Abstract
Retrotransposons, specifically Alu and L1 elements, have been especially successful in their expansion throughout primate genomes. While most of these elements integrate through an endonuclease-mediated process termed target primed reverse transcription, a minority integrate using alternative methods. Here we present evidence for one such mechanism, (which we term internal priming) and demonstrate that loci integrating through this mechanism are qualitatively different from “classical” insertions. Previous examples of this mechanism are limited to cell culture assays, which show that reverse transcription can initiate upstream of the 3′ polyA tail during retrotransposon integration. To detect whether this mechanism occurs in vivo as well as in cell culture, we have analyzed the human genome for internal priming events using recently integrated L1 and Alu elements. Our examination of the human genome resulted in the recovery of twenty events involving internal priming insertions, which are structurally distinct from both classical TPRT-mediated insertions and non-classical insertions. We suggest two possible mechanisms by which these internal priming loci are created and provide evidence supporting a role in staggered DNA double-strand break repair. Also, we demonstrate that the internal priming process is associated with inter-chromosomal duplications and the insertion of filler DNA.
Keywords: L1 elements, Alu elements, target primed reverse transcription, double-strand break repair
1. Introduction
L1 elements and Alu elements are highly successful and ubiquitous retrotransposons in primate genomes that are actively involved in shaping the genomic architecture. A full length L1 element is approximately 6kb in length and consists of a 5′ UTR containing an internal RNA polymerase II promoter, two open reading frames (ORFs) separated by an intergenic spacer, and a 3′ UTR region encompassing the poly-A tail (Kazazian and Moran, 1998). ORF1 codes for an RNA-binding protein with nucleic acid chaperone activity and ORF 2 codes for reverse transcriptase (RT) and endonuclease (EN) activities (Mathias et al., 1991; Feng et al., 1996; Kolosha and Martin, 1997). The L1 retrotransposon enzymatic machinery is used by the non-autonomous ~300bp Alu element, which does not code for any proteins, but carries an internal RNA polymerase III promoter (Fuhrman et al., 1981). Generally these elements mobilize by a “copy and paste” mechanism in their host genomes via a process termed retrotransposition. L1s and Alu elements are thought to insert into the genome through a mechanism described as target primed reverse transcription (TPRT), first reported in Bombyx mori (Luan et al., 1993)(Fig. 1a).
Figure 1. Alternative mechanisms of retrotransposon integration.
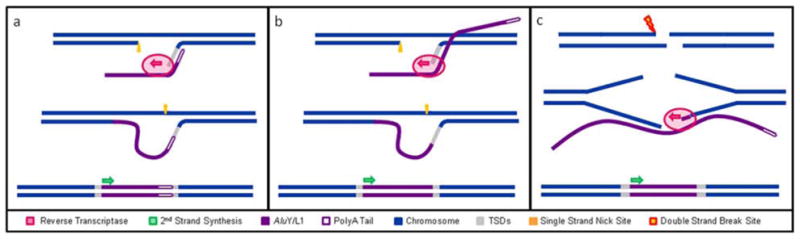
(a) Classical TPRT-mediated L1 or Alu insertion into the host primate genome. L1 EN creates a nick in the first strand (orange arrow) at the 5′-TTTT/A-3′ consensus and the retrotransposon mRNA (purple line) anneals to the genomic DNA (blue line) using its polyA tail (purple outline). L1 RT (pink oval) synthesizes the retrotransposon mRNA to complete insertion and the TSDs (grey) are filled in. (b) TPRT variant-mediated retrotransposon insertion. L1 RT internally primes on the L1 or Alu mRNA and the break is filled using classical TPRT machinery. (c) Staggered DSB repair with 5′ overhangs. A staggered DSB (lightning bolt) occurs and RT (pink oval) internally primes on the mobile element mRNA (purple line) that bridges the gap by binding to either end. Subsequent cDNA synthesis fills the break with a copy of the truncated element.
During TPRT, L1 EN makes a single nick at one of the preferred motifs (e.g. 5′-TTAAAA-3′) and the L1 or Alu element mRNA anneals to the nick site using its 3′ poly-A tail, following which the L1 RT initiates reverse transcription using the mRNA as a template and the second strand nick occurs downstream of the initial cleavage site. This process creates staggered breaks which are later filled in by direct repeats on either side of the element, termed target site duplications (TSDs) (Feng et al., 1996; Szak et al., 2002). The final two steps entail the integration of the newly synthesized single-stranded mobile element cDNA and synthesis of the second strand; the chronological order in which this happens is still unclear. If it proceeds to completion unhindered, TPRT results in the creation of characteristic structural features including intact TSDs and a variable length poly-A tail (Luan et al., 1993; Gilbert et al., 2005). Integration of retrotransposons using classical TPRT has been implicated in the disruption of gene function, deletions at the insertion site, termination of transcription and in the creation of certain disease states (e.g. neurofibromatosis, hemophilia) (Batzer and Deininger, 2002; Goodier and Kazazian, 2008). Though the majority of genomic Alu and L1 elements integrate using this method, a detectable minority integrate into the genome using alternative pathways and variants upon TPRT (Morrish et al., 2002; Callinan et al., 2005; Gilbert et al., 2005; Babushok et al., 2006; Sen et al., 2007; Srikanta et al., 2009).
A recent analysis of L1 elements reported a variation of the “classical” TPRT model of mobile DNA integration (Kulpa and Moran, 2006). This analysis involved an assay to detect ORF2p activity, and provided in vitro evidence that L1 RT preferentially acts upon its own template, as well as Alu elements. Sequencing of the resulting transcripts led to the discovery that RT had occasionally initiated transcription within and upstream of the poly-A tail, (as opposed to “classical” TPRT, where transcription begins at 3′ end of the poly-A tail), similar to a previous study of tRNA-derived retropseudogenes (Schmitz et al., 2004; Callinan et al., 2005; Kulpa and Moran, 2006).. To explore that a similar integration mechanism is active in vivo, we scanned the human genome for truncated Alu and L1 elements with TSDs ≥6bp (Szak et al., 2002). This mechanism of insertion, which we term Internal Priming (IP), appears to be an opportunistic alternative pathway for L1 and Alu mobilization and may play a role in repairing DNA double-strand breaks.
