless ribosome assembly on the internal ribosome entry site of cricket paralysis virus. J. Mol. Biol. 2002;324:889–902. doi: 10.1016/S0022-2836(02)01099-9. [DOI] [PubMed] [Google Scholar]
- 13.Pestova TV, Lomakin IB, Hellen CU. Position of the CrPV IRES on the 40S subunit and factor dependence of IRES/80S ribosome assembly. EMBO Rep. 2004;5:906–913. doi: 10.1038/sj.embor.7400240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Merrick WC. Assays for eukaryotic protein synthesis. Methods Enzymol. 1979;60:108–123. doi: 10.1016/S0076-6879(79)60011-3. [DOI] [PubMed] [Google Scholar]
- 15.Novac O, Guenier AS, Pelletier J. Inhibitors of protein synthesis identified by a high throughput multiplexed translation screen. Nucleic Acids Res. 2004;32:902–915. doi: 10.1093/nar/gkh235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bordeleau ME, et al. Functional characterization of IRESes by an inhibitor of the RNA helicase eIF4A. Nat. Chem. Biol. 2006;2:213–220. doi: 10.1038/nchembio776. [DOI] [PubMed] [Google Scholar]
- 17.Pestova TV, et al. Molecular mechanisms of translation initiation in eukaryotes. Proc. Natl. Acad. Sci. USA. 2001;98:7029–7036. doi: 10.1073/pnas.111145798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kozak M. Primer extension analysis of eukaryotic ribosome-mRNA complexes. Nucleic Acids Res. 1998;26:4853–4859. doi: 10.1093/nar/26.21.4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Komarova AV, et al. Rabies virus matrix protein interplay with eIF3, new insights into rabies virus pathogenesis. Nucleic Acids Res. 2007;35:1522–1532. doi: 10.1093/nar/gkl1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lyles DS. Cytopathogenesis and inhibition of host gene expression by RNA viruses. Microbiol. Mol. Biol. Rev. 2000;64:709–724. doi: 10.1128/MMBR.64.4.709-724.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sato H, et al. Measles virus N protein inhibits host translation by binding to eIF3-p40. J. Virol. 2007;81:11569–11576. doi: 10.1128/JVI.00570-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson SR, Sarnow P. Regulation of host cell translation by viruses and effects on cell function. Curr. Opin. Microbiol. 2000;3:366–370. doi: 10.1016/S1369-5274(00)00106-5. [DOI] [PubMed] [Google Scholar]
- 23.Everly DN, Jr., Read GS. Site-directed mutagenesis of the virion host shutoff gene (UL41) of herpes simplex virus (HSV): analysis of functional differences between HSV type 1 (HSV-1) and HSV-2 alleles. J. Virol. 1999;73:9117–9129. doi: 10.1128/jvi.73.11.9117-9129.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glaunsinger B, Ganem D. Lytic KSHV infection inhibits host gene expression by accelerating global mRNA turnover. Mol. Cell. 2004;13:713–723. doi: 10.1016/S1097-2765(04)00091-7. [DOI] [PubMed] [Google Scholar]
- 25.Rowe M, et al. Host shutoff during productive Epstein-Barr virus infection is mediated by BGLF5 and may contribute to immune evasion. Proc. Natl. Acad. Sci. USA. 2007;104:3366–3371. doi: 10.1073/pnas.0611128104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith TJ, Morrison LA, Leib DA. Pathogenesis of herpes simplex virus type 2 virion host shutoff (vhs) mutants. J. Virol. 2002;76:2054–2061. doi: 10.1128/jvi.76.5.2054-2061.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smiley JR. Herpes simplex virus virion host shutoff protein: immune evasion mediated by a viral RNase? J. Virol. 2004;78:1063–1068. doi: 10.1128/JVI.78.3.1063-1068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taddeo B, Zhang W, Roizman B. The UL41 protein of herpes simplex virus 1 degrades RNA by endonucleolytic cleavage in absence of other cellular or viral proteins. Proc. Natl. Acad. Sci. USA. 2006;103:2827–2832. doi: 10.1073/pnas.0510712103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elgadi MM, Hayes CE, Smiley JR. The herpes simplex virus vhs protein induces endoribonucleolytic cleavage of target RNAs in cell extracts. J. Virol. 1999;73:7153–7164. doi: 10.1128/jvi.73.9.7153-7164.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kwong AD, Frenkel N. Herpes simplex virus-infected cells contain a function(s) that destabilizes both host and viral mRNAs. Proc. Natl. Acad. Sci. USA. 1987;84:1926–1930. doi: 10.1073/pnas.84.7.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Colon-Ramos DA, et al. Direct ribosomal binding by a cellular inhibitor of translation. Nat. Struct. Mol. Biol. 2006;13:103–111. doi: 10.1038/nsmb1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martinez-Salas E, Pacheco A, Serrano P, Fernandez N. New insights into internal ribosome entry site elements relevant for viral gene expression. J. Gen. Virol. 2008;89:611–626. doi: 10.1099/vir.0.83426-0. [DOI] [PubMed] [Google Scholar]
- 33.Doma MK, Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006;440:561–564. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang F, Schoenberg DR. Endonuclease-mediated mRNA decay involves the selective targeting of PMR1 to polyribosome-bound substrate mRNA. Mol. Cell. 2004;14:435–445. doi: 10.1016/j.molcel.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 35.Doma MK, Parker R. RNA quality control in eukaryotes. Cell. 2007;131:660–668. doi: 10.1016/j.cell.2007.10.041. [DOI] [PubMed] [Google Scholar]
- 36.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 37.Gorbalenya AE, Enjuanes L, Ziebuhr J, Snijder EJ. Nidovirales: evolving the largest RNA virus genome. Virus Res. 2006;117:17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang TH, Rijnbrand RC, Lemon SM. Core protein-coding sequence, but not core protein, modulates the efficiency of cap-independent translation directed by the internal ribosome entry site of hepatitis C virus. J. Virol. 2000;74:11347–11358. doi: 10.1128/JVI.74.23.11347-11358.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ikegami T, Peters CJ, Makino S. Rift valley fever virus nonstructural protein NSs promotes viral RNA replication and transcription in a minigenome system. J. Virol. 2005;79:5606–5615. doi: 10.1128/JVI.79.9.5606-5615.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Banerjee S, An S, Zhou A, Silverman RH, Makino S. RNase L-independent specific 28S rRNA cleavage in murine coronavirus-infected cells. J. Virol. 2000;74:8793–8802. doi: 10.1128/JVI.74.19.8793-8802.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ostareck DH, Ostareck-Lederer A, Shatsky IN, Hentze MW. Lipoxygenase mRNA silencing in erythroid differentiation: The 3′UTR regulatory complex controls 60S ribosomal subunit joining. Cell. 2001;104:281–290. doi: 10.1016/S0092-8674(01)00212-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures 1–3 (PDF 84 kb)


