Summary
CXCL12 provides a chemotactic signal directing leukocyte migration and regulates metastatic behavior of tumor cells. We conducted a population-based case-control study to test the hypothesis that common genetic variation in CXCL12 (single SNP alleles and haplotypes) is associated with the risk of cervical carcinoma. Cases (N=917) were women diagnosed with invasive squamous cell cervical carcinoma (SCC), adenocarcinoma or adenosquamous carcinoma or adenocarcinoma in situ (ACIS) of the cervix, while residents of western Washington State. Control participants (N=849) were identified from the source population by random digit telephone dialing and frequency matched to cases on county and age. Nine CXCL12 tagSNPs chosen from the SeattleSNPs database were genotyped. The minor allele of intronic SNP rs266085 was inversely associated with cervical cancer under a recessive genetic effects model (OR=0.74, 95% C.I. 0.56–0.98). Among the ten common haplotypes inferred from the 9 tagSNPs, one haplotype defined by minor alleles at 5’ flanking SNP rs17885289 and rs266085, and common alleles at the other 7 SNPs occurred among 7.8% of cases and 10.6% of controls (dominant model OR=0.72, 95% C.I. 0.56–0.93; recessive model OR=0.35, 95% C.I. 0.12–0.97; and log additive model OR=0.72, 95% C.I. 0.57–0.90). A stepwise procedure identified rs17885289, rs266085, and 3’ UTR SNP rs266093 as the most parsimonious subset of SNPs necessary to define the haplotype inversely associated with cervical cancer risk in our study. A 3’ UTR SNP, rs1801157, previously found to be related to HIV pathogenesis, was not associated with cervical cancer risk. Further population-based studies are warranted to confirm these associations between genetic variation in CXCL12 and cervical cancer risk.
Keywords: cervical cancer, HPV epidemiology, CXCL12, single nucleotide polymorphism
Introduction
Cervical cancer is the second most common cancer among women worldwide, and the leading cause of cancer death among women in developing countries (Parkin, Bray et al. 2005). Infection with oncogenic human papillomavirus (HPV) is necessary to the development of cervical cancer (Bosch and de Sanjose 2003; IARC 2007), and oncogenic HPV types are found in >99% of cervical tumors (Walboomers, Jacobs et al. 1999). Although nearly 30% of sexually active women become infected with genital HPV soon after sexual debut (Winer, Lee et al. 2003), the large majority of women clear their infections at a young age; thus, only a small minority go on to develop the persistent infections that lead to cervical cancer and its precursor lesions (Bosch and de Sanjose 2003). Most women clear HPV infections through activation of cellular immunity. In addition to the central role of HPV, other environmental and genetic factors increase the risk of cervical cancer. For example, studies have linked variation in genes involved in the immune response with cervical cancer risk (e.g.,(Carreon, Martin et al. 2005; Carrington, Wang et al. 2005; Mehta, Jordanova et al. 2007; Madeleine, Johnson et al. 2008).
The chemokine CXCL12, also known as stromal-cell derived factor 1 (SDF-1), directs leukocyte migration through interactions with its receptor CXCR4 (Bleul, Fuhlbrigge et al. 1996). Rare mutations in the CXCR4 gene that increase cellular responsiveness to CXCL12 have been shown by linkage analysis to underlie WHIM (warts, hypogammaglobulinemia, immunodeficiency, and myelokatexis) syndrome, a rare immunodeficiency characterized by immune defects and extensive HPV-induced warts (Hernandez, Gorlin et al. 2003). This syndrome has been attributed to a deficit of mature leukocytes in the periphery, which may lead to HPV tolerance (Diaz 2005).
CXCL12 has been widely studied because its receptor CXCR4 is used by strains of HIV-1 to enter cells (Bleul, Farzan et al. 1996). Prior studies have identified altered risks of HIV infection and AIDS progression associated with genetic variation in CXCL12, focused largely on a common G->A single nucleotide polymorphism (SNP) (rs1801157) in the 3’ untranslated region (UTR) (Hendel, Henon et al. 1998; Modi, Scott et al. 2005; Petersen, Glashoff et al. 2005; Vidal, Peraire et al. 2005; Vissoci Reiche, Ehara Watanabe et al. 2006).
The CXCL12-CXCR4 axis also regulates metastatic behavior in tumor cell lines (Libura, Drukala et al. 2002; Kucia, Jankowski et al. 2004). Prior studies have found a positive association between the minor allele at rs1801157 and risk of carcinoma of the breast (Zafiropoulos, Crikas et al. 2004; Razmkhah, Talei et al. 2005), lung (Razmkhah, Doroudchi et al. 2005), prostate (Hirata, Hinoda et al. 2007), and colon (Hidalgo-Pascual, Galan et al. 2007).
We conducted a population-based case-control study to test the hypothesis that common genetic variation in CXCL12 is associated with cervical cancer risk. In contrast with prior genetic association studies involving CXCL12, we genotyped multiple SNPs in the CXCL12 region, allowing the measurement of disease risks associated with single SNP alleles as well as common CXCL12 haplotypes.
