Abstract
The study of protein interactions with DNA is important to gain a fundamental understanding of how numerous biological processes occur, including recombination, transcription, repair, etc. In this study, we use the EcoRII restriction enzyme, which employs a three-site binding mechanism in order to catalyze cleavage of a single recognition site. Using high-speed atomic force microscopy (HS-AFM) to image single-molecule interactions in real time, we were able to observe binding, translocation, and dissociation mechanisms of the EcoRII protein. The results show that the protein can translocate along DNA to search for the specific binding site. Also, once specifically bound at a single site, the protein is capable of translocating along the DNA to locate the second specific binding site. Furthermore, two alternative modes of dissociation of the EcoRII protein from the loop structure were observed, which result in the protein stably bound as monomers to two sites or bound to a single site as a dimer. From these observations, we propose a model in which this pathway is involved in the formation and dynamics of a catalytically active three-site complex.
The formation of synaptic protein-DNA complexes is central to many biological processes which require communication between two or more DNA regions, including recombination (1, 2), replication (3), transcriptional regulation (4), repair (5), transposition (6), and restriction (7, 8). Restriction enzymes serve as useful models to study mechanisms by which the intracellular protein machinery functions on DNA, including synapsis. Restriction enzymes (REases), which require binding to two or more cognate recognition sites in order to be catalytically active are widely spread (9). A multi-site mechanism suggests that restriction enzymes serve as evolutionary precursors to many DNA regulatory factors in the cell (10, 11). Such a mechanism could also serve an inhibitory function to prevent rare unmethylated recognition sites in the host genome from undergoing restriction (12). In addition to systems involving interactions of two DNA helices, interactions of three or more DNA molecules may occur (13-17).
EcoRII is a dimer which recognizes the sequence 5′-CCWGG-3′. It is generally known as a type IIE restriction enzyme. In general, the definition of typeIIE REases is that they bind two DNA recognition sites in order to cleave one of the sites (18). However, recent evidence suggests that the EcoRII protein actually requires three sites to concertedly cleave both strands of one recognition site. This formation of a triple synaptic complex (TSC) or two-loop complex was proposed based on kinetic studies with a plasmid containing three recognition sites, which showed that concerted cleavage of a single site occurred much more quickly than with a one site fragment (19). In addition, direct AFM imaging of the complexes confirmed that the EcoRII protein and a DNA fragment containing three sites could form TSCs (8). The model proposed based on the crystal structure suggests that this occurs by an autoinhibition mechanism (12, 20). The noncatalytic N-terminal binding domains occupy the catalytic C-terminal domain in the unbound protein. In order for DNA to interact with the catalytic site, the N-terminal sites must first bind to a DNA recognition site and undergo a conformational change to expose the catalytic site of the protein for binding.
There is much interest in how site specific proteins search for their cognate recognition site on DNA. It has been shown that proteins are able to ‘find’ their recognition site 100-1000 times faster than what would be expected for random diffusion using a two-step binding process where the protein first interacts nonspecifically with DNA, then undergoes a translocation process to it’s specific binding sequence (21). In 1981, Berg, Winter, and von Hippel proposed mathematical models of four possible site search mechanisms: macroscopic dissociation-reassociation (random collision), microscopic dissociation-reassociation (hopping), intersegmental transfer, or sliding (22, 23). Various approaches utilizing both bulk techniques (24-26) and single molecule techniques (27-29) have been applied to determine the translocation mechanisms employed by different proteins. Total Internal Reflection Fluorescence Microscopy (TIRFM) studies that have been popular for studying protein translocation along DNA (reviewed in (30)). This technique is useful because it allows for the direct visualization of a fluorescently labeled protein moving along the DNA, however the DNA is stretched prior to experiments, which may cause some limitations in what events can be seen. For instance, intersegmental transfer mechanisms which require the formation of DNA loops in order for the protein to be transferred to another site on the DNA would be problematic to observe with this technique.
For this study, we used high speed atomic force microscopy (HS-AFM) to directly image single molecule dynamics of the protein-DNA complexes formed by EcoRII restriction enzyme. It has been used previously to visualize looping and translocation mechanisms of the type III restriction enzyme EcoP15I (31). This HS-AFM relies on a small cantilever design based on the original design by T. Ando (32). This technique has the ability to observe molecular dynamics on a timescale that is 100 times faster than conventional AFM technology (31). On average, we collected images at a rate of 2 frames per second. With this experimental setup, we observed dynamics of specifically bound looped EcoRII complexes. We were able to observe EcoRII dissociation, interaction, and translocation. We observed that the protein can dissociate from a loop in two ways, resulting in stable binding of a dimer to a single site or of a monomer bound to each site. Also, the protein could be observed translocating along nonspecific DNA from one region to another. In addition, the complex was seen to move from one specifically bound looped complex to another site, revealing a possible translocation mechanism where the protein may bind specifically to one site, then bind nonspecifically to the second strand and translocate along the DNA to form the looped complex specifically bound at two sites. The observation of these events helps us to develop a model of how the protein moves along and interacts with DNA in order to carry out its catalytic functions and also demonstrates the utility of this technique for observing mechanisms of protein-DNA interactions.
