Abstract
The genomic cis-regulatory systems controlling regulatory gene expression usually include multiple modules. The regulatory output of such systems at any given time depends on which module is directing the function of the basal transcription apparatus, and ultimately on the transcription factor inputs into that module. Here we examine regulation of the S. purpuratus tbrain gene, a required activator of the skeletogenic specification state in the lineage descendant from the embryo micromeres. Alternate cis-regulatory modules were found to convey skeletogenic expression in reporter constructs. To determine their relative developmental functions in context, we made use of recombineered BAC constructs containing a GFP reporter, and of derivatives from which specific modules had been deleted. The outputs of the various constructs were observed spatially by GFP fluorescence and quantitatively over time by QPCR. In the context of the complete genomic locus, early skeletogenic expression is controlled by an intron enhancer plus a proximal region containing a HesC site as predicted from network analysis. From ingression onward, however, a dedicated distal module utilizing positive Ets1/2 inputs contributes to definitive expression in the skeletogenic mesenchyme. This module also mediates a newly-discovered negative Erg input which excludes non-skeletogenic mesodermal expression.
Keywords: tbrain gene, Gene regulatory network, cis-Regulatory analysis, Skeletogenic micromere lineage, Recombinant BAC
Introduction
The sea urchin regulatory gene tbrain (tbr) is zygotically expressed in the skeletogenic mesoderm (SM) of the cleavage and blastula stage embryo (Croce et al., 2001; Oliveri et al., 2002), and its expression is required for the postgastrular formation of the larval spicules (Fuchikami et al., 2002). Through transcriptional activation of a target gene, erg,tbr establishes an erg-hex-tgif-alx1 positive feedback circuit that maintains the regulatory state of the skeletogenic mesoderm (SM) domain from early in development, and eventually, together with other regulators, serves as a transcriptional driver of an initial set of differentiation genes (Oliveri et al., 2008). The tbr gene thus has essential roles, first in specification of the SM and then in definitive larval skeletogenesis. Yet these roles, and the circuitry underlying them, are evolutionarily derived traits, since only modern sea urchins precociously segregate a SM lineage. In the sister group to the echinoids, the sea cucumbers, tbr is expressed in the developing endomesoderm (Maruyama, 2000). This is the pleisiomorphic function of the tbr gene in embryogenesis, since it is also expressed in endomesoderm in the more distant sea star outgroup (Hinman and Davidson, 2007; Hinman et al., 2003; Shoguchi et al., 2000). Thus from an evolutionary standpoint the tbrcis-regulatory system is of particular interest since it must be at least partly “new”, and since it is a key mechanistic component of the skeletogenic micromere specification network: this, as a whole, is in itself a derived embryonic feature of the modern sea urchins (euechinoids).
Despite the simple pattern of tbr expression, which is confined entirely to the SM lineage throughout embryonic development, the cis-regulatory system of the tbr gene is anything but simple. Typically for regulatory genes (c.f. Davidson, 2006), tbr is controlled by multiple cis-regulatory modules. Regulatory modules were identified in an intron as well as proximally in the closely related (actually congeneric) strongylotrotid known as Hemicentrotus pulcherrimus (Ochiai et al., 2008). A different, also completely specific skeletogenic cis-regulatory module exists some distance upstream of the gene in S. purpuratus, as we describe below. A major objective of this work was to resolve the various roles of these modules. Gene regulatory network analysis had shown that tbr lies under control of a double negative gate (Oliveri et al., 2002; Oliveri et al., 2003; Oliveri et al., 2008; Revilla-i-Domingo et al., 2007). Thus the early zygotically expressed micromere repressor Pmar1 acts to prevent transcription in micromeres of the hesC gene, which encodes a dedicated repressor zygotically expressed everywhere in the embryo except in micromeres expressing the pmar1 gene. Among the targets of HesC repression is tbr, along with a small number of other initial founders of the SM regulatory state. The double negative gate thus results in derepression of the tbr gene in the SM lineage. The putative site of HesC interaction in tbrcis-regulatory DNA had been identified (Ochiai et al., 2008), but there was little detailed information as to hesC effects on the tbrcis-regulatory system. In addition cis-regulatory mutations as well as other evidence indicated that some member(s) of the Ets family of transcription factors are required for tbr expression (Fuchikami et al., 2002; Ochiai et al., 2008). On the other hand, it had also been reported that morpholino-substituted antisense oligonucleotides (MASO) directed against the S. purpuratus Ets family members SpErg, SpEts1/2, and SpTel had no very significant effect (i.e., caused <3-fold change) on tbr expression up to 24hpf (Oliveri et al., 2008). The role of Ets factors in tbr regulation altogether was clearly in need of further investigation. An additional mystery was that by late mesenchyme blastula stage hesC expression disappears from the non-skeletogenic mesenchyme (NSM) (Smith and Davidson, 2008b), and ets expression spreads to include the NSM (Rizzo et al., 2006); yet tbr expression does not expand, remaining confined to the SM. Thus there appeared to be a need for either an additional yet unidentified NSM repressor of tbr expression, or a spatially-dedicated SM activator of tbr in later stages.
These issues are resolved in the cis-regulatory analyses described in this paper. The approach we have taken differs from the conventional in that we have attempted to examine cis-regulatory modular function in the context of the complete genomic tbr locus. To this end we utilized recombineered BAC reporters bearing module deletions or site mutations. Thus we have been able to establish the sequence of module deployment as well as determine the functionality of key transcription factor target sites. Perhaps not surprisingly, some of the insights we obtained as to module function in context proved invisible from the vantage point of the usual minimal expression constructs.
Materials and methods
BAC homologous recombination
Deletions of the γ(2), B, and C modules from an SpTbrain GFP knock-in BAC (Damle et al., 2006) by homologous recombination were performed as described by Lee et al. (2007). The parental BAC is referred to as tbr::GFP BAC in the following. To produce a targeting cassette with homology to the regions bordering each module, a kanamycin resistance gene flanked by frt sites was amplified with the following primer pairs:
Δγ (2) module-Forward: 5′-GACATAGGTATTTCCTTATACATCGTCATGATTATGGTTACACTCTCTAGATAACTGATCAGCTT-3′
Δγ (2) module-Reverse: 5′-ATATATCTATAATTATATGGAATAAATTCCATGAAATCTCATGTGGAGCTATTCCAGAAGTAGTGA-3′
ΔB module-Forward: 5′-GGTAGTCACAAAGCCCAAATACCTTACAAGCTCCTCTTTTATGTCGGAGTATCTTAAGTACTCTTTGTAAAGCTGTCTAATTTTCCTGATTCTAGATAACTGATCAGCTT-3′
ΔB module-Reverse: 5′-AAATTCGTACGTTACTTTGAAATGAACCGACAATGCGGATTATAAGAGCTATTCCAGAAGTAGTGA-3′
ΔC module-Forward: 5′-CAGCTTAGGCACTTTAACAAAAAAAGAGTCTTTAGAATTCTTTGATCTAGATAACTGATCAGCTT-3′
ΔC module-Reverse: 5′-GAGCAAATCCTACATGATATCTACAGACATCATCAGATGCTTCAGGAGCTATTCCAGAAGTAGTGA-3′
Underlined sequences are homologous to the targeting cassette. Correct integration of the cassette into tbr::GFP BAC was confirmed by sequencing and diagnostic PCR. After removal of kanR by induction of flippase, a 125bp artifact of the cassette remained in the former location of each module.
To avoid undesired flippase recombination with a frt site at the GFP insertion site, mutation of the HesC binding site on tbr::GFP BAC was performed using a GalK positive/negative selection method (Warming et al., 2005). A targeting cassette containing galK was amplified with the following primers to introduce homology to the region flanking the HesC binding site:
HesCmut-cassette-Forward: 5′-CAGACTATTTTTTCTTCTTCGTCGTCGTCTAAATGTTATTTCGAGTCGCCTGTTGACAATTAATCATC-3′
HesCmut-cassette-Reverse: 5′-GGGCTACCAGACAATGGAGAGTCGCGCGTTGATTGGCCGCCAGGGAGGTCAGCACTGTCCTGCTCCTT-3′.
Underlined sequences are homologous to the targeting cassettes. After proper insertion was confirmed by sequencing and diagnostic PCR, the cassette was replaced with the mutated HesC site through homologous recombination with the following annealed oligonucleotides:
HesCmut-cassette-removal-Forward: 5′-CAGACTATTTTTTCTTCTTCGTCGTCGTCTAAATGTTATTTCGAGTCGGACTCCTCCCT GGCGGCCAATCAACGCGCGACTCTCGATTGTCTGGTAGCCC-3′
HesCmut-cassette-removal-Reverse: 5′-GGGCTACCAGACAATCGAGAGTCGCGCGTTGATTGGCCGCCAGGGAGGAGTCCGAC TCGAAATAACATTTAGACGACGACGAAGAAGAAAAAATAGTCTG-3′.
Generation of cis-regulatory reporter constructs
The γ(2)::EpGFP construct, in which the γ(2) regulatory region drives GFP expression from the endo16 basal promoter (Yuh and Davidson, 1996; Yuh et al., 1996), was produced by fusion PCR (Yon and Fried, 1989). The γ(2) fragment was amplified from SptbrBAC (clone 31;J08 from arrayed library) using γ(2)-Forward and γ(2)-EpGFP-Reverse primers. EpGFP was amplified from the EpGFPII expression vector (Cameron et al., 2004) using γ(2)-EpGFP-Forward and GFP-Reverse primers. The fusion product was amplified using both fragments and the γ(2)-Forward and GFP-Reverse primers. The resulting fragment was cloned into Promega pGEM-T Easy vector (Catalog #A1360) and fully sequenced to confirm proper fusion. The γ(2)::EpGFP reporter construct was then amplified from the plasmid using γ(2)-Forward and GFP-Reverse to produce linear fragments for injection.
