Abstract
During development, cascades of regulatory genes act in a hierarchical fashion to subdivide the embryo into increasingly specified body regions. This has been best characterized in Drosophila, where genes encoding regulatory transcription factors form a network to direct the development of the basic segmented body plan. The pair-rule genes are pivotal in this process as they are responsible for the first subdivision of the embryo into repeated metameric units. The Drosophila pair-rule gene fushi tarazu (ftz) is a derived Hox gene expressed in and required for the development of alternate parasegments. Previous studies suggested that Ftz achieves its distinct regulatory specificity as a segmentation protein by interacting with a ubiquitously expressed cofactor, the nuclear receptor Ftz-F1. However, the downstream target genes regulated by Ftz and other pair-rule genes to direct segment formation are not known. In this study, we selected candidate Ftz targets by virtue of their early expression in Ftz-like stripes. This identified two new Ftz target genes, drumstick (drm) and no ocelli (noc), and confirmed that Ftz regulates a serotonin receptor (5-HT2). These are the earliest Ftz targets identified to date and all are coordinately regulated by Ftz-F1. Engrailed (En), the best-characterized Ftz/Ftz-F1 downstream target, is not an intermediate in regulation. The drm genomic region harbors two separate 7-stripe enhancers, identified by virtue of predicted Ftz-F1 binding sites and these sites are necessary for stripe expression in vivo. We propose that pair-rule genes, exemplified by Ftz/Ftz-F1, promote segmentation by acting at different hierarchical levels, regulating first, other segmentation genes; second, other regulatory genes that in turn control specific cellular processes such as tissue differentiation; and, third, 'segmentation realizator genes' that are directly involved in morphogenesis.
Keywords: Segmentation, Hox genes, pair-rule genes, Ftz, Ftz-F1
INTRODUCTION
The development of multicellular organisms from single fertilized egg cells is a hierarchical process in which cell fates are increasingly specified as development proceeds. Nowhere have we learned more about this process than from genetic screens carried out in Drosophila melanogaster (Nusslein-Volhard et al., 1985). These screens identified sets of regulatory genes that sequentially subdivide the Drosophila embryo into increasingly specified body parts along the anterior-posterior axis of the egg, culminating in the formation of the body segments that are the basis of the insect body plan. The segmentation genes identified in these screens were found to participate in a complex regulatory cascade: the overlapping and staggered expression patterns of maternal and gap genes are required for striped expression of pair-rule genes, which in turn regulate segmental expression of segment polarity genes and region-specific expression of homeotic genes. Together, these genes form an integrated network of regulatory genes that control early embryonic development (reviewed in Nasiadka et al., 2002; Schroeder et al., 2004).
The pair-rule genes are the first genes to be expressed in repeated patterns in the early embryo in sets of transverse stripes in the primordia of alternating segmental units. Each pair-rule gene is expressed in the primordia of the alternating metameric region missing in embryos carrying mutations in that pair-rule gene. For example, the pair-rule gene fushi tarazu (ftz) is expressed in the primordia of the even numbered parasegments which are missing in ftz embryos (Carroll and Scott, 1985; Hafen et al., 1984). Elegant studies have explained how pair-rule stripes can be generated by the combinatorial action of activating and repressing maternal and gap transcription factors which act on stripe-specific cis-regulatory elements (reviewed in Arnosti et al., 1996; Small et al., 1996). However, less is known about how the pair-rule genes, once expressed in striped patterns, impact segment morphogenesis. We have begun to address this by identifying target genes of the pair-rule segmentation protein Ftz. Ftz contains a DNA-binding homeodomain and activates transcription (Florence et al., 1991; Han et al., 1989; Nelson and Laughon, 1990; Pick et al., 1990; Winslow et al., 1989; Yu et al., 1999). However, like other Hox proteins, Ftz binding to DNA is weak and promiscuous, with >14 million predicted binding sites in the Drosophila genome (Bowler et al., 2006). The specificity of Ftz target site selection is achieved by Ftz interaction with a specific cofactor, the orphan nuclear receptor Ftz-F1, whose DNA binding specificity is much more stringent than that of Ftz (Florence et al., 1997; Guichet et al., 1997; Yu et al., 1997); reviewed in (Pick et al., 2006). Ftz and Ftz-F1 form a stable complex in vivo and bind cooperatively to DNA (Yu et al., 1997; Yussa et al., 2001). Because Ftz-F1 DNA binding specificity is greater than Ftz DNA binding specificity, the predominant determinant of Ftz/Ftz-F1 target genes is the Ftz-F1 binding site. In fact, when Ftz is overexpressed, interaction with Ftz-F1 obviates the need for Ftz DNA binding, and the anti-ftz phenotype can be generated by a Ftz protein lacking its DNA binding homeodomain (Copeland et al., 1996; Fitzpatrick et al., 1992; Guichet et al., 1997).
The best-characterized Ftz/Ftz-F1 target genes are ftz itself (autoregulation) and the downstream target engrailed (en). The Ftz/Ftz-F1-dependent enhancers that regulate ftz and en each contain composite Ftz/Ftz-F1 binding sites that are necessary for gene expression in vivo (Florence et al., 1997; Han et al., 1998; Pick et al., 1990; Schier and Gehring, 1993a). The composite binding sites in the ftz and en enhancers differ in spacing and orientation; in the ftz enhancer, the Ftz binding site is 5' to the Ftz-F1 site, with 7 nucleotides between them while in the en enhancer, the Ftz-F1 site is 5' to the Ftz site and the sites are separated by 11 nucleotides (see Bowler et al., 2006). A previous study utilized these ftz-like and en-like binding site configurations, along with a compilation of all experimentally verified Ftz and Ftz-F1 binding sequences, to identify additional Ftz/Ftz-F1 targets in the Drosophila genome. This study identified apontic (apt) and Sulfated (Sulf1) as downstream targets of Ftz/Ftz-F1 but most of the genes harboring multiple ftz-like or en-like Ftz/Ftz-F1 binding sites were not in fact regulated by Ftz/Ftz-F1 (Bowler et al., 2006). This suggested that other approaches would be required to identify Ftz/Ftz-F1 targets genes.
Here, we have made use of the availability of an in situ database from the Berkeley Drosophila Genome Project (Tomancak et al., 2002) to identify candidate Ftz target genes that are expressed in striped patterns — 'stripy genes.' We characterized three genes that are expressed in stripes that overlap the Ftz stripes. Each genomic region contains multiple potential Ftz-F1 binding sites and each gene requires Ftz as well as Ftz-F1 for striped expression. These genes were 5-HT2, previously shown to require Ftz for expression in stripes (Colas et al., 1995); noc, a zinc finger transcription factor; and drm, which is expressed in an en-like pattern of 14 stripes, 7 of which require Ftz and Ftz-F1. We identified two regulatory elements for drm that direct expression in Ftz-like stripes, each containing predicted Ftz-F1 binding sites that were shown to be required for expression of drm-lacZ fusion genes in vivo. Together, these results suggest that Ftz and Ftz-F1 coordinately regulate downstream target genes that function at different levels of a gene hierarchy, with some target genes encoding regulatory proteins and others encoding products directly involved in morphogenesis.
MATERIALS AND METHODS
Fly Stocks
Flies were maintained at 25°C on a standard diet. The ftz mutant was ftz9H34 balanced over TM3, hb-lacZ to identify mutant embryos. Embryos derived from ftz-f1 germline clones (referred to as ftz-f1 mutants) were generated with the autosomal FLP-DFS technique (Chou and Perrimon, 1992; Chou and Perrimon, 1996; Chou et al., 1993) using ftz-f119(Broadus et al., 1999; Fortier et al., 2003; Pick et al., 2006). Ftz was ectopically expressed throughout blastoderm embryos by mating UAS-myc-ftz males (Lohr and Pick, 2005) with females homozygous for an NGT40/GAL4 driver (Tracey et al., 2000). Expression was examined for two en alleles, en1, Df(2R)42 and enE, each balanced over CyO, hb-lacZ to identify mutants. Transgenic fly lines were generated by Rainbow Transgenic Flies, CA. Multiple independent lines were established for each construct, maintained over balancer chromosomes or as homozygotes.
Analysis of embryonic expression patterns
Standard protocols were followed for in situ hybridization (Kosman and Small, 1997; Tautz and Pfeifle, 1989) and antibody staining (Gutjahr et al., 1994). Digoxigenin-labeled RNA probes were made with cDNA clones LD26791 (drm), LD28078 (noc), and RH04788 (5-HT2) from the Drosophila Genome Resource Center. Protein/RNA double staining followed standard in situ hybridization protocols, including the Proteinase K treatment followed by addition of primary antibodies, rat anti-Ftz (1:200) (Kosman et al., 1998) and sheep anti-digoxigenin (1:1000, Roche). Secondary antibodies, anti-rat Alexa Fluor 488 and anti-sheep Alexa Fluor 555 (Invitrogen), were used at 1:600. After washes, including a final wash in PBST overnight at 4ºC, and rinses with PBS, embryos were mounted in 90% glycerol, 0. 1M Tris-HCl, pH 7.9. For double RNA in situs, embryos were incubated simultaneously with digoxigenin-labeled lacZ or target RNA and biotinylated ftz RNA probes, followed by detection with mouse anti-biotin (Roche) and anti-digoxigenin antibodies as described above. For drm reporter constructs, embryos were stained with anti-β-galactosidase antibody (Cappel, 1:1000) (Gutjahr et al., 1993). Expression patterns shown here were observed in at least 3–5 independent lines for each construct. For drm5-lacZ, 5 independent lines were analyzed and in only one of these was expression detected. Expression was early in 7 stripes and developed into 14-stripes during germ band extension. Although the initial 7 stripes appeared to overlap with Ftz, the fact that this expression was detected in only one drm5-lacZ line, which was PCR-verified, suggests that it was due to a position effect and not enhancer sequences in the drm5 fragment. In situ hybridizations were visualized using a LeicaDMRB microscope. Fluorescent staining was captured using the Zeiss LSM 510 confocal microscope with a 16X Zeiss objective with oil immersion. Multiple fluorescent images were captured with multitrack switching. Alexa 488 antibodies were excited with a 488nm laser and detected at 505–530nm. Alexa 555 antibodies were excited with a 543nm laser and detected above 560nm.