In this analysis we report twenty mobile element insertions that resulted from the internal priming pathway for integration into the human genome. For each locus, we verified the pre-insertion sequence with PCR or cycle-sequencing of DNA from an outgroup primate genome. We confirmed that each had the hallmarks of internal priming (TSDs and 3′ truncation). We suggest that this mechanism of retrotransposon insertion, which has not been described before in the human genome, may constitute a third pathway (after TPRT and NCLI (non-classical L1 insertion (EN-independent)/NCAI (non-classical Alu insertion (EN-independent)) of integration for Alu and L1 element family insertions.
2. Materials and Methods
2.1 Computational extraction and manual authentication of putative IP loci
Alu element and L1 insertions used in this study were identified based on specific differences from both classical TPRT-mediated and non-classical insertion criteria. Characteristics of classical TPRT-mediated insertions include the presence of TSDs, variable length poly-A tails and “preferred” L1 EN-cleavage sites (Morrish et al., 2002); non-classical insertions lack TSDs, polyA tails and use EN-independent insertion sites. Putative internal priming (IP) events are 3′ truncated (lacking the poly-A tail and are ≤276bp for Alu elements, ≤6135bp for L1 elements), have TSDs no shorter than 6bp and do not appear to preferentially insert using preferred L1 EN-cleavage sites (Szak et al., 2002; Sen et al., 2007; Srikanta et al., 2009). Elements selected for this study were less than 2% diverged from the consensus sequence. These structural characteristics are similar to those described in Kulpa et al (2006).
To identify putative IP loci, we revised the method outlined in Sen et al (2007) and Srikanta et al (2009) for detecting non-classical retrotransposon insertions. The L1 and Alu element data were downloaded using whole-chromosome annotation files tabulating all mobile elements on each chromosome (available at http://hgdownload.cse.ucsc.edu/downloads.html#human) for the human (hg18) genome. We filtered the files to retain only Alu and L1 elements. Next, to scan for truncated Alu and L1 elements missing the poly-A tail used during classical TPRT-mediated integration, we used a Perl script to locate those Alu elements which had 3′ truncations to positions numbering 276 or less, along the 312bp AluY consensus sequence used by the RepeatMasker (RM) software package in its default settings (Smit et al., 1996), and those L1 elements which were 6135bp or less as described in Sen et al (2007).
Manual inspection of computationally detected loci involved extracting the putative truncated Alu or L1 element sequence with 5000bp of flanking sequence on both sides of each locus. Next, this sequence was used to query the chimpanzee (panTro2) and rhesus macaque (rheMac2) genomes using the BLAT software suite (http://www.genome.ucsc.edu/cgi-bin/), and a triple alignment of the locus was created to analyze the local pre-insertion and post-insertion sequence architecture. In particular, we scanned for the presence of TSDs longer than 6bp and for any target-site deletions present in the pre-insertion sequence, but absent following the Alu or L1 insertion. To ascertain whether the element was truly young and truncated (and thereby reduce the likelihood of finding false positives), we investigated the Alu and L1 elements within the context of 5000bp sequence flanking either side of the insertion. We discarded all elements >2% diverged from their respective consensus sequences according to the RM algorithm to limit our results to relatively recent integration events with easily reconstructed pre-insertion sequence architecture, using the chimpanzee and rhesus genomes.
We chose loci for experimental validation that matched the following four criteria: 3′ truncation as specified above, presence of TSDs ≥6bp in length, absence of a poly-A tail, and verifiable pre-insertion sequence structure in two other primate genomes. We cross-checked our putative IP loci against the orthologous pre-insertion sites in the other genomes to confirm there was no extraneous sequence between the starting points of the upstream and downstream matching flanking regions in the post-insertion genome (Table 1). To further confirm that loci fitting all the criteria described above were indeed atypical Alu and L1 insertions and not artifacts arising from sequence assembly errors, we PCR-amplified all loci from a panel of primate genomes and resequenced all ambiguous loci. To differentiate between Alu and L1 IP events we have labeled them AIP for Alu Internal Priming and L1IP for L1 element Internal Priming events.
Table 1. IP loci and insertion site characteristics.
In the column for lineage, H represents Human-specific loci, while HCG represents loci shared between subtribe Hominina, and HCGO represents loci shared between tribe Hominini.