Methods
EcoRII sample preparation
The EcoRII protein and PCR3 fragment were prepared in the laboratory of Dr. Virginijus Siksnys as described earlier (8). All complexes were assembled in eppendorf tubes prior to deposition onto a mica surface for imaging. Reactions contained 1.4 ng/μL PCR3 DNA fragment (810 bp), 3 nM EcoRII protein, 40 mM HEPES (pH 8.4), and 5 mM CaCl2 in a total volume of 10 μL. Mg2+ ions were replaced by Ca2+ ions, which are known to allow binding but prevent cleavage for many restriction enzymes(33). The interactions of the DNA are mediated by the Ca2+ ions acting as a bridge between the DNA and the negatively charged mica. Reactions were incubated at ambient temperature for 15 min. before depositing 3 μL onto a 1 mm mica disc for ~1 min and washing with buffer for imaging.
Fast-AFM Imaging
All images were acquired with the HS-AFM microscope in Dr. Takeyasu’s laboratory as described previously (31). Scan rates were 1-3 images per second. For visualization of the complexes without cleavage, the imaging buffer was the same as the binding buffer used to assemble the complexes.
Analysis
All length measurements were made using the segmented line tool to manually trace the DNA backbone using Image J, a free image-processing software available from the NIH website (http://rsb.info.nih.gov/ij/). The AFM scan software in Dr. Takeyasu’s laboratory (31) was used to generate the real time movies, as well as the volume analysis. All volume measurements were made by measuring the width of the protein in the x plane (a) and in the y plane (b), as well as the height of the protein in the z direction (h). The volume (V) was obtained using the equation for a segment of a sphere (34).
All errors are reported as the standard error of the mean (SEM). The Windows Movie Maker software was also used extensively for image viewing, as well as for image compression. The supplementary videos and images were rendered in adobe photoshop 7.0 to show only the molecule(s) of interest (see Fig. S1).
Results
Imaging of EcoRII complexes
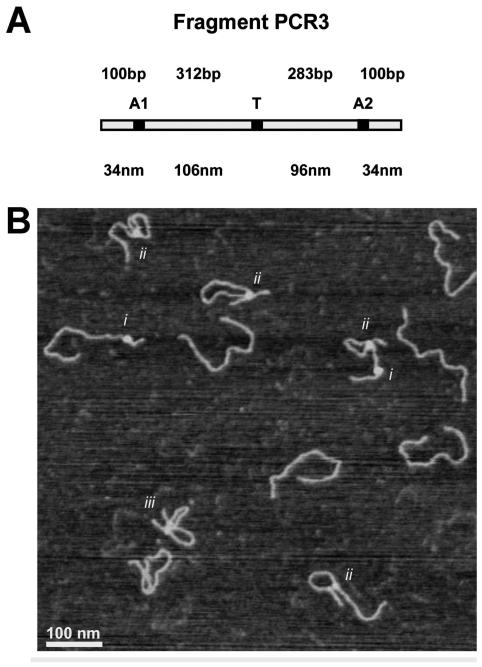
The fragment design used in this experiment is the same as reported previously (8) for characterization the three site binding behavior of the EcoRII restriction enzyme in dry AFM images (Fig. 1A). This fragment is 810 base pairs and contains three recognition sites located almost symmetrically along the DNA fragment. The recognition sites are separated by 283 and 312 bps and are flanked by 100 bp ‘arms’.
Figure 1.
Experimental design (A) PCR3 fragment design showing the location of the three 5 bp EcoRII recognition sites along an 810 bp fragment. (B) Types of EcoRII DNA complexes in dry AFM images: (i) one-site interactions, (ii) two-site interactions, and (iii) three site interactions.
A representative image is shown in Fig. 1B demonstrating the various types of complexes seen. The EcoRII protein can be seen interacting with a single recognition site (i), forming a looped structure with two recognition sites (ii), or forming a double loop structure interacting with all three sites (iii). These results are in line with previous AFM characterization of EcoRII complexes utilizing the same DNA fragment (8), which have shown the occurrence of one-site complexes (43%), two-site complexes (55%), and three site complexes(2%).
