Abstract
Conventional development of multi-gene expression models (GEMs) predicting therapeutic response of cancer patients are based on analysis of patients treated with specific regimens, which limits generalization to different or novel drug combinations. We overcome this limitation by developing GEMs based on in vitro drug sensitivities and microarray analyses of the NCI-60 cancer cell line panel. These GEMs were evaluated in blind fashion as predictors of tumor response and/or patient survival in seven independent cohorts of patients with breast (N=275), bladder (N=59), and ovarian (N=143) cancer treated with multi-agent chemotherapy, of which 233 patients were from prospectively-enrolled clinical trials. In all studies, GEMs effectively stratified tumor response and patient survival independent of established clinical and pathologic tumor variables. In bladder cancer patients treated with neoadjuvant MVAC (Methotrexate, Vinblastine, Doxorubicin, Cisplatin), the 3-year overall survival for those with favorable GEM scores was 81% vs. 33% for those with less favorable scores (p=0.002). GEMs for breast cancer patients treated with FAC (Fluorouracil, Doxorubicin, Cyclophosphamide) and ovarian cancer patients treated with platinum-containing regimens also stratified patient survival (5-year overall survival 100% vs. 74% (p=0.05) and 3-year overall survival 68% vs. 43% (p=0.008), respectively. Importantly, clinical prediction using our in vitro GEM was superior to that of conventionally-derived GEMs. We demonstrate a facile yet effective approach to GEM derivation that identifies patients most likely to benefit from selected multi-agent therapy. Use of such in vitro-based GEMs may provide a robust and generalizable approach to personalized cancer therapy.
Keywords: Combination Chemotherapy, Co-expression Extrapolation, Gene Expression-based Prediction Models
INTRODUCTION
Most cancer patients with advanced tumors undergo combination pharmacotherapy. A challenge in treating these patients is predicting which individuals will respond to a specific therapy. Recent studies have demonstrated the potential that tumor response to chemotherapeutics can be predicted using multivariate Gene Expression-based Models (GEMs).(1, 2) Such GEMs are developed using a training microarray set from tumors of human patients with known clinical responses to the drug(s), allowing selection of optimal marker genes and respective expression levels to construct such models.(3, 4) These are then evaluated in separate patient cohorts to determine their predictive ability. While validation is straightforward, formulation of such GEMs is lengthy, expensive, and requires human tumor tissue from patients treated with the specific drug regimen for which the GEM prediction is desired. In addition, this approach does not permit a priori prediction of responses to single or combination therapy that has never been clinically used, limiting the pairing of novel agents to their optimal disease types, thus hindering identification of novel applications of established agents.
To overcome these limitations, two recent studies derived GEMs based on biomarkers of in vitro drug sensitivity of cancer cell lines in the NCI-60 panel (5, 6) and demonstrated that such GEMs can predict the clinical outcome in patients who received systemic therapy. While promising, these reports did not provide validation of unaltered GEMs on multiple independent patient data sets. If such GEMs can maintain a robust prediction in this setting, this approach has the potential to revolutionize personalized cancer therapy and drug development since forecasting the clinical effectiveness of a compound could be undertaken very early in the drug development lifecycle.(7)
Here, we develop and apply GEMs derived from in vitro data and demonstrate the ability of these to predict clinical responses to combination pharmacotherapy regimens in 477 patients with three common cancer types reported in retrospective studies or prospective clinical trials. Importantly, the patient cohorts examined came from ethnically and geographically distinct regions.