Identification of drm transcription start site and enhancer construct design
RNA was extracted from 0–9 hr. D. melanogaster w1118 embryos with TRIzol (Invitrogen, CA) and a Qiagen RNA Extraction Kit (Qiagen, CA). The TSS was mapped using 5’ RLM-RACE (Ambion, TX). Briefly, RNA was treated with CIP and TAP to select full-length mRNAs. The RACE adapter was ligated to the 5’ end, and cDNA was made using reverse transcriptase. Two rounds of nested PCR resulted in a PCR product with the 5’ transcription start site immediately downstream of the adapter. The drm gene contains a perfect match to an INR, an almost perfect Downstream Promoter Elemet (DPE), and lacks an apparent TATA box. To construct drm enhancer-reporter transgenes, genomic fragments were isolated by PCR (primer sequences available upon request). Genomic location of fragments is: drm1, −606–2314; drm2, +513–1511; drm 34, +3198–4469; drm5, +11,900-12,714. Site-specific mutagenesis was carried out as described (Lohr and Pick, 2005). For expression in Drosophila, fragments were inserted directionally into the P-element vector pX28, upstream of a basal hsp70 promoter and lacZ reporter gene (Bowler et al., 2006; Segalat et al., 1994).
RESULTS
Identification of stripy genes as candidate Ftz targets
To identify candidate targets of Ftz in promoting segmentation, we searched the BDGP in situ expression database (Release 2, http://fruitfly.org:9005/cgi-bin/ex/insitu.pl; (Tomancak et al., 2002) for genes expressed early enough to be possible direct targets of pair-rule genes and with expression patterns that are modulated along the A-P axis. This identified 95 genes, corresponding to 6.7% (95/1403) of the genes in the database. Many of these are expressed in seven or fourteen stripes ('stripy genes'). Some stripy genes show additional modulation, such as restriction of stripes to the ventral side, restriction to the posterior abdominal regions of the embryo, or additional expression in the head (data not shown). These stripy genes are likely to be regulated by one or more of the seven pair-rule transcription factors expressed in different registers along the anterior-posterior axis. To narrow down potential Ftz targets among these genes, we searched computationally for genes harboring Ftz/Ftz-F1 binding sites with spacing matching those of the Ftz/Ftz-F1 binding sites in either the ftz or en enhancers (Introduction). Thirty of the 95 genes contain composite Ftz/Ftz-F1 sites within 20 kb of their annotated TSS (Bowler, 2004). Analysis of the expression of these 30 genes in wild type and ftz-f1 mutants identified 10 genes in addition to the previously characterized direct targets en, apt and Sulf1, that appeared to be responsive to Ftz-F1. These were: 5-HT2, noc, drm, CG12094, ImpL2, Sema-5c, Ama, Cyt-b5, danr, and RhoGAP71E. The remaining genes in the list of 95 stripy genes may be targets of other segmentation genes that control gene expression in different registers.
ftz-f1, as well as ftz, is required for 5-HT2 expression
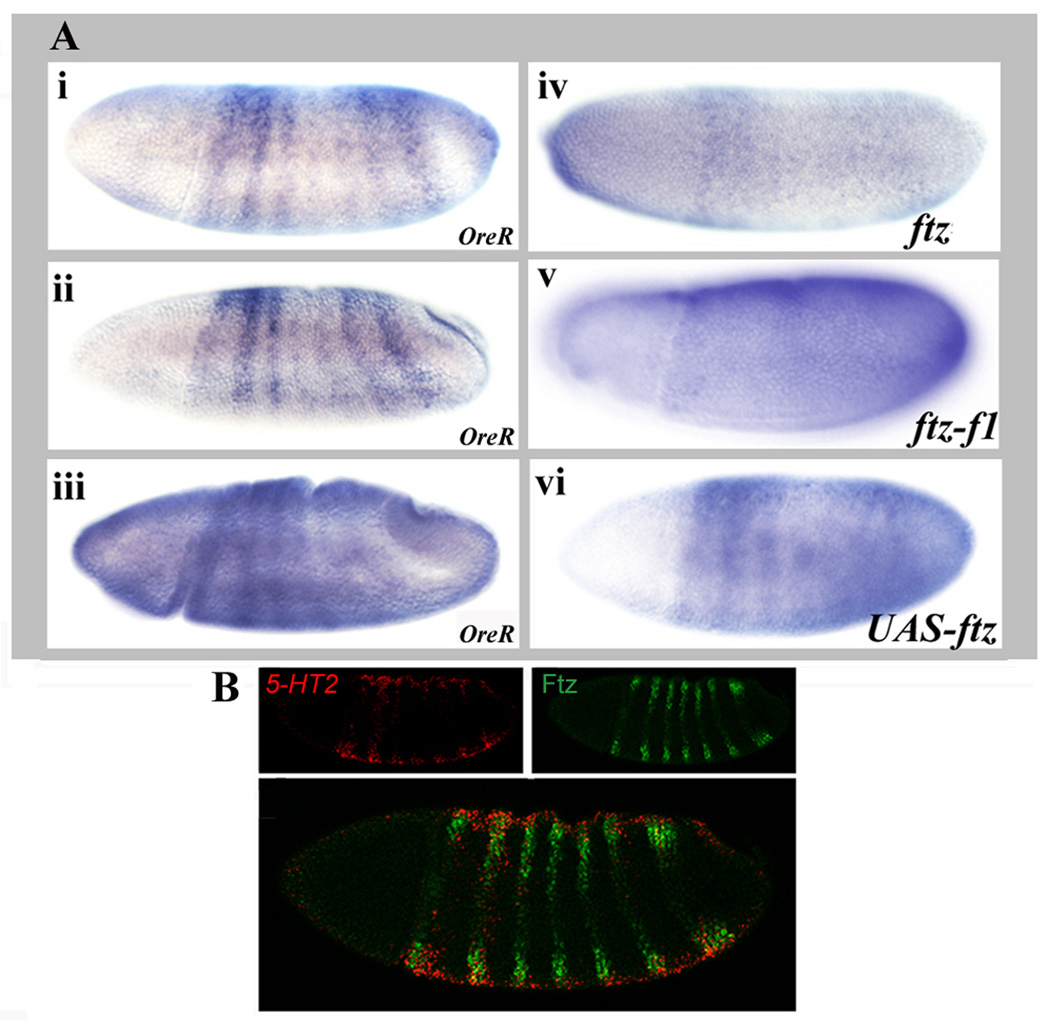
Among the earliest candidate target genes to be expressed during development is the 5-HT2 gene. As shown previously by Colas et al. (1995), 5-HT2 is expressed in stripes in the early embryo. Expression was first observed in seven stripes in blastoderm embryos. The first two and last two stripes were stronger than the middle three stripes (Fig. 1Ai). 5-HT2 RNA continued to be detectable in seven stripes during gastrulation, with the 4th and 5th stripe remaining weaker than the others (Fig. 1Aii). Stripes began to fade during germ band extension (Fig. 1Aiii). The 5-HT2 stripes overlap Ftz stripes in the ectodermal primordia (Fig. 1B, 5-HT2 red, Ftz green; and see (Colas et al., 1995). The overlap was apparent as early as the cellular blastoderm stage and during gastrulation. This is earlier than the overlap between Ftz and its other characterized downstream target genes, en, apt and Dsulf1 (Bowler et al., 2006; Lawrence et al., 1987). In ftz mutant embryos, 5-HT2 expression levels were reduced and stripes were no longer distinct (Fig. 1Aiv). Similarly, at later stages, low levels of 5-HT2 mRNA were detectable but distinct stripes were lost. These patterns are similar to those previously reported (Colas et al., 1995). As was the case for ftz mutants, in ftz-f1 mutants, low-level diffuse expression was seen at blastoderm and during germ band extension (Fig. 1Av). When Ftz was ectopically expressed throughout blastoderm embryos using the UAS/GAL4 system with an NGT40 driver (Tracey et al., 2000), changes in 5-HT2 expression were evident as early as cellular blastoderm, when the first two stripes were fused and the five posterior stripes were expanded (Fig. 1Avi). The fusing continued to be apparent during gastrulation and germ band extension when the first two stripes appeared as one broad stripe and the remaining stripes were expanded beyond their normal registers. The genomic region surrounding the 5HT-2 coding region contains 6 Ftz-F1 binding sites predicted computationally using experimentally verified Ftz-F1 binding sites to search the Drosophila genome, as done previously (Bowler et al., 2006). Each of these predicted Ftz-F1 sites is surrounded by multiple potential Ftz binding sites that could mediate cooperative interactions with Ftz-F1. In addition, four matches to a 7mer sequence, which is thought to act as a general enhancer for zygotic transcription via the zinc-finger transcription factor Zelda (De Renzis et al., 2007; Liang et al., 2008) are located in the 5HT-2 genomic region. Together, these results demonstrate that Ftz and its partner Ftz-F1 are necessary for the spatial patterning of 5-HT2. Future studies will determine whether the predicted Ftz/Ftz-F1 binding sites directly regulate 5HT-2 gene expression.
Figure 1. Ftz and its partner Ftz-F1 are necessary for 5-HT2 stripe expression.