Materials & Methods
Study participants, data, and specimens for this study were derived from a long-term population-based case-control study of anogenital cancer conducted in western Washington State that has been described in detail previously (Daling, Madeleine et al. 1996). Briefly, eligible cases were women diagnosed between 1986 and 2004 with histologically confirmed incident primary invasive squamous cell carcinoma (SCC, ICD-O 8010-8081), invasive adenocarcinoma (ICD-O 8140-8480), in situ adenocarcinoma (ACIS, ICD-O 8140-8480), or invasive adenosquamous carcinoma of the cervix (ICD-O 8560) and identified through the Cancer Surveillance System (CSS), a population-based cancer registry that is part of the National Cancer Institute’s Surveillance, Epidemiology, and End Results (SEER) program (Hankey, Ries et al. 1999). All invasive cervical carcinoma cases were residents of King, Pierce, and Snohomish counties at diagnosis: ACIS cases were residents of these three counties plus the remaining 10 counties in western Washington State that are covered by the CSS. Additionally, recruitment of ACIS cases was limited to diagnoses from 1990 through 2004. Residents of the 13 counties were identified and selected as controls using random digit telephone dialing and frequency matched to cases on age and county as previously described (Daling, Madeleine et al. 1996). All participants were aged 18–74 years on the date of diagnosis (if a case) or on a corresponding reference date (if a control).
Cases and controls were interviewed in person using a structured questionnaire administered by trained, professional interviewers. Interview items included information on smoking history, demographic characteristics, and reproductive, sexual, and birth control histories, as described previously (Daling, Madeleine et al. 1996). Blood was drawn from subjects who consented, and oral rinse samples were collected for participants who chose not to provide a blood sample. DNA was extracted from peripheral blood lymphocytes and buccal cells using phenol-chloroform or salting-out methods.
A total of 1384 cervical cancer cases were interviewed. The response proportion was 66%. Reasons for non-response included those who died prior to contact (7%), failed to participate due to doctor refusal (5%), and refusal to participate or loss to follow up (22%). Of cases interviewed, 91.5% provided a peripheral blood or oral rinse sample. The present analyses included 376 SCC, 170 invasive adenocarcinoma, 339 ACIS, and 32 adenosquamous carcinoma cases. These numbers represent all recruited cases for whom DNA samples were available at the time that assays were performed.
In total, 1636 controls with an intact uterus were recruited, with a response proportion of 67.4%. Of these controls, 1372 (83.9%) had a blood sample or oral rinse sample collected at interview. We selected 851 control samples for this study based on a random sample of the recruited controls with available DNA at the time the assays were performed. One control reporting no sex partners was dropped from the analysis, and another control was dropped due to genotyping failure at every CXCL12 SNP, leaving 849 controls for this analysis. Three study participants were excluded from haplotype inference because they were missing genotype data at more than four SNPs.
tagSNP Selection and Genotyping
The SeattleSNPs Project (http://pga.mbt.washington.edu) identified 118 SNPs in the CXCL12 gene region (18.5 kb on chromosome 10 spanning the CXCL12 coding region, introns, and upstream and downstream untranslated regions), based on resequencing of 7 individuals of European descent. Eleven tagging SNPs (tagSNPs) were chosen for genotyping based on three criteria: SNP minor allele frequency (MAF) greater than 5%; genotype correlation r2 threshold of 0.64 in each bin grouped by the LD_Select algorithm (Carlson, Eberle et al. 2004); and potential functional significance. When multiple possible tagSNPs were available for a given bin, SNPs were selected in the following order according to their location in the CXCL12 gene: coding region; UTR region; 5’ or 3’ region flanking the gene; and intronic region. These criteria were balanced against the feasibility of developing TaqMan® (Applied Biosystems Inc., Foster City, CA) genotyping assays. Of the 11 tagSNPs, two were not included in this analysis: rs169097 was excluded after it showed no variation in early genotyping results and rs2839682 was excluded due to assay failure. The 9 remaining tagSNPs (Figure 1 and Supplemental Table 1) include three located in the 5’ flanking region, which contains several partially characterized transcription factor binding motifs (Garcia-Moruja, Alonso-Lobo et al. 2005), three intronic SNPs, and three SNPs in the 3’ UTR including one (rs1801157) that has been associated with altered HIV infection and AIDS progression (Hendel, Henon et al. 1998; Modi, Scott et al. 2005; Petersen, Glashoff et al. 2005; Vissoci Reiche, Ehara Watanabe et al. 2006). DNA was genotyped using a TaqMan® platform as described previously (Hussain, Madeleine et al. 2008). Specimens were organized so that 10% replicate DNA aliquots were distributed throughout the reaction plates.
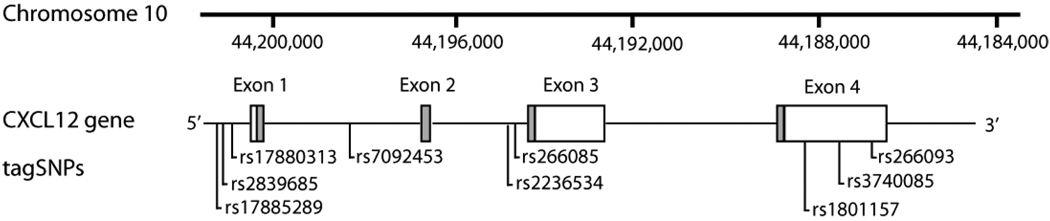
Figure 1.
Physical map and structure of the CXCL12 gene on chromosome 10, and the location of tagSNPs genotyped in this study. Coordinates refer to National Center for Bioinformatics (NCBI) (http://www.ncbi.nlm.nih.gov) chromosome 10 build 36.3. Coding regions of the CXCL12 gene are indicated as shaded boxes and transcribed non-coding regions as white boxes.