MATERIALS AND METHODS
Patient Data Sets for Gene Expression Model (GEM) Development and Evaluation
In vitro drug activity and microarray data of the NCI-60 cancer cell panel were previously described elsewhere.(6) Microarray gene expression data without associated treatment or clinical outcome information from three human tumor sample cohorts were used for GEM development including bladder cancer (N=89) (8), ovarian cancer (N=99) (9), and breast cancer (N=251).(10) These comprise the Training set (Figure 1). For GEM evaluation, through our own efforts and collaborations, we acquired and used seven independent Test sets in which data from microarray gene expression could be linked to clinical outcome. Test set studies included two bladder cancer (N=59), two ovarian cancer (N=143), and three breast cancer (N=275) sets of patients treated with multi-agent pharmacotherapy (Table 1). Of these, 233 patients were prospectively enrolled in clinical trials (BR-TX and BR-Ger). Training and test microarray data sets are publicly available or were obtained directly from the original authors. When survival or tumor response data (complete (CR), partial (PR), or down-grade/stage) were not publicly available, this information was obtained from the original investigators. Note that these test set studies included geographically and ethnically diverse populations with various tumor stages and tumor sampling methodologies and did not share any patients or clinical locations with the sets used for GEM development (Table 1 & Figure 1).
Figure 1.
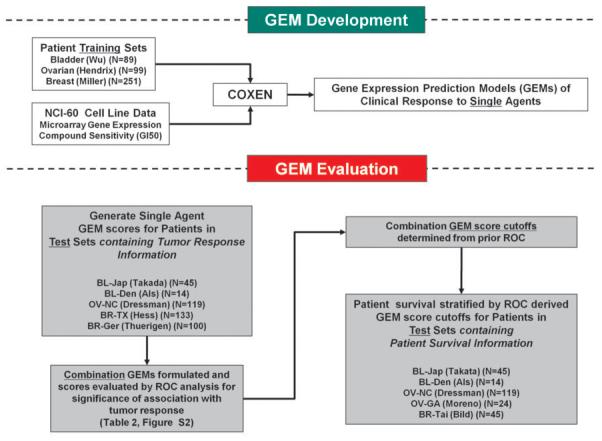
Schematic Overview of GEM development and evaluation.
Table 1.
Cell line and Patient Tumor Datasets for Gene Expression Models (GEMs) derivation and evaluation.
| Cancer Type |
Patient Population |
Study Name |
Microarray Type |
No. Patients |
Chemo Regimen |
Treatment Course |
Tumor Response* |
Survival Data** |
Reference |
|---|---|---|---|---|---|---|---|---|---|
|
Training Sets | |||||||||
| Mixed | (Cell lines) | NCI-60 | HG-U133A | 60 | None | n/a | n/a | n/a | Lee et al (6) |
| Bladder | New York | BL-89 | HG-U133A | 89* | None | n/a | n/a | n/a | Wu et al (8) |
| Ovarian | Michigan | OV-99 | HG-U133A | 99 | None | n/a | n/a | n/a | Hendrix et al (9) |
| Breast | Sweden | BR-251 | HG-U133A | 251 | None | n/a | n/a | n/a | Miller et al (10) |
|
Test Sets | |||||||||
| Bladder | Japan | BL-Jap | cDNA | 45 | MVAC | Neoadjuvant | Down- stage (R 23, N 22) |
Overall Survival |
Takata et al (15) |
| Denmark | BL-Den | HG-U133A | 14 | MVAC | No Surgery | WHO criteria (R 3, N 11) |
Overall Survival |
Als et al (16) |
|
| Ovarian | North Carolina, Florida |
OV-NC | HG-U133A | 119 | Platinum | Adjuvant | Pathologic Response (R 85, N 34) |
Overall Survival |
Dressman et al (3) |
| Georgia | OV-GA | HG-U95A | 24 | CT | Neoadjuvant | No | Overall Survival (S12, D 12) |
Moreno et al (19) |
|
| Breast | Texas / Peru |
BR-TX^ | HG-U133A | 133 | TFAC | Neoadjuvant | Pathologic Response (R 34, N 99) |
No | Hess et al (20) |
| North Carolina, Taiwan |
BR-Tai | HG-U95A | 45 | FAC | Adjuvant | No | Overall Survival (S 38, D 7) |
Bild et al (21) |
|
| Germany | BR-Ger^ | DKFZ/Opern Human Oligo |
100 | GED | Neoadjuvant | Pathologic Response (R 24, N 76) |
No | Thuerigen et al (22) |
|
MVAC (Methotrexate, Vinblastine, Adriamycin, Cisplatin), CT (Carboplatin, Taxol), Platinum (Carboplatin or Cisplatin and additional unspecified chemotherapeutic compounds), TFAC (Taxol, 5-FU, Adriamycin, Cyclophosphamide), FAC (same as TFAC without Taxol), GED (Gemcitabine, Epirubicin, Docetaxel).