| Alu | Coordinates | TSD | Alu bp_ins | Non-Alu bp_ins | bp_del | Lineage | Intragenic | Non Alu seq | subfamily | EN cleavage Site |
|---|---|---|---|---|---|---|---|---|---|---|
| AIP13 | chr6:141586702–141586851 | 13 | 150 | 3 | 1 | H | - | 3′ | Yg6 | 5′CTTA/A |
| AIP17 | chr7:44390521–44400651 | 15 | 133 | 0 | 0 | H | NUDCD3 | - | Y | 5′TTTG/G |
| AIP9 | chr4:177948302–177958449 | 15 | 97 | 100 | 0 | HCGO | - | 3′ | Y | 5′TTCT/T |
| AIP10 | chr4:46764433–46764462 | 7 | 30 | 57 | 0 | HCGO | GABRB1 | both | Yc3 | 5′TATA/A |
| AIP16 | chr7:157111896–157122003 | 14 | 120 | 0 | 2 | HCGO | PTPRN2 | - | Y | 5′AGTG/G |
| AIP29 | chrX:10054799–10064887 | 6 | 89 | 1 | 1 | HCGO | KIAA1280/WWC3 | 3′ | Y | 5′GAAA/A |
| L1 | TSD | L1 bp_ins | Non-L1 bp_ins | bp_del | Lineage | Intragenic | Non L1 seq | subfamily | ||
| L1IP8 | chr17:22510334–22515547 | 13 | 1932 | 3 | 0 | H | - | both | L1PA2 | 5′AAAA/A |
| L1IP16 | chr11:104044065–104057190 | 11 | 1371 | 594 | 0 | H | - | 5′ | L1HS | 5′TCAA/A |
| L1IP21 | chr1:102542356–102562760 | 16 | 405 | 57 | 0 | H | - | 5′ | L1PA2 | 5′ATGC/C |
| L1IP36 | chr4:142312647–142333603 | 7 | 954 | 0 | 0 | H | - | - | L1P1 | 5′GCTC/C |
| L1IP26 | chr2:229148117–229168169 | 15 | 51 | 36 | 0 | HCGO | - | 5′ | L1P2 | 5′GATT/T |
| L1IP27 | chr2:233940011–233960152 | 9 | 142 | 54 | 1 | HCGO | DGKD | 5′ | L1PA5 | 5′ATTT/T |
| L1IP28 | chr3:8809713–8829820 | 15 | 106 | 259 | 0 | HCGO | - | 5′ | L1P1 | 5′TCAA/A |
| L1IP31 | chr14:32591217–32611248 | 13 | 33 | 26 | 0 | HCG | NPAS3 | 5′ | L1PA2 | 5′TTGC/C |
| L1IP42 | chr7:117312394–117332534 | 14 | 141 | 31 | 1 | HCGO | - | 5′ | L1P1 | 5′CTCT/T |
| L1IP43 | chr8:23977525–23997785 | 10 | 253 | 0 | 0 | HCGO | - | - | L1P2 | 5′AATA/A |
| L1IP49 | chr11:81616736–81636875 | 16 | 140 | 85 | 0 | HCGO | - | 5′ | L1PA5 | 5′TTCC/C |
| L1IP54 | chr12:116338013–116358130 | 9 | 118 | 24 | 1 | HCGO | - | 5′ | L1PA3 | 5′AAAA/A |
| L1IP68 | chrX:113986811–114007431 | 16 | 621 | 259 | 0 | HCG | HTR2C | 5′ | L1P1 | 5′ATGT/T |
| L1IP159 | chr21:17169254–17189433 | 15 | 180 | 33 | 0 | HCG | - | 5′ | L1PA3 | 5′CATT/T |
2.2 Validation of loci through PCR amplification and resequencing
We designed primers for each locus using the Primer3 utility (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) and performed PCR in 25μl reactions using 15–25ng genomic DNA, 0.28μM primer, 200μM dNTPs in 50mM KCl, 1.5mM MgCl2, 10mM Tris-HCl (pH 8.4), and 2.5 units Taq DNA polymerase. Thermocycler programs were as follows: 95°C for 2 min (1 cycle), [95°C for 30sec, optimal annealing temperature for 30 sec, 72°C for 1 min] (35 cycles), 72°C for 10 min (1 cycle). PCR products were visualized on 2% agarose gels stained with ethidium bromide. For PCR fragments with expected lengths larger than 1.5kb, ExTaq™ (Takara) was used according to the manufacturer’s specified protocol. All loci were amplified from the following genomes: Homo sapiens (HeLa; cell line ATCC CCL-2), Pan troglodytes (common chimpanzee “Clint”; cell line NS06006B), Gorilla gorilla (Western lowland gorilla; cell line Coriell Cell Repositories AG05251), Pongo pygmaeus (orangutan; cell line GM04272A), Macaca mulatta (rhesus macaque; cell line NG07109), and Chlorocebus aethiops (African green monkey; cell line ATCC CCL70) (Fig. 2). Primer sequences are available from the Publications section of the Batzer laboratory website (http://batzerlab.lsu.edu).
Figure 2. A schematic detailing IP locus investigation.
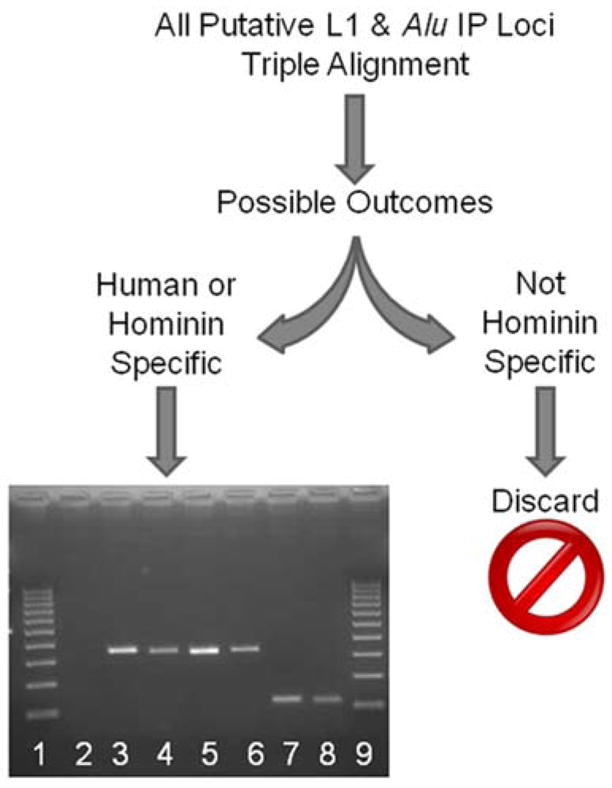
All computationally derived candidate loci were triple-aligned (human, chimpanzee, and rhesus macaque), and those loci found to be human- or hominin-specific were kept for wet bench verification. Gel chromatograph of PCR products from a phylogenetic analysis of a hominin-specific AIP locus (AIP 9). The numbers indicate the DNA template used: 1 & 9, 100bp ladder; 2, negative control (H2O); 3, human; 4, chimpanzee; 5, gorilla; 6, orangutan; 7, rhesus macaque; 8, green monkey.
Loci were sequenced directly from the PCR amplicons after cleanup using Wizard® gel purification kits (Promega Corporation) or ExoSAP-IT® (USB Corporation). Samples that could not be sequenced directly from PCR products were cloned into vectors using the TOPO TA (fragments <1kb) cloning kit (Invitrogen). Sequencing results were obtained using an ABI3130XL automated DNA sequencer and analyzed using the SeqMan, BioEdit and EditSeq utilities from the DNAStar® V.5 package. GC content was calculated using GEECEE (http://mobyle.pasteur.fr/cgi-bin/MobylePortal/portal.py?form=geecee) for both the flanking regions and the insertion. Close inspection of the flanking sequence and the results of the PCR and sequence analyses confirmed the pre-insertion loci from two outgroup genomes (Fig. 2).
