Abstract
We describe the identification and characterization of Trim43a, Trim43b, and Trim43c genes, whose expression are restricted to preimplantation stages and peak at the 8-cell to morula stage. We identified a 5 kb DNA fragment that covers upstream region of Trim43a as a putative promoter, which can drive the expression of mStrawberry fluorescent protein in a manner similar to endogenous Trim43 genes. Trim43 genes will be useful stage-specific markers for the study of preimplantation embryos.
Keywords: mouse preimplantation, gene expression, 8-cell, morula, fluorescent proteins, asymmetric PCR, promoter analysis
1. Results and Discussion
1.1. Rational for the study and selection of candidate genes
Mouse preimplantation embryogenesis is marked by three waves of gene expression: massive degradation of maternal RNAs, zygotic genome activation (ZGA) at the 2 cell stage, and mid-preimplantation gene activation (MGA) (Hamatani et al., 2004). Global expression profiles of mouse preimplantation embryos by microarrays have been reported recently (Hamatani et al., 2004; Wang et al., 2004; Wang et al., 2005; Zeng et al., 2004), providing a great opportunity to search for genes that show specific expression patterns during preimplantation development. We previously identified Zscan4 as a ZGA gene that is expressed uniquely in the 2-cell embryos (Falco et al., 2007). Here, we sought MGA genes, which should be expressed uniquely in the later stages of preimplantation development.
To identify genes whose expression peaks at the 8-cell or morula stage, we first used both EST frequency (Quackenbush et al., 2001; Sharov et al., 2003) and DNA microarray data (Hamatani et al., 2004; Zeng et al., 2004). We found 108 candidate genes and then narrowed them down to 6 candidate genes (Gpbp1l1, Zfyve27, Ranbp10, Zfp791, Zfp821, and EG547109) by discarding genes encoding enzymes and other well-known genes. We used qRT-PCR analysis to confirm that a gene model named EG547109 showed peak expression at the 8-cell stage (see the section 1.3) and decided to analyze this gene further.
1.2. Analysis of genome sequences
We detected three closely related genes corresponding to EG547109 by public database analysis (UCSC database, July 2007). These three gene models were annotated as EG547109, EG666731, and EG666747, which were mapped within 510 kb region on Chromosome 9 (Supplemental Fig. S1A). Based on the genome sequences, we predicted three cDNAs and open reading frames (Fig. 1A). Domain prediction analysis (htpp://smart.embl.de/) of the protein encoded by EG547109 revealed that this gene belongs to the TRIM/RBCC proteins, which are defined by the presence of the tripartite motif composed of a RING domain, one or two B-box motifs and a coiled-coil region. It is known that these proteins self–associate, resulting in the formation of large protein complexes that are involved in a broad range of biological processes (Meroni and Diez-Roux, 2005; Reymond et al., 2001; Sardiello et al., 2008; Torok and Etkin, 2001).
Figure 1.
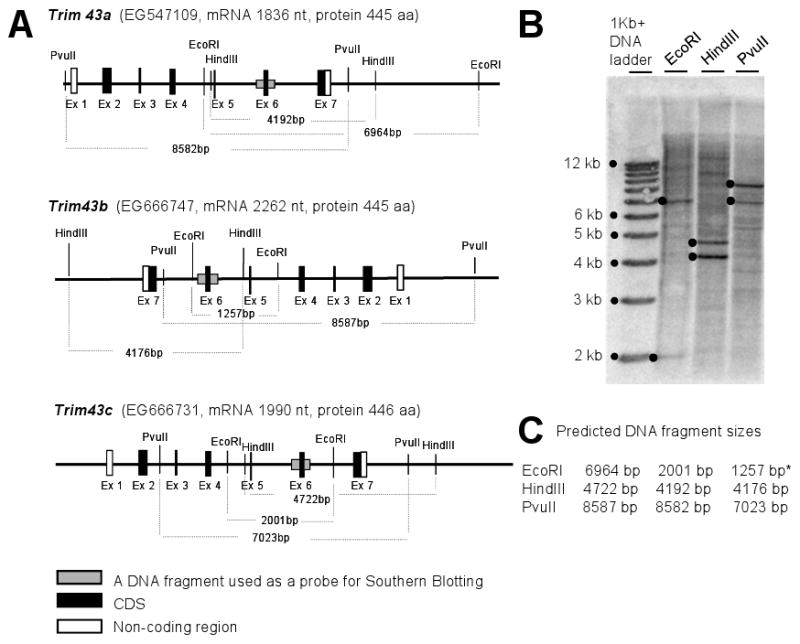
(A) Exon-Intron structures of three Trim43 genes. Black boxes represent the protein-coding region of cDNAs. New gene symbols we proposed are shown in bold italics with the current gene symbols, mRNA sizes (nucleotides), and protein sizes (amino acids). (B, C) Southern blot analysis of C57BL/6J genomic DNAs digested with EcoRI, HindIII, or PvuII restriction enzymes. Sizes of all DNA fragments hybridized with a Trim43 probe matched with those of predicted DNA fragments. A fragment with * is not shown in this figure, but was also detected in another Southern Blot (data not shown).