(A)5-HT2 expression is dependent upon ftz and ftz-f1. In situ hybridization of 5-HT2 mRNA: (i,ii,iii) OreR, at cellular blastoderm, gastrulation and early germ band extension; (iv) ftz9H34 mutant embryo; (v) ftz-f119 mutant embryo; and (vi) UAS-myc-ftz/NGT40 embryo. In either ftz or ftz-f1 mutants, 5HT-2 stripes were replaced by diffuse, low level expression. Ectopic ftz expression induced ectopic 5HT-2 expression. (B)5-HT2 and Ftz are coexpressed. Confocal images of embryos stained for 5-HT2 mRNA and Ftz protein. Stage 7 embryo: 5-HT2 RNA (red), Ftz protein (green), 5-HT2 RNA and Ftz protein merged. Note that overlapping expression in stripes does not produce a yellow color because Ftz protein is nuclear and 5-HT2 RNA is cytoplasmic.
noc is responsive to ftz and ftz-f1
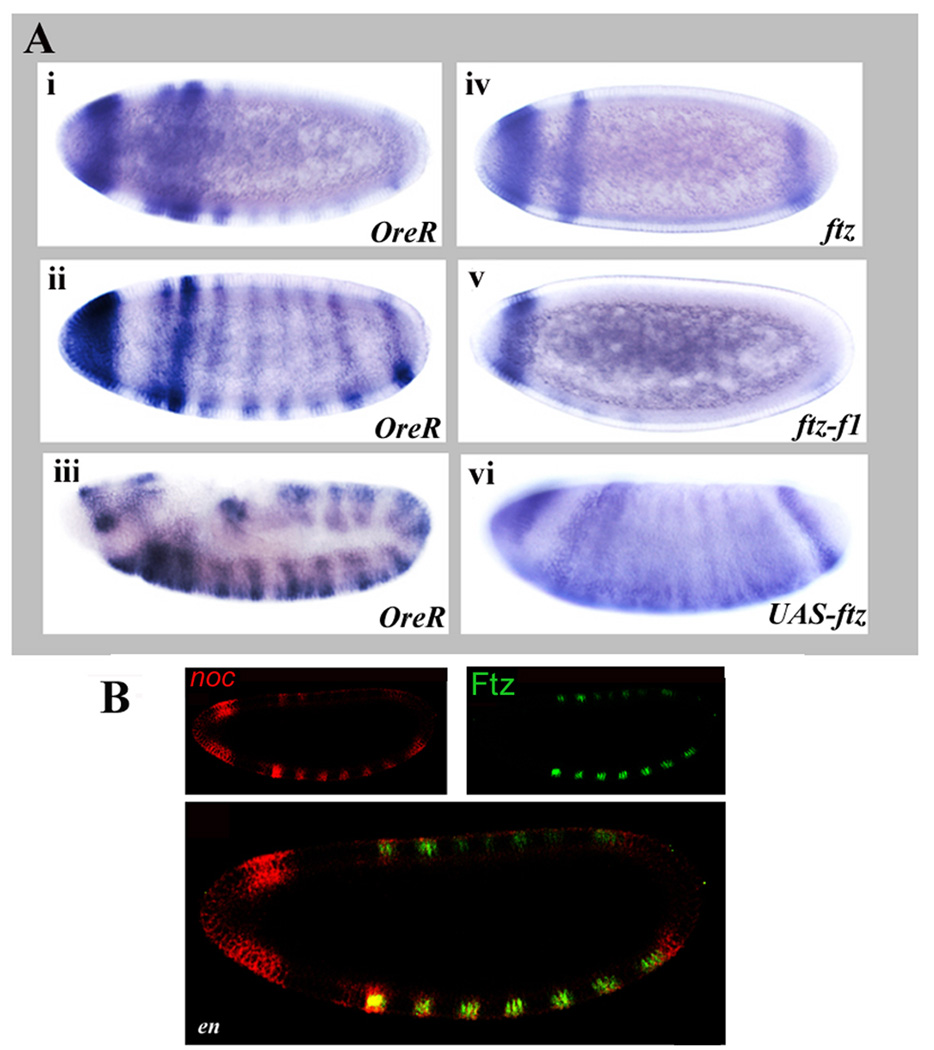
noc is expressed in a striped pattern in early embryos. noc transcripts were first detected at the cellular blastoderm stage, in the procephalic ectoderm, and one stripe (noc stripe 1). Subsequently, 7 additional stripes arose, with noc stripe 2 stronger than the other stripes (Fig. 2Ai). Stripes appeared stronger ventrally. Stripe 8 and stripes 3–7 became stronger as development progressed, with expression in the head remaining strong (Fig. 2Aii). Near the end of germ band extension, stripes in the central body region developed into doublets (Fig. 2Aiii), reminiscent of segment polarity gene expression. In ftz mutant embryos, noc stripes 2–7 were not detectable (Fig. 2Aiv). Expression in the head, and in noc stripe 1, which do not overlap with ftz (Fig. 2B and data not shown), were unaffected. In addition, noc stripe 8, which only partially overlaps ftz stripe 7 (Fig. 2B) was detected in ftz mutant embryos. During late germband extension, some stripes appeared in weak doublet configurations in the central region of the embryo suggesting that, at later stages, additional factors regulate noc. However, these stripes were diffuse and lacked the clear pattern seen in the wild type (data not shown). Similar expression patterns were seen in ftz-f1 mutants (Fig. 2Av), although it appeared that loss of ftz-f1 function also affected noc stripes 1 and 8. As for the ftz mutants, a diffuse, partial stripy pattern was evident at late germband extension. When Ftz was ectopically expressed throughout blastoderm embryos, an extra stripe just prior to the terminal stripe appeared at blastoderm, and soon after this stripes became diffuse, covering much of the central region of the embryo (Fig. 2Avi). Stripes continued to appear fused or expanded during germband extension. Together, these results demonstrate that Ftz and Ftz-F1 are required to establish the striped pattern of noc in the central region of the embryo and that ectopic expression of Ftz results in rapid ectopic activation of noc. In keeping with potential direct regulation, 7 potential Ftz-F1 binding sites were found within a region of 10 kb upstream of the annotated noc TSS (data not shown). Each of these Ftz-F1 sites is surrounded by multiple Ftz binding sites that could potentially mediate cooperative interaction. However, since Ftz and Ftz-F1 regulate en and En is itself a transcriptional regulator thought to mediate effects of Ftz and other pair-rule genes (DiNardo and O'Farrell, 1987; Howard and Ingham, 1986; Lawrence et al., 1987), we asked whether Ftz/Ftz-F1 effects on noc could be indirect via en. In fact, expression patterns in en mutants (Fig. 2B) were indistinguishable from wild type animals: Expression of noc stripes 2–7 overlapped precisely with the seven stripes of Ftz at cellular blastoderm and this overlap in register persisted through later stages (Fig. 2B and data not shown, noc red, Ftz green). noc stripe 8 overlapped with, but extended slightly posterior of ftz stripe 7. No overlap with ftz was seen in the head or with noc stripe 1, as expected. In sum, the early expression of noc in stripes, the dependence of stripes on both Ftz and Ftz-F1, the rapid responsiveness to ectopic Ftz expression, and the presence of potential Ftz/Ftz-F1 binding sites, together suggest that Ftz and Ftz-F1 regulate noc expression and that En is not an intermediate in this regulation.
Figure 2. ftz and ftz-f1 regulate noc expression.
(A) noc expression is dependent upon ftz and ftz-f1. In situ hybridization of noc mRNA: (i,ii,iii) OreR, at cellular blastoderm and late germ band extension stages show early expression in the head and in eight stripes, developing into a 14 stripe pattern; (iv) ftz9H34 mutant embryo; (v) ftz-f1ex19 mutant embryo; and (vi) UAS-myc-ftz/NGT40 embryo. In ftz mutant embryos, the six central noc stripes are missing while ectopic Ftz induced ectopic noc expression. (B) Ftz regulates noc expression independent of en. Confocal images of embryos stained for noc mRNA (red) or Ftz protein, as indicated, in an en1 mutant embryo: noc RNA (red), Ftz protein (green). Note that Ftz protein is nuclear and noc RNA is cytoplasmic, so cellular colocalization does not produce a yellow color. No change was observed in the noc expression pattern in en mutants, as compared to wild type controls.
drm is regulated by Ftz and Ftz-F1
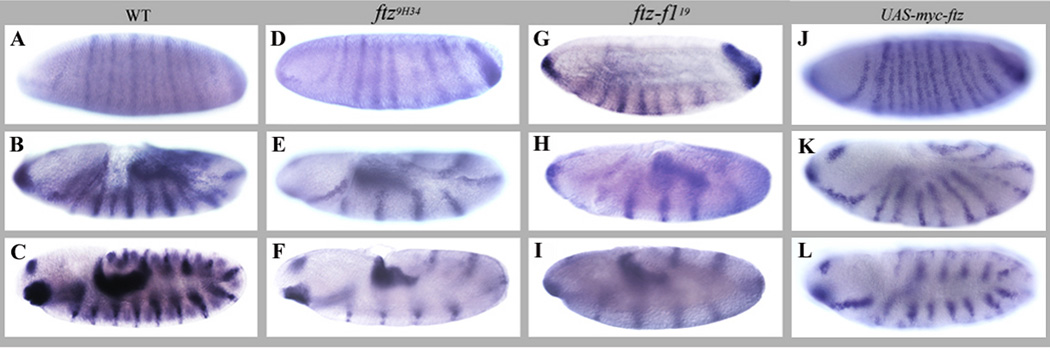
drm encodes a zinc finger transcription factor involved in differentiation, morphogenesis and cell movement during gut morphogenesis (reviewed in (Lengyel and Iwaki, 2002). We found that drm is expressed in a seven-stripe pattern at the cellular blastoderm stage (drm primary stripes). A weaker set of seven secondary stripes appeared during gastrulation (drm secondary stripes) resulting in a fourteen-stripe pattern, reminiscent of segment polarity genes (Fig. 3A). These stripes persisted through gastrulation and early germ band extension, with the primary stripes thicker and stronger than the secondary stripes (Fig. 3B). Additional expression in the head in the proventricular and hindgut primordia was evident at early stages and became stronger as development proceeded. During late germ band extension, the stripes became more equal in intensity and the expression in the hindgut primordium increased (Fig. 3C). In ftz mutant embryos, half of the stripes were missing while hindgut expression was unaffected, as expected. The loss of alternate stripes was apparent as early as stage 6, and persisted through late germ band extension (Fig. 3D–F). This pattern was also observed in ftz-f1 mutants (Fig. 3G–I). When Ftz was ectopically expressed, the spacing of drm stripes became irregular (Fig. 3J, K) and at late germ band extension the stripes adopted a doublet configuration (Fig. 3L) reminiscent of the en response to ectopic Ftz expression (Ish-Horowicz et al., 1989).
Figure 3. Alternate drm stripes require Ftz and Ftz-F1.