3. Results and Discussion
3.1 Characterization of putative internal priming mechanisms
Based on our analyses, we suggest that two alternatives to TPRT may be responsible for the internal priming structures observed (Fig. 1). The first is an opportunistic mechanism wherein a first strand nick is created by the L1 EN and instead of annealing by its polyA tail as in “classical” TPRT, the retrotransposon mRNA attaches to the host genome using a limited number of complementary bases at a site within the mobile element upstream of the polyA tail). RT activity (albeit possibly at reduced fidelity) fills in the break with a single-stranded copy of the element, and the other steps of the integration proceed as with classical TPRT (Fig 1b), i.e. a second strand nick then occurs, the entire break is filled, and TSDs form as in classical TPRT. In the second mechanism, retrotransposon mRNA attaches to both ends of a preexisting staggered double-strand break in the genome using complementary base pairing at sequences within the length of the element (as opposed to the 5′ or 3′ ends). RT activity begins at the 3′ binding site, with subsequent cDNA synthesis joining the ends of the DSB with a copy of the truncated element (Fig 1c) (Lin et al., 1999; Lin and Waldman, 2001; Ostertag and Kazazian, 2001; Valerie and Povirk, 2003; Haber, 2006; Haber, 2008; Lieber et al., 2008). Due to the staggered nature of the break, TSDs are formed, filling in the cleavage sites. Low levels of microhomology found only at the 3′ ends of these insertions could provide further support for the opportunistic nature of mobile element recruitment to the break site. Termed “Internal Priming” (IP), these insertions differ from those found during classical TPRT and NCI events in that they are truncated elements with intact TSDs (Kulpa and Moran, 2006).
3.2 Investigation of human genomic internal priming events
Using a combination of computational data mining and wet-bench verification, we analyzed the human genome for evidence of an internal priming mechanism of retrotransposition, specifically Alu and L1 elements. We excluded all classical TPRT-mediated insertions through a stringent manual inspection of putative IP loci following a triple alignment of the three genomes at each locus and PCR analyses (Fig. 2). A total of twenty IP insertions from the hg18 assembly were verified in this manner, six human-specific loci (two AIP and four L1IP) and fourteen loci (4 AIP and 10 L1IP) that were shared among the hominin genomes (i.e., human, chimpanzee, gorilla, and orangutan), with the pre-insertion architecture confirmed via PCR-assay and sequencing (Table 1). Along with the truncated Alu and L1 elements, we found approximately 1.63kb of non-retrotransposon sequence inserted at experimentally confirmed IP loci, with ~163bp associated with Alu elements and ~1.47kb associated with L1s (Table 1).
3.3 Sequence composition of IP loci and alignment to the full-length consensus sequence
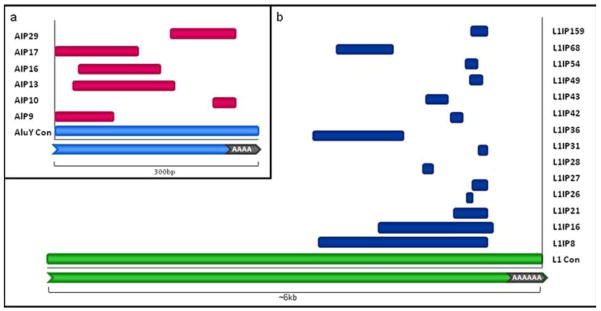
Alu internal priming (AIP) loci ranged in size from 30bp to 150bp, with an average AIP length of ~103bp, in contrast to full-length Alu elements, which are ~300bp in length. The L1 internal priming (L1IP) loci ranged from 33bp to 1.9kb in length as compared to a consensus L1 sequence, which is ~6kb in length, with an average L1IP length of ~460bp (Fig. 3). A multiple alignment of AIP and LIP loci with their respective full-length sequences revealed that the AIP loci had a slight tendency to cluster towards the 5′end whereas L1IP loci had a tendency to cluster towards the 3′ end of their consensus sequences (Fig. 3). Of 20 total insertions, only 2 are 5′ intact, and both are AIP loci (AIP 17 & 9), which can be explained by the short insertion length of Alu elements. As full-length Alu elements are only ~300bp in length, when RT internally primes somewhere within the Alu element, it is more likely to reach the 5′ end of the Alu mRNA, whereas a full-length L1 element is much longer and may be more likely to be 5′ truncated. None of the L1IP insertions were 5′ intact. L1IP loci showed at least 3.5kb 5′ truncation and four AIP showed at least 35bp of 5′ truncation. Two AIP loci had intact middle polyA rich regions and one AIP was truncated within the middle polyA rich region, whereas only one L1IP locus (L1IP 36) had an intact intergenic spacer region. A common feature of classical TPRT insertions is the creation of target site deletions. In our data only six of the twenty IP loci had target site deletions associated with their insertions. Fourteen loci lacked target site deletions, only 7bp total were deleted whereas ~8.7kb mobile element and non-mobile elements sequences were inserted. These findings are consistent with the theory that IP events arise as a consequence of a DSB repair mechanism.
Figure 3. Alignment of IP loci to their respective consensus sequences.
(a) AIP fragments juxtaposed with a representation of a full-length Alu element consensus sequence. The Alu fragments are pink and the consensus sequence is light blue. Two AIP loci are 5′ intact and overall AIP loci align to the consensus sequence with no bias. (b) L1IP fragments juxtaposed with a representation of a full-length L1 element consensus sequence The L1 fragments are. dark blue and the consensus sequence is green. L1IP loci show an alignment bias for the 3′ end of the consensus sequence.
It is theoretically possible that post-insertion 3′ truncation events would mimic the unique local sequence architecture of IP events. In this analysis, we tried to minimize such errors using two different methods. First, we compared the orthologous flanking sequence in all three primate genomes to confirm that post-insertion random genomic deletions did not delete the portion of the element immediately upstream of the 3′ TSD, creating a truncated structure that could mimic the AIP or LIP structure; we assume that the probability of post-insertion 3′ truncation occurring independently at exactly the same position in three separate primate genomes is negligible. Second, we further confirmed that 3′ truncation events were not created by “private” deletions in the reference human genome mimicking IP events by PCR amplification of all loci on a population panel consisting of 80 individuals from four different geographic ancestral origins: African Americans, South Americans, Europeans and Asians. Gel electrophoresis of the PCR amplicons showed no variation in the expected size, and DNA sequencing also confirmed the PCR amplicons contained only the truncated element and no individual had a full length Alu or L1 element.
