Recently a C>T SNP at position −169 (rs7528684) in the promoter region of the FCRL3 gene was shown to be associated with systemic lupus erythematosus (SLE), rheumatoid arthritis (RA), and other autoimmune disorders in a Japanese population (1, 2). The −169C allele, which was overrepresented in SLE patients confers a more orthodox consensus binding site for the NF-kB transcription factor, increasing promoter activity in vitro and upregulating FCRL3 mRNA transcription. However, the relationship between −169CT alleles and FCRL3 expression on the B cell surface has remained unexplored. Since there is a strong association between the −169CT alleles and FCRL3 mRNA expression, we used a newly developed anti-FCRL3 monoclonal antibody (3D2) (3) to explore the relationship between the −169CT alleles and FCRL3 protein expression on B lymphocytes from disease-free individuals and SLE patients with inactive disease.
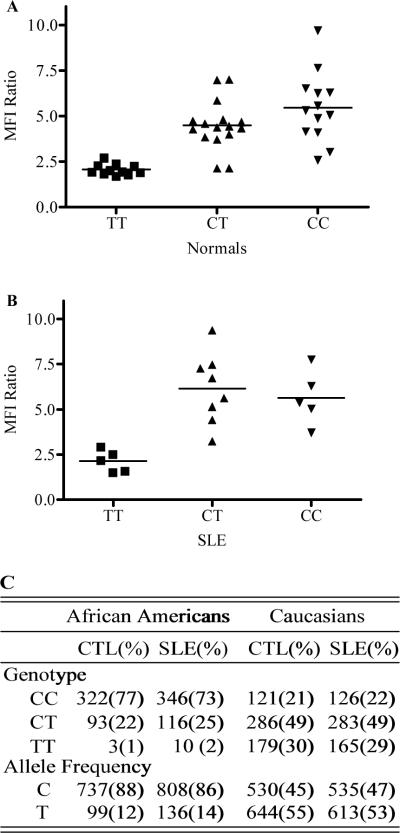
To investigate whether FCRL3 surface expression varies according to the allele status of the −169CT polymorphism, venous blood samples from 40 normal donors were stained using 3D2 and an isotype-matched antibody control for analysis by flow cytometry. Figure 1A demonstrates that CD19+ B cells from CC homozygotes expressed significantly higher levels of FCRL3 than B cells from TT homozygotes (p < 0.0001), with CT heterozygotes expressing intermediate levels (p < 0.0001, TT vs CT). These results are consistent with previous mRNA transcription data showing that CC homozygotes expressed higher levels of FCRL3 mRNA than TT homozygotes (1). Importantly, these data indicate that FCRL3 protein levels may be regulated by NF-kB in an allele-dependent fashion. In addition, our study group included both African American and European American donors. Since expression levels did not segregate by ethnicity, this suggests that FCRL3 expression levels do not vary across these ethnic study groups.
Figure 1.
Association of FCRL3 expression with −169CT alleles. African American and European American SLE patients that met the ACR 1997 revised criteria, and matched healthy controls were recruited by the faculty of the Division of Clinical Immunology and Rheumatology at UAB. A, MFI Ratio is the ratio of the mean fluorescence intensity staining of the anti-FCRL3 mAb (3D2) and a mIgG isotype-matched control. For TT vs CC and TT vs CT p < 0.0001 (Student's t-test). B, for TT vs CC and TT vs CT, p < 0.0015. C, for genotype distribution in controls vs SLE, p = 0.142 (African Americans) and p = 0.755 (European Americans) (2 × 3 Chi Square). For allele frequency distribution, p = 0.122 (African Americans) and p = 0.505 (European Americans) (Fisher's Exact Test).
We also examined FCRL3 surface expression on B cells from 18 SLE patients with quiescent disease. Our results showed no significant variation of expression between SLE patients and healthy volunteers (Figure 1B). As with normal donors, B cells from SLE CC homozygotes expressed higher levels of FCRL3 than TT homozygotes (p < 0.0015). However, there was no significant difference in expression between SLE and normal homozygotes of the same genotype (Figures 1A and 1B).