In situ hybridization of drm mRNA to embryos at late cellular blastoderm, early germ band extension and full germ band extended stages: (A, B, C) OreR; (D, E, F) ftz9H34 mutant; (G, H, I) ftz-f119 mutant; and (J, K, L) UAS-myc-ftz/ NGT40 embryos. drm is expressed in seven strong primary stripes, with a set of weaker secondary stripes becoming stronger as development proceeds. In ftz or in ftz-f1 mutant embryos, half of the stripes seen in the wild type are missing, while ectopic Ftz expression alters the pattern of drm stripes.
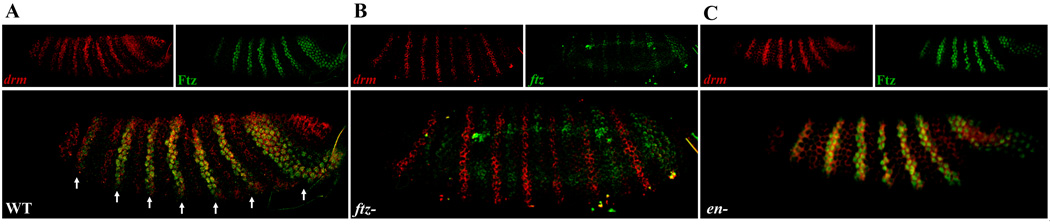
Ftz protein and drm co-localized in the primary drm stripes (white arrows, Fig. 4A,drm RNA red; Ftz green). Expression in the head region, hindgut primordium, and the secondary stripes did not overlap with Ftz. Quantitation of expression based upon fluorescence confirmed that the primary drm stripes that overlap Ftz are stronger than the secondary stripes (data not shown). To determine which set of drm stripes was lost in ftz embryos, we made use of the strong ftz allele, ftz9H34 that expresses ftz RNA but no protein (Furukubo-Tokunaga et al., 1992); Fig. S1). As shown in Fig. 4B, the set of seven drm stripes remaining in ftz mutants was out of register with ftz (drm RNA red, ftz RNA green), demonstrating that the missing set of stripes in these mutants are the primary drm stripes that overlap with ftz in wild type (Ftz-dependent drm stripes). These Ftz-dependent stripes require both Ftz and Ftz-F1 for expression. However, as for noc (see above), it was possible that regulation of drm was mediated via the activation of En by Ftz/Ftz-F1. To test this, drm expression was examined in en mutants (Fig. 4C). The Ftz-dependent drm stripes were present and still overlapped with Ftz stripes in en mutant embryos. Thus, En is not a necessary intermediate in the regulation of the primary drm stripes by Ftz/Ftz-F1. It appeared that the secondary drm stripes were weaker in en mutants, suggesting that En may contribute to the regulation of the Ftz-independent drm stripes (data not shown). Additionally, it was reported that drm expression levels were lower in hh mutants at later stages (Hatini et al., 2005).
Figure 4. ftz is required for expression of the primary drm stripes independent of en.
Confocal images of embryos double stained for drm mRNA and Ftz protein (A,C) or ftz RNA (B). (A)Ftz and drm overlap in the primary drm stripes. OreR, stage 6 embryo: drm RNA (red), Ftz protein (green). (B)Primary drm stripes are lost in ftz mutants. ftz9H34 stage 5 embryo: drm RNA (red), ftz RNA, (green). The remaining drm stripes in the ftz mutant embryo are out of register with the ftz stripes. (C)Primary drm stripes do not require en. Stage 6 en embryo: drm RNA (red), Ftz protein (green). No change in the Ftz-dependent drm stripes was observed in en mutants.
In sum, drm is expressed in a pattern of 7 primary and 7 secondary stripes, in addition to expression in the head and hindgut primordia. Ftz overlaps with drm in the primary drm stripes. ftz and ftz-f1 are each required for expression of these alternate stripes and ectopic Ftz expression alters the pattern of drm expression. The Ftz-dependent drm stripes are not dependent upon en, demonstrating that En is not an intermediate in the regulation of the primary drm stripes by Ftz/Ftz-F1.
drm contains independent stripe enhancers
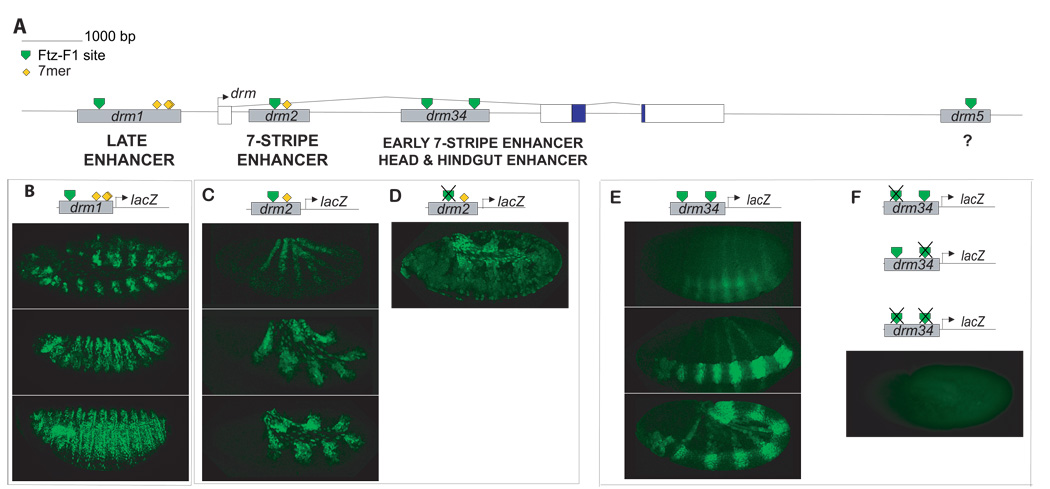
To ask whether Ftz/Ftz-F1 directly regulate drm expression, the major embryonic drm TSS was mapped (Materials and Methods) and potential Ftz-F1 binding sites were identified computationally based upon a position list for experimentally identified Ftz-F1 binding sites, BSAAGGHYRHH (Bowler et al., 2006). Within a 20kb drm genomic region, there are five potential Ftz-F1 sites (Fig. 5A, green triangles). Each of these has multiple potential Ftz binding sites within 25 bases of the core Ftz-F1 site (not shown). To determine whether these predicted Ftz/Ftz-F1 binding sites are components of functional enhancers, fragments of ∼1kb spanning them were inserted upstream of a basal promoter and a lacZ reporter gene in the P-element vector pX28. Multiple independent transgenic lines were generated for each construct and expression was analyzed with anti-β-galactosidase antibody (Figure 5B). drm1 contains the only predicted Ftz-F1 binding site located upstream of the TSS, at – 1.9 kb. This site is flanked by 5 potential Ftz binding sites. Two constructs were generated: the first, drm1-lacZ, spans 1 kb and does not contain any 7mer sequences. The second, drm1–7mer, was generated because no expression was detected for drm1. This construct is extended by ∼ 700 bp to contain five 7mer sequences located 3' of the end of drm1 (Fig. 5A, yellow diamonds). drm1–7mer-lacZ expression was not detected before late germ band extension. At this time, it was expressed in fourteen lines or patches that extended only partially around the embryo (Fig. 5B). This pattern evolved into a complex segmental pattern that closely resembled expression of endogenous drm at stages 9–12 (Tomancak et al., 2002). These results indicate that drm1–7mer harbors an enhancer that directs drm expression at late stages of development (drm Late Enhancer, Fig. 5A). Although clearly not a strict test of 7mer function, the finding that drm1-lacZ was not expressed is consistent with a role for the 7mer sequences in facilitating expression directed by a region-specific late drm enhancer.
Figure 5. Multiple stripe enhancers control drm expression.
(A) Schematic of the structure of the drm gene. The drm gene includes three exons, and two introns, with the coding region initiating in exon 2. Five potential Ftz-F1 binding sequences (green triangles) and multiple 7mers (of a total of eighteen) are indicated. Putative cis-regulatory elements for drm were identified by virtue of the presence of potential Ftz-F1 binding sites. Fragments containing these sites (gray rectangles, drm1-7mer, drm2, drm34, drm5, as indicated, were inserted upstream of a basal promoter and lacZ reporter gene in the P-element vector pX28. Enhancers identified in this analysis are indicated below the line. No reproducible expression pattern was obtained for drm5-lacZ (B–F)Expression of drm-lacZ transgenes. (B)drm1-7mer-lacZ expression at the end of germ band extension and in a complex segmental pattern through germ band retraction. (C) Expression of drm2-lacZ in seven stripes at early, mid and late germ band extension stages. (D) Expression of drm2M-lacZ, in which the predicted Ftz-F1 binding site was mutated, in seven weak stripes at germ band extension. Amnioserosa expression appears to be due to vector sequences. (E) Expression of drm34-lacZ in seven stripes at the cellular blastoderm stage and during gastrulation. Striped expression persisted through germ band extension. Expression in the head and hindgut was detected from gastrulation through germ band extension. (F) Mutation of the drm3, drm4 or both drm3 and 4 predicted Ftz-F1 binding sites abolished expression of lacZ fusion genes.