Based on the local genomic architecture of these insertions and an analysis of the L1 EN cleavage sites of the loci, we suggest the preferred model for IP may be a rare variant of TPRT or another more opportunistic mechanism, staggered double-strand break repair (Fig. 1b, 1c). Four loci contained only the L1 or Alu element while sixteen IP loci had non-mobile element DNA associated with them; in some cases this could represent “filler DNA” (Roth et al., 1989). The twenty IP loci described could be the products of template jumping activity, which has been previously documented for reverse transcriptase (Cost et al., 2002; Kulpa and Moran, 2006; Kurzynska-Kokorniak et al., 2007; Eickbush and Jamburuthugoda, 2008). Four out of six AIP insertions and three of fourteen L1IP insertions occurred in intragenic regions; though there are only twenty loci, this may suggest an internal priming repair mechanism using available mRNA from nearby actively transcribed elements. Both Alu and L1 elements are mobilizing in the genome, and we suggest a variant reverse transcriptase-mediated pathway that operates opportunistically.
The search criteria used in this analysis were quite stringent and loci that could potentially have represented IP insertions may have been culled. We were only able to find those germline events that have been successfully inherited; many more germline events are likely to have occurred, but were lost. There could also be many somatic events, but these would remain mainly unrecoverable by our analysis. RepeatMasker has difficulty correctly discerning insertions under 30bp in length, even when using the most sensitive setting, and can miscall ambiguous repetitive elements. By sampling from only one genome, our analysis will not recover many low-frequency polymorphic human loci that could be present in the species (Hedges et al., 2004; Callinan et al., 2005). This study was made even more conservative by discarding all elements >2% diverged from their consensus sequences and keeping only those loci with unambiguous TSDs ≥6bp and in which the pre-insertion sequence could be authenticated through triple-alignment and wet-bench verification. There are potentially many more IP loci, and this analysis is by no means comprehensive, but the loci presented here provide evidence of an opportunistic, non-standard pathway involving internal priming of reverse transcriptase.
3.4 IP microhomology and endonuclease cleavage site analyses
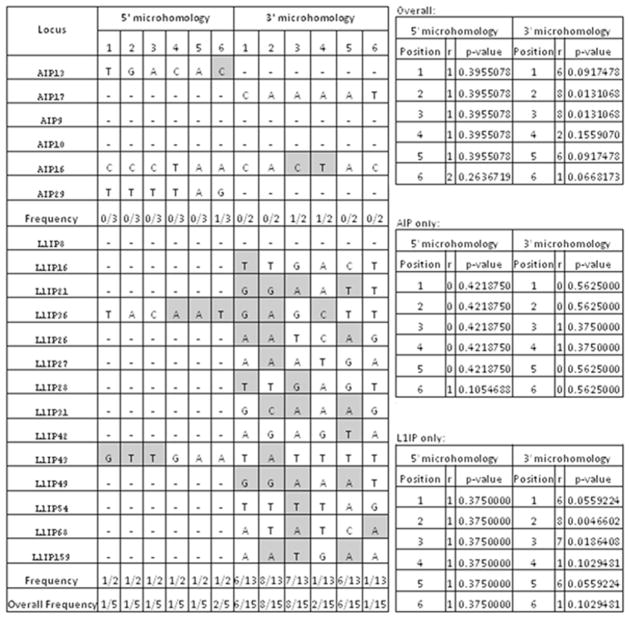
To attempt to distinguish between our two hypothetical mechanisms accounting for IP events, we performed two analyses to determine the independent nature of the insertion site as well as attachment at the insertion site. Using the method outlined in Srikanta et al (2009), microhomology analyses were performed on AIP and L1IP loci separately and combined (Fig. 4) (Zingler et al., 2005). We compared 6bp stretches at both ends of the insert, using only those loci whose 5′ and 3′ ends did not include non-L1 or Alu sequence. Of the 20 loci possible for this analysis, five loci at the 5′ end (3 AIP and 2 L1IP) and fifteen loci at the 3′ end (2 AIP and 13 L1IP) included Alu or L1 sequence. Our results indicate a slightly increased level of microhomology at the 3′ insertion junctions using L1IP data alone, and in the combined data set (Fig. 4). This suggests that the microhomology at the 3′ insertion junction of L1IP events may mediate attachment to the break site. The small number of AIP events does not provide enough support to draw conclusions about whether the same might be true for Alu elements.
Figure 4. Combined AIP & L1IP microhomology analysis.
Complementary nucleotide positions are counted in opposite directions at the 5′ and 3′ ends of the respective consensus sequences. Bases are highlighted in grey if they are complementary to the corresponding nucleotide on the L1 or Alu RNA. The number of matches at each position (r) and the corresponding p-values indicate the likelihood of obtaining the observed numbers of matches by chance alone. Using a binomial probability distribution, we calculated p-values assuming the chance of success (i.e. complimentary pairing) was 1/4 and the chance of failure was 3/4 at each position.
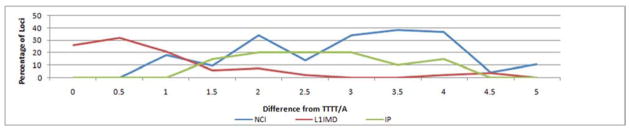
If the IP insertions occurred through a variant of TPRT that retains the dependence on the L1 EN to create the nicks in the host genome, few differences from the typical L1 EN cleavage site (5′TTTT/A) would be expected. To test this hypothesis, we inspected the sequence at the insertion sites of the loci in this analysis and find that there is substantial deviation from both the preferred and atypical cleavage sites for L1 EN (Morrish et al., 2002; Han et al., 2005). Using a previously described analysis system for L1 EN dependence (Han et al., 2005), IP loci were compared to a combined analysis of non-classical Alu and L1 insertions and a recent analysis of TPRT-mediated insertions (Fig. 5). These comparisons indicate that the cleavage sites of IP events differ from typical EN cleavage sites, but not as substantially as non-classical insertion cleavage sites. These findings are more consistent with the hypothesis that mobile elements are opportunistically integrating into genomic lesions as a mechanism for repairing staggered DSBs using an internal priming mechanism, as opposed to the TPRT variant mechanism (Fig. 1, 4 & 5).
Figure 5. IP insertion site divergence from the preferred L1 endonuclease cleavage site sequence.
Loci generated by three different insertion studies (L1IMD, NCI and IP) were analyzed for presence or absence of the preferred L1 EN cleavage site motif. The red line indicates loci analyzed for L1IMD events, which occur via classical TPRT; the blue line indicates NCI events, which are L1 EN-independent; and the green line indicates IP events. The results indicate increased divergence from the preferred motif used by L1 EN-mediated classical TPRT, suggesting that IP events use a mechanism more similar to NCI than L1IMD. These findings are consistent with an opportunistic mechanism.