After consulting with the International Committee on Standardized Genetic Nomenclature for Mice, we named the mouse EG547109 Trim43a, EG666731 Trim43b, and EG666747 Trim43c, primarily based on the following sequence similarity searches. To search for the human ortholog of EG547109, we carried out a BLASTP (Altschul et al., 1990) analysis of the mouse protein against all the human proteins. Top two hits were humanTRIM43 and a protein similar to human TRIM17; both proteins shared 41% identities and were mapped to Chromosome 2. The third hit was human TRIM11 that shared 36% identities and was mapped to Chromosome 11. When the human TRIM43 protein was searched against all the mouse proteins, EG666747 (TRIM43C), EG547109 (TRIM43A), and EG666731 (TRIM43B) were among the top hits, sharing 42% (TRIM43C) and 41% (TRIM43A and TRIM43B) identities with the human TRIM43 protein. The domain sequences of mouse TRIM43A protein showed a high conservation in RING (48.7%), Bbox (54.7%), and SPRY (50%) of the human TRIM43 protein.
Although it was clear from the sequence alignments to mouse genome (Supplemental Fig. S1A) that Trim43a, Trim43b, and Trim43c are different genes, their transcript sequences and predicted amino acid sequences were very similar (Supplemental Fig. S2, S3). At the nucleotide level, transcripts show 97% identity between each other. At the protein level, identities are 95% (TRIM43A vs TRIM43B), 92% (TRIM43A vs TRIM43C), and 94% (TRIM43B vs TRIM43C). Southern blot analysis confirmed that Trim43a, Trim43b, and Trim43c are indeed present in the mouse genome (Fig. 1B, C).
1.3. Expression patterns of Trim43 genes
EST frequency analysis detected the expression of Trim43 genes only in preimplantation embryos (Supplemental Fig. S1B, C). Analysis of Trim43 genes by RT-PCR also did not detect their expression in major organs (Fig. 2A). We then carried out qRT-PCR analysis of preimplantation embryos and found that Trim43 transcripts were detected at a low level by the late 2-cell stage, but showed the highest level of transcripts in the 8-cell and morula stages (Fig. 2B). Because Zscan4 — another preimplantation embryo-specific gene that we reported recently (Falco et al., 2007) is also expressed in mouse embryonic stem (ES) cells, we examined the expression of Trim43 genes in ES cells, but no expression was found. (Fig. 2C).
Figure 2.
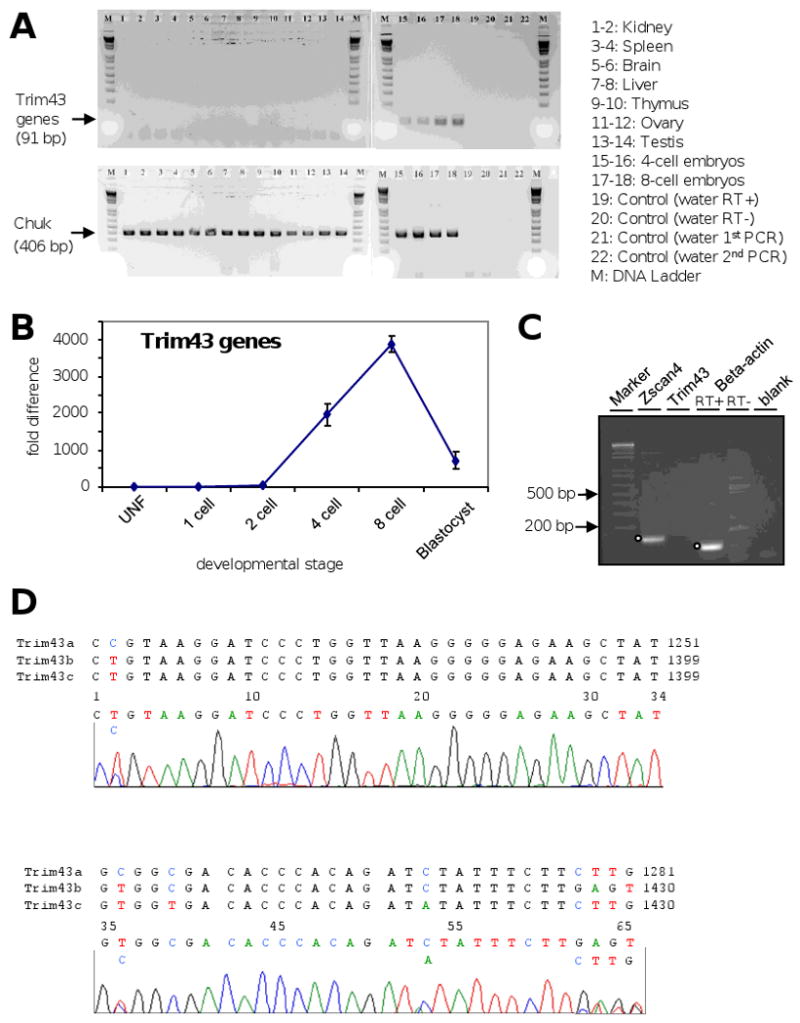
(A) Analysis of Trim43 expression in mouse tissues by RT-PCR. Chuk was used as a control. (B) Expression profile of Trim43 during preimplantation development by qRT-PCR analysis. Three sets of 10 pooled embryos were collected from each stage (UNF, MetaII oocyte; 1-cell embryo; 2-cell embryo; 4-cell embryo; 8-cell embryo; and blastocyst) and used for qRT-PCR analysis. Expression levels of Trim43 were normalized by those of control Chuk, and then the averaged expression at each stage was represented as a fold change compared to the expression level in oocytes. (C) Analysis of Trim43 expression in ES cells. Zscan4, used as a positive control together with Beta-actin, shows a clear band, whereas no band is detected with Trim43 primers. (D) Electropherograms of Trim43 cDNA sequences amplified from embryos at the morula stage.