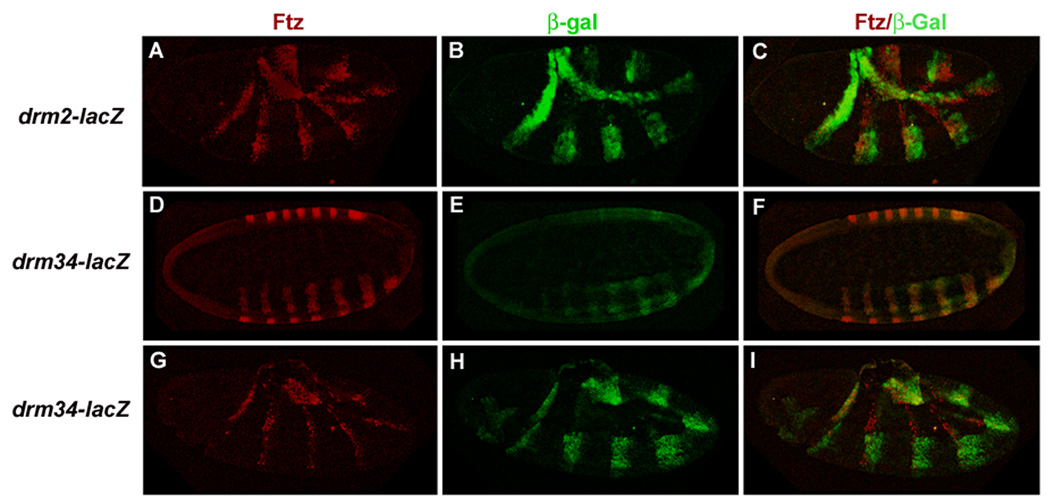
drm2 contains one potential Ftz-F1 binding site, located in the first drm intron. This Ftz-F1 site is flanked by 7 potential Ftz binding sites and one 7mer. drm2-lacZ was expressed in seven strong stripes (Fig. 5C). These stripes were evident at late gastrulation and became more prominent during early germ band elongation. Expression in stripes remained strong during mid- and late- germ band extension stages. Additional expression in the amnioserosa was detected, which may be a result of vector sequences. The drm2-lacZ stripes are in register with Ftz (Fig. 6A–C) and thus represent the primary drm stripes. This stripe pattern, directed by the drm 7-Stripe Enhancer, is reminiscent of ftz-lacZ fusion genes that are directly responsive to Ftz/Ftz-F1 (Han et al., 1998; Pick et al., 1990; Yussa et al., 2001).
Figure 6. Two independent drm enhancers direct expression in Ftz-like stripes.
Immunohistochemical staining of drm-lacZ transgenic embryos with anti-Ftz antibody (red) and anti-β-galactosidase antibody (green). (A–C) Expression of drm2-lacZ in seven stripes was detected during germ band elongation. Ftz and β-galactosidase overlap. Note that Ftz stripes have become thinner by this stage, but the stability of β-galactosidase results in thick stripes for the drm-lacZ transgene. (D–F) A confocal cross-section through a drm34-lacZ transgenic embryo at cellular blastoderm. Ftz and β-galactosidase expression overlap exactly. As the Ftz stripes thin during germ band elongation (G–I), Ftz and drm34-lacZ stripes continue to overlap but β-galactosidase expression is broader, because of the stability of β-galactosidase protein, resembling the pattern seen for ftz autoregulatory elements that are direct Ftz targets (Pick et al., 1990).
drm34 contains 2 Ftz-F1 potential binding sites, located about 800 bp apart in the first drm intron. The predicted Ftz-F1 binding site in drm3 is flanked by 6 potential Ftz binding sites and in drm4 by 7 potential Ftz sites. drm34-lacZ was also expressed in a strong 7-stripe pattern that was detectable as early as the cellular blastoderm stage (Fig. 5E; Early 7-Stripe Enhancer, Fig. 5A). During gastrulation and germ band extension, expression in the 7 stripes increased and additional expression in the proventriculus and hindgut primordia became apparent. Expression in seven stripes, the head and hindgut persisted at least through the end of germ band extension. Surprisingly, these stripes also overlapped with Ftz, as did the drm2-lacZ stripes. Complete overlap between Ftz and β-galactosidase expression was evident at the cellular blastoderm stage (Fig. 6D–F) and continued through germ band elongation (Fig. 6G–I) indicating that the drm34 Early 7-Stripe Enhancer directs expression in the primary drm stripes.
drm5 is located downstream of the drm coding region. This potential Ftz-F1 binding site is flanked by 5 potential Ftz binding sites. No reproducible pattern of expression was observed, suggesting that it either does not harbor a drm enhancer element or that it also harbors repressors that mask functional enhancer sequences.
In sum, this analysis identified 3 independent enhancers of drm (Fig. 5A). Each directs expression in portions of the endogenous drm pattern. The upstream drm1 region harbors a Late Enhancer. The first intron harbors two independent enhancers: the drm2 7-Stripe Enhancer directs expression in 7 stripes that overlap Ftz. The drm34 Early 7-Stripe Enhancer directs expression in 7 stripes that also overlap Ftz but which initiate earlier than drm2-directed stripes. The drm34 enhancer also directs expression in the proventriculus and hindgut. The two 7-stripe enhancers each direct expression in the primary, Ftz-dependent drm stripes.
Predicted Ftz-F1 binding sites are functional in vivo
To ask whether the computationally identified Ftz-F1 binding sites in the two 7-stripe enhancers are functional in vivo, point mutations were generated in the core Ftz-F1 binding sequences (AAGG to AGAT) to generate a sequence known to abolish binding of purified protein Ftz-F1 and Ftz-F1 protein in Drosophila nuclear extracts (Han et al., 1998). Fragments carrying Ftz-F1 site mutations were inserted upstream of a basal promoter and a lacZ reporter gene in the P-element vector pX28, as above, and multiple independent transformant lines were generated. Expression of drm2M-lacZ, containing a mutation in the single predicted Ftz-F1 binding site in this 7-stripe enhancer, was drastically decreased (Fig. 5D). In two independent transformant lines, no expression was detected; in one line, reasonably strong stripes were found; and in two independent lines, very weak striped expression was observed, as shown in the figure. We interpret this result as indicating that the predicted Ftz-F1 binding site is necessary for full expression of this enhancer, but that other factors are also able to generate weak stripes in the absence of Ftz-F1.
Expression of drm34-lacZ fusion genes with mutations in either the drm3 or drm4 predicted Ftz-F1 binding sites or simultaneous mutations in both the drm3 and drm4 sites was abolished (Fig. 5F, one example shown). Expression of drm3M4-lacZ, drm34M-lacZ, or drm3M4M-lacZ was undetectable in 5/5 independent transgenic lines for each of these constructs. These results demonstrate that each of the Ftz-F1 sites in this drm 34 Early 7-Stripe Enhancer is necessary for striped expression.
DISCUSSION
The regulatory transcription factors that direct Drosophila development have been studied in great detail. These transcription factors interact in a largely linear hierarchy of maternal, gap, pair-rule, segment-polarity and homeotic genes, with cross-regulation occurring at each level of the hierarchy. This network of transcription factors provides a blueprint for the development and differentiation of body segments. How this blueprint of regulatory or instructive information is translated into morphology is of considerable interest. Although some progress has been made in understanding control of morphology by homeotic selector genes (reviewed in (Pearson et al., 2005), it is less clear if genes acting earlier in the hierarchy impact morphology or function solely to establish the expression patterns of segment polarity and homeotic genes, which then impact morphology. In particular, there is considerable debate as to whether the pair-rule transcription factors are purely pre-patterning genes that regulate solely other selector genes in the hierarchy, or if they are involved in regulating segment formation independent of the segment polarity and homeotic genes, by controlling genes more intimately involved with segment formation and morphogenesis. We have begun to address this issue by identifying downstream targets of the pair-rule transcription factors Ftz and Ftz-F1, regulators that direct formation of alternate parasegments in the Drosophila embryo. Our findings support those of others (see below) in suggesting that the pair-rule genes do not participate in a strictly linear hierarchy, regulating only other selector genes to indirectly control segmentation, but that they control the expression of a range of different classes of genes, thereby providing branch points in a linear hierarchy that amplify the information provided by striped pair-rule expression patterns.
Ftz and Ftz-F1 coordinately regulate the expression of multiple target genes
This study identified Ftz targets based upon a search for genes expressed in striped patterns in the early Drosophila embryo (Fig. 1–Fig. 4). Each of these Ftz-dependent genes is also regulated by Ftz-F1, an orphan nuclear receptor previously shown to interact with Ftz in vitro and in vivo (Guichet et al., 1997; Yu et al., 1997). Unlike Ftz, which is expressed in a striped pattern in the Drosophila blastoderm, Ftz-F1 is expressed ubiquitously, in all somatic cells at the blastoderm stage (Yussa et al., 2001). The finding here that all three additional Ftz-dependent genes, identified by virtue of their striped expression patterns, require Ftz-F1 for expression in stripes lends support to the model that interaction with Ftz-F1 is the key to Ftz functional specificity as a segmentation protein. The three genes characterized in this study, 5-HT2, noc and drm, are the earliest identified downstream targets of Ftz. Expression in stripes was observed at the cellular blastoderm stage when Ftz-F1 is highly expressed throughout the embryo and the 7 Ftz stripes are at their peak levels. These early target gene stripes were lost in ftz and also in ftz-f1 mutants. In addition, ectopic expression was observed at early stages when Ftz was ectopically expressed throughout the embryo. En, long thought to be a major mediator of Ftz function in segmentation, is expressed later than these target genes and we verified that En is not required for the Ftz-dependent stripe expression of noc or drm. These findings suggest that Ftz and Ftz-F1 directly regulate expression of these three target genes. This new study brings to seven the targets of Ftz that appear to be directly co-regulated by Ftz and Ftz-F1: ftz itself (Han et al., 1998; Hiromi and Gehring, 1987; Pick et al., 1990; Schier and Gehring, 1993a; Schier and Gehring, 1993b; Yussa et al., 2001), en (Florence et al., 1997; Kassis, 1990), apt, Dsulf1(Bowler et al., 2006), 5HT-2, noc and drm. For each gene, multiple potential Ftz-F1 binding sites were found within a 15–20 kb genomic region. In all cases, multiple potential Ftz binding sites surround the Ftz-F1 binding sites that could mediate cooperative interactions between Ftz and Ftz-F1. Many of these sites have been maintained during evolution and are present in distant Drosophila species (Bowler et al., 2006; Maier et al., 1990); data not shown). Other Ftz targets, such as Ubx (Muller and Bienz, 1992), prd, odd (Nasiadka and Krause, 1999) and tsh (Core et al., 1997) are also likely to be co-regulated by Ftz-F1.