Given the impact of the FCRL3 −169CT SNP on receptor protein expression in B cells, its location in an SLE-linked genetic locus, and its association with SLE in Japanese, we investigated whether these alleles are associated with SLE in African Americans and European Americans (4–6). 472 African American SLE patients and 418 matched controls, as well as 574 European American SLE patients and 586 matched controls were genotyped by pyrosequencing. Data analysis indicated no significant difference in genotypes or allele frequency distribution between SLE and control donors in either ethnic group (Figure 1C). An investigation of other SNP genotypes in the FCRL3 promoter (rs945635 and rs225828), and in the coding region (rs944627) also failed to show an association with SLE in our cohorts (data not shown). Therefore, these results suggest that the FCRL3 −169CT alleles are not associated with SLE in our African American and European American cohorts. Since the results of individual studies may be explained by a lack of sufficient power to detect an effect, we performed a meta-analysis that included data from the four published studies of the −169CT alleles in SLE (1, 7–9). Using a random effects model our analysis failed to show any significant effect in Caucasians (European Americans and Spanish; N = 2,220; OR = 1.0478, p = 0.45; 95% CI 0.9304–1.180) (8, 10) or Asians (Japanese, Koreans, Chinese; N = 4,267; OR = 1.0616, p = 0.412; 95% CI 0.9204–1.2244) (1, 7, 9). Therefore, given the significant association seen in the Japanese study, our analysis suggests that the genetic effect of the −169CT SNP does not extend to the pan-Asian group examined or other ethnic groups.
Although its ligand(s) remains unknown, it is clear that the −169CT NF-kB site in the FCRL3 promoter significantly alters surface expression levels of FCRL3 in B cells suggesting that, along with its tyrosine-based functional potential, this polymorphism could have an important influence on immune regulation (11). However, in contrast to previous results indicating an association between −169CT alleles and SLE in Japanese, our data show no association between these SNP alleles and SLE in African Americans or European Americans. Similar variation in genetic effects across different ethnic groups have been observed in many studies and likely reflect the involvement of multiple immunologic pathways characteristic of complex genetic traits (for example, PADI4 found only in Japanese RA, and PTPN22 found only in Caucasian RA and SLE) (12–16). Moreover, recent studies of SLE and the −169CT alleles in other ethnic groups failed to show a genetic effect (7–9). While individual studies may have lacked sufficient power to detect an effect, our meta-analysis combining several study groups also failed to show an effect in Asians and Caucasians.
In conclusion, our data indicate that the FCRL3 −169CT SNP induces differential protein expression levels in healthy individuals and SLE patients, but does not appear to be an SLE susceptibility factor in African Americans or European Americans.
Acknowledgments
This work was supported by NIH grants PO1 AR49084 and RO1 AR42476, and the Dana Foundation Program in Human Immunology (RSD).