Statistical Analyses
Estimates of linkage disequilibrium (r2 and |D’|) between tagSNPs were calculated among controls using Haploview (Barrett, Fry et al. 2005). We estimated univariate associations between cervical carcinoma (all histologic types combined) and genotypes of individual tagSNPs using unconditional logistic regression in Stata 10.0 (StataCorp LP, College Station, TX) to calculate odds ratio (OR) estimates and 95% confidence intervals. OR estimates and 95% confidence intervals were also calculated under dominant, recessive, and log additive genetic effects models. We then used an estimating equations approach, implemented in the software package HPlus (Li, Khalid et al. 2003), to infer haplotypes and diplotypes and calculate OR estimates and 95% confidence intervals under recessive, dominant, and log additive relationships with the risk of cervical cancer. We calculated histologic type specific associations by each technique for SCC and all adenocarcinomas combined, as well as for invasive and in situ adenocarcinoma. Differences in associations with genotypes between histologic types were assessed by polytomous logistic regression using a likelihood ratio test to compare a model in which risk estimates were constrained to be equal between cervical cancer histology groups with an unconstrained model in which risk estimates were allowed to vary.
All analyses were adjusted for age at diagnosis (18 to 34, 35 to 44, 45 to 54, 55 to 64, and 65 to 74) or reference date and race (categorized as white or nonwhite). Sub-analyses excluded nonwhites and were compared to analyses including all subjects.
Forward, backward, and hybrid stepwise modeling approaches (Yang, Li et al. 2008), were used to identify the minimum number of SNPs required for imputation of haplotypes with potentially altered risk. Briefly, the stepwise procedure adds or removes one SNP at a time and asks whether an observed disease association is altered (Yang, Li et al. 2008).
Results
The mean ages of cases (SCC, invasive adenocarcinoma, ACIS, and adenosquamous carcinoma combined) and controls were 40.3 years and 41.5 years, respectively. Whites comprised approximately 90% of each group (Table 1). Cases tended to have lower educational level and income than controls, and were more likely to be current smokers and have a higher number of lifetime sex partners. Among cases, women with SCC tended to be older than women with adenocarcinoma (AC, including invasive adenocarcinoma, ACIS, and adenosquamous carcinoma combined unless otherwise noted) cases. Cases and controls with SNP genotype data did not differ substantially in behavioral and demographic characteristics from those without genotype information (data not shown).
Table 1.
Characteristics of cervical cancer cases and controls with CXCL12 genotyping data available, Seattle-Puget Sound Region, 1986 – 2004
| Controls | Cases | |||||||
|---|---|---|---|---|---|---|---|---|
| Squamous Cell Cervical Carcinoma |
Adenocarcinomaa | All Histological Types |
||||||
| Characteristic | N=849 | (%) | N=376 | (%) | N=541 | (%) | N=917 | (%) |
| Age (years) | ||||||||
| 18–34 | 268 | (31.6) | 107 | (28.5) | 221 | (40.9) | 328 | (35.8) |
| 35–44 | 274 | (32.3) | 115 | (30.6) | 191 | (35.3) | 306 | (33.4) |
| 45–54 | 189 | (22.3) | 86 | (22.9) | 91 | (16.8) | 177 | (19.3) |
| 55–64 | 69 | (8.1) | 42 | (11.2) | 24 | (4.4) | 66 | (7.2) |
| 65–74 | 49 | (5.8) | 26 | (6.9) | 14 | (2.6) | 40 | (4.4) |
| Race | ||||||||
| White | 767 | (90.3) | 312 | (83.0) | 492 | (90.9) | 804 | (87.7) |
| Black | 23 | (2.7) | 15 | (4.0) | 6 | (1.1) | 21 | (2.3) |
| Native American or Alaskan Inuit | 4 | (0.5) | 6 | (1.6) | 7 | (1.3) | 13 | (1.4) |
| Asian or Pacific Islander | 35 | (4.1) | 27 | (7.2) | 21 | (3.9) | 48 | (5.2) |
| More than one, other, or unknown | 20 | (2.4) | 16 | (4.3) | 15 | (2.8) | 31 | (3.4) |
| Education | ||||||||
| High school or less | 212 | (25.0) | 139 | (37.7) | 137 | (25.5) | 276 | (30.4) |
| Some college or greater | 636 | (75.0) | 230 | (62.3) | 401 | (74.5) | 631 | (69.6) |
| Unknown | 1 | (0.1) | 7 | (1.9) | 3 | (0.6) | 10 | (1.1) |
| Annual household income | ||||||||
| <$15K | 70 | (8.3) | 69 | (18.7) | 54 | (10.1) | 123 | (13.6) |
| $15–30K | 159 | (18.9) | 99 | (26.8) | 100 | (18.7) | 199 | (22.0) |
| $30–45K | 155 | (18.4) | 75 | (20.3) | 112 | (20.9) | 187 | (20.7) |
| 45K+ | 459 | (54.4) | 126 | (34.1) | 270 | (50.4) | 396 | (43.8) |
| Unknown | 6 | (0.7) | 7 | (1.9) | 5 | (0.9) | 12 | (1.3) |
| Lifetime number of sex partners | ||||||||
| 1 | 193 | (22.8) | 39 | (10.4) | 67 | (12.4) | 106 | (11.6) |
| 2–4 | 255 | (30.2) | 121 | (32.3) | 149 | (27.5) | 270 | (29.5) |
| 5–14 | 288 | (34.1) | 161 | (42.9) | 228 | (42.1) | 389 | (42.5) |
| 15 or greater | 109 | (12.9) | 54 | (14.4) | 97 | (17.9) | 151 | (16.5) |
| Unknown | 4 | (0.5) | 1 | (0.3) | 0 | (0.0) | 1 | (0.1) |
| Cigarette smoking at reference date | ||||||||
| Never | 455 | (53.6) | 156 | (41.5) | 293 | (54.2) | 449 | (49.0) |
| Former | 219 | (25.8) | 98 | (26.1) | 132 | (24.4) | 230 | (25.1) |
| Current | 175 | (20.6) | 122 | (32.4) | 116 | (21.4) | 238 | (26.0) |
| Number of live or still births | ||||||||
| 0 | 229 | (27.0) | 71 | (18.9) | 182 | (33.6) | 253 | (27.6) |
| 1 | 139 | (16.4) | 61 | (16.2) | 93 | (17.2) | 154 | (16.8) |
| 2 | 248 | (29.2) | 106 | (28.2) | 163 | (30.1) | 269 | (29.3) |
| 3+ | 233 | (27.4) | 138 | (36.7) | 103 | (19.0) | 241 | (26.3) |
Adenocarcinoma includes in situ adenocarcinoma (n=339), invasive adenocarcinoma (n=170) and adenosquamous carcinoma (n=32).