The seven Ftz/Ftz-F1 target genes identified to date play diverse roles in segmentation and act at different levels of the embryonic hierarchy. First, Ftz acts in a cross-regulatory fashion to modulate expression of other pair-rule genes: it interacts with Ftz-F1 in autoregulation and also has been shown to regulate the pair-rule genes prd, odd and slp (Baumgartner and Noll, 1990; Gutjahr et al., 1994; Nasiadka and Krause, 1999). Second, Ftz/Ftz-F1 directly regulate components of the segment polarity system: first, they activate en expression in alternate stripes, and, second, they regulate Dsulf1, thought to modulate Wg activity (Lai et al., 2002). Ftz has also been shown to repress wg expression (Ingham et al., 1988; Nasiadka and Krause, 1999). Ftz/Ftz-F1 thus indirectly control compartment border formation, via regulation of En and Wg. Third, Ftz/Ftz-F1 regulate transcription factors that in turn control the differentiation of specific cell types: apt, noc, drm. drm encodes an odd-skipped family zinc finger transcription factor that it is required for patterning the dorsal epidermis, thus regulating the differentiation of specific cell types (Hatini et al., 2005). noc plays a role in trachael morphogenesis with mutants displaying defects in branch migration and expanded expression of trachael-specific genes (Dorfman et al., 2002). Similarly, apt is involved in this process as a regulator of the migration of trachael precursor cells (Bowler et al., 2006; Liu et al., 2003). Finally, Ftz/Ftz-F1 regulate a target gene more directly involved in morphogenesis, 5HT-2. 5-HT2 encodes a serotonin receptor that demonstrates specific ligand binding in transfected cells and in Drosophila embryo extracts (Colas et al., 1995). Phenotypic analysis suggested a role for 5-HT2 and other genes involved in serotonin biosynthesis in morphogenetic movements during gastrulation: deficiency embryos lacking 5HT-2 displayed delayed and incomplete movements during germband extension accompanied by mislocalization of Armadillo protein, suggestive of abnormalities in adherens junctions (Colas et al., 1999a; Colas et al., 1999b; Schaerlinger et al., 2007). It will be of interest in the future to determine whether other pair-rule genes direct expression of additional cell surface proteins that coordinate these processes.
drm is regulated by multiple stripe enhancers
We have identified enhancers of drm by combining bioinformatics with enhancer-reporter gene expression analysis in vivo (Fig. 5,Fig. 6). Fragments chosen for the in vivo analysis contained one or more matche(s) to a Ftz-F1 binding site. Three of the four fragments directed expression in drm-like patterns in vivo (Fig. 5). The upstream fragment, drm1, harbors a late stage enhancer that directs segmental expression of drm. drm2 directed expression in seven strong stripes. drm34 harbors enhancers for the region-specific expression of drm in the proventriculus and hindgut, expression that is important for the development of the fore- and hindgut (Johansen et al., 2003), as well as an early 7-stripe enhancer. We have not investigated whether any of these enhancers also direct expression in the leg imaginal discs (Hao et al., 2003). Two of the fragments tested here, drm2 and drm34, directed expression in 7-stripe patterns. Surprisingly, for each of them, the set of seven stripes is in register with Ftz, suggesting that both regulate expression of the drm-primary stripes (Fig. 6). Although unexpected, this phenomenon has been observed in other cases where it was suggested that enhancers directing the same or similar expression patterns function as shadow enhancers to enhance the precision of expression patterns and facilitate the rapid evolution of cis-regulatory sequences (Hong et al., 2008). Point mutations of either or both of the predicted Ftz-F1 binding sites in the drm34 Early 7-Stripe Enhancer abolished expression of lacZ fusion genes (Fig. 5F). Stripe expression was decreased but not completely abolished by mutation of the single predicted Ftz-F1 binding site in the drm2 7-Stripe Enhancer (Fig. 5D), suggesting additional inputs into regulation of the drm primary stripes by this enhancer. Together, these results suggest that Ftz-F1 activates expression in the primary drm stripes via the drm34 Early 7-Stripe Enhancer. We speculate that following this initial activation, autoregulation by Drm may augment Ftz-F1 activation of stripes via the drm2 7-Stripe Enhancer to raise levels of transcription in drm primary stripes.
The role of pair-rule patterning in Drosophila segmentation
Drosophila ftz is a typical pair-rule gene: ftz mutant embryos die lacking even-numbered body segments (Wakimoto et al., 1984). How this wild type function of ftz, and other pair-rule genes, is executed is not yet known. As for other segmentation mutants, the pair-rule mutant phenotype results from cell death (Magrassi and Lawrence, 1988; Pazdera et al., 1998). However, this cell death appears to be an indirect effect (Hughes and Krause, 2001). Similarly, pair-rule genes regulate segment border formation indirectly, via activation of the segment polarity genes such as en and wg (Carroll, 1990; Carroll et al., 1988; DiNardo and O'Farrell, 1987; Howard and Ingham, 1986; Ingham et al., 1988; Jaynes and Fujioka, 2004; Lawrence and Johnston, 1989; Lawrence et al., 1987; Lawrence and Pick, 1998). In addition to this, segment-polarity-independent roles for the pair-rule genes in morphogenesis have been revealed by careful studies from the Wieschaus lab (reviewed in (Dawes-Hoang and Wieschaus, 2001; Wieschaus et al., 1991). For example, it was found that cell intercalation and germ band extension are regulated by the pair-rule genes, independent of segment polarity genes (Irvine and Wieschaus, 1994). Similarly, cellular studies defined two subtle morphogenetic processes that occur before gastrulation – one, controlled by the pair-rule gene paired (Blankenship and Wieschaus, 2001). More recently, studies have shown that the planar polarity and organization of intercalating cells during germ band extension are controlled by the striped expression patterns of eve and runt (Blankenship et al., 2006; Zallen and Wieschaus, 2004) and that the longitudinal division of cells during germ band extension is controlled by eve (da Silva and Vincent, 2007). These studies are suggestive of direct roles for the pair-rule system in cell shape changes and rearrangements during germ band extension (reviewed in Pilot and Lecuit, 2005; Zallen and Blankenship, 2008). Together, these studies support the notion that combinatorial expression of early patterning genes assigns unique identities in the blastoderm at a single cell level, as originally proposed by Gergen et al., 1986; Scott and O'Farrell, 1986). Here we have shown that the pair-rule gene ftz regulates target genes prior to and independent of En. These findings support the model that the stripes of pair-rule genes play active roles in patterning the embryo rather than serving solely as intermediary patterns whose function is to produce the segmental stripes of segment polarity genes. One role for these pair-rule stripes may be to establish differential adhesiveness to groups of cells in the blastoderm embryo (Irvine and Wieschaus, 1994) and see above). Future work identifying additional pair-rule targets will be required to explain the fundamental biological roles of pair-rule patterning and to understand how the assignment of positional identities by pair-rule genes, prior to morphogenesis, translates into the development and differentiation of body segments.
Supplementary Material
(A–B) Cuticular preparations of control w1118 and ftz9H34 embryos. Note that the ftz9H34 embryos display strong pair-rule phenotypes. (C) Confocal image of in situ hybridization of ftz RNA to ftz9H34 stage 5 embryo. ftz RNA was expressed in a pattern indistinguishable from wild type, including the apical localization of the RNA, in ftz9H34 mutants.
ACKNOWLEDGEMENTS
We thank Rootvij Patel for help with fly stocks, Caroline Li for comments on the manuscript and the Bloomington Fly Center for fly stocks. This work was supported by grants to L.P. from the National Institutes of Health (HD 27937) and National Science Foundation (IBN 0641717).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Arnosti DN, Barolo S, Levine M, Small S. The eve stripe 2 enhancer employs multiple modes of transcriptional synergy. Development. 1996;122:205–214. doi: 10.1242/dev.122.1.205. [DOI] [PubMed] [Google Scholar]
- Baumgartner S, Noll M. Network of interactions among pair-rule genes regulating paired expression during primordial segmentation of Drosophila. Mech Dev. 1990;33:1–18. doi: 10.1016/0925-4773(90)90130-e. [DOI] [PubMed] [Google Scholar]
- Blankenship JT, Backovic ST, Sanny JS, Weitz O, Zallen JA. Multicellular rosette formation links planar cell polarity to tissue morphogenesis. Dev Cell. 2006;11:459–470. doi: 10.1016/j.devcel.2006.09.007. [DOI] [PubMed] [Google Scholar]
- Blankenship JT, Wieschaus E. Two new roles for the Drosophila AP patterning system in early morphogenesis. Development. 2001;128:5129–5138. doi: 10.1242/dev.128.24.5129. [DOI] [PubMed] [Google Scholar]
- Bowler T. Downstream targets of Fushi Tarazu, Ph.D. Thesis. Mount Sinai Medical School of New York University; 2004. [Google Scholar]