References
- 1.Kochi Y, Yamada R, Suzuki A, Harley JB, Shirasawa S, Sawada T, et al. A functional variant in FCRL3, encoding Fc receptor-like 3, is associated with rheumatoid arthritis and several autoimmunities. Nat Genet. 2005;37(5):478–85. doi: 10.1038/ng1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davis RS, Dennis G, Jr., Odom MR, Gibson AW, Kimberly RP, Burrows PD, et al. Fc receptor homologs: newest members of a remarkably diverse Fc receptor gene family. Immunol Rev. 2002;190:123–36. doi: 10.1034/j.1600-065x.2002.19009.x. [DOI] [PubMed] [Google Scholar]
- 3.Li FJ, Ding S, Pan J, Shakhmatov MA, Kashentseva E, Wu J, et al. FCRL2 expression predicts IGHV mutation status and clinical progression in chronic lymphocytic leukemia. Blood. 2008;112(1):179–87. doi: 10.1182/blood-2008-01-131359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davis RS, Wang YH, Kubagawa H, Cooper MD. Identification of a family of Fc receptor homologs with preferential B cell expression. Proc Natl Acad Sci U S A. 2001;98(17):9772–7. doi: 10.1073/pnas.171308498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edberg JC, Langefeld CD, Wu J, Moser KL, Kaufman KM, Kelly J, et al. Genetic linkage and association of Fcgamma receptor IIIA (CD16A) on chromosome 1q23 with human systemic lupus erythematosus. Arthritis Rheum. 2002;46(8):2132–40. doi: 10.1002/art.10438. [DOI] [PubMed] [Google Scholar]
- 6.Croker JA, Kimberly RP. Genetics of susceptibility and severity in systemic lupus erythematosus. Curr Opin Rheumatol. 2005;17(5):529–37. doi: 10.1097/01.bor.0000169360.15701.27. [DOI] [PubMed] [Google Scholar]
- 7.Choi CB, Kang CP, Seong SS, Bae SC, Kang C. The −169C/T polymorphism in FCRL3 is not associated with susceptibility to rheumatoid arthritis or systemic lupus erythematosus in a case-control study of Koreans. Arthritis Rheum. 2006;54(12):3838–41. doi: 10.1002/art.22248. [DOI] [PubMed] [Google Scholar]
- 8.Sanchez E, Callejas JL, Sabio JM, de Haro M, Camps M, de Ramon E, et al. Polymorphisms of the FCRL3 gene in a Spanish population of systemic lupus erythematosus patients. Rheumatology (Oxford) 2006;45(8):1044–6. doi: 10.1093/rheumatology/kel160. [DOI] [PubMed] [Google Scholar]
- 9.You Y, Wang Z, Deng G, Hao F. Lack of association between Fc receptor-like 3 gene polymorphisms and systemic lupus erythematosus in Chinese population. J Dermatol Sci. 2008 doi: 10.1016/j.jdermsci.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 10.Fleiss JL. The statistical basis of meta-analysis. Stat Methods Med Res. 1993;2(2):121–45. doi: 10.1177/096228029300200202. [DOI] [PubMed] [Google Scholar]
- 11.Davis RS. Fc receptor-like molecules. Annual Review of Immunology. 2007;25:525–560. doi: 10.1146/annurev.immunol.25.022106.141541. [DOI] [PubMed] [Google Scholar]
- 12.Barton A, Bowes J, Eyre S, Spreckley K, Hinks A, John S, et al. A functional haplotype of the PADI4 gene associated with rheumatoid arthritis in a Japanese population is not associated in a United Kingdom population. Arthritis Rheum. 2004;50(4):1117–21. doi: 10.1002/art.20169. [DOI] [PubMed] [Google Scholar]
- 13.Suzuki A, Yamada R, Chang X, Tokuhiro S, Sawada T, Suzuki M, et al. Functional haplotypes of PADI4, encoding citrullinating enzyme peptidylarginine deiminase 4, are associated with rheumatoid arthritis. Nat Genet. 2003;34(4):395–402. doi: 10.1038/ng1206. [DOI] [PubMed] [Google Scholar]
- 14.Begovich AB, Carlton VE, Honigberg LA, Schrodi SJ, Chokkalingam AP, Alexander HC, et al. A missense single-nucleotide polymorphism in a gene encoding a protein tyrosine phosphatase (PTPN22) is associated with rheumatoid arthritis. Am J Hum Genet. 2004;75(2):330–7. doi: 10.1086/422827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ikari K, Momohara S, Inoue E, Tomatsu T, Hara M, Yamanaka H, et al. Haplotype analysis revealed no association between the PTPN22 gene and RA in a Japanese population. Rheumatology (Oxford) 2006;45(11):1345–8. doi: 10.1093/rheumatology/kel169. [DOI] [PubMed] [Google Scholar]
- 16.Kyogoku C, Langefeld CD, Ortmann WA, Lee A, Selby S, Carlton VE, et al. Genetic association of the R620W polymorphism of protein tyrosine phosphatase PTPN22 with human SLE. Am J Hum Genet. 2004;75(3):504–7. doi: 10.1086/423790. [DOI] [PMC free article] [PubMed] [Google Scholar]