Defined as reported by the original authors (R: responder, N: non-responder).
Used absolute survival as a surrogate of clinical response if no clinical response data available (S: survivor, D: deceased).
Prospective Clinical Trial.
Statistical Analysis
GEM Development
The COXEN algorithm (6), originally demonstrated to predict single-agent chemosensitivity for a cell line panel and small breast cancer sets, was expanded and refined to derive GEMs and subsequent GEM-predicted chemosensitivity probabilities (named “GEM Scores”) in patients with bladder, ovarian, and breast cancer (Figure 1; Supplementary Methods). Briefly, each microarray data set was first standardized within each gene for consistent GEM development and evaluation. Chemosensitivity biomarkers for each compound were then identified in the NCI-60 microarray dataset by comparing each gene’s differential expression between the sensitive and resistant cells for each compound. These biomarkers were next triaged based on the COXEN coefficient which represents the degree of concordance of expression between the NCI-60 set and one of three cancer patient microarray data sets used for development---BR-251, BL-89, or OV-99.(6) This resulted in a panel of 15~95 gene probe sets per compound that were used for the GEM training of statistical multivariate classification modeling between the NCI-60 sensitive and resistant cell lines for that compound. Individual compound GEMs were then combined to generate the prediction model for relevant combination pharmacotherapy, assuming the modes of action of individual compounds in combination chemotherapy were independent. Importantly, this model development and training did not use any clinical information or microarray data from the test sets used for GEM evaluation, thus maintaining strict independence between training and test data sets.(11)
GEM Evaluation
We compared the GEM scores between clinical responders and non-responders as reported in five studies for which tumor response data after pharmacotherapy was available (Figure 1). The significance of this score difference was evaluated using the two-sample t-test between the responder and non-responder patient groups. In the two studies for which tumor response data were not available, OV-GA and BR-Tai, we compared GEM scores between actual surviving and deceased patients---the only available information of patient outcomes in these two studies, using the absolute call of patients’ survival as a surrogate metric for tumor response. Patient survival would have depended not only on chemotherapeutic response but also on many other clinical factors but we simply tested whether the survivor group was more enriched with predicted responders to chemotherapy. Once a GEM was derived for a particular agent or combination of agents in a cancer type, it was used without any alteration in all subsequent evaluations. Where information on patient survival time was available, we performed Kaplan-Meier (KM) analyses on five cohorts stratified by the GEM scores from the analyses of tumor response. In order to classify predicted responders from predicted non-responders a priori, a Receiver Operating Characteristic (ROC) curve was plotted (Figure S1). The optimal cutoff on the ROC curve was determined by maximizing the so-called Youden index (=sensitivity+specificity−1), at which sensitivity, specificity, positive predictive value, (PPV), and negative predictive value (NPV) were evaluated for each combination GEM in stratifying patients’ clinical response.(12)