γ(2)-Forward:5′-GTCTCTAGCAAGATATGTTACT-3′
γ(2)-EpGFP-Reverse: 5′-ACAGTTTAACCCGGGAGATCTACTCTATAAACCACTACTGTACTCTA-3′
γ(2)-EpGFP-Forward: 5′-TAGAGTACAGTAGTGGTTTATAGAGTAGATCTCCCGGGTTAAACTGT-3′ GFP-Reverse:5′-ACTGGGTTGAAGGCTCTCAA-3′
The Stratagene QuikChange Mutagenesis Kit (catalog #200518) was used to mutate or delete putative transcription factor binding sites on the γ(2)::EpGFP plasmid. The resulting plasmids were sequenced to confirm introduction of the mutation. The following primer pairs were used to produce the otxmut γ(2)::EpGFP and complex Δγ(2)::EpGFP plasmids:
Otxmut γ(2)::EpGFP-Forward: -CTGGTGATCGGTCAACTGATTCCTTCCGGTTGGACGTGAA-3′
Otxmut γ(2)::EpGFP-Reverse: 5′-TTCACGTCCAACCGGAAGGAATCAGTTGACCGATCACCAG-3′
Complex Δγ(2)::EpGFP-Forward: 5′-TGTGCGTGCTTTACACCTGTCTGGTGATCG-3′
Complex Δγ(2)::EpGFP-Reverse: 5′-CGATCACCAGACAGGTGTAAAGCACGCACA-3′
Complex Δγ (2)::EpGFP-Reverse: 5′-CGATCACCAGACAGGTGTAAAGCACGCACA-3′. The mutated clones were checked by sequencing.
Ets and bHLH binding site mutations were introduced into γ(2)::EpGFP by fusion PCR. “Left” fragments (produced using γ(2)-Forward and the mutation's reverse primer) and “right” fragments (amplified with GFP-Reverse and the mutation's forward primer) were mixed to produce a megaprimer template for fusion PCR with γ(2)-Forward and GFP-Reverse primers. The resulting full-length fragment was gel-purified and ligated into pGEM-T Easy vector for sequencing and amplification. The etsmut1+2 γ(2)::EpGFP construct was produced using etsmut1 γ(2)::EpGFP as a template for fusion PCR with the etsmut2 mutation primers.
Etsmut1-Forward: 5′-GTCATTGACCTCAGATAGTCTGGTGATCG-3′
Etsmut1-Reverse: 5′-TCACCAGACTATCTGAGGTCAATGACCGCTT-3′
Etsmut2-Forward: 5′-CGAAGTTCACGTCCAAGAATCTGGGATTAGTT-3′
Etsmut2-Reverse: 5′-GTCAACTAATCCCAGATACTTGGACGTGAA-3′
bHLHmut-Forward: 5′-CATTGACCTCTTCCTGCTACATGATCGGTCA-3′
bHLHmut-Reverse: 5′-AGTTGACCGATCATGTAGCAGGAAGAGGT-3′
The γ(2) module was located by reiterative reporter assays as described (Smith and Davidson, 2008a). εδγβα::GFP was produced by fusion PCR between the 5′ intergenic region of tbr (amplified from SptbrBAC using TbrA-Forward and TbrA-Reverse primers) and GFP amplified with primers homologous to the basal promoter of tbr (εδγβα-GFP-Forward and GFP-Reverse). εδγβα::GFP was obtained by the same scheme using a different GFP forward primer, εδγβα-GFP-Forward. PCR fragments were cloned into pGEM-T Easy vector and sequenced. The εδγβα::GFP, γβα::GFP, βα::GFP, and α::GFP reporter constructs were produced from εδγβα::GFP using GFP-Reverse and the corresponding forward primers. γα::GFP, γ::EpGFP, and γ(2)α::GFP were generated by fusion PCR using an analogous method.
TbrA-Forward: 5′-GGAACGATACGAAAACTTTG-3′
TbrA-Reverse:5′-CTTAGGACCGTGTTATATAC -3′
εδγβα-Forward:5′-CAGACAATCTAGATTGCCTA-3′
γβα-Forward:5′-TATAGGACCGTGTTATATACCTC-3′
βα-Forward:5′-TATGTGTGCATGACTTTGCTT-3′
α-Forward: 5′-AGATGGTTATTCTTCCAGACTA-3′
Shortened fragments of γ(2)::EpGFP were produced by PCR amplification of γ(2)::EpGFP using GFP-Reverse as a reverse primer and the following forward primers: γ(2.1)-Forward, γ(2.2)-Forward, γ(2.3)-Forward, γ(2.4)-Forward. These fragments were cloned into pGEM-T Easy vector and sequenced. The reporter fragment γ(2.2-3)::EpGFP was produced by fusion PCR between the amplified region between the primers γ(2.2)-Forward and γ(2.3)-Forward (using γ(2.2)-Forward and γ(2.3)-Reverse) as well as amplified EpGFP with homology to γ(2.3) produced by amplification with GFP-Reverse and γ(2.3)-EpGFP-Forward.
γ(2.1)-Forward:5′-CATTTATTCGATCATCGA-3′
γ(2.2)-Forward:5′-TGCTTTACAGTGATAACA-3′
γ(2.3)-Forward:5′-TTGGACGTGAACTTCGA-3′
γ(2.4)-Forward:5′-CCATATAATTATAGATATATGA-3′
γ(2.3)-Reverse:5′-GGGAGATCTACTCCGGAAGGGATTAG-3′
γ(2.3)-EpGFP-Forward:5′-CCTTCCGGAGTAGATCTCCCGGGT-3′
Quantitative PCR
Embryos injected with recombineered BACs or reporter constructs were collected at the indicated timepoints. DNA and RNA were extracted using the Qiagen AllPrep DNA/RNA Mini kit (catalog #80204). Reverse transcriptase PCR was performed on the extracted RNA using the Biorad iScript cDNA synthesis kit (catalog #170-8890). BAC/reporter construct incorporation number and expression level were quantified by quantitative real-time PCR performed on extracted DNA and cDNA, respectively (Revilla-i-Domingo et al., 2004). The single-copy gene foxA and two genes of well-characterized expression, Spz12 (Wang et al., 1995) and ubiquitin (ubq) (Oliveri et al., 2002; Ransick et al., 2002), were also quantified for comparison. The number of transcripts per embryo was determined by multiplying the fold difference in construct expression level (relative to Spz12 or ubq) by the number of Spz12 or ubq transcripts present at that timepoint, adjusting for GFP construct incorporation relative to foxA (Materna and Oliveri, 2008). Spz12 and ubq standardizations gave consistent results; graphs shown are standardized relative to Spz12. The QPCR primers used are available online at: http://sugp.caltech.edu/SUGP/resources/methods/q-pcr.php.
Culture, microinjection, and fluorescence visualization
Culture and microinjection were performed as described (Flytzanis et al., 1985; McMahon et al., 1985) with the following modifications: eggs were not filtered prior to dejellying, and no BSA was added to dejellied eggs. Zygotes were injected with 10pL of solution containing 150 molecules/pL of reporter construct or 40 molecules/pL of BAC and 120 mM KCl. HindIII fragment carrier DNA (4nM) was added to injection solutions containing small reporter constructs. All BACs were linearized with AscI prior to injection.
Translation and splice-blocking morpholino antisense oligonucleotides (MASO) were designed by GeneTools. For coinjections, MASO was added to the injection solution at the indicated concentrations. Embryos injected with a randomized mixture of morpholinos (IUPAC sequence: N25) served as a mock-knockdown control.
| Elk trans MASO:5′CGCTTCCGACATTGTGATGATTCTG-3′ | 400μM | |
| Ets1/2 trans MASO:5′-GAACAGTGCATAGACGCCATGATTG-3′ | 500μM | |
| Ets4 splicing MASO: 5′- GCAAACTTCGCCAGTTGAGAACATG -3′ | 400μM | |
| Erg trans MASO*:5′-GCATATAACAAATTGAGGAACACTG-3′ | 200μM | |
| Erg splicing MASO*:5′-GGCCACTTCCTGCAAAAACGAAC-3′ | 200μM | |
| HesC trans MASO:5′-GTTGGTATCCAGATGAAGTAAGCAT-3′ | 500μM | |
| Tel trans MASO:5′-CCTGTCTGGTAGAGGCCGGGTCCAT-3′ | 400μM |
Equal amounts of Erg trans and splicing MASOs were combined for Erg MASO injections (Oliveri et al., 2008).
pmar1 and ets1/2 mRNA were obtained by plasmid transcription as described (Oliveri et al., 2002). Injection solution for mRNA co-injections contained 200ng/μL ets1/2 mRNA or 10ng/μL pmar1 mRNA. The final concentration of injected transcript did not exceed the maternal (for ets1/2, (Rizzo et al., 2006) or early blastula (for pmar1, (Oliveri et al., 2002) transcript number by more than tenfold, as recommended to maintain binding specificity (Materna and Oliveri, 2008).