The presence of three similar copies of Trim43 on the mouse genome prompted us to examine which copies are indeed expressed in mouse preimplantation embryos. To this end, we PCR-amplified and sequenced the CDS of Trim43 from cDNA mixtures of embryos from the morula stage. We were able to distinguish each copy of Trim43 by examining the sequence chromatograph, when it showed multiple nucleotide peaks at the same nucleotide position. Fig. 2D shows an example of such analyses, which indicate that all three copies were expressed in morula. For example, transcripts of Trim43a was present, because a peak of “C” (2 n.t. position in the chromatograph) was detected. Similarly, Trim43b was present, because peaks of GAGT (62 - 65 n.t.) were detected (Fig. 2D). The presence of a small peak “A” (53 n.t.) indicates the presence of Trim43c transcripts, but the level of expression was low, which is also consistent with a lack of a peak “T” (39 n.t.). Because primers used to amplify a region of Trim43 cDNAs 100% matched to all three copies, it is most likely that abundance of each transcript in PCR products represents the abundance of each transcripts in cDNA mixture. We, therefore, estimated the abundance of transcripts for each gene in the morula cDNA mixture using peak height information of each nucleotide at multiple locations: Trim43b seems to be the most highly expressed, followed by Trim43a, while Trim43c shows the lowest expression level.
1.4. Attempts to suppress Trim43 transcript level in preimplantation development
As a first step to characterize the function of Trim43, we explored a possibility to knockdown all three copies (Trim43a, Trim43b, Trim43c) using siRNA technology and injected a mixture of four oligonucleotide siRNAs (obtained from Dharmacon) into 1-cell mouse embryos. However, we could not obtain a down-regulation of Trim43 expression, even after trying different concentration of siRNAs (5, 10, and 20 μM). Considering the late expression timing during preimplantation stages, we also injected the mixture of siRNAs into both blastomeres of 2-cell embryos but failed to suppress the Trim43 expression. It is possible that siRNA mixtures were diluted as the embryo cleaves and the level of siRNA mixtures were too low to affect the expression of Trim43, which reaches very high expression levels by the morula stage.
1.5. Analysis of Trim43 promoter activity in preimplantation embryos
To investigate whether the defined DNA region of Trim43 promoter can drive the expression of a marker gene in the same manner as the endogenous Trim43, we PCR-amplified a 5 kb DNA fragment that covers a region from ATG codon to 5 kb upstream of the ATG codon of Trim43a gene (containing the first and second exons) and fused it to a green fluorescent protein (GFP) variant - Emerald (green) or mStrawberry (red) (Shaner et al., 2005) and 3′UTR of Trim43 gene containing the polyA signal (Supplemental Fig S4). These DNA fragments were named pTrim43a(ATG)-Emerald-3′UTR and pTrim43a(ATG)-mStrawberry-3′UTR, respectively. The promoter sequences of Trim43a, Trim43b and Trim43c share 98% similarity. A total 6.1 kb of this expression unit was purified and microinjected into a male pronucleus or both pronuclei of 1-cell embryos. A fluorescent signal was detected by the late 2-cell stage but dramatically increased in the later stages with the highest signal intensity at the morula stage (Fig. 3). The results confirm the specific expression pattern of Trim43 obtained by qRT-PCR (Fig. 2B), microarray, and EST frequency analysis (Supplemental Fig. S1). A low level of fluorescent signal was still present even at the blastocyst stage, but it disappeared after hatching. Essentially the same results were obtained with two additional constructs we tested: pTrim43a(ATG)-Emerald-tkPolyA [5 kb upstream region from ATG – Emerald - HSV tk polyA] and pTrim43a(1stExon)-Emerald-3′UTR [5 kb upstream region from the first exon – Emerald - 3′-UTR of Trim43] (data not shown). In contrast, no fluorescent signal was detected when a 500 bp DNA fragment that covers a region between ATG and 500 bp upstream from ATG (named pTrim43a(500 bp)-Emerald-3′UTR) was used as a promoter (Fig. 3B).