The distributions of CXCL12 SNP genotypes among the white controls were consistent with Hardy-Weinberg equilibrium. All of the SNPs in our study had call rates greater than 95% with the exception of rs3740085, which was genotyped to 94.6% completion. Among all SNPs, there was 99.8% consistency between blinded replicate pairs.
There was no statistically significant association between any of the SNP alleles under dominant or log additive genetic effects models and cervical cancer (Table 2). The minor allele at intronic SNP rs266085 was associated with a lower risk of cervical cancer under the recessive (OR=0.74, 95% C.I. 0.56–0.98) model. In the codominant model, an unconstrained genetic model in which heterozygotes and minor allele homozygotes are compared separately to common allele homozygotes, having two copies of the minor allele at rs266085 was significantly associated with a lower risk of cervical cancer (OR=0.72, 95% C.I. 0.53–0.97) (Supplemental Table 2). The minor allele at 3’ UTR SNP rs266093 was associated with a weakly increased risk of cervical cancer (dominant model OR=1.11, 95% C.I. 0.92–1.35, recessive model OR=1.23, 95% C.I. 0.94–1.63, and log additive model OR=1.12, 95% C.I. 0.97–1.28), although confidence intervals included the null value for OR estimates. Genotype analyses restricted to Caucasians yielded results that were similar to those including all races. No differences in risk estimates between invasive and non-invasive cervical cancer, or between SCC and AC were statistically significant.
Table 2.
Association of CXCL12 SNP alleles with cervical cancer risk, Seattle-Puget Sound Region, 1986 – 2004
| Genetic Effects Model | ||||||
|---|---|---|---|---|---|---|
| Frequency | Dominant | Recessive | Log Additive | |||
| SNP | Allele | Casesa | Controls | OR (95% CI)b | OR (95% CI)b | OR (95% CI)b |
| N = 917 | N=849 | |||||
| rs17885289 | G | 0.725 | 0.707 | |||
| A | 0.275 | 0.293 | 0.92 (0.76–1.11) | 0.88 (0.62–1.24) | 0.93 (0.80–1.08) | |
| rs2839685 | C | 0.865 | 0.865 | |||
| T | 0.149 | 0.143 | 1.05 (0.85–1.30) | 1.07 (0.47–2.40) | 1.05 (0.86–1.27) | |
| rs17880313 | C | 0.878 | 0.870 | |||
| T | 0.128 | 0.125 | 1.05 (0.84–1.32) | 0.95 (0.45–2.02) | 1.04 (0.85–1.28) | |
| rs7092453 | A | 0.806 | 0.813 | |||
| G | 0.202 | 0.191 | 1.09 (0.89–1.33) | 1.24 (0.76–2.00) | 1.09 (0.92–1.29) | |
| rs2236534 | C | 0.821 | 0.815 | |||
| A | 0.195 | 0.195 | 1.01 (0.82–1.23) | 0.94 (0.60–1.50) | 1.00 (0.84–1.18) | |
| rs266085 | G | 0.666 | 0.637 | |||
| A | 0.350 | 0.377 | 0.89 (0.74–1.08) | 0.74 (0.56–0.98) | 0.88 (0.76–1.00) | |
| rs1801157 | G | 0.821 | 0.797 | |||
| A | 0.189 | 0.193 | 0.90 (0.74–1.10) | 1.17 (0.67–2.03) | 0.94 (0.79–1.11) | |
| rs3740085 | G | 0.738 | 0.719 | |||
| C | 0.247 | 0.242 | 1.02 (0.84–1.24) | 0.94 (0.64–1.39) | 1.01 (0.86–1.18) | |
| rs266093 | C | 0.620 | 0.638 | |||
| G | 0.388 | 0.362 | 1.11 (0.92–1.35) | 1.23 (0.94–1.63) | 1.12 (0.97–1.28) | |
Abbreviations: SNP, single nucleotide polymorphism; OR, odds ratio, CI, confidence interval.
Bold indicates significance level (P-value) below 0.05.
Cases include squamous cell carcinoma (n=376), invasive adenocarcinoma (n=170), adenocarcinoma in situ (n=339), and adenosquamous carcinoma (n=32).
Risk estimates include all races, adjusted for age (categorically, as presented in Table 1) and race (white versus nonwhite).
CXCL12 haplotypes were imputed from all nine tagSNPs genotyped and the ten haplotypes that occurred at a frequency of 1% or greater in cases or controls are shown in Table 3. One haplotype defined by minor alleles at the 5’ flanking SNP rs17885289 and the intronic SNP rs266085, and common alleles at the other 7 SNPs occurred among 7.8% of case chromosomes and 10.6% of control chromosomes and was inversely associated with cervical cancer risk under dominant (OR=0.72, 95% C.I. 0.56–0.93), recessive (OR=0.35, 95% C.I. 0.12–0.97), and log additive (OR=0.72, 95% C.I. 0.57– 0.90) models.