- Bowler T, Kosman D, Licht JD, Pick L. Computational Identification of Ftz/Ftz-F1 target genes. Developmental Biology. 2006;299:78–90. doi: 10.1016/j.ydbio.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Broadus J, McCabe JR, Endrizzi B, Thummel CS, Woodard CT. The Drosophila βFTZ-F1 orphan nuclear receptor provides competence for stage-specific responses to the steroid hormone ecdysone. Mol. Cell. 1999;3:143–149. doi: 10.1016/s1097-2765(00)80305-6. [DOI] [PubMed] [Google Scholar]
- Carroll SB. Zebra patterns in fly embryos: activation of stripes or repression of interstripes? Cell. 1990;60:6–16. doi: 10.1016/0092-8674(90)90711-m. [DOI] [PubMed] [Google Scholar]
- Carroll SB, DiNardo S, O'Farrell PH, White RAH, Scott MP. Temporal and spatial relationships between segmentation and homeotic gene expression in Drosophila embryos: distributions of the fushi tarazu, engrailed, Sex combs reduced, Antennapedia, and Ultrabithorax proteins. Genes Dev. 1988;2:350–360. doi: 10.1101/gad.2.3.350. [DOI] [PubMed] [Google Scholar]
- Carroll SB, Scott MP. Localization of the fushi tarazu protein during Drosophila embryogenesis. Cell. 1985;43:47–57. doi: 10.1016/0092-8674(85)90011-x. [DOI] [PubMed] [Google Scholar]
- Chou T-B, Perrimon N. Use of a yeast site-specific recombinase to produce female germline chimeras in Drosophila. Genetics. 1992;131:643–653. doi: 10.1093/genetics/131.3.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou T-B, Perrimon N. The autosomal FLP-DFS technique for generating germline mosaics in Drosophila melanogaster. Genetics. 1996;144:1673–1679. doi: 10.1093/genetics/144.4.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou TB, Noll E, Perrimon N. Autosomal P[ovoD1] dominant female-sterile insertions in Drosophila and their use in generating germ-line chimeras. Development. 1993;119:1359–1369. doi: 10.1242/dev.119.4.1359. [DOI] [PubMed] [Google Scholar]
- Colas J-F, Launay J-M, Vonesch J-L, Hickel P, Maroteaux L. Serotonin synchronises convergent extension of ectoderm with morphogenetic gastrulation moevements in Drosophila. Mech. Dev. 1999a;87:77–91. doi: 10.1016/s0925-4773(99)00141-0. [DOI] [PubMed] [Google Scholar]
- Colas JF, Launay JM, Kellermann O, Rosay P, Maroteaux L. Drosophila 5-HT2 serotonin receptor: coexpression with fushi-tarazu during segmentation. Proc Natl Acad Sci U S A. 1995;92:5441–5445. doi: 10.1073/pnas.92.12.5441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colas JF, Launay JM, Maroteaux L. Maternal and zygotic control of serotonin biosynthesis are both necessary for Drosophila germband extension. Mech Dev. 1999b;87:67–76. doi: 10.1016/s0925-4773(99)00140-9. [DOI] [PubMed] [Google Scholar]
- Copeland WR, Nasiadka A, Dietrich BH, Krause HM. Patterning of the Drosophila embryo by a homeodomain-deleted Ftz polypeptide. Nature. 1996;379:162–165. doi: 10.1038/379162a0. [DOI] [PubMed] [Google Scholar]
- Core N, Charroux B, McCormick A, Vola C, Fasano L, Scott MP, Kerridge S. Transcriptional regulation of the Drosophila homeotic gene teashirt by the homeodomain protein Fushi tarazu. Mech Dev. 1997;68:157–172. doi: 10.1016/s0925-4773(97)00144-5. [DOI] [PubMed] [Google Scholar]
- da Silva SM, Vincent JP. Oriented cell divisions in the extending germband of Drosophila. Development. 2007;134:3049–3054. doi: 10.1242/dev.004911. [DOI] [PubMed] [Google Scholar]
- Dawes-Hoang RE, Wieschaus EF. Cell and developmental biology--a shared past, an intertwined future. Dev Cell. 2001;1:27–36. doi: 10.1016/s1534-5807(01)00020-x. [DOI] [PubMed] [Google Scholar]
- De Renzis S, Elemento O, Tavazoie S, Wieschaus EF. Unmasking activation of the zygotic genome using chromosomal deletions in the Drosophila embryo. PLoS Biol. 2007;5:e117. doi: 10.1371/journal.pbio.0050117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiNardo S, O'Farrell P. Establishment and refinement of segmental patterning in Drosophila embryos: spatial control of engrailed expression by pair-rule genes. Genes & Dev. 1987:1212–1225. doi: 10.1101/gad.1.10.1212. [DOI] [PubMed] [Google Scholar]
- Dorfman R, Glazer L, Weihe U, Wernet MF, Shilo BZ. Elbow and Noc define a family of zinc finger proteins controlling morphogenesis of specific tracheal branches. Development. 2002;129:3585–3596. doi: 10.1242/dev.129.15.3585. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick VD, Percival-Smith A, Ingles CJ, Krause HM. Homeodomain-independent activity of fushi tarazu polypeptide in Drosophila embryos. Nature. 1992;356:610–612. doi: 10.1038/356610a0. [DOI] [PubMed] [Google Scholar]
- Florence B, Guichet A, Ephrussi A, Laughon A. Ftz-F1 is a cofactor in Ftz activation of the Drosophila engrailed gene. Development. 1997;124:839–847. doi: 10.1242/dev.124.4.839. [DOI] [PubMed] [Google Scholar]
- Florence B, Handrow R, Laughon A. DNA-binding specificity of the fushi tarazu homeodomain. Mol. Cell Biol. 1991;11:3613–3623. doi: 10.1128/mcb.11.7.3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortier TM, Vasa PP, Woodard CT. Orphan nuclear receptor betaFTZ-F1 is required for muscle-driven morphogenetic events at the prepupal-pupal transition in Drosophila melanogaster. Dev Biol. 2003;257:153–165. doi: 10.1016/s0012-1606(03)00036-8. [DOI] [PubMed] [Google Scholar]
- Furukubo-Tokunaga K, Muller M, Affolter M, Pick L, Kloter U, Gehring WJ. In vivo analysis of the helix-turn-helix motif of the fushi tarazu homeo domain of Drosophila melanogaster. Genes & Dev. 1992;6:1082–1096. doi: 10.1101/gad.6.6.1082. [DOI] [PubMed] [Google Scholar]
- Gergen JP, Coulter D, Wieschaus E. Gametogenesis and the Early Embryo. Alan R. Liss, Inc; 1986. Segmental Pattern and Blastoderm Cell Identities; pp. 195–220. [Google Scholar]
- Guichet A, Copeland JWR, Erdelyi M, Hlousek D, Zavorszky P, Ho J, Brown S, Percival-Smith A, Krause HM, Ephrussi A. The nuclear receptor homologue Ftz-F1 and the homeodomain protein Ftz are mutually dependent cofactors. Nature. 1997;385:548–552. doi: 10.1038/385548a0. [DOI] [PubMed] [Google Scholar]
- Gutjahr T, Frei E, Noll M. Complex regulation of early paired expression: initial activation by gap genes and pattern modulation by pair-rule genes. Development. 1993;117:609–623. doi: 10.1242/dev.117.2.609. [DOI] [PubMed] [Google Scholar]
- Gutjahr T, Vanario Alonso CE, Pick L, Noll M. Multiple regulatory elements direct the complex expression patterns of the Drosophila segmentation gene paired. Mech. Dev. 1994;48:119–128. doi: 10.1016/0925-4773(94)90021-3. [DOI] [PubMed] [Google Scholar]
- Hafen E, Kuroiwa A, Gehring WJ. Spatial distribution of transcripts from the segmentation gene fushi tarazu during Drosophila embryonic development. Cell. 1984;37:833–841. doi: 10.1016/0092-8674(84)90418-5. [DOI] [PubMed] [Google Scholar]
- Han K, Levine MS, Manley JL. Synergistic activation and repression of transcription by Drosophila homeobox proteins. Cell. 1989;56:573–583. doi: 10.1016/0092-8674(89)90580-1. [DOI] [PubMed] [Google Scholar]
- Han W, Yu Y, Su K, Kohanski RA, Pick L. A binding site for multiple transcriptional activators in the fushi tarazu proximal enhancer is essential for gene expression in vivo. Mol. Cell. Biol. 1998;18:3384–3394. doi: 10.1128/mcb.18.6.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao I, Green RB, Dunaevsky O, Lengyel JA, Rauskolb C. The odd-skipped family of zinc finger genes promotes Drosophila leg segmentation. Dev Biol. 2003;263:282–295. doi: 10.1016/j.ydbio.2003.07.011. [DOI] [PubMed] [Google Scholar]
- Hatini V, Green RB, Lengyel JA, Bray SJ, Dinardo S. The Drumstick/Lines/Bowl regulatory pathway links antagonistic Hedgehog and Wingless signaling inputs to epidermal cell differentiation. Genes Dev. 2005;19:709–718. doi: 10.1101/gad.1268005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiromi Y, Gehring WJ. Regulation and function of the Drosophila segmentation gene fushi tarazu. Cell. 1987;50:963–974. doi: 10.1016/0092-8674(87)90523-x. [DOI] [PubMed] [Google Scholar]
- Hong JW, Hendrix DA, Levine MS. Shadow enhancers as a source of evolutionary novelty. Science. 2008;321:1314. doi: 10.1126/science.1160631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard K, Ingham P. Regulatory interactions between the segmentation genes fushi tarazu, hairy and engrailed in the Drosophila blastoderm. 1986;44:949–957. doi: 10.1016/0092-8674(86)90018-8. [DOI] [PubMed] [Google Scholar]
- Hughes SC, Krause HM. Establishment and maintenance of parasegmental compartments. Development. 2001;128:1109–1118. doi: 10.1242/dev.128.7.1109. [DOI] [PubMed] [Google Scholar]
- Ingham PW, Baker NE, Martinez-Arias A. Regulation of segment polarity genes in the Drosophila blastoderm by fushi tarazu and even skipped. 1988;331:73–75. doi: 10.1038/331073a0. [DOI] [PubMed] [Google Scholar]
- Irvine KD, Wieschaus E. Cell intercalation during Drosophila germband extension and its regulation by pair-rule segmentation genes. Development. 1994;120:827–841. doi: 10.1242/dev.120.4.827. [DOI] [PubMed] [Google Scholar]
- Ish-Horowicz D, Pinchin SM, Inghma PW, Gyurkovics HG. Autocatalytic ftz activation and metameric instability induced by ectopic ftz expression. 1989;57:223–232. doi: 10.1016/0092-8674(89)90960-4. [DOI] [PubMed] [Google Scholar]
- Jaynes JB, Fujioka M. Drawing lines in the sand: even skipped et al. and parasegment boundaries. Dev Biol. 2004;269:609–622. doi: 10.1016/j.ydbio.2004.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen KA, Green RB, Iwaki DD, Hernandez JB, Lengyel JA. The Drm-Bowl-Lin relief-of-repression hierarchy controls fore- and hindgut patterning and morphogenesis. Mech Dev. 2003;120:1139–1151. doi: 10.1016/j.mod.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Kassis JA. Spatial and temporal control elements of the Drosophila engrailed gene. Genes Dev. 1990;4:433–443. doi: 10.1101/gad.4.3.433. [DOI] [PubMed] [Google Scholar]