Figure 3.
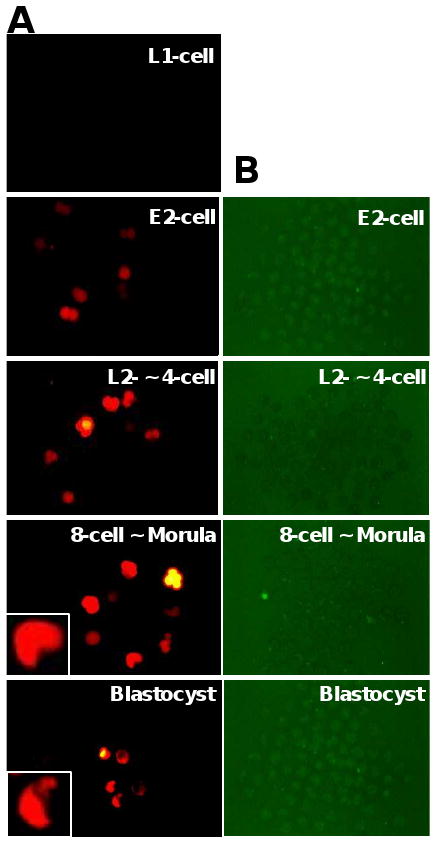
(A) Representative images of embryos cultured after microinjecting a DNA fragment pTrim43a(ATG)-mStrawberry-3′UTR into the pronucleus of 1-cell embryos. Fluorescent signals were detected by the late 2 cell stage but dramatically increased at the later stage with the highest peak at the 8-cell ∼ morula stage. See Supplemental Table S1 for the detail. (B) Representative images of embryos cultured after microinjecting a DNA fragment pTrim43a(500 bp)-Emerald-3′UTR into the pronucleus of 1-cell embryos. No fluorescent signal was detected through preimplantation stages.
Interestingly, in most cases we observed a fluorescent signal in only one of the two blastomeres at the late 2-cell stage. These embryos subsequently developed into the morula and blastocyst, where fluorescent signals were observed in only about 50% of blastomeres (Fig. 4A). At this stage, the fluorescence was present in both the inner cell mass (ICM) and the trophectoderm (TE). This unexpected marking of one of the 2-cell blastomeres and seemingly unbiased distribution of fluorescence at the blastocyst stage may thus support that notion that both blastomeres at the 2-cell stage give rise to both ICM and TE (Hiiragi et al., 2006).
Figure 4.
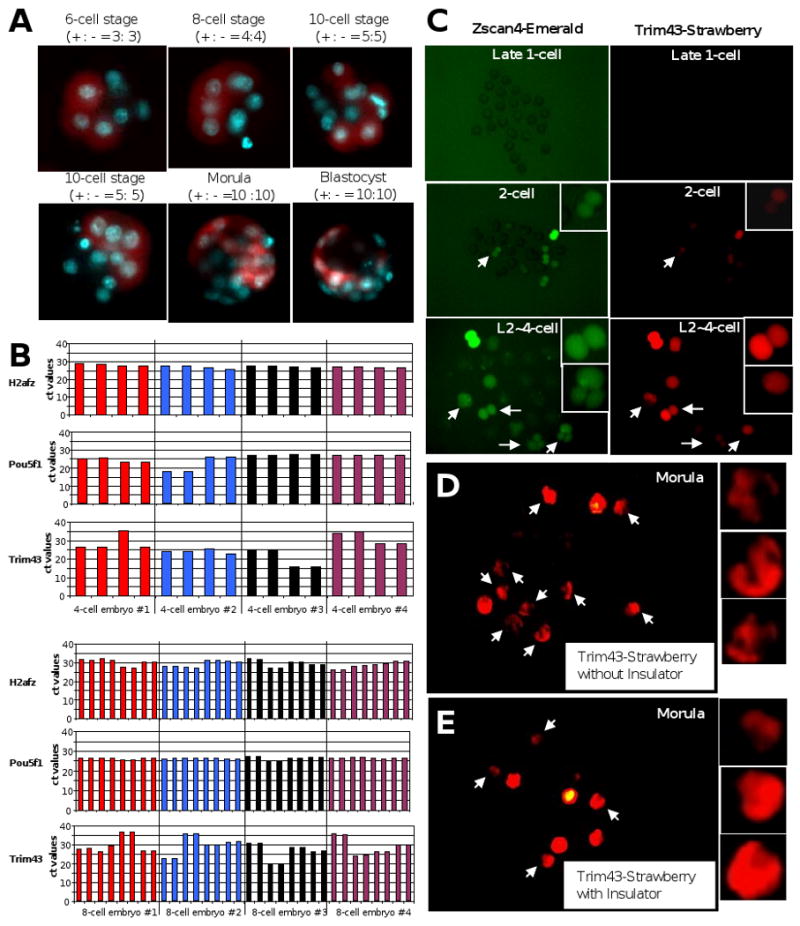
(A) Representative images of embryos cultured after microinjecting a plasmid vector pTrim43a(ATG)-Emerald-tkPolyA into the pronucleus of 1-cell embryos. Emerald fluorescence is shown as red color in this case. Numbers of blastomeres of Emerald positive (+) vs Emerald negative (-) are also shown. See Supplemental Table S2 for the detail. (B) Analysis of Trim43 expression of single 4-cell and 8-cell embryo blastomeres by qRT-PCR. Control genes - H2afz and Pou5f1 - show similar ct values among the blastomeres of both 4-cell and 8-cell stage embryos. In contrast, Trim43 shows lower level of expression (ct values around 35 cycles) in 3/16 blastomeres of 4-cell stage embryos and in 6/32 blastomeres of 8-cell stage embryos. (C) Representative images of embryos cultured after co-microinjecting a pTrim43a(ATG)-mStrawberry-3′UTR and a Zscan4 promoter-Emerald construct (reported in Falco et al., 2007) into 1-cell embryos. An asymmetric signal was only observed for Trim43, starting at the 2 cell stage. See Supplemental Table S3 for the detail. (D) Representative images of embryos cultured after injecting a pTrim43a(ATG)-mStrwaberry-3′UTR into 1-cell embryos. See Supplemental Table S4 for the detail. (E) Representative images of embryos cultured after injecting a pTrim43a(ATG)-mStrwaberry-3′UTR-Insulator into 1-cell embryos. See Supplemental Tables S4 and S5 for the detail.
The asymmetric distribution of weak fluorescent signals at the 2-cell stage was not observed in our earlier study using a Zscan4 promoter construct (Falco et al., 2007). To test whether the asymmetric expression of the Trim43 promoter is a reflection of that of endogenous Trim43 gene, we carried out qRT-PCR analyses of single blastomeres in 4-cell and 8-cell embryos. We used four embryos for both stages, isolated individual blastomeres from each embryo, and tested Trim43, Pou5f1 (control), and H2afz (control) for each