GFP expression pattern was evaluated at the indicated timepoints on an epifluorescence Axioscope 2 Plus microscope (Zeiss, Hallbergmoos, Germany). Images were recorded with an AxioCam MRm (Zeiss) and fluorescence overlays produced in Adobe Photoshop CS 3.
Electrophoretic mobility gel shift assays and probe preparation
Gel shifts were performed using 12h embryonic nuclear extract as described (Yuh et al., 1994). Double-stranded oligonucleotides were annealed and 32P-labeled with Klenow DNA polymerase by the end-fill reaction. Underlined sequence represents overhang serving as a template for Klenow labeling.
H-Forward:5′-GAGAGCCCCTGTGCGTGCTTTACAGTGATAACAC-3′
H-Reverse:5′-GAGAGTGTTATCACTGTAAAGCACGCACAGGGGC-3′
I-Forward:5′-GAGAGTGCTTTACAGTGATAACACAAAGCGGTCA-3′
I-Reverse:5′-GAGATGACCGCTTTGTGTTATCACTGTAAAGCAC-3′
J-Forward:5′-GAGAGTGATAACACAAAGCGGTCATTGACCTCTT-3′
J-Reverse: 5′-GAGAAAGAGGTCAATGACCGCTTTGTGTTATCAC-3′
K-Forward: 5′-GAGAAAAGCGGTCATTGACCTCTTCCTGTCTGGT-3′
K-Reverse:5′-GAGAACCAGACAGGAAGAGGTCAATGACCGCTTT-3′
L-Forward:5′-GAGATTGACCTCTTCCTGTCTGGTGATCGGTCAA-3′
L-Reverse:5′-GAGATTGACCGATCACCAGACAGGAAGAGGTCAA-3′
M-Forward: 5′-GAGACCTGTCTGGTGATCGGTCAACTAATCCCTT-3′
M-Reverse:5′-GAGAAAGGGATTAGTTGACCGATCACCAGACAGG-3′
N-Forward:5′-GAGAGATCGGTCAACTAATCCCTTCCGGTTGGAC-3′
N-Reverse:5′-GAGAGTCCAACCGGAAGGGATTAGTTGACCGATC-3′
O-Forward:5′-GAGACTAATCCCTTCCGGTTGGACGTGAACTTCG-3′
O-Reverse:5′-GAGACGAAGTTCACGTCCAACCGGAAGGGATTAG-3′
P-Forward:5′-GAGACCGGTTGGACGTGAACTTCGACCGCTGGTT-3′
P-Reverse:5′-GAGAAACCAGCGGTCGAAGTTCACGTCCAACCGG-3′
Results
Spatial and temporal expression pattern of recombineered tbr::GFP BAC
Abundant and ubiquitously distributed maternal transcript obscures the early zygotic expression pattern of the endogenous tbr gene. To visualize zygotic transcription we used a recombinant BAC, in which the coding region of GFP had been inserted at the start codon of tbr exon1 (Damle et al., 2006). Figure 1B shows an expression time-course generated by quantifying GFP transcripts produced by this expression construct, tbr::GFP BAC, in embryos collected at 6-48 hours after fertilization (hpf) and injection. GFP transcript number was normalized to the number of BAC DNA molecules incorporated per embryo. This was determined in QPCR measurements by comparing the incorporated genomic GFP coding sequence content to that of a known single copy gene, foxa.
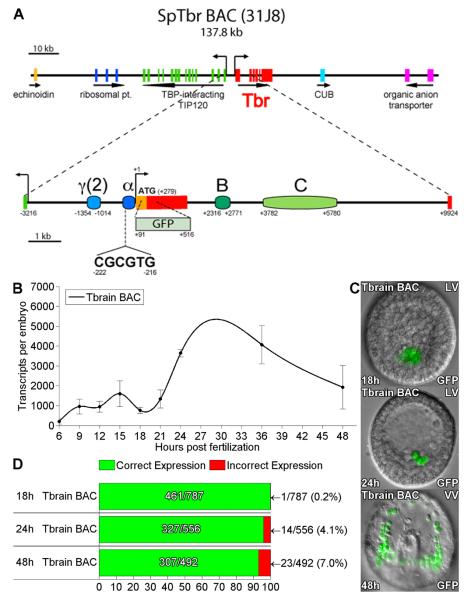
Fig. 1.
Accurate skeletogenic expression of tbr::GFP BAC. (A) Map of the gene-rich tbrain BAC. The GFP coding sequence was recombined into tbr exon 1 as previously described (Damle et al, 2006). Positions of relevant cis-regulatory modules are indicated relative to the transcription start site (bent arrow) and exons (red boxes). (B) QPCR measurements of GFP mRNA in embryos bearing tbr::GFP BAC, 6-48hpf. Transcript levels were normalized to measured BAC molecules in each sample, in this and all subsequent time-courses shown (see Methods). Error bars indicate SEM in repetitions of the same experiments. (C) GFP fluorescence image overlays of tbr::GFP BAC-injected embryos at 18, 24, and 48 hpf. Expression is limited to the skeletogenic cells at all stages: LV, lateral view; VV,ventral view. (D) Summary of spatial expression statistics. Green and red bars indicate the fraction of embryos expressing GFP that showed fluorescence restricted to the SM cells (correct expression) vs. partially or completely ectopic fluorescence (incorrect expression), respectively. The total number of embryos injected is given in the denominators of these fractions.
Expression begins between 6 and 9hpf, coincident with the disappearance of transcript encoding HesC, the predicted tbr repressor, from the micromeres between 8 and 12 hpf (Revilla-i-Domingo et al., 2007). There were ~1000 GFP transcripts/embryo between 9 and 21 hpf, increasing three-fold by 24 hpf, and remaining high at 36 and 48 hpf. This pattern of expression is consistent with previous time-courses for endogenous tbr transcript (Oliveri et al., 2008); and additional unpublished data). The spatial expression pattern of tbr::GFP BAC was visualized in injected embryos by fluorescence microscopy at the blastula (18hpf), mesenchyme blastula (24 hpf), and late gastrula (48 hpf) stages, as illustrated in Fig. 1C. Expression was highly specific to the SM lineage; the percentage of injected embryos displaying fluorescence anywhere else was ≤ 7% at all stages, and essentially zero at 18 h (Fig. 1D). The tbr::GFP BAC construct recapitulates both the spatial and temporal expression pattern of the endogenous gene with high fidelity.
The tbr gene is strikingly up-regulated by pmar1 mRNA injection (Oliveri et al., 2002) and by hesC morpholino antisense oligonucleotide (MASO) injection (Revilla-i-Domingo et al., 2007), as required by the double negative gate architecture. So indeed is the tbr::GFP BAC. Embryos coinjected with this construct and with pmar1 mRNA, with hesC MASO, or with a random (N) MASO control were visualized by fluorescence microscopy at 18, 24, and 48 hpf. Both pmar1 mRNA and hesC MASO injection resulted in increased amount of expression and grossly ectopic fluorescence relative to the control (Fig. S2A,C,E; Table S1). The tbr::GFP construct thus includes the genomic sequence required for these known regulatory inputs into the gene.
Ectopic GFP expression following HesC binding site mutation
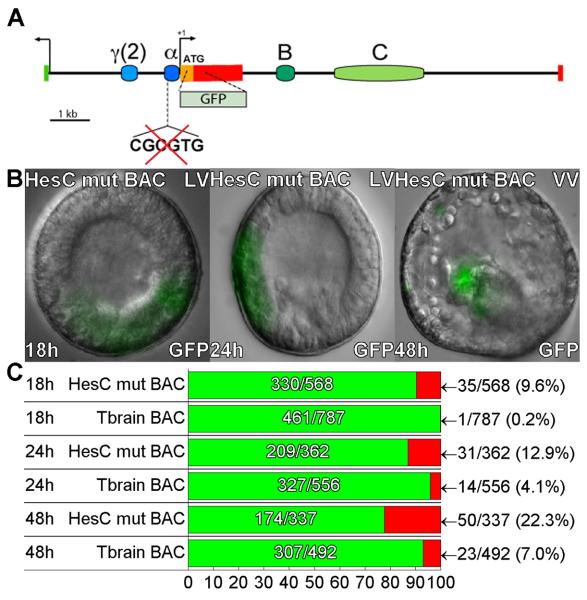
A class C bHLH factor binding site (Iso et al., 2003) near the basal promoter is necessary for repression of a Hemicentrotus tbr construct outside of the SM territory (Ochiai et al., 2008), and was thought to be a binding site for the HesC repressor implicated by gene network analysis in the control of tbr spatial expression in S. purpuratus (Oliveri et al., 2002; Revilla-i-Domingo et al., 2007). This sequence (CGCGTG) is conserved in the S. purpuratus tbr gene at −222 −217 relative to the transcription start site (Fig. 2A). To determine whether mutation of this single site would suffice to induce ectopic expression in the complete genomic context, a 4bp mutation was introduced on the tbr::GFP BAC by means of homologous recombination. The mutation resulted in a significant increase in ectopic GFP expression relative to the tbr::GFP BAC control, while GFP expression in the SM lineage was unaffected (Fig. 2B,C). However, GFP misexpression was observed in only 10%, 13%, and 23% of embryos at 18h blastula, 24h mesenchyme blastula, and 48h prism stages. This suggested that there could be additional undiscovered HesC sites: thus, by comparison, pmar1 mRNA, which works by shutting down hesC expression, produced 49% ectopic expression by mesenchyme blastula stage, and the hesC MASO treatment used in these particular experiments 24% (Table S1). Computational analysis of the whole tbr regulatory apparatus identifies several other potential HesC sites here not investigated; however, most of these lie in non-conserved regions of the sequence. Alternatively, this difference in misexpression rate caused by the mutation and that caused by pmar1 mRNA and hesC MASO could be due to an indirect effect: both pmar mRNA and hesC MASO injection cause the ectopic expression of ets1/2, an activator of tbr (see Discussion). In addition, we note that in a MASO injection the antisense oligo must be in excess to block the translation of the continuously transcribed hesC, which is not always attained, while the pmar1 MOE produces enough transcriptional repressor to completely turn off the hesC gene.