3.5 Features of IP loci are consistent with a model of DNA double-strand break repair
IP loci possess distinct characteristics that set them apart from both classical TPRT-mediated insertions and non-classical (EN-independent) insertions. We propose that this internal priming mechanism can act as an alternative integration pathway for retrotransposons in primate genomes and may occasionally be involved in repair of staggered DNA double-strand breaks. Both microhomology and EN cleavage site analyses provide support for an opportunistic mechanism that bridges breaks neatly, resulting in little loss or gain of genomic material.
Also in contrast to classical TPRT-mediated insertions, sixteen out of twenty IP loci (4 AIPs and 12 L1IPs) had non-mobile element DNA inserted along with the retrotransposon insertion (Table 1). These fragments ranged in size from 1bp to 594bp and were generally found 5′ of the mobile element in the insertion site (Roth et al., 1989). Of the four AIP loci with non-Alu inserted sequence, one had non-Alu sequence on both sides of the truncated Alu (AIP 10) and the other three had 3′ non-Alu inserted sequence (AIPs 13, 9, 29). Of the twelve L1IP loci with non-L1 inserted sequence, only one had non-LI inserted sequence (L1IP 8) on both sides of the truncated L1, while eleven loci had non-L1 sequence inserted 5′ of the truncated L1 (L1IPs 16, 21, 26, 27, 28, 31, 42, 49, 54, 68, 159), as opposed to 3′ as was observed in the AIP events. Both L1IP 28 (found on chromosome 3) and L1IP 68 (found on the X chromosome) appear to have 5′ transduced sequence. The transduced sequences are 245bp and 206bp, respectively, and share more than 94% sequence homology with different non-repetitive sequence on chromosome 8. L1IPs 49, 54 and 159 appear to have included sequence from unknown locations while the majority of non-Alu or L1 sequence inserted with the IP loci is in the form of simple or low-complexity repeats suggesting that the internal priming process could play a role in creating new simple and low complexity repeats (Ovchinnikov et al., 2001; Mirkin, 2006; Sen et al., 2007).
Three IP loci were characterized with either AT or CA-rich repeats at the 3′ or 5′ ends. Both Alu elements and L1 elements have previously been associated with the expansion and formation of microsatellites; however, as these microsatellites may have expanded or contracted over time, it is difficult to determine the exact sequence at the time of insertion (Arcot et al., 1995). Along with simple and low complexity repeats, we found evidence for capture of extra L1 RNA at one locus (L1IP 16). The non-mobile element inserted sequence did not have significant matches when searches were performed in BLAT and BLAST. Eight L1IP events showed a polyT repeat at the 5′ end of their insertions (L1IPs 16, 27, 31, 42, 49, 54, 69, 159). These stretches ranged from 7bp to 37bp and are not the complementary sequence to the polyA tail of a retrotranspositionally-competent L1. Such polyT stretches have been suggested to cause instability and act as recombination hotspots (Chambeyron et al., 2002; Wallace et al., 2008).
3.6 Evidence for non-traditional mobilization in primate genomes
We have provided evidence for the existence of an alternative integration mechanism for L1 and Alu elements in primate genomes. With this analysis, we have shown an integration mechanism that differs from both classical TPRT and EN-independent insertion activity. The structural features of the loci discussed in this study leave little doubt that the internal priming-based integration mechanism we report is distinct from classical TPRT and constitutes a non-preferred method of Alu and L1 mobilization. Previous in vitro systems have shown the existence of internal priming for L1s (Kulpa and Moran, 2006); however, our analysis confirms that this mechanism is active in vivo as well.
While overall, the large proportion of TPRT-mediated L1 and Alu insertions in primate genomes are essentially neutral, individual loci may be associated with disruption of gene function and creation of local genomic instability, largely due to the “active” role of the L1 EN in creating DSBs (Hedges and Deininger, 2007; Goodier and Kazazian, 2008). In contrast to such insertions, the “passive” role IP events seem to be playing in the fortuitous repair of genomic lesions gives them a role (albeit minor) in maintaining genomic stability through an RNA-mediated DNA repair mechanism. L1 and Alu elements make up a significant portion of primate genomes and have been implicated in a number of mechanisms that have led to lineage-specific evolutionary changes. The relatively conservative estimate of the number of IP events in hominins that we present here is due to the methods we used: restricting our computational search to the human genome, RepeatMasker limitations described in section 3.3, host genome tolerance, and the ≤2% divergence from the consensus sequence we allowed in order to filter for the youngest elements. This estimate undoubtedly represents only a fraction of the total number of IP events possible in primate genomes. The human genome contains ~1.2 million Alu elements and ~0.5 million L1s (Batzer and Deininger, 2002; Goodier and Kazazian, 2008; Comeaux et al., 2009). Using the BLAT Tables utility (Kent, 2002), and filtering for Alu elements showing divergence of 2% or less from the consensus sequence, we found 572 young inserts in the human genome. We also found 706 L1 elements using the same criteria. Out of twenty IP loci, we had six human-specific events, two were Alu element-based and four were L1-based. Employing a similar analysis approach to that was used in Srikanta et al 2009, our data suggest a rate of insertions among young elements by this internal priming pathway in the human genome to be ~0.35% for Alu elements and ~0.57% or ~0.6% for L1 elements. Two percentages are given here for L1 element insertions as we calculated this rate using two different estimates of the number of L1s, all L1s versus only those in the L1PA1 and L1HS subfamilies (Khan et al., 2006; Giordano et al., 2007). Since the beginning of the radiation of the primate lineage (~65 million years), as few as 3680 and as many as 4196 Alu elements may have inserted using this pathway (1 AIP insertion per ~15,000–18,000 years). A similar extrapolation with L1 elements suggests that anywhere between 2833 and 3125 L1 elements have inserted in this fashion (1 L1IP insertion per ~20,000–23,000 years).