blastomere. The control genes (H2afz and Pou5f1) showed similar transcript levels among blastomeres (Fig. 4B). However, Trim43 showed more variability in transcript levels among blastomeres in both 4-cell embryos and 8-cell embryos (Fig. 4B). At the 4-cell stage, one or two blastomeres showed much lower level of Trim43 than other blastomeres. At the 8-cell stage, in three of four embryos, two blastomeres showed much lower level of Trim43 than other blastomeres (Fig. 4B). However, due to the observed embryo-to-embryo variation of transcript levels, the results are not conclusive at this point.
To further test whether the asymmetric expression is specific to Trim43 promoter, we co-injected Trim43 promoter-Strawberry construct and Zscan4 promoter-Emerald construct into 1-cell embryos. The majority of embryos showed Emerald expression in both blastomeres, but an asymmetric expression signal was observed only for Strawberry (Fig. 4C). This supports the notion that the variegation is specifically related to Trim43 promoter. According to the earlier report (Saveliev et al., 2003), the presence of repeats in a transgene leads to chromosomal position independent variegation. We then carefully inspected Trim43 promoter sequence and found a block of repetitive sequences. We suspected that this asymmetric expression was indeed caused by the variegated expression phenomenon (Frazar et al., 2003). To test this possibility, we added a chicken beta-globin 5′ HS4 insulator element at the 5′-end of the expression unit (see Experimental procedures for further details). A total 8672 bp DNA fragment was injected into the pronuclei of mouse 1-cell embryo. We observed that the asymmetric expressions were attenuated with the addition of the insulator sequences, although some asymmetry remained (Fig. 4D, E).
In this paper, we have reported a cloning and characterization of novel genes that show specific expression in mouse preimplantation embryos. Identification of promoter sequences that can drive the expression of marker genes (e.g., GFP) in a stage-specific manner will provide a useful tool for the molecular analysis of preimplantation embryo development.
2. Experimental procedures
2.1. Collection of eggs and preimplantation embryos
B6D2F1 mice aged 4–5 weeks were superovulated by injections of 5 IU pregnant mare serum gonadotropin (PMS, Sigma, St. Louis, MO), followed 46–47 hr later by 5 IU human chorionic gonadotropin (hCG, Sigma). Unfertilized eggs and embryos were harvested as previously described (Falco et al., 2006). Briefly, after removing cumulus cells by incubation in M2 medium (Specialty Media, NJ) with 300 μg/ml hyaluronidase (Sigma), unfertilized eggs and 1-cell embryos were thoroughly washed, selected for good morphology, and collected. The fertilized eggs (1-cell embryos) were further cultured in KSOM medium (Specialty Media, NJ) at 37 °C in an atmosphere of 5% CO2. Embryos were harvested at 23, 43, 55, 66, 80 and 102 hours post hCG respectively for 1-cell, late 2-cell, 4-cell, 8-cell, morula, and blastocyst. Good-looking embryos were transferred in PBT 1× (Phosphate Buffered Saline (PBS) + Tween20 0.1%) solution into three subsets of pooled (10) embryos and stored in liquid nitrogen.