Relationship between GEM and Clinical Parameters
We examined whether our multi-gene expression-based GEM scores are correlated with conventional clinical parameter information such tumor stage, grade, well-known marker types e.g., ER, PR, Her2 in breast cancer . We first calculated simple univariate (rank-based Spearman or binary) correlations between the GEM scores and each of clinical parameters. Next, in order to see the relationship between GEM scores and combined clinical information, we measured the correlation between the GEM scores and a published clinical variable-based breast cancer nomogram that predicts pCR after preoperative chemotherapy based on patient age, tumor size, ER-status, tumor grade and the type and number of chemotherapy treatments.(13)
RESULTS
GEM stratification of tumor response and absolute patient survival
Bladder cancer
Using prospectively-predicted and outcome-blinded GEM scores the single-drug GEMs of adriamycin and cisplatin provided scores of responder patients which were significantly higher than those of non-responders (adriamycin GEM score with two-sample t-test p-value=0.049 and 0.061 and cisplatin GEM score with p-value=0.007 and 0.012) both in the neoadjuvant (BL-Jap) and advanced tumor (BL-Den) studies (Table 2). The GEM score for methotrexate and vinblastine was significantly different only in the BL-Jap study (p-value<0.001 and =0.002). The single drug GEMs of methotrexate, vinblastine, adriamycin, and cisplatin were then combined to formulate the combination GEM for MVAC response. Using this combination GEM, a significant difference in scores was observed between responders and non-responders in both studies (p-value=0.002 and 0.033). This combination GEM provided sensitivity 83%, specificity 64%, PPV 71%, and NPV 78% at the cutoff value maximizing the Youden index; the overall significance of ROC curve against random classification was also extremely significant (Wilcoxen test p-value<0.001, Supplementary Figure S1). Note again that each of these GEMs was simultaneously applied and evaluated unaltered on the Japan (BL-Jap) and Europe (BL-Den) sets. We further examined whether the GEM scores were correlated or overlapped with the information from conventional clinical parameters of patients. Using a univariate analysis of rank-based correlation on the BL-Jap cohort, we found that the GEM for MVAC response was independent of available clinical parameters such as tumor stage, grade, age, or gender (Spearman correlation −0.14 ~ 0.07). We also found the GEM score to be the only significant predictor of MVAC response on this cohort by a multivariate analysis that was performed together with the GEM score and the above clinical and tumor pathological variables (logistic regression GEM p-value=0.03; Table 3).(14) The BL-Den study was not large enough for a multivariate analysis.
Table 2.
Comparison of GEM prediction probabilities of chemosensitivity to actual tumor response and patient survival.
| Cancer Type | Study Name | Agent or Combination | GEM Score probability Responders (mean+/− 95% CI) |
GEM Score probability Non-Responders (mean+/− 95% CI) |
p-value† |
|---|---|---|---|---|---|
| Responder vs. | Non-Responder | ||||
| Bladder | BL-Jap | Methotrexate | 0.625 +/− 0.071 | 0.371 +/− 0.071 | <0.001 |