Dynamic EcoRII-DNA interactions
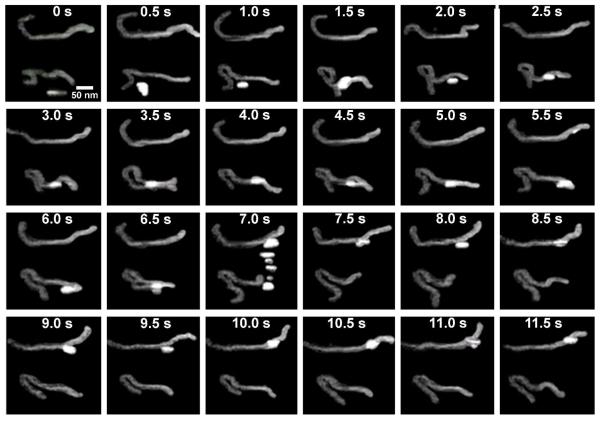
Next, the protein imaged in aqueous buffer conditions in order to obtain dynamic images of EcoRII-DNA interactions. Fig. 2 displays 24 frames acquired at a rate of 2 frames/s demonstrating various aspects of the complex dynamics. Supplementary video 1 also displays this event. Two instances of protein translocation can be seen in this event, first from 4-6 s, and then from 7-11.5 s. Although the protein is seen to ‘track’ the DNA, it appears to follow an interesting path where it is seen to completely overlap the DNA in some frames (as seen at 10-10.5 s), but lay next to the DNA fragment in other frames (as seen at 9-9.5 s). In addition, the protein appeared to briefly interact with two sites to form a transient loop also, as can be seen at 3.5 s and at 6.5 s. At 7 s, the protein can be seen to transfer to another DNA fragment. Multiple blips of the protein motion can be observed in this frame due to the raster scan pattern of the tip, in which the protein is moving towards the other fragment as the tip is scanning, causing the protein to be captured multiple times in the same image. The contribution of the tip to the molecules structure and dynamics is impossible to eliminate. However, in this particular case the protein translocation occurred in the direction perpendicular to the scanning direction. Therefore, the observed dynamics very likely relates to the protein translocation rather than the protein motion exerted by scanning tip. More examples of 1D diffusion can be observed in Fig. S2 and Supplementary videos 1-5.
Figure 2.
EcoRII translocation. Consecutive frames of EcoRII movement over DNA with a time resolution of 500 ms. The protein is seen to bind and form loops. In addition, the protein is seen to bind apparent nonspecific regions of the DNA and translocate down the DNA (5.0 s to 6.0 s and 7.5 s to 11.5 s). The protein is seen to ‘come away’ from and rebind the protein (8.0s and 9.5 s). Macrohopping to an adjacent DNA strand is seen as well (7.0 s). The smearing effect is due to the raster scan pattern of the microscope in which the tip is tracking the protein as it moves. This event can be seen in supplementary video 1.
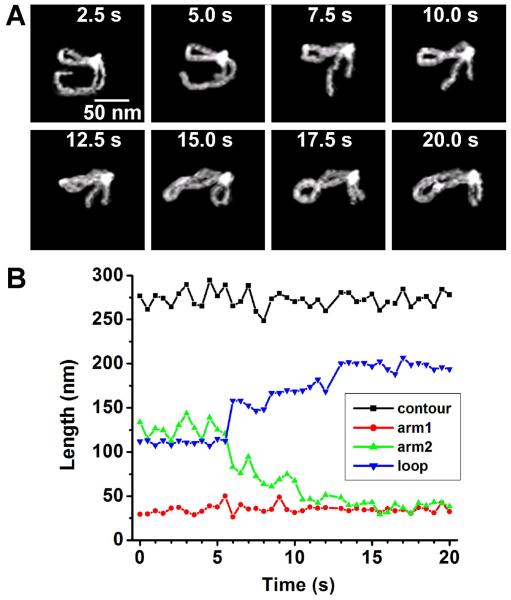
In addition to simple translocation of the protein interacting with a single DNA site, translocation was observed on looped DNA as well. The protein remains bound at one recognition site, while the adjacent strand is observed to gradually translocate from one recognition site to another. Fig. 3A displays 8 frames of this event spanning 20 s. The full event, imaged at a rate of 2 frames/s, can be observed in supplementary video 6. To verify that the protein was interacting with its specific binding sites, the change in loop, arms, and contour length were measured over time (Fig. 3B). The contour length stays close to the expected value of 275 nm for an 810 bp fragment showing that the dynamics of the molecule do not interfere considerably with the measurements. The short arm1 also stays at a value close to the expected 34 nm for specific binding 100 bp from the end throughout the imaging period, which shows that the protein remains specifically bound at this recognition site the entire time. The measurements for arm 2 are expected to correspond to either 388 bp (132 nm) or 417 bp (142 nm) when bound at the middle site. This is seen for the most part, although some increased dynamics of the molecule for this arm at the beginning part of the imaging period cause the value to fall below this value from time to time. These fluctuations are also reflected in the contour length, as many of the ‘peaks’ and ‘valleys’ match, suggesting that the end of the DNA becomes detached from the surface intermittently causing the DNA to appear shorter. The starting value for the loop also corresponds closely to the expected value for either 283 bp (96 nm) or 312 bp (106 nm). At about 6 s, the loop length begins to increase as the arm length decreases, showing translocation occurs along one ‘arm’ of the DNA. After 10 s, translocation stops with the protein bound at another specific DNA site. This is validated by the length measurements which show that arm 2 stops at the expected length of 34 nm expected for binding 100 bp from the end, and that the loop stops at the expected value of 207 nm which corresponds to the large 610 bp loop. The change in the loop length occur over a period of about 10 seconds covering a distance of about 300 bp (102 nm) until is stops at another recognition site. This means translocation occurs at a rate of about 30 bp/s (10.2 nm/s).