Table 3.
Risk of cervical cancer associated with common CXCL12 haplotypes, Seattle-Puget Sound Region, 1986 – 2004
| Frequency | Genetic Effects Model | ||||
|---|---|---|---|---|---|
| Dominant | Recessive | Log Additive | |||
| Haplotypea | Caseb Chromosomes | Control Chromosomes |
OR (95% CI)c | OR (95% CI)c | OR (95% CI)c |
| N = 1830 | N=1696 | ||||
| 000001100 | 0.188 | 0.200 | 0.88 (0.73–1.08) | 1.06(0.62–1.82) | 0.92 (0.77–1.09) |
| 000010010 | 0.192 | 0.190 | 1.01 (0.83–1.23) | 1.11 (0.69–1.79) | 1.02 (0.86–1.20) |
| 010000001 | 0.142 | 0.137 | 1.06 (0.86–1.31) | 1.00 (0.43–2.32) | 1.05 (0.86–1.28) |
| 101100001 | 0.123 | 0.119 | 1.10 (0.87–1.38) | 0.92 (0.43–2.00) | 1.07 (0.87–1.32) |
| 100001000 | 0.078 | 0.106 | 0.72 (0.56–0.93) | 0.35 (0.12–0.97) | 0.72 (0.57–0.90) |
| 000001000 | 0.078 | 0.066 | 1.16 (0.88–1.54) | 1.21 (0.27–5.45) | 1.15 (0.88–1.51) |
| 100100001 | 0.072 | 0.063 | 1.14 (0.86–1.51) | 1.95 (0.46–8.33) | 1.15 (0.88–1.50) |
| 000000010 | 0.062 | 0.059 | 1.11 (0.82–1.50) | 0.88 (0.26–2.90) | 1.09 (0.82–1.44) |
| 000000001 | 0.041 | 0.035 | 1.13 (0.78–1.62) | - | 1.14 (0.80–1.64) |
| 000000000 | 0.008 | 0.010 | 1.02 (0.47–2.19) | - | 1.02 (0.47–2.18) |
Abbreviations: SNP, single nucleotide polymorphism; OR, odds ratio, CI, confidence interval.
Odds ratios omitted where numbers were insufficient for calculation.
Bold indicates significance level (P-value) below 0.05.
Haplotypes defined by SNP alleles in chromosomal order from 5’ to 3’ across the CXCL12 gene region (rs17885289, rs2839685, rs17880313, rs7092453, rs2236534, rs266085, rs1801157, rs3740085, rs266093), where zero (0) indicates the common allele and one (1) indicates minor allele.
Cases include squamous cell carcinoma (n=376 subjects), invasive adenocarcinoma (n=169 subjects), adenocarcinoma in situ (n=339 subjects), and adenosquamous carcinoma (n=31 subjects).
Risk estimates include all races, adjusted for age (categorically, as presented in Table 1) and race (white versus nonwhite).
In diplotype analysis the reference diplotype occurred among 6.7% of cases and 8.2% of controls and consisted of one haplotype with minor alleles at rs266085 and rs1801157 and a second haplotype with minor alleles at rs2236534 and rs3740085. There was a decreased risk associated with having two copies of the haplotype with minor alleles at rs17885289 and rs266085, which occurred among 0.6% of cases and 1.6% of controls (OR=0.36, 95% C.I. 0.13–1.04). No other haplotypes or diplotypes, including two others containing the minor allele at rs266085, were associated with significantly altered risks of cervical cancer.
A stepwise procedure was used to identify the most parsimonious subset of SNPs associated with cervical cancer risk (Yang, Li et al. 2008). Using this subset of SNPs (5’ flanking SNP rs17885289, intronic SNP rs266085, and 3’ UTR SNP rs266093), the inferred minimal haplotype associated with risk was defined by minor alleles at rs17885289 and rs266085, and the common allele at rs266093 (Supplemental Table 3). Linkage analysis showed that the other 6 SNPs were in high linkage disequilibrium with these three haplotype-defining SNPs in our control population (Supplemental Table 4).
Discussion
This study is the first report of the association between genetic variation in the chemokine CXCL12 and risk of cervical carcinoma. In the single SNP analysis, the minor allele at intronic SNP rs266085 was associated with a significantly lower risk of cervical cancer (OR=0.74, 95% C.I. 0.56–0.98) under the recessive model. Haplotype inference based on the nine tagSNPs genotyped in this study identified ten haplotypes that were present in at least 1% of cases or controls, including three that contained the rare allele at rs266085. Only one of these haplotypes (characterized by minor alleles at SNPs rs17885289 and rs266085 and common alleles at all other tagSNPs) was inversely associated with risk of cervical cancer, suggesting that other factors, such as SNPs in linkage with rs266085 but not genotyped in our study, may contribute to cervical cancer risk. Additional studies should be conducted to test whether the association between rs266085 and cervical cancer risk is robust, and whether this association is limited to certain haplotypic backgrounds, as we found.
The SNP rs266085 was chosen to tag a bin containing one other SNP (rs266087) with which rs266085 is in perfect linkage disequilibrium (|D'|=1.0) according to SeattleSNPs. Neither rs266085 nor rs266087 has been reported to have disease associations or functional consequences known to us. rs266085 is located between exons 2 and 3 of CXCL12, and rs266087 is located between exons 3 and 4. CXCL12 has multiple splice variants that differ in the fourth exon (Gleichmann, Gillen et al. 2000; Yu, Cecil et al. 2006), and it is possible that rs266087 (or others variants such as rs266085) influence the relative accumulation of different CXCL12 isoforms (Kimura, Nishioka et al. 2005). SNPs in the 5’ or 3’ UTRs or flanking regions of CXCL12 could also affect isoform accumulation or overall expression levels. The various isoforms of CXCL12 differ in tissue localization (Yu, Cecil et al. 2006), and this could affect immune surveillance or other anti-carcinogenic processes mediated by CXCL12 or its receptor CXCR4, such as angiogenesis or apoptosis.