| Vinblastine | 0.594 +/− 0.071 | 0.358 +/− 0.079 | 0.002 | ||
| Adriamycin | 0.552 +/− 0.078 | 0.402 +/− 0.090 | 0.049 | ||
| Cisplatin | 0.570 +/− 0.076 | 0.364 +/− 0.080 | 0.007 | ||
| MVAC* | 0.944 +/− 0.022 | 0.818 +/− 0.054 | 0.002 | ||
| BL-Den | Methotrexate | 0.534 +/− 0.122 | 0.506 +/− 0.168 | 0.436 | |
| Vinblastine | 0.602 +/− 0.192 | 0.490 +/− 0.166 | 0.331 | ||
| Adriamycin | 0.756 +/− 0.121 | 0.441 +/− 0.154 | 0.061 | ||
| Cisplatin | 0.783 +/− 0.075 | 0.442 +/− 0.148 | 0.012 | ||
| MVAC* | 0.996 +/− 0.002 | 0.871 +/− 0.104 | 0.033* | ||
| Ovarian | OV-NC^ | Carboplatin | 0.715 +/− 0.074 | 0.563 +/− 0.141 | 0.034 |
| Taxol | 0.429 +/− 0.087 | 0.223 +/− 0.114 | 0.003 | ||
| CT** | 0.826 +/− 0.061 | 0.638 +/− 0.132 | 0.007 | ||
| Breast | BR-TX | Taxol | 0.568 +/− 0.130 | 0.345 +/− 0.072 | 0.002 |
| 5-FU | 0.549 +/− 0.119 | 0.472 +/− 0.077 | 0.146 | ||
| Adriamycin | 0.326 +/− 0.135 | 0.164 +/− 0.061 | 0.019 | ||
| Cyclophosphamide | 0.340 +/− 0.138 | 0.218 +/− 0.071 | 0.064 | ||
| FAC^ | 0.721 +/− 0.116 | 0.576 +/− 0.072 | 0.021 | ||
| BR-Ger | Gemcitabine | 0.219 +/− 0.046 | 0.154 +/− 0.033 | 0.014 | |
| Epirubicin | 0.430 +/− 0.113 | 0.324 +/− 0.057 | 0.055 | ||
| Docetaxel | 0.804 +/− 0.076 | 0.726 +/− 0.042 | 0.044 | ||
| GED^^ | 0.919 +/− 0.034 | 0.859 +/− 0.023 | 0.003 | ||
| Survivor vs. | Deceased | ||||
| Ovarian | OV-GA | Carboplatin | 0.498 +/− 0.253 | 0.202 +/− 0.212 | 0.047 |
| Taxol | 0.397 +/− 0.227 | 0.114 +/− 0.136 | 0.025 | ||
| CT** | 0.721 +/− 0.213 | 0.310 +/− 0.224 | 0.008 | ||
| Breast | BR-Tai | 5-FU | 0.514 +/− 0.112 | 0.301 +/− 0.155 | 0.024 |
| Adriamycin | 0.211 +/− 0.107 | 0.036 +/− 0.042 | 0.002 | ||
| Cyclophosphamide | 0.263 +/− 0.118 | 0.114 +/− 0.124 | 0.052 | ||
| FAC^ | 0.611 +/− 0.120 | 0.392 +/− 0.177 | 0.033 | ||
Combination GEM for BL-Jap and BL-Den was made with the single-drug GEMs of Methotrexate, Vinblastin, Adriamycin and Cisplatin.
Combination GEM for OV-NC and OV-GA was made with the single-drug GEMs of Carboplatin and Taxol.
Combination GEM for BR-TX and BR-Tai was made with the single-drug GEMs for 5-FU, Adriamycin, and Cyclophosphamide.
Combination GEM for BR-Ger was made with the single-drug GEMs of Gemcitabine, Epirubicin, Docetaxel.
P-value calculated by two- sample t-test.
Table 3.
Multivariate analysis on GEM and clinical variables for stratifying tumor response.
| Variable | Estimate | Std. Error | z value | Pr(>|z|) |
|---|---|---|---|---|
|
Bladder (BL-Jap) | ||||
| GEM (MVAC) | 2.872 | 1.326 | 2.165 | 0.030 |
| Gender | 0.118 | 0.755 | 0.156 | 0.875 |
| Age | −0.024 | 0.051 | −0.482 | 0.630 |
| Stage | −0.242 | 0.817 | −0.297 | 0.767 |
| Grade | 0.091 | 0.558 | 0.164 | 0.869 |
|
Ovarian (OV-NC) | ||||
| GEM (CT) | −1.425 | 0.626 | −2.276 | 0.022 |
| Stage | 0.421 | 0.579 | 0.728 | 0.466 |
| Grade | −0.009 | 0.378 | −0.025 | 0.980 |
|
Breast (BR-TX) | ||||
| GEM (TFAC) | 1.517 | 0.710 | 2.137 | 0.032 |
| Age | −0.051 | 0.026 | −1.935 | 0.053 |
| Race | −0.306 | 0.187 | −1.640 | 0.101 |
| Grade | 0.320 | 0.657 | 0.487 | 0.626 |
| ER | −2.205 | 0.634 | −3.481 | <0.001 |
| Her2 | 0.656 | 0.548 | 1.197 | 0.231 |
| PR | −0.682 | 0.615 | −1.110 | 0.267 |
Logistic regression analysis was performed using both GEM and other clinical and pathological parameters for each of BL-Jap, OV-NC, and BR-TX. P-values by the significance of each parameter estimate compared to its error from each regression model.