Figure 3.
EcoRII translocation (A) Individual frames are shown every 2.5 s. Supplementary video 6 shows this event at 2 frames/s. (B) the change in DNA length over a time period of 20s measured in 0.5s intervals. As the long arm gradually gets shorter, the loop length gradually increases. The contour lengths of the entire molecule and the short arm have consistent values over the entire timescale. The translocation over the length of 300 base pairs occurs within 10 s.
EcoRII dissociation
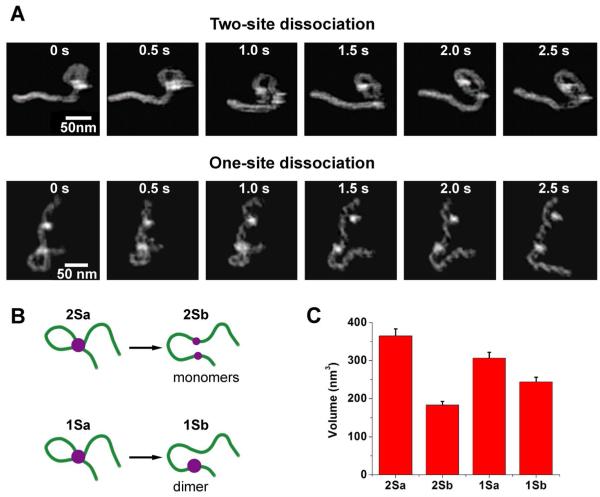
For the next portion of the analysis, the dissociation of the EcoRII protein from the DNA was examined. For the dissociation of single loop complexes, it was observed that there were two distinct modes of dissociation (Fig. 4A and Supplementary videos 7-10). A model is shown in Fig. 4B demonstrating the two pathways, resulting in either a dimer or two monomers which stably interact with a single site. Out of 31 observed loop dissociation events, 18 complexes (58%) were seen to dissociate to an intermediate complex interacting with a single site, and 13 complexes (42%) were seen to dissociate by the breaking apart of protein subunits, resulting in a subunit left interacting with both sites.
Figure 4.
EcoRII dissociation (A) Images showing the two modes of dissociation by the EcoRII protein, one which results in two protein monomers bound to either site (supplementary video 7) and another resulting in a dimer bound to a single site (supplementary video 9). (B) Models illustrating the two routes of possible dissociation of the protein with the protein dissociation into the monomers (top diagram) and without the protein dissociation (bottom diagram). (C) The volume analysis for both types of events. For the dissociation to two sites, protein volume is cut in half, as expected if a dimer is breaking into monomers. For the dissociation to one site, the volume is smaller after dissociation, presumably because there is a contribution of the second DNA strand into the volume measurement.
To gain further insight into the protein stoichiometry in the dissociated complexes, the volume was measured before and after loop dissociation (Fig. 4C). The dissociation to two site bound complexes resulted in an average volume that was about half of the volume (183.2 ± 19.0 nm3) of the pre-dissociation complex (365.1 ± 29.9 nm3), providing evidence that the protein is, in fact dissociating into its constituent subunits. When the protein dissociates to bind one site, there is also a slight reduction in volume (244.0 ± 20.1 nm3) from the pre-dissociation complex (306.5 ± 28.5 nm3), which could reflect the loss of the contribution of the second DNA strand to the volume of the complex.
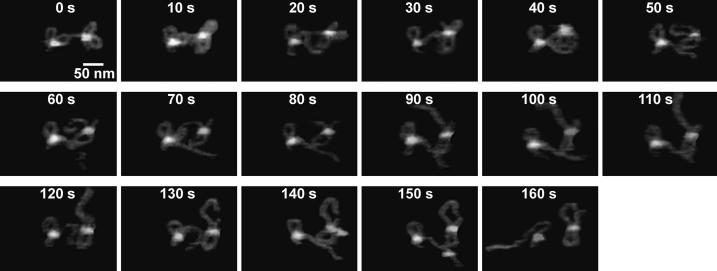
In addition to single loop complexes, 13 double loop complexes were observed to dissociate (Fig. 5 and Supplementary videos 11-12). Of those 13 events, 6 (46%) dissociated to a small loop, 4 (31%) dissociated to a big loop, and 2 (15%) seemed to spontaneously dissociate without a looped intermediate. In the last event of Supplementary video 2, the two-loop complex actually appeared to dissociate into a big loop, then possibly reform into the two loop complexes once again, and then dissociate to a small loop intermediate (see Fig. S4). These results show that two site binding is an intermediate complex formed during the dissociation process. Furthermore, it also shows no clear geometrical bias for the mode of double loop dissociation.
Figure 5.
Frames showing two-loop dissociation every 10 s. The triple synaptic complex (TSC) on the left dissociates to a single loop at 20 s, followed by the TSC on the right dissociating to a single loop at 50 s. At 160 s, the loop on the left dissociates to a one-site complex. These events, originally imaged at 2 frames/s can be seen as movies in Supplementary video 11.