Fig. 2.
Ectopic expression of tbr::GFP BAC following mutation of a HesC site. (A) Map of tbr locus. The location of the HesC binding site (CGCGTG) in the α region is indicated. (B) Examples of ectopic expression in ectoderm, endoderm, and NSM of tbr::GFP BAC in which this site had been mutated, at 18, 24, and 48hpf. (C) Expression statistics for mutant and control BAC constructs as in figure 1D.
Deletion of conserved intronic regions from tbr::GFP BAC
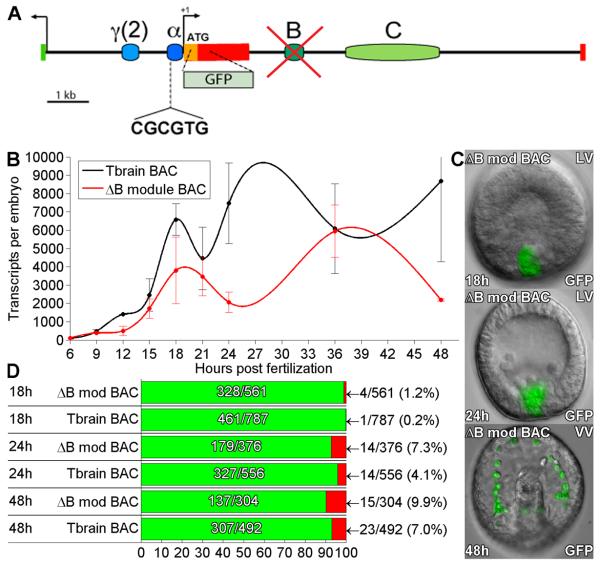
Ochiai et al. (2008) reported that Snail family consensus binding sites in a conserved intronic cis-regulatory module were necessary for repression of ectopic expression in a Hemicentrotus tbr reporter construct. The corresponding region, here identified as the B module (Fig. 3A), was deleted from the S. purpuratus tbr::GFP BAC by homologous recombination. Quantification of GFP transcripts revealed no very significant differences in temporal expression pattern in the ΔB module BAC relative to the control, though there may be a transient depression of the level of activity soon after ingression (Fig. 3B). More importantly, there was no change whatsoever in the accuracy of expression caused by deletion of B module (Fig. 3C). Thus in S. purpuratus, the putative Snail binding site of B module has no detectable repressive spatial function when measured in complete genomic context.
Fig. 3.
Effects of deletion of B module from tbr::GFP BAC. (A) Map of tbrain locus. The B module was deleted by recombination (see Methods). (B) QPCR timecourse of GFP mRNA levels generated by the B module deletion, and by the parental tbr::GFP BAC. In this and the following comparisons of diverse constructs data were obtained in experiments in which both constructs were injected into the same batches of embryos; i.e., the controls of each set of experiments were those of that comparison. Transcript levels were not adjusted for DNA incorporation rate of each construct. Error bars indicate SEM. No very large differences in expression level are observed in the B module deletion. (C) Examples of GFP fluorescence image overlays showing embryos expressing the B module deletion, at 18, 24, and 48hpf. Expression is confined to SM cells. (D) Expression statistics for mutant and control BAC constructs as in figure 1D.
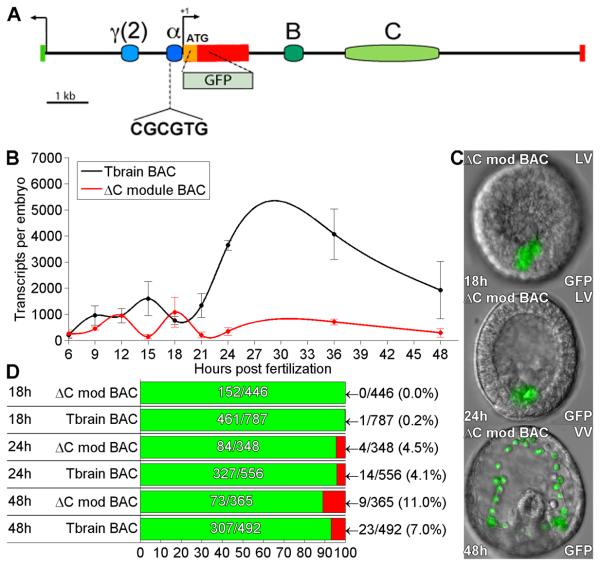
An additional conserved region in the first intron of the Tbrain gene was identified as an enhancer in Hemicentrotus (Ochiai et al., 2008). When this region, here the C module, was deleted from the tbr::GFP BAC (Fig. 4A), a very significant decrease in GFP transcript levels was observed at all time-points examined (Fig. 4B). Although the analogous deletion from a 7kb HpTbrain reporter construct caused an increase in ectopic expression (Ochiai et al., 2008), we could detect no difference in the amount of ectopic expression produced by the ΔC module BAC vs. the control tbr::GFP BAC (Fig. 4C,D). Thus in S. purpuratus, C module in the context of the complete system appears to act as a quantitative enhancer of expression, but is not required for spatial accuracy of expression.
Fig. 4.
Effects of deletion of C module from tbr::GFP BAC. (A) Map of tbrain locus. The C module was deleted by recombination (see Methods). (B) QPCR timecourse of GFP mRNA levels generated by the C module deletion, and by the parental tbr::GFP BAC. Sharply decreased expression is observed relative to the control tbr::GFP BAC. Error bars indicate SEM of repeated experiments. (C) Examples of GFP fluorescence image overlays showing embryos expressing the B module deletion, at 18, 24, and 48hpf. (D) Expression statistics for mutant and control BAC constructs as in figure 1D.
γ(2) module drives expression after ingression of the SM cells
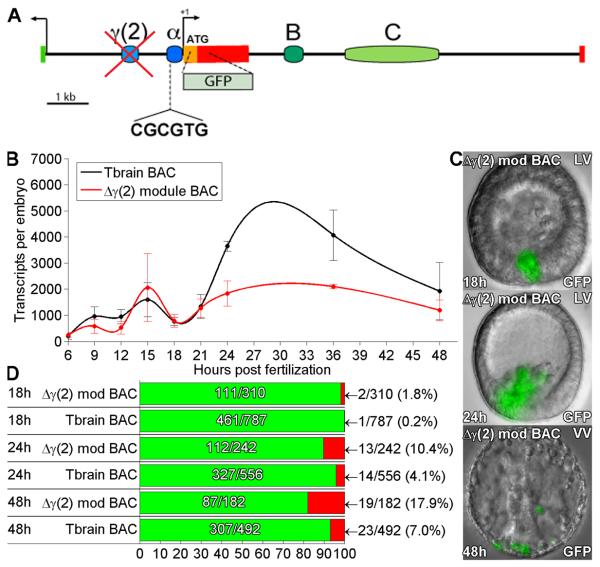
A novel cis-regulatory module, γ(2), which also mediates skeletogenic expression, was identified in the 5′ intergenic region of the tbr locus (Fig.5A). It was found by means of iterative deletions from a large expression construct that included the whole intergenic region between tbr and the next gene upstream (Fig. S1). Successive deletions and results are shown in Fig. S3 and Table S2. To determine the function of γ(2) module in the context of the whole genomic regulatory system, this module was specifically deleted from the tbr::GFP BAC by homologous recombination. Study of the expression of this deletion construct revealed that it is expressed quite normally temporally and spatially until the time of ingression, but between 24 and 48h a major decrease in expression levels is seen; this result is shown in Fig. 5B-D. In addition the γ(2) deletion produced a minor but significant increase in ectopic expression during this period, typically in the non-skeletogenic mesoderm. Thus in genomic context, γ(2) module functions after ingression. Since as shown in Fig. 4 C module also functions during this period, we conclude that these two non-contiguous cis-regulatory modules collaborate in generating the definitive expression of the tbr gene in differentiated skeletogenic cells.
Fig. 5.
Effects of deletion of γ(2) module from tbr::GFP BAC. (A) Map of tbrain locus. The γ(2) module was deleted by recombination (see Methods). (B) QPCR timecourse of GFP mRNA levels generated by the γ(2) module deletion, and by the parental tbr::GFP BAC. Significantly decreased expression is seen after 18hpf. . Error bars indicate SEM of repeated experiments. (C) Examples of GFP fluorescence image overlays showing embryos expressing the γ(2) module deletion, at 18, 24, and 48hpf. Increased ectopic GFP fluorescence was observed in the NSM cells of embryos bearing this deletion at 48hpf, but not at 18hpf. (D) Expression statistics for mutant and control BAC constructs as in figure 1D.