In the only study to have evaluated the functional impact of genetic variation in CXCL12, Kimura et al. (2005) genotyped 14 SNPs (including five SNPs also genotyped in this study: rs2839685, rs2236534, rs266085, rs1801157, and rs266093) in cell lines from Indonesian individuals, and found that cell lines with certain reconstructed haplotypes showed stronger CXCL12 induction (measured by mRNA accumulation) than others (Kimura, Nishioka et al. 2005). Although the haplotypes containing the minor allele at rs266085 were associated with strong CXCL12 induction, neither rs1788529 nor a correlated SNP were genotyped by Kimura et al., prohibiting us from determining whether the haplotype we found to be in association with cervical cancer risk had a functional effect in that study (Kimura, Nishioka et al. 2005). Even had rs1788529 been genotyped, it is possible that the haplotype structure in Indonesians is sufficiently different from our study population to make comparison difficult.
CXCL12 genotypes have been previously observed to be associated with risk of disease including HIV infection and progression to AIDS (Hendel, Henon et al. 1998; Modi, Scott et al. 2005; Petersen, Glashoff et al. 2005; Vissoci Reiche, Ehara Watanabe et al. 2006), and cancers of the breast (Zafiropoulos, Crikas et al. 2004; Razmkhah, Talei et al. 2005), lung (Razmkhah, Doroudchi et al. 2005), prostate (Hirata, Hinoda et al. 2007), and colon (Hidalgo-Pascual, Galan et al. 2007). Associations with HIV and AIDS have been inconsistent. A large cohort study by Modi et al. found that SNP genotypes containing minor alleles at rs266085 and rs1801157 were more common among high-risk exposed uninfected study participants compared to seroconverters (p=0.06 and p=0.01, respectively) (Modi, Scott et al. 2005). These two SNPs were the only SNPs genotyped in both their study and ours. Increased risks of both lung and breast cancer have been associated with the rs1801157 A allele (Zafiropoulos, Crikas et al. 2004; Razmkhah, Doroudchi et al. 2005; Razmkhah, Talei et al. 2005); this SNP was not related to cervical cancer in this study.
There are several limitations to this study. First, study participants genotyped were only a subset of the total eligible population; some cases died prior to recruitment, and some chose not to participate in the study. In this study, 7% of cases died prior to recruitment and 22% of eligible cases refused to participate. Given the putative role of CXCL12 in cancer metastasis (Koizumi, Hojo et al. 2007), including cervical cancer (Zhang, Lu et al. 2007), nucleotide variation in this gene may be plausibly related to prognosis (Burger and Kipps 2006). Thus, if certain CXCL12 genotypes or haplotypes are associated with death from cervical cancer or being too sick to participate, those genotypes would be under-represented among participating cases, creating an inverse association with risk (if the true association were null), or hindering the detection of positive associations between genotypes and risk.
Another potential limitation to this study is that our tagSNP selection scheme may not have captured all relevant genetic variation in CXCL12 due to potential differences in allele frequencies or haplotype structure between our study population and the SeattleSNPs source population. This limitation would be especially relevant if we missed SNPs that occurred with MAF <5% in SeattleSNPs. Furthermore, one linkage bin of SNPs identified in SeattleSNPs was unsampled in our study due to genotyping failure of the SNP we selected from this bin. This bin contained 12 SNPs with an average MAF of 18%. Lastly, our study lacked power to detect weak associations.
We did not adjust statistical tests for multiple comparisons because most of the comparisons in this study were not independent, yet shared different types of dependence, and it is therefore not clear what method of adjustment would be appropriate. For example, the statistical significance of a risk estimate for a single SNP under the dominant genetic effects model is likely to be dependent on the estimate for the same SNP under recessive and log additive models, but in different ways from how it relates to the risk estimate for a haplotype containing that SNP. Even tests of associations with individual SNPs under a single genetic effects model are dependent because of linkage disequilibrium. Applying a P-value cutoff of 0.05, 1/20 false positives are expected. In comparison, our results showed one significant association out of nine tagSNPs tested, and one significant haplotype association out of 10 common haplotypes tested. We considered subanalyses such as those limited by histology or race to be exploratory analyses that need to be replicated in other studies.
Our study differed from most prior genetic association studies of CXCL12 by including multiple SNPs that captured a substantial amount of the common variation in the gene. We provide evidence for an association between genetic variation in CXCL12 and cervical cancer risk that joins a growing field of research suggesting that genetic variation in cytokines may contribute to oncogenic outcomes of viral infection (eg., (Cordano, Lake et al. 2005; Deshpande, Nolan et al. 2005; Tseng, Lin et al. 2006). Further population-based studies are warranted to test these results. The minimal haplotype associated with an altered risk of cervical cancer in our study was defined by SNPs located in the 5’ UTR (rs17885289), between exons 2 and 3 (rs266085), and in the 3’ UTR (rs266093). These SNP locations are consistent with multiple possible etiologic mechanisms including regulation of CXCL12 induction and alternative splicing. Laboratory experiments similar to those performed by Kimura et al. (2005) would be needed to examine the effect of SNP genotypes on CXCL12 induction and the distribution of CXCL12 splice variants in cell culture (Kimura, Nishioka et al. 2005). Furthermore, planned future work includes investigating associations between genotypes of the receptor CXCR4 and cervical cancer risk, both alone and in combination with CXCL12 genotypes.