2.2. RNA isolation and cDNA preparation
A freeze/thaw step has been done to facilitate mechanical rupture of pooled eggs/embryos (collected in PBT 1×) that were used to obtain RNA as a template for cDNA preparation. The Ovation System RNA Amplification (Nugen Technologies, San Carlos, CA, USA) was used in order to prepare amplified cDNA for gene expression analysis.
2.3. qPCR primer design, Quantitative Real-time PCR, and PCR on mouse preimplantation embryos
PCR primer pairs were designed by Vector NTI software (Invitrogen, Carlsbad, CA, USA) and were tested using ovary cDNA with SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA) as described in (Falco et al., 2006). Primer pairs used for qPCR are reported in Supplemental Fig. S4. The cDNAs obtained from oocytes and embryos with the Ovation kit were diluted to 1:25 in a total of 1000 μl and 2 μl were used as a template. Quantitative Real-Time PCR was performed as previously described, and data were normalized by Chuk with the ΔΔCt method (Falco et al., 2006; Livak and Schmittgen, 2001). To understand which copy of Trim43 genes are expressed during preimplantation development, cDNA was synthesized from a set of 10 pooled embryos at 8-cell stage using primers amplifying the entire CDS. The cDNAs obtained were sequenced using a 3100ABI Automatic Sequencer and four-color fluorescent dye chemistry.
2.4. Southern blot
A probe, containing exon 6, was designed and amplified from mouse DNA extracted from ES cells. The primers used are reported in Supplemental Fig. S4. Reactions were set up in a 50 μl final volume using Titanium taq PCR Kit (Clontech) and using 200 ng of DNA as a template. Thermal cycling was initiated with a denaturation step for 3 min at 95°, followed by 30 cycles at 95° for 30 sec, 60° for 30 sec, 70° for 1 min with a final extension of 10 min at 70°.
The PCR product was purified using GFX PCR DNA and Gel band purification kit (GE Healthcare) following the instructions of the kit. Fifteen μg of genomic DNA, extracted from mouse ES cells, was digested to completion with EcoRI, HindIII, PvuII restriction enzymes, fractionated on a 0.8% (w/v) agarose gel, transferred and immobilized to nitrocellulose membranes in 20× SSC (SSC = 3M sodium chloride/0.3M-sodium citrate pH 7.0) and cross-linked to the membrane by baking blots at 80°C for 30 minutes. Blots were hybridized under standard conditions with random-primed 32P-labeled DNA probes. Membranes were subjected to 3× washes of 30 min each: 1-SSC 2× /0.1% (w/v) SDS at room temperature, 2- SSC 0.5× /0.1% (w/v) SDS at 42°C, 3- SSC 0.1× /0.1% (w/v) SDS concentrations at room temperature and autoradiographed finally for 48 hours at -80°C.
2.5. RT-PCR on ES cells
RNA was extracted by ES cells with the Trizol method, and cDNA was synthesized using SuperScript III Reverse Transcriptase (Invitrogen).
2.6. RT-PCR on mouse organs, 4-cell embryos, and 8-cell embryos
Total RNAs were extracted from tissues using the Trizol; a pool of 10 embryos at 4-cell and 8-cell stages were collected as already described above, and cDNA was synthesized using SuperScript III Reverse Transcriptase (Invitrogen). For tissues, 50 ng of total RNAs were used. A nested-PCR was used to amplify Trim43 in tissues and embryos at the 4-cell and 8-cell stages. A high fidelity Taq polymerase (Platinum, Invitrogen) was used with the following thermal cycling conditions: an initial denaturation step of 95°C for 5 min followed by 20 cycles at 95°C 15 sec, 55°C 1.30 min and 72°C 7 min (1st PCR). A 2nd PCR was performed using 5μl of product from the 1st PCR as a template. The amplification conditions used are the following: an initial denaturation step of 95°C for 5 min followed by 35 cycles at 95°C 15 sec, 55°C 1 min, 72°C 1 min and 72°C 7 min. PCR product was visualized under a shortwave length UV on a Gel Logic 1500 Imaging System (Kodak).