Expression of a short γ(2) module construct lacking any other regulatory sequence
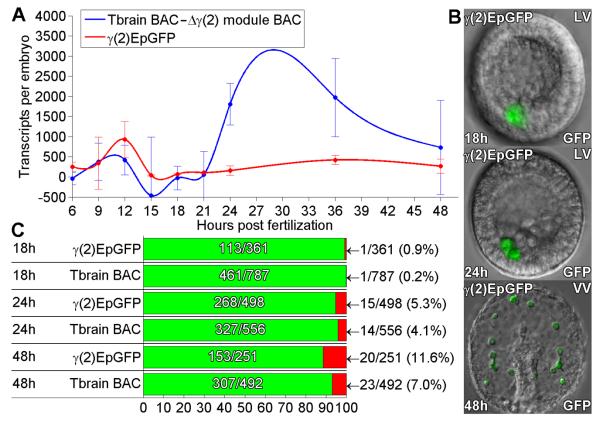
A standard minimal expression construct was created by fusing the γ(2) module (Fig. 6 and S3) to the endo16 basal promoter::GFP reporter (construct “γ(2)::EpGFP”). On its own this basal promoter has no specific intrinsic spatial or temporal regulatory activity, but it mediates transcription in any domain of the embryo if provided with an exogenous cis-regulatory module active in that domain (Yuh and Davidson, 1996). In a head-to-head comparison the short γ(2)::EpGFP construct is expressed just as accurately as is tbr::GFP BAC (Fig. 6B,C). We then compared the quantitative expression of this construct across developmental time to that of the tbr::GFP BAC from which γ(2) module had been deleted, as for the experiments of Fig. 5. The simplest case we can consider is that the activity of the whole system is just the sum of the activities of its individual cis-regulatory modules. In this case the activity of the short construct should match the calculated difference between the activities of the tbr::GFP BAC and the tbr::GFP γ(2) deletion BAC. This comparison is plotted in Fig.6A.
Fig. 6.
Expression of a minimal γ(2) module construct. A short construct consisting of the isolated γ(2) module associated with our standard endo16-GFP expression vector was constructed as described in Methods (construct γ(2)::EpGFP). (A) QPCR timecourse of GFP mRNA levels generated by γ(2)::EpGFP (red), and co-plotted with the calculated difference between the timecourse produced by the control tbr::GFP BAC and that produced by tbr::GFP BAC from which γ(2) module had been deleted (blue). All data are after normalization to the numbers of copies of the respective vectors incorporated at each timepoint, as above. . Error bars indicate SEM. (B) Examples of GFP fluorescence image overlays showing embryos expressing γ(2)::EpGFP, at 18, 24, and 48hpf. (C) Expression statistics for γ(2)::EpGFP together with control tbr::GFP BAC as in figure 1D.
There are two interesting aspects of the result. First, and most obviously, γ(2)::EpGFP does not generate nearly as much activity per incorporated construct, in the period after 24 h, as is lost from the complete system when the γ(2) module is deleted. To test whether this might be due to the exogenous endo16 promoter used in this construct, we generated a construct in which the γ(2) module was associated only with the endogenous tbr promoter, denoted in the maps shown in Fig. S3 as “α” (construct “γ(2)α::GFP”). This construct was expressed spatially with the same accuracy as γ(2)::EpGFP, and quantitatively at exactly the same level (Table S2; Fig. S3). Promoter strength or identity is therefore not the explanation for the weak expression per incorporated molecule of the short construct. There is some other reason, as discussed below, that the short construct functions far less efficiently in isolation than does the very same cis-regulatory module in context.
The second interesting aspect of the comparison in Fig. 6A is that in the period earlier than 21 h, the short construct is expressed at the same level, and also in the same skeletogenic cells as is tbr::GFP BAC. In other words, in the context of the whole system, Fig. 5B shows that γ(2) module plays no role whatsoever prior to ingression, but in isolation, as shown in Fig. 6A, it is capable of generating apparently normal spatial expression prior to ingression.
Given its accurate expression, we tested whether γ(2)::EpGFP would respond similarly to tbr::GFP BAC in perturbations of the upstream regulators. And indeed, injection of both pmar1 mRNA and hesC MASO caused gross ectopic expression of the γ(2)::EpGFP construct (Fig. S2; Table S1).
Ets family transcription factors regulate γ(2) module
To identify the transcriptional activator(s) of the γ(2) module, and to determine whether HesC is a direct or indirect regulator, a gel shift analysis was carried out using nuclear extract from 12 h embryos. We found a 71 bp subregion of γ(2) module (Fig. 7A) which drove GFP expression specifically in the SM, though less strongly than does the full γ(2) module when incorporated in an expression construct (γ(2.2-3)::EpGFP; Fig. S3a; Table S3). As Fig. 7B shows, there are three putative kinds of DNA-protein complex in this region, which are found respectively in oligonucleotides containing Ets family consensus binding sites (Consales and Arnone, 2002), oligonucleotides containing an Otx family consensus binding site (Mao et al., 1994), and an oligonucleotide that included a 30bp upstream region which produced an unresolved additional set of complexes. The activities of the γ(2)::EpGFP construct and of derivatives in which each of these putative binding sites were mutated are given in the chart in Fig. 7C. Mutation of the putative Otx binding site had minor effect (from 38.4% in WT to 29.1% when mutated), while deletion of the 30 bp sequence (which partially overlapped an Ets binding site) decreased the level of GFP expression and the number of injected embryos visibly expressing GFP. Mutation of either Ets binding site significantly reduced the number of GFP-expressing embryos, more strongly for site 1 than for site 2, and when both Ets binding sites were mutated, GFP expression was abolished. But none of these mutations produced any ectopic expression (e.g., Fig. S4a-g). Although no corresponding DNA-protein complex was observed, a consensus bHLH binding site in this region was also considered as a candidate HesC binding site. However, mutation of this site in γ(2)::EpGFP affected neither quantitative nor ectopic expression (Fig. 7c; Fig. S4g).
Fig. 7.
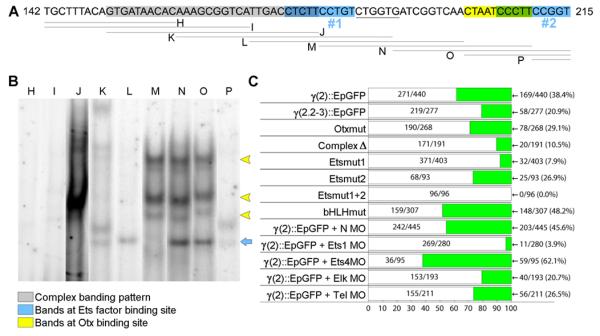
Transcription factor binding sites in a subregion of γ(2) module. (A) Map of a 71bp subregion of γ(2) module, showing the positions of oligonucleotides (H-P) used in the electrophoretic gel mobility shift assay. The inferred locations of putative target sites are indicated: Ets, blue; Otx, yellow; a possible bHLH site is underlined. (B) Gel shift results with oligonucleotides (H-P). Similar banding patterns are observed on oligonucleotides containing a putative Otx binding site (M-O, yellow arrowheads) and Ets factor binding sites (K-L and N-O, blue arrows). (C) Summary of expression results obtained with indicated site mutations and MASO treatments. Data are for 24h embryos. All constructs, if they expressed at all, expressed accurately, and only quantitative expression results are shown. Embryos with ectopic GFP expression were rare and omitted from this figure for clarity; a full tally is provided in Table S.3. Mutation of the putative Otx site had a minor effect and mutation of the bHLH sites had no effect on expression of γ(2)::EpGFP. However, mutation of either Ets binding site or deletion of the 30bp region overlapping an Ets site (gray in (A)) dramatically decreased the number of embryos expressing GFP without affecting the spatial expression pattern. Mutation of both Ets sites eliminated almost all expression. Coinjection of γ(2)::EpGFP with a random MASO (N MO) or MASOs directed against Ets family members Ets4 or Tel had no effect on expression, but −ets1/2 MASO and elk MASO dramatically decreased the number of embryos expressing GFP.
There are five genes of the Ets family expressed in the SM by mesenchyme blastula stage, viz. erg, ets1/2, ets4, elk, and tel (Kurokawa et al., 1999; Rizzo et al., 2006). MASO directed against each of these Ets family members was co-injected with γ(2)::EpGFP. The results, also summarized in Fig. 7C, reveal that Ets1/2 (and possibly Elk, which had a weak effect) are required for normal levels of expression of γ(2)::EpGFP. This raised the possibility that the spatial control of this short construct by HesC could be indirect, since the ets1/2 gene is itself controlled by the pmar1/hesC double negative gate. To test this, ets1/2 mRNA was co-injected with γ(2)::EpGFP or with tbr::GFP BAC. There was a striking difference in the early expression (18hpf) outcome: γ(2)::EpGFP was now expressed ectopically all over the embryo but the tbr::GFP BAC was not (Fig. S2g,h: Table S1). Thus the complete system encompassed in the tbr::GFP BAC is subject to dominant repression by HesC as shown above, whereas the short construct is regulated only by Ets1/2. In contrast, at later stages, when the γ(2) module is functional, both tbr::GFP BAC and γ(2)::EpGFP are ectopically expressed in ets1/2 mRNA co-injection. This distinction in behavior excludes the possibility that γ(2) module is literally redundant with the rest of the regulatory system.