Supplementary Material
Acknowledgements
Funding for this work was provided by training grants (R25CA094880) from the National Institutes for Health and (T32HG00035) from the National Human Genome Research Institute to SNM, research and program project grants (R01CA112512, P01CA04279) and contract N01-PC-35412 from the National Cancer Institute, the University of Washington Center for Ecogenetics and Environmental Health (P30ES07033), and institutional funds from the Fred Hutchinson Cancer Research Center.
References
- Barrett JC, Fry B, et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Bleul CC, Farzan M, et al. The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry. Nature. 1996;382(6594):829–833. doi: 10.1038/382829a0. [DOI] [PubMed] [Google Scholar]
- Bleul CC, Fuhlbrigge RC, et al. A highly efficacious lymphocyte chemoattractant, stromal cell-derived factor 1 (SDF-1) J Exp Med. 1996;184(3):1101–1109. doi: 10.1084/jem.184.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch FX, de Sanjose S. Chapter 1: Human papillomavirus and cervical cancer--burden and assessment of causality. J Natl Cancer Inst Monogr. 2003;(31):3–13. doi: 10.1093/oxfordjournals.jncimonographs.a003479. [DOI] [PubMed] [Google Scholar]
- Burger JA, Kipps TJ. CXCR4: a key receptor in the crosstalk between tumor cells and their microenvironment. Blood. 2006;107(5):1761–1767. doi: 10.1182/blood-2005-08-3182. [DOI] [PubMed] [Google Scholar]
- Carlson CS, Eberle MA, et al. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74(1):106–120. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreon JD, Martin MP, et al. Human leukocyte antigen class I and II haplotypes and risk of cervical cancer. Tissue Antigens. 2005;66(4):321–324. doi: 10.1111/j.1399-0039.2005.00478.x. [DOI] [PubMed] [Google Scholar]
- Carrington M, Wang S, et al. Hierarchy of resistance to cervical neoplasia mediated by combinations of killer immunoglobulin-like receptor and human leukocyte antigen loci. J Exp Med. 2005;201(7):1069–1075. doi: 10.1084/jem.20042158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordano P, Lake A, et al. Effect of IL-6 promoter polymorphism on incidence and outcome in Hodgkin's lymphoma. Br J Haematol. 2005;128(4):493–495. doi: 10.1111/j.1365-2141.2004.05353.x. [DOI] [PubMed] [Google Scholar]
- Daling JR, Madeleine MM, et al. The relationship of human papillomavirus-related cervical tumors to cigarette smoking, oral contraceptive use, and prior herpes simplex virus type 2 infection. Cancer Epidemiol Biomarkers Prev. 1996;5(7):541–548. [PubMed] [Google Scholar]
- Deshpande A, Nolan JP, et al. TNF-alpha promoter polymorphisms and susceptibility to human papillomavirus 16-associated cervical cancer. J Infect Dis. 2005;191(6):969–976. doi: 10.1086/427826. [DOI] [PubMed] [Google Scholar]
- Diaz GA. CXCR4 mutations in WHIM syndrome: a misguided immune system? Immunol Rev. 2005;203:235–243. doi: 10.1111/j.0105-2896.2005.00226.x. [DOI] [PubMed] [Google Scholar]
- Garcia-Moruja C, Alonso-Lobo JM, et al. Functional characterization of SDF-1 proximal promoter. J Mol Biol. 2005;348(1):43–62. doi: 10.1016/j.jmb.2005.02.016. [DOI] [PubMed] [Google Scholar]
- Gleichmann M, Gillen C, et al. Cloning and characterization of SDF-1gamma, a novel SDF-1 chemokine transcript with developmentally regulated expression in the nervous system. Eur J Neurosci. 2000;12(6):1857–1866. doi: 10.1046/j.1460-9568.2000.00048.x. [DOI] [PubMed] [Google Scholar]
- Hankey BF, Ries LA, et al. The surveillance, epidemiology, and end results program: a national resource. Cancer Epidemiol Biomarkers Prev. 1999;8(12):1117–1121. [PubMed] [Google Scholar]
- Hendel H, Henon N, et al. Distinctive effects of CCR5, CCR2, and SDF1 genetic polymorphisms in AIDS progression. J Acquir Immune Defic Syndr Hum Retrovirol. 1998;19(4):381–386. doi: 10.1097/00042560-199812010-00009. [DOI] [PubMed] [Google Scholar]
- Hernandez PA, Gorlin RJ, et al. Mutations in the chemokine receptor gene CXCR4 are associated with WHIM syndrome, a combined immunodeficiency disease. Nat Genet. 2003;34(1):70–74. doi: 10.1038/ng1149. [DOI] [PubMed] [Google Scholar]
- Hidalgo-Pascual M, Galan JJ, et al. Analysis of CXCL12 3'UTR G>A polymorphism in colorectal cancer. Oncol Rep. 2007;18(6):1583–1587. doi: 10.3892/or.18.6.1583. [DOI] [PubMed] [Google Scholar]
- Hirata H, Hinoda Y, et al. CXCL12 G801A polymorphism is a risk factor for sporadic prostate cancer susceptibility. Clin Cancer Res. 2007;13(17):5056–5062. doi: 10.1158/1078-0432.CCR-07-0859. [DOI] [PubMed] [Google Scholar]
- Hussain SK, Madeleine MM, et al. Cervical and vulvar cancer risk in relation to the joint effects of cigarette smoking and genetic variation in interleukin 2. Cancer Epidemiol Biomarkers Prev. 2008;17(7):1790–1799. doi: 10.1158/1055-9965.EPI-07-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IARC. Human papillomaviruses. IARC Monogr Eval Carcinog Risks Hum. 2007;90:1–636. [PMC free article] [PubMed] [Google Scholar]