4. Conclusion
In conclusion, using a combination of computational data mining and experimental verification, we have established that the retrotransposon internal priming events seen in cell culture also occur in vivo. Recent analyses provide evidence supporting alternative pathways to integration for mobile elements (Morrish et al., 2002; Gilbert et al., 2005; Babushok et al., 2006; Kulpa and Moran, 2006; Sen et al., 2007; Srikanta et al., 2009). Internal priming events may play a role in genomic stability by repairing genomic lesions. This mechanism is distinct from classical TPRT and an EN-independent pathway, as distinguished by inspection of the pre-insertion and post-insertion features of the sequence architecture. Internal priming events seem to have occurred at a much lower frequency than either TPRT or NCI events. This is consistent with the results of in vitro assays which demonstrated that priming upstream of the 3′ poly-A tail results in reduced retrotransposition (Kulpa and Moran, 2006). Internal priming is an inefficient pathway, suggesting the mechanism of insertion is occurring in trans. While the internal priming mechanism could be explained as a variant of TPRT that we term TPRT variant, the characteristic features of these loci are more indicative of a random integration mechanism occasionally resulting in the repair of DSBs, which would otherwise be deleterious to the genome (Lin and Waldman, 2001; Rudin and Thompson, 2001; Hagan et al., 2003; Brugmans et al., 2007; Helleday et al., 2007; Ichiyanagi et al., 2007; Wallace et al., 2008). Overall, growing evidence from recent analyses of such non-deleterious roles for both the L1 and Alu families is providing support for a role for TEs in maintaining genomic stability, illuminating yet another aspect to the biology of non-LTR retrotransposons in primate genomes.
Acknowledgments
The authors would like to thank all members of the Batzer laboratory for their support and feedback. They would especially like to thank J.A. Walker, K. Han and M. Konkel for suggestions and advice. They are grateful to T.J. Meyer, C. Faulk and J. Huang for their useful comments during the preparation of the manuscript. This research was supported by LSU Biograds #08-10 (D. S.), National Science Foundation grant BCS-0218338 (M.A.B.), and National Institutes of Health RO1 GM59290 (M.A.B.).
Abbreviations
- IP
internal priming
- HS
human-specific
- DSBs
double-strand breaks
- TPRT
target primed reverse transcription
- PCR
polymerase chain reaction
- NHEJ
non-homologous end-joining
- EN
endonuclease
- RT
reverse transcriptase
- SINE
short interspersed element
- RM
RepeatMasker
- TSD
target site duplication
- NCLI
non-classical L1 insertion (EN-independent)
- NCAI
non-classical Alu insertion (EN-independent)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arcot SS, Wang Z, Weber JL, Deininger PL, Batzer MA. Alu repeats: a source for the genesis of primate microsatellites. Genomics. 1995;29:136–144. doi: 10.1006/geno.1995.1224. [DOI] [PubMed] [Google Scholar]
- Babushok DV, Ostertag EM, Courtney CE, Choi JM, Kazazian HH., Jr L1 integration in a transgenic mouse model. Genome Res. 2006;16:240–250. doi: 10.1101/gr.4571606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nat Rev Genet. 2002;3:370–379. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- Brugmans L, Kanaar R, Essers J. Analysis of DNA double-strand break repair pathways in mice. Mutat Res. 2007;614:95–108. doi: 10.1016/j.mrfmmm.2006.01.022. [DOI] [PubMed] [Google Scholar]
- Callinan PA, Wang J, Herke SW, Garber RK, Liang P, Batzer MA. Alu Retrotransposition-mediated Deletion. J Mol Biol. 2005;348:791–800. doi: 10.1016/j.jmb.2005.02.043. [DOI] [PubMed] [Google Scholar]
- Chambeyron S, Bucheton A, Busseau I. Tandem UAA repeats at the 3′-end of the transcript are essential for the precise initiation of reverse transcription of the I factor in Drosophila melanogaster. J Biol Chem. 2002;277:17877–17882. doi: 10.1074/jbc.M200996200. [DOI] [PubMed] [Google Scholar]
- Comeaux MS, Roy-Engel AM, Hedges DJ, Deininger PL. Diverse cis factors controlling Alu retrotransposition: What causes Alu elements to die? Genome Res. 2009 doi: 10.1101/gr.089789.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cost GJ, Feng Q, Jacquier A, Boeke JD. Human L1 element target-primed reverse transcription in vitro. Embo J. 2002;21:5899–5910. doi: 10.1093/emboj/cdf592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickbush TH, Jamburuthugoda VK. The diversity of retrotransposons and the properties of their reverse transcriptases. Virus Res. 2008;134:221–234. doi: 10.1016/j.virusres.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Moran JV, Kazazian HH, Jr, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- Fuhrman SA, Deininger PL, LaPorte P, Friedmann T, Geiduschek EP. Analysis of transcription of the human Alu family ubiquitous repeating element by eukaryotic RNA polymerase III. Nucleic Acids Res. 1981;9:6439–6456. doi: 10.1093/nar/9.23.6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert N, Lutz S, Morrish TA, Moran JV. Multiple fates of L1 retrotransposition intermediates in cultured human cells. Mol Cell Biol. 2005;25:7780–7795. doi: 10.1128/MCB.25.17.7780-7795.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano J, Ge Y, Gelfand Y, Abrusan G, Benson G, Warburton PE. Evolutionary History of Mammalian Transposons Determined by Genome-Wide Defragmentation. PLoS Comput Biol. 2007;3:e137. doi: 10.1371/journal.pcbi.0030137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier JL, Kazazian HH., Jr Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell. 2008;135:23–35. doi: 10.1016/j.cell.2008.09.022. [DOI] [PubMed] [Google Scholar]
- Haber JE. Chromosome breakage and repair. Genetics. 2006;173:1181–1185. doi: 10.1093/genetics/173.3.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber JE. Alternative endings. Proc Natl Acad Sci U S A. 2008;105:405–406. doi: 10.1073/pnas.0711334105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan CR, Sheffield RF, Rudin CM. Human Alu element retrotransposition induced by genotoxic stress. Nat Genet. 2003;35:219–220. doi: 10.1038/ng1259. [DOI] [PubMed] [Google Scholar]