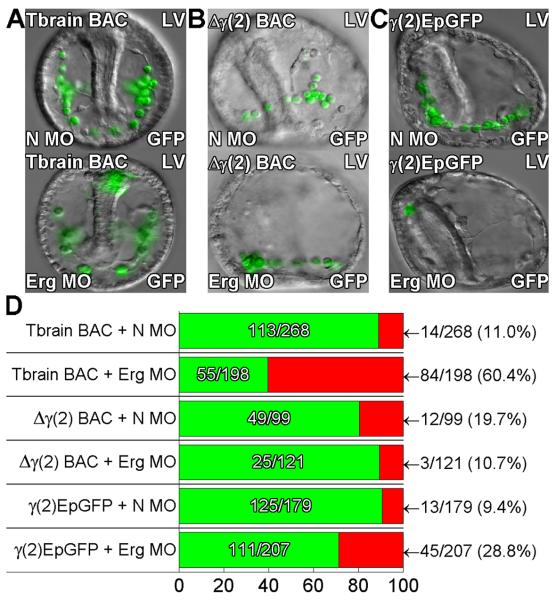
An unexpected and important result of these MASO experiments was that introduction of erg MASO caused expansion of expression of both tbr::GFP BAC (Fig. 8A,D) and γ(2)::EpGFP (Fig. 8C,D) into the NSM at 48hpf. However, the tbr::GFP BAC construct from which the γ(2) module had been deleted (Fig.8b,d) was immune to this effect. Thus another late role of the γ(2) module in the whole system is revealed: this function is to suppress transcription of the tbr gene in NSM in the gastrula stage embryo, a role necessitated by the expansion of ets1/2 expression to the NSM by this stage.
Fig. 8.
Effect of erg MASO on expression of recombinant tbr BACs including and lacking γ(2) module. (A) Examples of accurate spatial expression of control tbr::GFP BAC in presence of random MASO but ectopic expression in NSM cells in presence of erg MASO. (B) Example of accurate expression of tbr::GFP BAC from which the γ(2) module had been deleted even in presence of erg MASO. (C) Effects of erg MASO similar to those in (A) are obtained with γ(2)::EpGFP. (D) Spatial expression statistics for these experiments.
Discussion
The tbr gene lies at an essential node, high in the gene regulatory network subcircuit which establishes the initial lineage specific regulatory state of the future skeletogenic mesoderm (SM) (Oliveri et al., 2008). Network analysis predicts the key features of the genomic cis-regulatory code determining the transcriptional activity of this gene, and an initial motivation of this work was to explore these predictions. But it soon devolved that there are multiple components of this regulatory system: Ochiai et al (2008) identified several cis-regulatory modules in the tbr gene of a related species, while we had found a distinct tbr cis-regulatory module in a different region of the locus in S. purpuratus. Here we recount a system scale analysis that includes all known active modular units of the locus, based on recombineered BAC constructs which cover the complete locus and extend into the territories of the flanking genes on either side. The network prediction that tbr is a primary target of the pmar1-hesC double negative gate (Oliveri et al, 2002; 2008; Revilla-i-Domingo et al, 2007) was demonstrated true, and in this work we also solved the identity of the missing inferred control input that precludes tbr expression in the nonskeletogenic mesoderm (NSM). But in addition to resolving the functions of its various cis-regulatory inputs, we have gained unexpected insight into two other interesting aspects of the regulatory biology of the tbr gene. We discovered how different tbr cis-regulatory modules are deployed at different stages of development, and how, in this case, cis-regulatory inputs affect module choice. Not much is known about the subject of module choice, though it is obvious that the phenomenon is pervasive, as most regulatory genes appear to utilize multiple cis-regulatory modules (for review, Davidson, 2006). A related consequence, which has sharp implications for standard operating procedures in cis-regulatory analysis, was the demonstration that a “minimal enhancer” construct may display more functionality when introduced into an embryo than it actually executes in context, where what it does depends on whether it, rather than another module, is actually deployed. Finally the whole elaborate regulatory system we have revealed is cast into a particularly interesting light by the evolutionary novelty of this derived system, for as reviewed briefly in Introduction, only in echinoids is the tbr gene utilized at all in an embryonic SM cell lineage.
The early tbr control system
Disruption of the single HesC site in the α region of the tbr::GFP BAC produces a significant amount of ectopic expression in 18 and 24 h embryos, which though quantitatively minor is to be compared with the almost completely accurate expression of the parental BAC (Fig.2, Table S1). A higher rate of ectopic expression was produced at these times by treatment with hesC MASO, using the wild type tbr::GFP BAC. The hesC MASO is clearly active as shown in earlier work (Revilla-i-Domingo et al., 2007), and as noted below, it sufficed in this study to produce 100% ectopic expression from the short γ(2)::EpGFP construct later in development (Table S1). However, early in development when hesC is intensely transcribed everywhere in the embryo (except in the SM pmar1 domain), it may be relatively difficult to block the presence of all HesC protein. We were mainly concerned to test in full genomic context the function of the single α module HesC site discovered by Ochiai et al (2008), and as noted above it is very possible that additional functional HesC sites exist elsewhere in the tbr locus.
The positive early control system consists of modules α plus C, as shown in the BAC deletions of Figs. 4 and 5. However, Fig. 4C,D show that of these, C module is not required to produce accurate expression in the whole BAC. C module appears to contribute only a quantitative booster input since there is no increase in ectopic expression whatsoever when it is deleted, though there is a great decrease in level of expression (Fig. 4B). α module and its HesC site are able to do the job of ensuring that what expression remains is accurate. The location of any additional repressive HesC sites elsewhere in the locus would not have been tested in these deletions. Nonetheless, the significant destabilization of the very tight control executed by the early system operating in tbr::GFP BAC prior to ingression when the single known α module HesC site is destroyed, justifies the placement of this gene downstream of the pmar1-hesC double negative gate.
tbr regulation after ingression
As shown very clearly in Fig. 5, when the upstream γ(2) module is deleted from the complete system carried in tbr::GFP BAC, there is no effect of any kind on expression prior to ingression (21-22hpf), either quantitative or spatial. But thereafter, the level of expression is greatly compromised; and in addition ectopic expression increases significantly, particularly in NSM cells (examples in Figs. 5C, 24 and 48h embryos). The γ(2) module is thus a late acting driver of expression in cells executing active skeletogenesis. It does not act alone, however, and again module C functions as a booster. These two modules interact cooperatively, since the sum of the expression in the late phase when C is deleted plus when γ(2) is deleted does not equal the level of late expression when neither is deleted (Figs. 4,5).
The γ(2) module has two different regulatory inputs, which probably use the same target sites. The experiments in Fig. 7 and Table S.3 prove that the activating driver is indeed Ets1/2, interactions with which account entirely for its activity. We also demonstrated that the short γ(2) module construct, γ(2)::EpGFP, responds sharply to hesC MASO; in fact by late gastrula this treatment causes 100% of embryos to mis-express the GFP reporter (Table S.1). So also does global expression of the Ets1/2 driver (Table S.1). But γ(2) module has no functional HesC site, and the effect of HesC on its expression is indirect. We can understand this at once by reference to the network architecture, for the ets1/2 gene is itself a primary target of HesC repression immediately downstream of the pmar1 double negative gate. Thus HesC MASO causes global ectopic expression of Ets1/2 which in postgastrular embryos is normally confined to SM and NSM cells. That is why it causes global expression of γ(2)::EpGFP, the same effect on expression as direct injection into the egg of ets1/2 mRNA (Table S.1).
The experiments in Fig. 8 show that the reason the γ(2) module does not express in the NSM even though the Ets1/2 driver is present in these cells is that another NSM Ets family factor, Erg, acts to repress the activation potential of the module. After gastrulation erg is not transcribed in SM but continues to be expressed in NSM (Rizzo et al., 2006). Erg and Ets bind similar DNA target sites and so this is likely a case of competitive binding at the Ets sites, such that if the repressor Erg is present it wins. Thus erg MASO produces ectopic NSM expression of both the γ(2)::EpGFP short construct and of tbr::GFP BAC (Fig. 8). But, the additional striking result in Fig. 8 is that erg MASO produces no ectopic NSM expression in the derivative of tbr::GFP from which γ(2) module has been deleted. This reveals another late regulatory role of γ(2) module in its normal context: not only does it cooperatively (with module C) drive expression in the SM, but it also represses it in the NSM.
Minimal module illusions, and the mechanism of γ(2) module exclusion in early development
γ(2)::EpGFP is a typical “minimal” expression construct, consisting of only the module itself and a promiscuous basal promoter-reporter apparatus. It gave near perfect expression both early and late (Fig. 6C), though as pointed out above, the short construct is quantitatively much less active per copy relative to its function in context. This could be due to the much greater flexibility of the longer DNA “arm” separating the module from the promoter in the normal context, allowing a greater variety of productive contact conformations, or to a greater tendency of the individual construct units to interfere with one another in the incorporated concatenate, or to titration of activators by the large number of short construct copies, or to a combination of these. The main point is not this, but the shocking discovery that in context the γ(2) module apparently produces no output whatsoever prior to ingression, while when isolated in γ(2)::EpGFP it does function prior to ingression. We see immediately that in the short construct, where there is no other option, the basal promoter will use whatever it can get, so to speak. The short construct does not exactly “lie” about γ(2) module functionality; rather it “exaggerates”: only a part of what it displays may be utilized in context, because there is another layer of control, module choice. The fact that the complete system minus the γ(2) module functions the same in early development as when γ(2) module is present shows directly that γ(2) module provides no significant input while the early plus C module system is running. It operates differently, not redundantly with the α plus C module system, as shown by the strikingly different response to Ets overexpression in pre-ingression embryos. The interactions controlling γ(2) module revealed in this study can also explain why it is silent in the early embryo.