- Kimura R, Nishioka T, et al. Allele-specific transcript quantification detects haplotypic variation in the levels of the SDF-1 transcripts. Hum Mol Genet. 2005;14(12):1579–1585. doi: 10.1093/hmg/ddi166. [DOI] [PubMed] [Google Scholar]
- Koizumi K, Hojo S, et al. Chemokine receptors in cancer metastasis and cancer cell-derived chemokines in host immune response. Cancer Sci. 2007;98(11):1652–1658. doi: 10.1111/j.1349-7006.2007.00606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucia M, Jankowski K, et al. CXCR4-SDF-1 signalling, locomotion, chemotaxis and adhesion. J Mol Histol. 2004;35(3):233–245. doi: 10.1023/b:hijo.0000032355.66152.b8. [DOI] [PubMed] [Google Scholar]
- Li SS, Khalid N, et al. Estimating haplotype frequencies and standard errors for multiple single nucleotide polymorphisms. Biostatistics. 2003;4(4):513–522. doi: 10.1093/biostatistics/4.4.513. [DOI] [PubMed] [Google Scholar]
- Libura J, Drukala J, et al. CXCR4-SDF-1 signaling is active in rhabdomyosarcoma cells and regulates locomotion, chemotaxis, and adhesion. Blood. 2002;100(7):2597–2606. doi: 10.1182/blood-2002-01-0031. [DOI] [PubMed] [Google Scholar]
- Madeleine MM, Johnson LG, et al. Comprehensive analysis of HLA-A, HLA-B, HLA-C, HLA-DRB1, and HLA-DQB1 loci and squamous cell cervical cancer risk. Cancer Res. 2008;68(9):3532–3539. doi: 10.1158/0008-5472.CAN-07-6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta AM, Jordanova ES, et al. Genetic variation of antigen processing machinery components and association with cervical carcinoma. Genes Chromosomes Cancer. 2007;46(6):577–586. doi: 10.1002/gcc.20441. [DOI] [PubMed] [Google Scholar]
- Modi WS, Scott K, et al. Haplotype analysis of the SDF-1 (CXCL12) gene in a longitudinal HIV-1/AIDS cohort study. Genes Immun. 2005;6(8):691–698. doi: 10.1038/sj.gene.6364258. [DOI] [PubMed] [Google Scholar]
- Parkin DM, Bray F, et al. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55(2):74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- Petersen DC, Glashoff RH, et al. Risk for HIV-1 infection associated with a common CXCL12 (SDF1) polymorphism and CXCR4 variation in an African population. J Acquir Immune Defic Syndr. 2005;40(5):521–526. doi: 10.1097/01.qai.0000186360.42834.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razmkhah M, Doroudchi M, et al. Stromal cell-derived factor-1 (SDF-1) gene and susceptibility of Iranian patients with lung cancer. Lung Cancer. 2005;49(3):311–315. doi: 10.1016/j.lungcan.2005.04.014. [DOI] [PubMed] [Google Scholar]
- Razmkhah M, Talei AR, et al. Stromal cell-derived factor-1 (SDF-1) alleles and susceptibility to breast carcinoma. Cancer Lett. 2005;225(2):261–266. doi: 10.1016/j.canlet.2004.10.039. [DOI] [PubMed] [Google Scholar]
- Tseng LH, Lin MT, et al. Correlation of interleukin-10 gene haplotype with hepatocellular carcinoma in Taiwan. Tissue Antigens. 2006;67(2):127–133. doi: 10.1111/j.1399-0039.2006.00536.x. [DOI] [PubMed] [Google Scholar]
- Vidal F, Peraire J, et al. Lack of association of SDF-1 3'A variant allele with long-term nonprogressive HIV-1 infection is extended beyond 16 years. J Acquir Immune Defic Syndr. 2005;40(3):276–279. doi: 10.1097/01.qai.0000176653.89769.4d. [DOI] [PubMed] [Google Scholar]
- Vissoci Reiche EM, Ehara Watanabe MA, et al. The effect of stromal cell-derived factor 1 (SDF1/CXCL12) genetic polymorphism on HIV-1 disease progression. Int J Mol Med. 2006;18(4):785–793. [PubMed] [Google Scholar]
- Walboomers JM, Jacobs MV, et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999;189(1):12–19. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Winer RL, Lee SK, et al. Genital human papillomavirus infection: incidence and risk factors in a cohort of female university students. Am J Epidemiol. 2003;157(3):218–226. doi: 10.1093/aje/kwf180. [DOI] [PubMed] [Google Scholar]
- Yang Y, Li SS, et al. A systematic search for SNPs/haplotypes associated with disease phenotypes using a haplotype-based stepwise procedure. BMC Genet. 2008;9(1):90. doi: 10.1186/1471-2156-9-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Cecil J, et al. Identification and expression of novel isoforms of human stromal cell-derived factor 1. Gene. 2006;374:174–179. doi: 10.1016/j.gene.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Zafiropoulos A, Crikas N, et al. Significant involvement of CCR2-64I and CXCL12-3a in the development of sporadic breast cancer. J Med Genet. 2004;41(5):e59. doi: 10.1136/jmg.2003.013649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JP, Lu WG, et al. Study on CXCR4/SDF-1alpha axis in lymph node metastasis of cervical squamous cell carcinoma. Int J Gynecol Cancer. 2007;17(2):478–483. doi: 10.1111/j.1525-1438.2007.00786.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.