2.7 Promoter study
To construct DNA fragments pTrim43a(ATG)-Emerald-3′UTR and a pTrim43a(ATG)-mStrawberry-3′UTR, a 4,996 bp DNA fragment that covers a region between the ATG codon (located in the 2nd exon) and 5 kb upstream of the ATG codon of Trim43a gene was PCR amplified from RP23-46B12-BAC DNA using primers flanked by the MluI (upstream) restriction enzyme recognition sequence (Supplemental Fig. S4, S5). CDSs of green fluorescent protein (GFP) variants - Emerald and mStrawberry (a gift from Dr. Roger Tsien) - were amplified from vectors pcDNA 6.2/EmGFP and pRSET-B, respectively. The 3′UTR of Trim43, containing the polyA signal, was amplified from BAC RP23-46B12. The three fragments were ligated together by a modified protocol of the asymmetric PCR technique (Sparwasser et al., 2004). Briefly, the amplified DNA fragments were fused together by the use of single 50 nt, HPLC purified overlapping primers named Trim43 promoter/emerald (or strawberry) and emerald (or strawberry)/3UTR reported in Supplemental Fig. S4. A ratio of 100:1 between the external and the overlapping primers (named 1A-3B and 4A-4B respectively in Supplemental Fig. S5) was used in a PCR reaction with the 3 PCR amplified fragments as templates. A high fidelity Taq polymerase (Platinum, Invitrogen) was used with the following thermal cycling conditions: an initial denaturation step of 94° for 2.5 min followed by 5 cycles at 94°C 30 sec, 65°C 45 sec, 68°C 8 min and 30 cycles at 94°C 30 sec 60°C 45 sec, 68°C 8 min. The PCR product was run on 1% agarose gel and a band corresponding to the correct size (6145 bp) was purified using GFX PCR DNA and Gel band purification kit (GE Healthcare) following the instructions of the kit. The fragments corresponding to five kb upstream of ATG and upstream of the first exon were also cloned into pcDNA 6.2/EmGFP by TOPO cloning reaction, resulting in pTrim43a(ATG)-Emerald-tkPolyA [5 kb upstream region from ATG – Emerald - HSV tk polyA] and pTrim43a(1stExon)-Emerald-3′UTR [5 kb upstream region from the first exon – Emerald - 3′-UTR of Trim43] plasmids. Plasmids were subsequently cut with MluI and SmaI restriction enzymes, thus obtaining fragments containing Trim43 putative promoter, emerald CDS, and TK poly A signal. The digested fragments were run on 1% agarose gel and purified using GFX PCR DNA and Gel band purification kit (GE Healthcare) kit. The purified probes were microinjected into the male and into both pronuclei of 1-cell stage embryos.
For a construct with an insulator sequence, a 2.4 kb tandem insulator from chicken beta-globin was excised from pJC13-1 (a gift from Dr. Gary Felsenfeld) and cloned into a PTRE2 vector. The asymmetric PCR product containing Trim43 promoter, mStrawberry, and 3′UTR was re-amplified using external primers containing MluI and SalI restriction enzyme site and cloned into the aforementioned PTRE2 vector carrying the insulator. The vector was named pTrim43a(ATG)-mStrawberry-3′UTR-Insulator. We then digested the modified vector with AatII restriction enzyme (whose site is located just upstream of the insulator) and with SalI, thus obtaining a probe of 8672 bp that was subsequently injected into pronuclei of mouse 1-cell embryo.
2.8 Single blastomere quantitative RT-PCR
Mouse 4-cell and 8-cell embryos were incubated for 2-3 min in the acid Tyrode's solution (Sigma Aldrich, Inc. St. Louis, MO) to remove zona pellucida. Individual blastomeres were then disaggregated by gentle pipetting with a thin mouth pipette, collected in single tubes containing 1 μl of PBT 1×, and stored in liquid nitrogen. cDNA from each single blastomere was synthesized using SuperScript III Reverse Transcriptase (Invitrogen), and a first round of PCR was performed to prepare the template for a quantitative Real-Time PCR reaction. A high fidelity Taq polymerase (Platinum, Invitrogen) was used with the following thermal cycling conditions: an initial denaturation step of 95°C for 5 min followed by 18 cycles at 95°C 15 sec, 65°C 1.5 min and 72°C 7 min. Quantitative Real-Time PCR was performed as previously described (Falco et al., 2006; Livak and Schmittgen, 2001) and H2afz and Pou5f1 (see Supplemental Fig. S4 for oligos sequence) were used as controls. Trim43 expression was analyzed on 16 and 32 blastomeres derived from four 4-cell and 8-cell embryos.