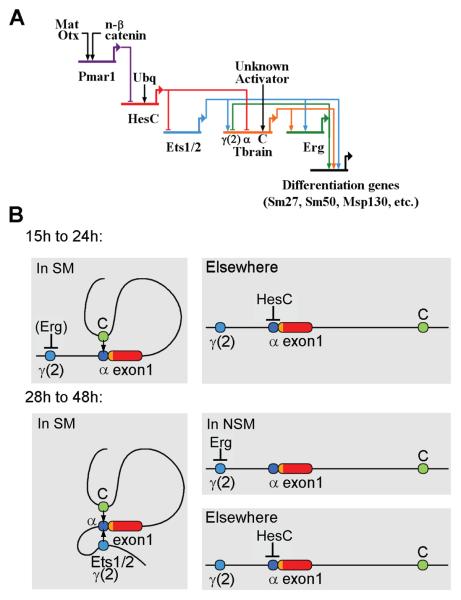
In the pre-ingression SM we believe that the same thing happens to γ(2) module as happens in the post-ingression NSM. As network analysis has shown (Oliveri et al., 2008), just downstream of the regulatory targets activated by the pmar1-hesC double negative gate (i.e., ets1/2, alx1, tel, and tbr), a positive feedback subcircuit is activated by inputs from these primary responders. The first gene in this subcircuit is none other than erg. It receives an input from tbr itself as well as from ets1/2, then forges interactions with hex and tgif, including a feedback onto erg from hex. As we have seen, in the context of the whole system the γ(2) module is dominantly repressed by Erg in the presence of Ets1/2, and so in the pre-ingression SM, once erg is turned on and kept on, γ(2) module should be inactive. This is a case of short range repression (Gray et al., 1994) since the gene is not silenced, only the γ(2) module. The circuitry, summarized in Fig. 9A, is fascinating. Essentially, tbr expression is the cause of γ(2) module repression, via the negative feedback from the tbr target erg. Or in other words the tbr gene itself ends up controlling which regulatory module will be deployed actively, and the exclusion of γ(2) module activation potential is probably the cause of deployment of the α-C module system that operates in the early embryo rather than γ(2) module. Later when erg expression is extinguished in the SM (for reasons not yet known, as this occurs later than our comprehensive network analysis so far extends), γ(2) module is called into action, also in collaboration with C. The alternative conformations implied by these deployments are diagrammed in Fig. 9B. This is our preferred model, but it is also possible that an insulator contributes to silencing γ(2) module in the complete construct, since we observed that interposition of a large stretch of upstream sequence in γ(2) expression constructs prevents expression (Table S2; Fig. S3).
Fig. 9.
Regulatory interactions of the tbr control system. (A) An updated tbrain regulatory subcircuit showing the inputs into the γ(2) module from Ets1/2 and Erg identified in this study. The diagram displays interactions that occur in the SM prior to ingression. During this period the γ(2) module is inactive (due to the negative feedback from the erg gene) and tbr expression is controlled by modules C and α, the site of HesC spatial repression (see text). Network diagram was constructed in BioTapestry (Longabaugh et al., 2009). (B) Schematics showing proposed interactions of tbrain cis-regulatory modules in different embryonic territories. This model is based on the now commonly assumed mechanism of interactions between distant cis-regulatory modules by DNA looping. Between 15 and about 21hpf (Smith et al, 2008), dominant repression by HesC prevents tbrain expression outside of the skeletogenic micromere lineage. While Ets1/2 is present in the SM and then NSM during this period, tbrain activation in the PMCs occurs primarily through the C module due to the accumulation of the Erg repressor of γ(2) module in the SM, as in (A). By ingression, HesC expression has disappeared in the NSM (Smith and Davidson, 2008b), and for unknown reasons expression of erg is extinguished in the SM cells; Erg and Ets1/2 are now both present in NSM but only Ets1/2 in the post-ingression SM. The result is that the γ(2) module can now respond to SM Ets1/2 and contribute to tbr expression along with the module C booster, while γ(2) module is shut off in the NSM by Erg and the expression of tbr elsewhere continues to be excluded by HesC.
There are at least two possible reasons that the short γ(2)::EpGFP construct does not respond to Erg repression in the early SM: first, the Ets activator may have a competitive advantage when its target sites are brought into immediate proximity of the basal transcription apparatus, forming a stable activation complex; second, the γ(2)::EpGFP construct runs on an exogenous, promiscuous promoter from the endo16 gene, and Erg repression may require elements of the endogenous promoter. As usual, negative results are subject to various interpretations, and it is what the γ(2)::EpGFP construct does that is more informative than what it does not do.
Evolutionary considerations: how could all this have happened?
Almost all of the embryonic SM specification and differentiation gene regulatory network appears also to be utilized in the skeletogenic centers in which the initial spines and test plates of the adult body plan are constructed during mid-late larval life (Gao and Davidson, 2008). This includes the ets1/2 and alx genes, as well as the triple feedback erg, hex, tgif subcircuit genes, and downstream regulators as well. Since the same apparatus is evidently deployed in the skeletogenesis centers of the sea star larva (which has no embryonic skeletogenic mesoderm lineage whatsoever), all of these genes appear to be components of a pleisiomorphic echinoderm skeletogenic network (Gao and Davidson, 2008). This network was evidently linked in toto into the embryonic specification system defining the micromere lineage in the evolutionary branch leading to the euechinoids, the modern sea urchins which display a precociously-ingressing skeletogenic micromere lineage. But none of this pertains to the tbr gene, because this gene is not part of the adult skeletogenic apparatus in either sea urchins or sea stars (Gao and Davidson, 2008). As reviewed in Introduction, tbr is expressed in the embryonic endoderm in other echinoderm classes and in euechinoid embryos exclusively in the SM.
The acquisition of tbr by the embryonic skeletogenic control apparatus of the euechinoids is a classic case of co-option, here seen directly at the network level. The switch away from its pleisiomorphic endodermal function may have had nothing to do directly with the tbr co-option process, since many regulatory genes participate in multiple developmental processes. There is some evidence that a key role of tbr in sea star embryonic specification, to provide a necessary feed into the otx gene, an essential endoderm regulator, has been supplanted by a different gene in the euechinoids, viz. blimp1 (Hinman et al., 2007; Hinman et al., 2003). But this could have happened before, during or after tbr acquired its skeletogenic role. One essential step we can infer in the co-option process was placing tbr under control of the pmar1-hesC double negative gate, as pointed out earlier (Gao and Davidson, 2008). This gate is not part of the adult skeletogenic apparatus either, and it is the definitive initiator of micromere specification. The other three first tier regulators answering to the double negative gate also had to be placed under HesC control. Cis-regulatory studies on several double negative gate targets (Smith and Davidson, 2008b) and unpublished data) show that one or two HesC sites does the job, and this aspect of the co-option process is easy to imagine.
But there is something special about tbr co-option, just because this gene is not part of the pleisiomorphic skeletogenic network apparatus, and the characteristics of γ(2) module may hold the answer to the conundrum. The tbr gene has acquired several downstream targets in the SM, and so it is presumably useful as a differentiation driver. However unlike most others of these, tbr is never expressed in the NSM, as are ets1/2, erg, hex, etc. The reason, as we have seen, lies in the Erg repression function of the γ(2) module. SM and NSM regulatory states greatly overlap but, because of γ(2) module, tbr is an exception. In the evolutionary process leading to establishment of the embryonic euechinoid SM, γ(2) module thus provided a mechanism for building a unique, non-skeletogenic mesodermal regulatory state. It is not the only one, for there is one other regulatory gene just downstream of the double negative gate that is also never expressed in the NSM, viz. alx1. The evolutionary role of γ(2) module suggested here fits with its amazingly simple cis-regulatory construction, which depends essentially only on a couple of Ets1/2 target sites.
In summary, evolutionary co-option of tbr may have provided the special function of differentiating the SM from the NSM, just because the means of co-option included the appearance of γ(2) module. Two other parts of this same function were provided by the still unknown mechanism by which transcription of the erg repressor is shut off in the SM, and by the equally SM-specific cis-regulatory apparatus of the alx1 gene.