Ovarian cancer
In ovarian cancer, the unaltered carboplatin GEM also showed consistently significant results for patient response to pharmacotherapy in the OV-NC study (p-value=0.034) and for absolute patient survival in the OV-GA study (p-value=0.047) (Table 2). In the OV-GA study, patients received carboplatin and Taxol (paclitaxel) and a large proportion of patients in the OV-NC study were also reported to receive Taxol. We thus tested the ability of GEM prediction for this drug to stratify the outcome in the two ovarian studies. The scores of the Taxol GEM, in fact, correlated well with the clinical response in the OV-NC study (p-value=0.003) and with absolute patient survival in the OV-GA (p-value=0.025) study. When the carboplatin and Taxol (CT) combination GEM was applied, the differences in GEM scores between responders and non-responders in OV-NC was highly significant (p-value=0.007). Moreover, the same combination GEM significantly segregated scores between patients who survived and deceased patients in the OV-GA study (p-value=0.008). This combination GEM provided sensitivity 77%, specificity 56%, PPV 71%, and NPV 78% for the OV-NC cohort at the Youden cutoff value; the overall ROC curve was also extremely significant (p-value<0.001, Supplementary Figure S1). Together, results in ovarian cancer also showed that the identical GEMs of individual agents and the combination were successful in stratifying not only the measured tumor response in the OV-NC study but also absolute patient survival (survivor vs. deceased) in the OV-GA study. In the OV-NC study, for which other clinical parameters were available, the CT combination GEM score was found to be independent of other clinical and pathological variables such as tumor grade and stage (Spearman correlation −0.15, 0.04) and the only significant predictor of tumor response to pharmacotherapy (logistic regression GEM p-value=0.022; Table 3).
Breast cancer
The response to pharmacotherapy of patients in the neoadjuvant breast cancer study BR-TX and survival of patients entered on the adjuvant BR-Tai breast cancer study also correlated to our GEM scores (Table 2). For example, the identical single-drug GEMs for adriamycin and cyclophosphamide were consistently different in tumor response to pharmacotherapy in the BR-TX study (two-sample t-test p-value=0.019 and 0.064) and in absolute patient survival in the BR-Tai study (p-value=0.002 and 0.052). The GEM of 5-FU was significantly different only in BR-Tai (p-value=0.024). Nevertheless, the combination prediction model based on the three single-drug GEMs of 5-FU, adriamycin, and cyclophosphamide - compounds used in both studies - was significantly associated with patient chemotherapeutic responses in BR-TX (p-value=0.021) and survival status in BR-Tai (p-value=0.033). This combination GEM provided sensitivity 71%, specificity 53%, PPV 32%, and NPV 85% at the Youden cutoff value; the overall ROC curve was highly significant (p-value=0.007, Figure S1). For the neoadjuvant BR-Ger breast cancer study, all the single-drug GEMs of gemcitabine, epirubicin, and docetaxel (p-value=0.014, 0.055, and 0.044) and the combination prediction model based on the three single-drug GEMs were significantly different between responders and non-responders to pharmacotherapy (p-value=0.003) (Table 2). In the BR-TX study, the only breast cancer trial both with patients’ chemotherapeutic response and other clinical variable information, the GEM for TFAC response was independent from other clinical and pathological parameters (Spearman correlation −0.07 ~ 0.13). We also compared our GEM scores with a previously-published nomogram prediction of chemotherapeutic response based on the clinical characteristics of the patient and tumor.(13) We found that the Taxol GEM scores are weakly correlated with the nomogram (Pearson correlation = 0.24) and the FAC GEM scores are not correlated (Pearson correlation = 0.07). In multivariate prediction modeling, both ER status and GEM score were found to be significant predictors of tumor response in this cohort, implying potentially improved predictability by combining GEM and clinical parameter information (logistic regression p-value <0.001, =0.032; Table 3).