Discussion
With HS-AFM, we have demonstrated the visualization of single-molecule EcoRII-DNA complex dynamics at the nanoscale. One important finding is that the protein can perform the search for a second cognate site on the DNA while being bound to the first recognition site. This is an expected property of proteins that can bind to several recognition sites but has never been observed directly. The results obtained here also suggest that translocation along the DNA strand by either sliding and/or hopping mechanisms in search of the initial binding site and the second binding site. The current view of search mechanisms suggests that the optimal route for the protein to quickly locate its target site involves a combination of short range slides and hops with long range jumps (35). One study using computer simulations came to the conclusion that the most efficient search occurs when a protein is sliding along DNA 20% of the time, and hopping and jumping for the other 80% of the time (36). Generally hopping over DNA is considered to occur in increments of <10 bp (<3.4 nm) (35). These small-scale dissociation-reassociation events are difficult to observe with the resolution limits of HS-AFM which usually ranges from about 5-10 nm. Although the resolution limits make it difficult to make any definitive conclusions about the degree of hopping/sliding employed by this protein, comparison to a previous HS-AFM study using the same imaging system for the ATP-dependent type III restriction enzyme EcoP15I shows that the protein translocates over the DNA for long distances resulting in accumulated supercoiling (31). In contrast the EcoRII protein translocated along the DNA for an average distance of 74 nm (218 bp), ranging from 47 nm (138 bp) to 128 nm (376 bp) before dissociating from the DNA. This value is slightly larger than the previously suggested value of 100 bp (37), but is within reason because translocations over shorter distance may have been missed due to the time resolution of the microscope. If translocation occurred over 1-2 seconds, this event would last 4 frames or less, which makes it difficult to identify events which might have occurred in shorter timescales. Similar HS-AFM studies with different DNA-binding proteins may further our understanding of how proteins utilizing various search mechanisms behave during the imaging process.
It is interesting to consider how fast the protein is moving along the DNA. From our images, the Einstein equation yields values of 7.2×10-4 μm2/s for 1D diffusion and about 1.8×10-5 μm2/s for 2D diffusion (Fig. S3). The protein movement was obviously impaired for 2D diffusion due to interactions with the surface, but the influence of the surface or the tip on is not clear. A previous study using the same imaging system attempted to vary the scan rate to see if there was an effect on the measured diffusion rates and found that the diffusion coefficients obtained were not significantly changed (38). Currently, TIRF studies are the standard technique for the measurement of diffusion coefficients, but the measurement of looped translocations cannot be performed with such a technique because the DNA must be stretched over the surface (30, 39).
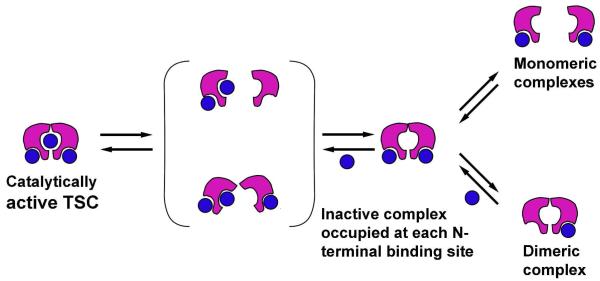
In this study, EcoRII was shown to have two distinct dissociation pathways resulting in either a dimer or two monomers, which remain stably bound to a single site following dissociation of the complexes. These observations support our previous assumption based on imaging of a bi-lobed shape of the protein in some looped complexes (8) and are in line with the results of gel shift experiments (40, 41). These observations prompted us to propose the following model of the EcoRII-DNA complex formation and dynamics (Fig. 6). The catalytically active triple synaptic complex loses one contact resulting in one-looped complexes bound at two recognition sites. We did not find a preference for either possible arrangement of the loops as both small and large loops were observed. In this scenario, the looped complex is shown bound to the N-terminal regions of the protein dimer, but we cannot exclude binding to N and C termini. The final step has two possible outcomes which were both observed in our AFM images. The protein either dissociates from one site with the dimer being bound to the other site, or the dimer can fall apart, forming monomeric complexes. Interestingly, free protein is primarily a dimer in solution (42), suggesting that dissociation into monomers is promoted by the protein-DNA interaction. The dissociation of the TSC may proceed via intermediate states shown in brackets. The crystal structure shows that the non-catalytic N-terminal domains block binding to the catalytic C-terminal domain in the native unbound crystal structure (20). If the two N-terminal domains are able to bind two strands, which subsequently break apart into monomers, this may expose the C-terminal site for binding. The third strand may then bind and the structure may then re-associate to form the active dimeric complex. The association of half complexes to form the active synaptic complexes is reminiscent of the mechanisms utilized by restriction enzymes SgrA1 (43, 44) and FokI (45). An alternative is dissociation-reassociation of the third strand via opening of the cleft, as has been proposed in previously(19). In this scenario, the formation of monomers may simply be a result of dissociation, and not directly observed as an intermediate for complex formation. The first model may lead to the formation of the looped complex mediated by a protein monomer. We did not directly observe these intermediates, so the model of the opening of the cleft for grabbing or dissociation of the third strand is still possible.