Supplementary Material
Acknowledgments
We thank Dr. Toshio Hamatani for the initial contribution to the work, Dr. Yuhki Nakatake for the discussion, Ms. Donna Tignor and Dr. Suresh Poosala for help in mouse husbandry, and Mr. Dawood Dudekula for help in DNA sequence analysis. We also thank Dr. Gary Felsenfeld for a generous gift of the insulator DNA, Dr. Roger Tsien for a generous gift of mStrawberry, and Ms. Lois Maltais for help in assigning gene symbols. This work was supported entirely by the Intramural Research Program of the National Institute on Aging, NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Falco G, Lee SL, Stanghellini I, Bassey UC, Hamatani T, Ko MS. Zscan4: a novel gene expressed exclusively in late 2-cell embryos and embryonic stem cells. Dev Biol. 2007;307:539–50. doi: 10.1016/j.ydbio.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falco G, Stanghellini I, Ko MS. Use of Chuk as an internal standard suitable for quantitative RT-PCR in mouse preimplantation embryos. Reprod Biomed Online. 2006;13:394–403. doi: 10.1016/s1472-6483(10)61445-9. [DOI] [PubMed] [Google Scholar]
- Frazar TF, Weisbein JL, Anderson SM, Cline AP, Garrett LJ, Felsenfeld G, Gallagher PG, Bodine DM. Variegated expression from the murine band 3 (AE1) promoter in transgenic mice is associated with mRNA transcript initiation at upstream start sites and can be suppressed by the addition of the chicken beta-globin 5′ HS4 insulator element. Mol Cell Biol. 2003;23:4753–63. doi: 10.1128/MCB.23.14.4753-4763.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamatani T, Carter MG, Sharov AA, Ko MSH. Dynamics of global gene expression changes during mouse preimplantation development. Dev Cell. 2004;6:117–31. doi: 10.1016/s1534-5807(03)00373-3. [DOI] [PubMed] [Google Scholar]
- Hiiragi T, Louvet-Vallee S, Solter D, Maro B. Embryology: does prepatterning occur in the mouse egg? Nature. 2006;442:E3–4. doi: 10.1038/nature04907. discussion E4. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Meroni G, Diez-Roux G. TRIM/RBCC, a novel class of ‘single protein RING finger’ E3 ubiquitin ligases. Bioessays. 2005;27:1147–57. doi: 10.1002/bies.20304. [DOI] [PubMed] [Google Scholar]
- Quackenbush J, Cho J, Lee D, Liang F, Holt I, Karamycheva S, Parvizi B, Pertea G, Sultana R, White J. The TIGR Gene Indices: analysis of gene transcript sequences in highly sampled eukaryotic species. Nucleic Acids Res. 2001;29:159–164. doi: 10.1093/nar/29.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L, Riganelli D, Zanaria E, Messali S, Cainarca S, Guffanti A, Minucci S, Pelicci PG, Ballabio A. The tripartite motif family identifies cell compartments. EMBO J. 2001;20:2140–51. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardiello M, Cairo S, Fontanella B, Ballabio A, Meroni G. Genomic analysis of the TRIM family reveals two groups of genes with distinct evolutionary properties. BMC Evol Biol. 2008;8:225. doi: 10.1186/1471-2148-8-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saveliev A, Everett C, Sharpe T, Webster Z, Festenstein R. DNA triplet repeats mediate heterochromatin-protein-1-sensitive variegated gene silencing. Nature. 2003;422:909–13. doi: 10.1038/nature01596. [DOI] [PubMed] [Google Scholar]
- Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Methods. 2005;2:905–9. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- Sharov AA, Piao Y, Matoba R, Dudekula DB, Qian Y, VanBuren V, Falco G, Martin PR, Stagg CA, Bassey UC, Wang Y, Carter MG, Hamatani T, Aiba K, Akutsu H, Sharova L, Tanaka TS, Kimber WL, Yoshikawa T, Jaradat SA, Pantano S, Nagaraja R, Boheler KR, Taub D, Hodes RJ, Longo DL, Schlessinger D, Keller J, Klotz E, Kelsoe G, Umezawa A, Vescovi AL, Rossant J, Kunath T, Hogan BL, Curci A, D'Urso M, Kelso J, Hide W, Ko MS. Transcriptome analysis of mouse stem cells and early embryos. PLoS Biol. 2003;1:E74. doi: 10.1371/journal.pbio.0000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparwasser T, Gong S, Li JY, Eberl G. General method for the modification of different BAC types and the rapid generation of BAC transgenic mice. Genesis. 2004;38:39–50. doi: 10.1002/gene.10249. [DOI] [PubMed] [Google Scholar]
- Torok M, Etkin LD. Two B or not two B? Overview of the rapidly expanding B-box family of proteins. Differentiation. 2001;67:63–71. doi: 10.1046/j.1432-0436.2001.067003063.x. [DOI] [PubMed] [Google Scholar]
- Wang QT, Piotrowska K, Ciemerych MA, Milenkovic L, Scott MP, Davis RW, Zernicka-Goetz M. A genome-wide study of gene activity reveals developmental signaling pathways in the preimplantation mouse embryo. Dev Cell. 2004;6:133–44. doi: 10.1016/s1534-5807(03)00404-0. [DOI] [PubMed] [Google Scholar]
- Wang S, Cowan CA, Chipperfield H, Powers RD. Gene expression in the preimplantation embryo: in-vitro developmental changes. Reprod Biomed Online. 2005;10:607–16. doi: 10.1016/s1472-6483(10)61668-9. [DOI] [PubMed] [Google Scholar]
- Zeng F, Baldwin DA, Schultz RM. Transcript profiling during preimplantation mouse development. Dev Biol. 2004;272:483–96. doi: 10.1016/j.ydbio.2004.05.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