Supplementary Material
Acknowledgments
We would like to thank Prof. Andrew Murray and anonymous reviewers for critical reading and helpful suggestions. Research was supported by the Caltech SURF program, the Camilla Chandler Frost Fellowship, the US Department of Defense NDSEG Fellowship Program, and NIH grants HD037105 and GM075089.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Cameron RA, Oliveri P, Wyllie J, Davidson EH. cis-Regulatory activity of randomly chosen genomic fragments from the sea urchin. Gene Expr. Patterns. 2004;4:205–13. doi: 10.1016/j.modgep.2003.08.007. [DOI] [PubMed] [Google Scholar]
- Consales C, Arnone MI. Functional characterization of Ets-binding sites in the sea urchin embryo: three base pair conversions redirect expression from mesoderm to ectoderm and endoderm. Gene. 2002;287:75–81. doi: 10.1016/s0378-1119(01)00891-5. [DOI] [PubMed] [Google Scholar]
- Croce J, Lhomond G, Lozano JC, Gache C. ske-T, a T-box gene expressed in the skeletogenic mesenchyme lineage of the sea urchin embryo. Mech. Dev. 2001;107:159–62. doi: 10.1016/s0925-4773(01)00470-1. [DOI] [PubMed] [Google Scholar]
- Damle S, Hanser B, Davidson EH, Fraser SE. Confocal quantification of cis-regulatory reporter gene expression in living sea urchin. Dev. Biol. 2006;299:543–50. doi: 10.1016/j.ydbio.2006.06.016. [DOI] [PubMed] [Google Scholar]
- Davidson EH. The Regulatory Genome: Gene Regulatory Networks in Development and Evolution. Academic Press; San Diego, CA: 2006. [Google Scholar]
- Flytzanis CN, McMahon AP, Hough-Evans BR, Katula KS, Britten RJ, Davidson EH. Persistence and integration of cloned DNA in postembryonic sea urchins. Dev. Biol. 1985;108:431–42. doi: 10.1016/0012-1606(85)90046-6. [DOI] [PubMed] [Google Scholar]
- Fuchikami T, Mitsunaga-Nakatsubo K, Amemiya S, Hosomi T, Watanabe T, Kurokawa D, Kataoka M, Harada Y, Satoh N, Kusunoki S, Takata K, Shimotori T, Yamamoto T, Sakamoto N, Shimada H, Akasaka K. T-brain homologue (HpTb) is involved in the archenteron induction signals of micromere descendant cells in the sea urchin embryo. Development. 2002;129:5205–16. doi: 10.1242/dev.129.22.5205. [DOI] [PubMed] [Google Scholar]
- Gao F, Davidson EH. Transfer of a large gene regulatory apparatus to a new developmental address in echinoid evolution. Proc Natl Acad Sci U S A. 2008;105:6091–6. doi: 10.1073/pnas.0801201105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray S, Szymanski P, Levine M. Short-range repression permits multiple enhancers to function autonomously within a complex promoter. Genes Dev. 1994;8:1829–38. doi: 10.1101/gad.8.15.1829. [DOI] [PubMed] [Google Scholar]
- Hinman VF, Davidson EH. Evolutionary plasticity of developmental gene regulatory network architecture. Proc. Natl. Acad. Sci. U S A. 2007;104:19404–9. doi: 10.1073/pnas.0709994104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinman VF, Nguyen A, Davidson EH. Caught in the evolutionary act: precise cis-regulatory basis of difference in the organization of gene networks of sea stars and sea urchins. Dev. Biol. 2007;312:584–95. doi: 10.1016/j.ydbio.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Hinman VF, Nguyen AT, Cameron RA, Davidson EH. Developmental gene regulatory network architecture across 500 million years of echinoderm evolution. Proc. Natl. Acad. Sci U S A. 2003;100:13356–61. doi: 10.1073/pnas.2235868100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iso T, Kedes L, Hamamori Y. HES and HERP families: multiple effectors of the Notch signaling pathway. J. Cell Physiol. 2003;194:237–55. doi: 10.1002/jcp.10208. [DOI] [PubMed] [Google Scholar]
- Kurokawa D, Kitajima T, Mitsunaga-Nakatsubo K, Amemiya S, Shimada H, Akasaka K. HpEts, an ets-related transcription factor implicated in primary mesenchyme cell differentiation in the sea urchin embryo. Mech. Dev. 1999;80:41–52. doi: 10.1016/s0925-4773(98)00192-0. [DOI] [PubMed] [Google Scholar]
- Lee PY, Nam J, Davidson EH. Exclusive developmental functions of gatae cis-regulatory modules in the Strongylocentrorus purpuratus embryo. Dev. Biol. 2007;307:434–45. doi: 10.1016/j.ydbio.2007.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longabaugh WJ, Davidson EH, Bolouri H. Visualization, documentation, analysis, and communication of large-scale gene regulatory networks. Biochim. Biophys. Acta. 2009;1789:363–74. doi: 10.1016/j.bbagrm.2008.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao CA, Gan L, Klein WH. Multiple Otx binding sites required for expression of the Strongylocentrotus purpuratus Spec2a gene. Dev. Biol. 1994;165:229–42. doi: 10.1006/dbio.1994.1249. [DOI] [PubMed] [Google Scholar]
- Maruyama YK. A Sea Cucumber Homolog of the Mouse T-Brain-1 is Expressed in the Invaginated Cells of the Early Gastrula in Holothuria leucospilota. Zoolog Sci. 2000;17:383–7. doi: 10.2108/jsz.17.383. [DOI] [PubMed] [Google Scholar]
- Materna SC, Oliveri P. A protocol for unraveling gene regulatory networks. Nat. Protoc. 2008;3:1876–87. doi: 10.1038/nprot.2008.187. [DOI] [PubMed] [Google Scholar]
- McMahon AP, Flytzanis CN, Hough-Evans BR, Katula KS, Britten RJ, Davidson EH. Introduction of cloned DNA into sea urchin egg cytoplasm: replication and persistence during embryogenesis. Dev. Biol. 1985;108:420–30. doi: 10.1016/0012-1606(85)90045-4. [DOI] [PubMed] [Google Scholar]
- Ochiai H, Sakamoto N, Momiyama A, Akasaka K, Yamamoto T. Analysis of cis-regulatory elements controlling spatio-temporal expression of T-brain gene in sea urchin, Hemicentrotus pulcherrimus. Mech. Dev. 2008;125:2–17. doi: 10.1016/j.mod.2007.10.009. [DOI] [PubMed] [Google Scholar]
- Oliveri P, Carrick DM, Davidson EH. A regulatory gene network that directs micromere specification in the sea urchin embryo. Dev. Biol. 2002;246:209–28. doi: 10.1006/dbio.2002.0627. [DOI] [PubMed] [Google Scholar]
- Oliveri P, Davidson EH, McClay DR. Activation of pmar1 controls specification of micromeres in the sea urchin embryo. Dev. Biol. 2003;258:32–43. doi: 10.1016/s0012-1606(03)00108-8. [DOI] [PubMed] [Google Scholar]
- Oliveri P, Tu Q, Davidson EH. Global regulatory logic for specification of an embryonic cell lineage. Proc. Natl. Acad. Sci. U S A. 2008;105:5955–62. doi: 10.1073/pnas.0711220105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransick A, Rast JP, Minokawa T, Calestani C, Davidson EH. New early zygotic regulators expressed in endomesoderm of sea urchin embryos discovered by differential array hybridization. Dev. Biol. 2002;246:132–47. doi: 10.1006/dbio.2002.0607. [DOI] [PubMed] [Google Scholar]
- Revilla-i-Domingo R, Minokawa T, Davidson EH. R11: a cis-regulatory node of the sea urchin embryo gene network that controls early expression of SpDelta in micromeres. Dev. Biol. 2004;274:438–51. doi: 10.1016/j.ydbio.2004.07.008. [DOI] [PubMed] [Google Scholar]
- Revilla-i-Domingo R, Oliveri P, Davidson EH. A missing link in the sea urchin embryo gene regulatory network: hesC and the double-negative specification of micromeres. Proc. Natl. Acad. Sci. U S A. 2007;104:12383–8. doi: 10.1073/pnas.0705324104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo F, Fernandez-Serra M, Squarzoni P, Archimandritis A, Arnone MI. Identification and developmental expression of the ets gene family in the sea urchin (Strongylocentrotus purpuratus) Dev. Biol. 2006;300:35–48. doi: 10.1016/j.ydbio.2006.08.012. [DOI] [PubMed] [Google Scholar]
- Shoguchi E, Satoh N, Maruyama YK. A starfish homolog of mouse T-brain-1 is expressed in the archenteron of Asterina pectinifera embryos: possible involvement of two T-box genes in starfish gastrulation. Dev. Growth Differ. 2000;42:61–8. doi: 10.1046/j.1440-169x.2000.00481.x. [DOI] [PubMed] [Google Scholar]
- Smith J, Davidson EH. A new method, using cis-regulatory control, for blocking embryonic gene expression. Dev. Biol. 2008a;318:360–5. doi: 10.1016/j.ydbio.2008.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J, Davidson EH. Gene regulatory network subcircuit controlling a dynamic spatial pattern of signaling in the sea urchin embryo. Proc. Natl. Acad. Sci. U S A. 2008b;105:20089–94. doi: 10.1073/pnas.0806442105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang DG, Britten RJ, Davidson EH. Maternal and embryonic provenance of a sea urchin embryo transcription factor, SpZ12-1. Mol. Mar. Biol. Biotechnol. 1995;4:148–53. [PubMed] [Google Scholar]
- Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 2005;33:e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yon J, Fried M. Precise gene fusion by PCR. Nucleic Acids Res. 1989;17:4895. doi: 10.1093/nar/17.12.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuh CH, Davidson EH. Modular cis-regulatory organization of Endo16, a gut-specific gene of the sea urchin embryo. Development. 1996;122:1069–82. doi: 10.1242/dev.122.4.1069. [DOI] [PubMed] [Google Scholar]
- Yuh CH, Moore JG, Davidson EH. Quantitative functional interrelations within the cis-regulatory system of the S. purpuratus Endo16 gene. Development. 1996;122:4045–56. doi: 10.1242/dev.122.12.4045. [DOI] [PubMed] [Google Scholar]
- Yuh CH, Ransick A, Martinez P, Britten RJ, Davidson EH. Complexity and organization of DNA-protein interactions in the 5′-regulatory region of an endoderm-specific marker gene in the sea urchin embryo. Mech. Dev. 1994;47:165–86. doi: 10.1016/0925-4773(94)90088-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.