Figure 6.
Dynamic model of the catalytically active triple synaptic complex (TSC). Circles in this scheme denote the DNA capable of binding to one of the three binding sites of the protein. An active TSC complex is formed after the two N-terminal binding sites are occupied and the third strand binds to C-terminal pocket. The scheme in brackets shows hypothetical transient forms of TSC complex illustrating that the complex can dissociate and reform with or without protein dissociation. The latter pathway is in line with available crystallographic data whereas the first pathway is consistent with the AFM volume measurements (8). The TSC complex breaks apart into two-site complexes followed by the dissociation into two monomeric complexes or a dimeric complex bound to a single binding site.
In summary, this study demonstrates the power of HS-AFM for the study of protein-DNA interactions. This technique demonstrates the ability to image multisite protein-DNA complexes with DNA in a relaxed conformation not amenable to other methods such as TIRF imaging techniques
Supplementary Material
Acknowledgements
We thank L. Shlyakhtenko and A. Lushnikov for the help in preparations for the HS-AFM experiments and useful comments on the manuscript.
The work was supported in part by grants GM 062235 (NIH), PHY 0615590 (NSF), Nebraska Research Imitative (NRI) (all to YLL); 0812853 (NSF EAPSI program) and the Structural biology and biophysics fellowship to JG provided by GAANN funding (U.S. Department of Education grant P200A060150), Grant-in-aid for Scientific Research (A) (19207001) and Priority Areas (16084203) to K.T., Grant-in-Aid for JSPS Fellows (21-5533) to Y.S.
Abbreviations
- HS-AFM
High Speed Atomic Force Microscopy
- TSC
Triple Synaptic Complex
- TIRFM
Total Internal Reflection Fluorescence Microscopy
References
- 1.Swanson P. The bounty of RAGs: recombination signal complexes and reaction outcomes. Immunol Rev. 2004;200:90–114. doi: 10.1111/j.0105-2896.2004.00159.x. [DOI] [PubMed] [Google Scholar]
- 2.Mouw K, Rowland S, Gajjar M, Boocock M, Stark W, Rice P. Architecture of a serine recombinase-DNA regulatory complex. Mol Cell. 2008;30:145–155. doi: 10.1016/j.molcel.2008.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moens P, Marcon E, Shore J, Kochakpour N, Spyropoulos B. Initiation and resolution of interhomolog connections: crossover and non-crossover sites along mouse synaptonemal complexe. J Cell Sci. 2007;120:1017–1027. doi: 10.1242/jcs.03394. [DOI] [PubMed] [Google Scholar]
- 4.Hoverter N, Waterman M. A Wnt-fall for gene regulation: repression. Sci Signal. 2008;1:pe43. doi: 10.1126/scisignal.139pe43. [DOI] [PubMed] [Google Scholar]
- 5.Weterings E, Chen D. The endless tale of non-homologous end-joining. Cell Res. 2008;18:114–124. doi: 10.1038/cr.2008.3. [DOI] [PubMed] [Google Scholar]
- 6.Vaezeslami S, Sterling R, Reznikoff W. Site-directed mutagenesis studies of tn5 transposase residues involved in synaptic complex formation. J Bacteriol. 2007;189:7436–7441. doi: 10.1128/JB.00524-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lushnikov A, Potaman V, Oussatcheva E, Sinden R, Lyubchenko Y. DNA strand arrangement within the SfiI-DNA complex: atomic force microscopy analysis. Biochemistry. 2006;45:152–158. doi: 10.1021/bi051767c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shlyakhtenko L, Gilmore J, Portillo A, Tamulaitis G, Siksnys V, Lyubchenko Y. Direct visualization of the EcoRII-DNA triple synaptic complex by atomic force microscopy. Biochemistry. 2007;46:11128–11136. doi: 10.1021/bi701123u. [DOI] [PubMed] [Google Scholar]
- 9.Halford S, Welsh A, Szczelkun M. Enzyme-mediated DNA looping. Annu Rev biophys Biomol Struct. 2004;33:1–24. doi: 10.1146/annurev.biophys.33.110502.132711. [DOI] [PubMed] [Google Scholar]
- 10.Mucke M, Kruger D, Reuter M. Diversity of type II restriction endonucleases that require two DNA recognition sites. Nucleic Acids Res. 2003;31:6079–6084. doi: 10.1093/nar/gkg836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mucke M, Grelle G, Behlke J, Kraft R, Kruger D, Reuter M. EcoRII: a restriction enzyme evolving recombination functions. EMBO J. 2002;21:5262–5268. doi: 10.1093/emboj/cdf514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Szczepek M, Mackeldanz P, Moncke-Buchner E, Alves J, Kruger D, Reuter M. Molecular analysis of restriction endonuclease EcoRII from Escherichia coli reveals precise regulation of its enzymatic activity by autoinhibition. Mol Microbiol. 2009;72:1011–1021. doi: 10.1111/j.1365-2958.2009.06702.x. [DOI] [PubMed] [Google Scholar]