- Han K, Sen SK, Wang J, Callinan PA, Lee J, Cordaux R, Liang P, Batzer MA. Genomic rearrangements by LINE-1 insertion-mediated deletion in the human and chimpanzee lineages. Nucleic Acids Res. 2005;33:4040–4052. doi: 10.1093/nar/gki718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges DJ, Callinan PA, Cordaux R, Xing J, Barnes E, Batzer MA. Differential alu mobilization and polymorphism among the human and chimpanzee lineages. Genome Res. 2004;14:1068–1075. doi: 10.1101/gr.2530404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges DJ, Deininger PL. Inviting instability: Transposable elements, double-strand breaks, and the maintenance of genome integrity. Mutat Res. 2007;616:46–59. doi: 10.1016/j.mrfmmm.2006.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helleday T, Lo J, van Gent DC, Engelward BP. DNA double-strand break repair: from mechanistic understanding to cancer treatment. DNA Repair (Amst) 2007;6:923–935. doi: 10.1016/j.dnarep.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Ichiyanagi K, Nakajima R, Kajikawa M, Okada N. Novel retrotransposon analysis reveals multiple mobility pathways dictated by hosts. Genome Res. 2007;17:33–41. doi: 10.1101/gr.5542607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian HH, Jr, Moran JV. The impact of L1 retrotransposons on the human genome. Nat Genet. 1998;19:19–24. doi: 10.1038/ng0598-19. [DOI] [PubMed] [Google Scholar]
- Kent WJ. BLAT--the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan H, Smit A, Boissinot S. Molecular evolution and tempo of amplification of human LINE-1 retrotransposons since the origin of primates. Genome Res. 2006;16:78–87. doi: 10.1101/gr.4001406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolosha VO, Martin SL. In vitro properties of the first ORF protein from mouse LINE-1 support its role in ribonucleoprotein particle formation during retrotransposition. Proc Natl Acad Sci U S A. 1997;94:10155–10160. doi: 10.1073/pnas.94.19.10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulpa DA, Moran JV. Cis-preferential LINE-1 reverse transcriptase activity in ribonucleoprotein particles. Nat Struct Mol Biol. 2006;13:655–660. doi: 10.1038/nsmb1107. [DOI] [PubMed] [Google Scholar]
- Kurzynska-Kokorniak A, Jamburuthugoda VK, Bibillo A, Eickbush TH. DNA-directed DNA polymerase and strand displacement activity of the reverse transcriptase encoded by the R2 retrotransposon. J Mol Biol. 2007;374:322–333. doi: 10.1016/j.jmb.2007.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber MR, Lu H, Gu J, Schwarz K. Flexibility in the order of action and in the enzymology of the nuclease, polymerases, and ligase of vertebrate non-homologous DNA end joining: relevance to cancer, aging, and the immune system. Cell Res. 2008;18:125–133. doi: 10.1038/cr.2007.108. [DOI] [PubMed] [Google Scholar]
- Lin Y, Lukacsovich T, Waldman AS. Multiple pathways for repair of DNA double-strand breaks in mammalian chromosomes. Mol Cell Biol. 1999;19:8353–8360. doi: 10.1128/mcb.19.12.8353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Waldman AS. Promiscuous patching of broken chromosomes in mammalian cells with extrachromosomal DNA. Nucleic Acids Res. 2001;29:3975–3981. doi: 10.1093/nar/29.19.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- Mathias SL, Scott AF, Kazazian HH, Jr, Boeke JD, Gabriel A. Reverse transcriptase encoded by a human transposable element. Science. 1991;254:1808–1810. doi: 10.1126/science.1722352. [DOI] [PubMed] [Google Scholar]
- Mirkin SM. DNA structures, repeat expansions and human hereditary disorders. Current Opinion in Structural Biology. 2006;16:351–358. doi: 10.1016/j.sbi.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Morrish TA, Gilbert N, Myers JS, Vincent BJ, Stamato TD, Taccioli GE, Batzer MA, Moran JV. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nat Genet. 2002;31:159–165. doi: 10.1038/ng898. [DOI] [PubMed] [Google Scholar]
- Ostertag EM, Kazazian HH., Jr Twin priming: a proposed mechanism for the creation of inversions in l1 retrotransposition. Genome Res. 2001;11:2059–2065. doi: 10.1101/gr.205701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovchinnikov I, Troxel AB, Swergold GD. Genomic Characterization of Recent Human LINE-1 Insertions: Evidence Supporting Random Insertion. Genome Res. 2001;11:2050–2058. doi: 10.1101/gr.194701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth DB, Chang XB, Wilson JH. Comparison of filler DNA at immune, nonimmune, and oncogenic rearrangements suggests multiple mechanisms of formation. Mol Cell Biol. 1989;9:3049–3057. doi: 10.1128/mcb.9.7.3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudin CM, Thompson CB. Transcriptional activation of short interspersed elements by DNA-damaging agents. Genes Chromosomes Cancer. 2001;30:64–71. [PubMed] [Google Scholar]
- Schmitz J, Churakov G, Zischler H, Brosius J. A novel class of mammalian-specific tailless retropseudogenes. Genome Res. 2004;14:1911–1915. doi: 10.1101/gr.2720104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen SK, Huang CT, Han K, Batzer MA. Endonuclease-independent insertion provides an alternative pathway for L1 retrotransposition in the human genome. Nucleic Acids Res. 2007;35:3741–3751. doi: 10.1093/nar/gkm317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit A, Hubley R, Green P. RepeatMasker Open-3.0 1996 [Google Scholar]
- Srikanta D, Sen SK, Huang CT, Conlin EM, Rhodes RM, Batzer MA. An alternative pathway for Alu retrotransposition suggests a role in DNA double-strand break repair. Genomics. 2009;93:205–212. doi: 10.1016/j.ygeno.2008.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szak ST, Pickeral OK, Makalowski W, Boguski MS, Landsman D, Boeke JD. Molecular archeology of L1 insertions in the human genome. Genome Biol. 2002;3:research0052. doi: 10.1186/gb-2002-3-10-research0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valerie K, Povirk LF. Regulation and mechanisms of mammalian double-strand break repair. Oncogene. 2003;22:5792–5812. doi: 10.1038/sj.onc.1206679. [DOI] [PubMed] [Google Scholar]
- Wallace NA, Belancio VP, Deininger PL. L1 mobile element expression causes multiple types of toxicity. Gene. 2008;419:75–81. doi: 10.1016/j.gene.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zingler N, Willhoeft U, Brose HP, Schoder V, Jahns T, Hanschmann KM, Morrish TA, Lower J, Schumann GG. Analysis of 5′ junctions of human LINE-1 and Alu retrotransposons suggests an alternative model for 5′-end attachment requiring microhomology-mediated end-joining. Genome Res. 2005;15:780–789. doi: 10.1101/gr.3421505. [DOI] [PMC free article] [PubMed] [Google Scholar]