- Kosman D, Small S. Concentration-dependent patterning by an ectopic expression domain of the Drosophila gap gene knirps. Development. 1997;124:1343–1354. doi: 10.1242/dev.124.7.1343. [DOI] [PubMed] [Google Scholar]
- Kosman D, Small S, Reinitz J. Rapid preparation of a panel of polyclonal antibodies to Drosophila segmentation proteins. Dev Genes Evol. 1998;208:290–294. doi: 10.1007/s004270050184. [DOI] [PubMed] [Google Scholar]
- Lai M, Ai X, Sun W, Emerson C, Standiford DM. Abstract: Regulation of Wg signaling by Drosophila sulfated. Dev. Biol. 2002;247:468. [Google Scholar]
- Lawrence PA, Johnston P. Pattern formation in the Drosophila embryo: allocation of cells to parasegments by even-skipped and fushi tarazu. 1989;105:761–767. doi: 10.1242/dev.105.4.761. [DOI] [PubMed] [Google Scholar]
- Lawrence PA, Johnston P, Macdonald P, Struhl G. Borders of parasegments are delimited by the fushi tarazu and even-skipped genes. Nature. 1987;328:440–445. doi: 10.1038/328440a0. [DOI] [PubMed] [Google Scholar]
- Lawrence PA, Pick L. How does the fushi tarazu gene activate engrailed in the Drosophila embryo? Dev Genet. 1998;23:28–34. doi: 10.1002/(SICI)1520-6408(1998)23:1<28::AID-DVG3>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Lengyel JA, Iwaki DD. It takes guts: the Drosophila hindgut as a model system for organogenesis. Dev Biol. 2002;243:1–19. doi: 10.1006/dbio.2002.0577. [DOI] [PubMed] [Google Scholar]
- Liang HL, Nien CY, Liu HY, Metzstein MM, Kirov N, Rushlow C. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature. 2008;456:400–403. doi: 10.1038/nature07388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu QX, Jindra M, Ueda H, Hiromi Y, Hirose S. Drosophila MBF1 is a co-activator for Tracheae Defective and contributes to the formation of tracheal and nervous systems. Development. 2003;130:719–728. doi: 10.1242/dev.00297. [DOI] [PubMed] [Google Scholar]
- Lohr U, Pick L. Cofactor-interaction motifs and the cooption of a homeotic Hox protein into the segmentation pathway of Drosophila melanogaster. Curr Biol. 2005;15:643–649. doi: 10.1016/j.cub.2005.02.048. [DOI] [PubMed] [Google Scholar]
- Magrassi L, Lawrence PA. The pattern of cell death in fushi tarazu, a segmentation gene of Drosophila. Development. 1988;3:447–451. doi: 10.1242/dev.104.3.447. [DOI] [PubMed] [Google Scholar]
- Maier D, Press A, Powell JR. Regulation of the segmentation gene fushi tarazu has been functionally conserved in Drosophila. EMBO J. 1990;9:3957–3966. doi: 10.1002/j.1460-2075.1990.tb07616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J, Bienz M. Sharp anterior boundary of homeotic gene expression conferred by the fushi tarazu protein. Embo J. 1992;11:3653–3661. doi: 10.1002/j.1460-2075.1992.tb05450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasiadka A, Dietrich BH, Krause HM. Anterior-posterior patterning in the Drosophila embryo. Advances in Developmental Biology and Biochemistry. 2002;12:156–204. [Google Scholar]
- Nasiadka A, Krause HM. Kinetic analysis of segmentation gene interactions in Drosophila embryos. Development. 1999;126:1515–1526. doi: 10.1242/dev.126.7.1515. [DOI] [PubMed] [Google Scholar]
- Nelson HB, Laughon A. The DNA binding specificity of the Drosophila fushi tarazu protein: a possible role for DNA bending in homeodomain recognition. New Biologist. 1990;2:171–178. [PubMed] [Google Scholar]
- Nusslein-Volhard C, Kluding H, Jurgens G. Genes affecting the segmental subdivision of the Drosophila embryo. Cold Spring Harbor Symp. Quant. Biol. 1985;50:145–154. doi: 10.1101/sqb.1985.050.01.020. [DOI] [PubMed] [Google Scholar]
- Pazdera TM, Janardhan P, Minden JS. Patterned epidermal cell death in wild-type and segment polarity mutant Drosophila embryos. Development. 1998;125:3427–3436. doi: 10.1242/dev.125.17.3427. [DOI] [PubMed] [Google Scholar]
- Pearson JC, Lemons D, McGinnis W. Modulating Hox gene functions during animal body patterning. Nature Reviews Genet. 2005 doi: 10.1038/nrg1726. in press. [DOI] [PubMed] [Google Scholar]
- Pick LAS, Affolter M, Schmidt-Glenewinkel T, Gehring WJ. Analysis of the ftz upstream element: germ layer-specific enhancers are independently autoregulated. Genes & Dev. 1990;4:1224–1239. doi: 10.1101/gad.4.7.1224. [DOI] [PubMed] [Google Scholar]
- Pick L, Shultz J, Anderson WR, Woodard CT. The Ftz-F1 family: orphan nuclear receptors regulated by novel protein-protein interactions. In: Taneja R, editor. Nuclear Receptors in Development. Elsevier; 2006. [Google Scholar]
- Pilot F, Lecuit T. Compartmentalized morphogenesis in epithelia: from cell to tissue shape. Dev Dyn. 2005;232:685–694. doi: 10.1002/dvdy.20334. [DOI] [PubMed] [Google Scholar]
- Schaerlinger B, Launay JM, Vonesch JL, Maroteaux L. Gain of affinity point mutation in the serotonin receptor gene 5-HT2Dro accelerates germband extension movements during Drosophila gastrulation. Dev Dyn. 2007;236:991–999. doi: 10.1002/dvdy.21110. [DOI] [PubMed] [Google Scholar]
- Schier AF, Gehring WJ. Analysis of a fushi tarazu autoregulatory element: multiple sequence elements contribute to enhancer activity. EMBO J. 1993a;12:1111–1119. doi: 10.1002/j.1460-2075.1993.tb05752.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schier AF, Gehring WJ. Functional specificity of the homeodomain protein fushi tarazu: The role of DNA-binding specificity. Proc. Natl. Acad. Sci. USA. 1993b;90:1450–1454. doi: 10.1073/pnas.90.4.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder MD, Pearce M, Fak J, Fan H, Unnerstall U, Emberly E, Rajewsky N, Siggia ED, Gaul U. Transcriptional control in the segmentation gene network of Drosophila. PLoS Biol. 2004;2:E271. doi: 10.1371/journal.pbio.0020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott MP, O'Farrell PH. Spatial programming of gene expression in early Drosophila embryogenesis. Ann. Rev. Cell Biol. 1986;2:49–80. doi: 10.1146/annurev.cb.02.110186.000405. [DOI] [PubMed] [Google Scholar]
- Segalat L, Berger G, Lepesant JA. Dissection of the Drosophila pourquoi-pas? promoter: complex ovarian expression is driven by distinct follicle cell- and germ line-specific enhancers. Mech Dev. 1994;47:241–251. doi: 10.1016/0925-4773(94)90042-6. [DOI] [PubMed] [Google Scholar]
- Small S, Blair A, Levine M. Regulation of two pair-rule stripes by a single enhancer in the Drosophila embryo. Dev. Biol. 1996;175:314–324. doi: 10.1006/dbio.1996.0117. [DOI] [PubMed] [Google Scholar]
- Tautz D, Pfeifle C. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- Tomancak P, Beaton A, Weiszmann R, Kwan E, Shu S, Lewis SE, Richards S, Ashburner M, Hartenstein V, Celniker SE, Rubin GM. Systematic determination of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-12-research0088. RESEARCH0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracey WD, Ning X, Klingler M, Kramer SG, Gergen JP. Quantitative analysis of gene function in the Drosophila embryo. Genetics. 2000;154:273–284. doi: 10.1093/genetics/154.1.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakimoto BT, Turner FR, Kaufman TC. Defects in embryogenesis in mutants associated with the antennapedia gene complex of Drosophila melanogaster. Dev Biol. 1984;102:147–172. doi: 10.1016/0012-1606(84)90182-9. [DOI] [PubMed] [Google Scholar]
- Wieschaus E, Sweeton D, Costa M. Convergence and extension during germband elongation in Drosophila embryos. In: Keller R, editor. Gastrulation. New York: Plenum Press; 1991. [Google Scholar]
- Winslow GM, Hayashi S, Krasnow M, Hogness DS, Scott MP. Transcriptional activation by the Antennapedia and fushi tarazu proteins in cultured Drosophila cells. Cell. 1989;57:1017–1030. doi: 10.1016/0092-8674(89)90340-1. [DOI] [PubMed] [Google Scholar]
- Yu Y, Li W, Su K, Han W, Yussa M, Perrimon N, Pick L. The nuclear hormone receptor FTZ-F1 is a cofactor for the Drosophila homeodomain protein Ftz. Nature. 1997;385:552–555. doi: 10.1038/385552a0. [DOI] [PubMed] [Google Scholar]
- Yu Y, Yussa M, Song J, Hirsch J, Pick L. A double interaction screen identifies positive and negative ftz gene regulators and Ftz-interacting proteins. Mech. Dev. 1999;83:95–105. doi: 10.1016/s0925-4773(99)00038-6. [DOI] [PubMed] [Google Scholar]
- Yussa M, Lohr U, Su K, Pick L. The nuclear receptor Ftz-F1 and homeodomain protein Ftz interact through evolutionarily conserved protein domains. Mech. Dev. 2001;107:39–53. doi: 10.1016/s0925-4773(01)00448-8. [DOI] [PubMed] [Google Scholar]
- Zallen JA, Blankenship JT. Multicellular dynamics during epithelial elongation. Semin Cell Dev Biol. 2008;19:263–270. doi: 10.1016/j.semcdb.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zallen JA, Wieschaus E. Patterned gene expression directs bipolar planar polarity in Drosophila. Dev Cell. 2004;6:343–355. doi: 10.1016/s1534-5807(04)00060-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A–B) Cuticular preparations of control w1118 and ftz9H34 embryos. Note that the ftz9H34 embryos display strong pair-rule phenotypes. (C) Confocal image of in situ hybridization of ftz RNA to ftz9H34 stage 5 embryo. ftz RNA was expressed in a pattern indistinguishable from wild type, including the apical localization of the RNA, in ftz9H34 mutants.