- 13.Watson N, Chaconas G. Three-site synapsis during Mu DNA transposition: A critical intermediate preceding engagement of the active site. Cell. 1996;85:435–445. doi: 10.1016/s0092-8674(00)81121-6. [DOI] [PubMed] [Google Scholar]
- 14.Kobryn K, Watson M, Allison R, Chaconas G. The Mu three-site synapse: a strained assembly platform in which delivery of the L1 transposase binding site triggers catalytic commitment. Mol Cell. 2002;10:659–669. doi: 10.1016/s1097-2765(02)00596-8. [DOI] [PubMed] [Google Scholar]
- 15.Merickel S, Haykinson M, Johnson R. Communication between Hin recombinase and Fis regulatory subunits during coordinate activation of Hin-catalyzed site-specific DNA inversion. Genes Dev. 1998;12:2803–2816. doi: 10.1101/gad.12.17.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heichman K, Moskowitz J, Johnson R. Configuration of DNA strands and mechanism of strand exchange in the Hin invertasome as revealed by analysis of recombinant knots. Genes Dev. 1991;5:1622–1634. doi: 10.1101/gad.5.9.1622. [DOI] [PubMed] [Google Scholar]
- 17.Merickel S, Johnson R. Topological analysis of Hin-catalysed DNA recombination in vivo and in vitro. Mol Microbiol. 2004;51:1143–1154. doi: 10.1046/j.1365-2958.2003.03890.x. [DOI] [PubMed] [Google Scholar]
- 18.Roberts R, Belfort M, Bestor T, Bhagwat A, Bickle T, Bitinaite J, Blumenthal R, Degtyarev S, Dryden K, Dybvig K, Firman K, Gromova E, Gumport R, Halford S, Hattman S, Heitman J, Hornby D, Janulaitus A, Jeltsch A, Josephsen J, Kiss A, Klaenhammer T, Kobayashi I, Kong H, Kruger D, Lacks S, Marinus M, Miyahara M, Morgan R, Murray N, Nagaraja V, Piekarowicz A, Pingoud A, Raleigh E, Rao D, Reich N, Repin V, Selker E, Shaw P, Stein D, Stoddard B, Szybalski W, Trautner T, Van Etten J, Vitor J, Wilson G, Xu S. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 2003;31:1805–1812. doi: 10.1093/nar/gkg274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tamulaitis G, Sasnauskas G, Mucke M, Siksnys V. Simultaneous binding of three recognition sites is necessary for a concerted plasmid DNA cleavage by EcoRII restriction endonuclease. J Mol Biol. 2006;358:406–419. doi: 10.1016/j.jmb.2006.02.024. [DOI] [PubMed] [Google Scholar]
- 20.Zhou X, Wang Y, Reuter M, Mucke M, Kruger D, Meehan E, Chen L. Crystal structure of type IIE restriction endonuclease EcoRII reveals an autoinhibition mechanism by a novel effector-binding fold. J Mol Biol. 2004;335:307–319. doi: 10.1016/j.jmb.2003.10.030. [DOI] [PubMed] [Google Scholar]
- 21.Riggs A, Bourgeois S, Cohn M. The lac repressor-operator interaction. 3. Kinetic studies. J Mol Biol. 1970;53:401–417. doi: 10.1016/0022-2836(70)90074-4. [DOI] [PubMed] [Google Scholar]
- 22.Berg O, Winter R, von Hippel P. Diffusion-Driven Mechanisms of protein Translocation on Nucleic Acids 1. Models and Theory. Biochemistry. 1981;20:6929–6948. doi: 10.1021/bi00527a028. [DOI] [PubMed] [Google Scholar]
- 23.Winter R, von Hippel P. Diffusion-Driven Mechanisms of protein Translocation on Nucleic Acids 2. The Echerichia coli repressor-Operator Interaction: Equilibrium Measurements. Biochemistry. 1981;20:6948–6960. doi: 10.1021/bi00527a029. [DOI] [PubMed] [Google Scholar]
- 24.Stanford N, Szczelkun M, Marko J, Halford S. One- and three-dimensional pathways for proteins to reach specific DNA sites. EMBO J. 2000;19:6546–6557. doi: 10.1093/emboj/19.23.6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gowers D, Halford S. Protein motion from non-specific to specific DNA by three-dimensional routes aided by supercoiling. EMBO J. 2003;22:1410–1418. doi: 10.1093/emboj/cdg125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gowers D, Wilson G, Halford S. Measurement of the contributions of 1D and 3D pathways to the translocation of a protein along DNA. Proc Natl Acad Sci USA. 2005;102:15883–15888. doi: 10.1073/pnas.0505378102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guthold M, Zhu X, Rivetti C, Yang G, Thomson N, Kasas S, Hansma H, Smith B, Hansma P, Bustamante C. Direct observation of one-dimensional diffusion and transcription by Escherichia coli RNA polymerase. Biophys J. 1999;77:2284–2294. doi: 10.1016/S0006-3495(99)77067-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sakata-Sogawa K, Shimamoto N. RNA polymerase can track a DNA groove during promoter search. Proc Natl Acad Sci USA. 2004;104:14731–14735. doi: 10.1073/pnas.0406441101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van den Broek B, Lomholt M, Kalisch S, Metzler R, Wuite G. How DNA coiling enhances target localization by proteins. Proc Natl Acad Sci USA. 2008;105:15738–15742. doi: 10.1073/pnas.0804248105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gorman J, Greene E. Visualizing one-dimensional diffusion of proteins along DNA. Nat Struct Mol Biol. 2008;15:768–774. doi: 10.1038/nsmb.1441. [DOI] [PubMed] [Google Scholar]
- 31.Crampton N, Yokokawa M, Dryden D, Edwardson J, Rao D, Takeyasu K, Yoshimura S, Henderson R. Fast-scan atomic force microscopy reveals that the type III restriction enzyme EcoP15I is capable of DNA translocation and looping. Proc Natl Acad Sci USA. 2007;104:12755–12760. doi: 10.1073/pnas.0700483104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ando T, Kodera N, Takai E, Maruyama D, Saito K, Toda A. A high-speed atomic force microscope for studying biological macromolecules. Proc Natl Acad Sci USA. 2001;98:12468–12472. doi: 10.1073/pnas.211400898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bellamy S, Kovacheva Y, Zulkipli I, Halford S. Differences between Ca2+ and Mg2+ in DNA binding and release by the SfiI restriction endonuclease: implications for DNA looping. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp569. DOI 10.1093/nar/gkp569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Henderson R, Schneider S, Li Q, Hornby D, White S, Oberleithner H. Imaging ROMK1 inwardly rectifying ATP-sensitive K+ channel protein using atomic force microscopy. Proc Natl Acad Sci USA. 1996;93:8756–8760. doi: 10.1073/pnas.93.16.8756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Halford S. An end to 40 years of mistakes in DNA-protein association kinetics? Biochem Soc Trans. 2009;37:343–348. doi: 10.1042/BST0370343. [DOI] [PubMed] [Google Scholar]
- 36.Givaty O, Levy Y. Protein sliding along DNA: Dynamics and Structural Characterization. J Mol Biol. 2008;385:108797. doi: 10.1016/j.jmb.2008.11.016. [DOI] [PubMed] [Google Scholar]
- 37.Halford S, JF M. How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res. 2004;32:3040–3052. doi: 10.1093/nar/gkh624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yokokawa M, Yoshimura S, Naito Y, Ando T, Yagi N, Sakai N, Takeyasu K. Fast-scanning atomic force microscopy reveals the molecular mechanism of DNA cleavage by ApaI endonuclease. IEE Proc Nanobiotechnol. 2006;153:60–66. doi: 10.1049/ip-nbt:20050018. [DOI] [PubMed] [Google Scholar]
- 39.Fazio T, Visnapuu M, Wind S, Greene E. DNA curtains and nanoscale curtain rods: high-throughput tools for single molecule imaging. Langmuir. 2008;24:10524–10531. doi: 10.1021/la801762h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karpova E, Kubareva E, Gromova E, Buryanov Y. Peculiarities of the binding of restriction endonuclease EcoRII to synthetic DNA duplexes. Biochem Mol Biol Int. 1993;29:113–121. [PubMed] [Google Scholar]
- 41.Karpova E, Kubareva E, Shabarova Z. A Model of EcoRII Restriction Endonuclease Action: The Active complex is Most Likely Formed by One Protein Subunit and One DNA Recognition Site. IUBMB Life. 1999;48:91–98. doi: 10.1080/713803460. [DOI] [PubMed] [Google Scholar]
- 42.Reuter M, Kupper D, Meisel A, Schroeder C, Kruger D. Cooperative Binding Properties of Restriction Endonuclease EcoRII with DNA Recognition Sites. J Biol Chem. 1998;273:8294–8300. doi: 10.1074/jbc.273.14.8294. [DOI] [PubMed] [Google Scholar]
- 43.Wood K, Daniels L, Halford S. Long-range communications between DNA sites by the dimeric restriction endonuclease SgrAI. J Mol Biol. 2005;350:240–253. doi: 10.1016/j.jmb.2005.04.053. [DOI] [PubMed] [Google Scholar]
- 44.Daniels L, Wood K, Scott D, Halford S. Subunit assembly for DNA cleavage by restriction endonuclease SgrAI. J Mol Biol. 2003;327:579–591. doi: 10.1016/s0022-2836(03)00143-8. [DOI] [PubMed] [Google Scholar]
- 45.Bitinaite J, Wah D, Aggarwal A, Schildkraut I. FokI dimerization is required for DNA cleavage. Proc Natl Acad Sci USA. 1998;95:10570–10575. doi: 10.1073/pnas.95.18.10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.