Utility of GEM scores in stratifying actuarial patient survival
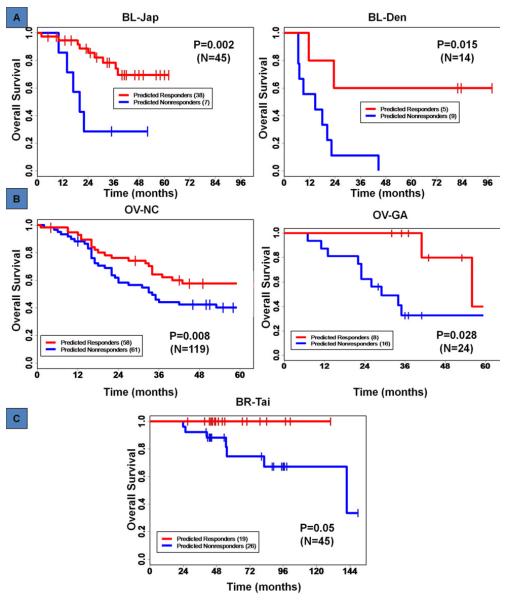
Unaltered GEMs evaluated above were then examined for their ability to stratify actuarial patient survival. Using the selected score cutoffs for the combination GEMs, patient survival time was stratified in the Kaplan-Meier analysis for all five studies with patient survival time information across the three tumor types. With the stratification by the combination GEM of MVAC in bladder cancer, the 3-year overall survival rates were 81% for predicted responders compared to 33% for predicted non-responders on the BL-Jap study (log-rank test p-value=0.002), and 61% vs. 16% on the BL-Den study (p-value=0.015), (Figure 2A). In ovarian cancer, the combination GEM of carboplatin and Taxol also effectively stratified patient survival time in both OV-NC (log-rank test p-value=0.008) and OV-GA (p-value=0.028) studies (Figure 2B). In this analysis, patient stratification by the GEM scores provided a difference in 3-year overall survival of 68% vs. 43% in OV-NC and >95% vs. 23% in OV-GA. In breast cancer, based on the combination GEM of FAC, we performed the Kaplan-Meier analysis on the BR-Tai study, the only breast cancer set with patient survival time information. This GEM stratification was also significant (p-value=0.05) with the 5-year overall survival rates ~100% vs. 74% between the predicted responders and predicted non-responders (Figure 2C).
Figure 2. GEM prediction of overall patient survival.
GEM score stratified Kaplan-Meier analysis. (A) for the BL-Jap and BL-Den studies, (B) for the OV-NC and OV-GA studies, and (C) for the BR-Tai study. P-values by log-rank test.
Predictive performance of in vitro COXEN based compared to classical GEMs
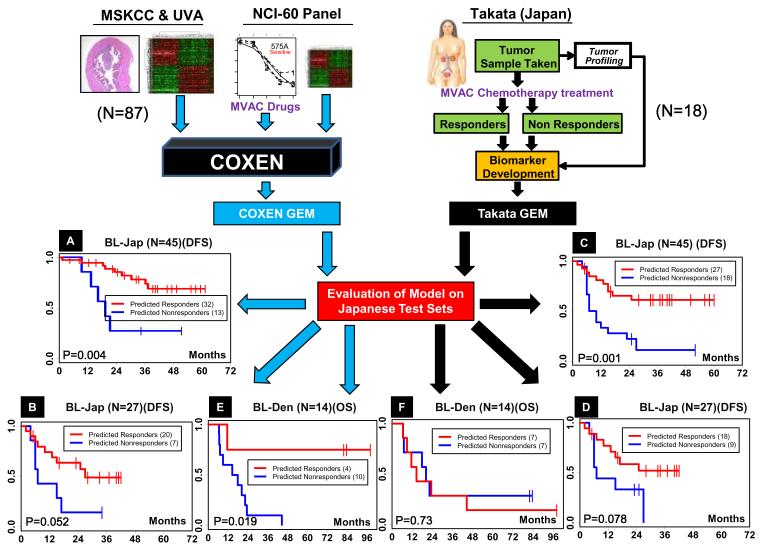
An important advantage of COXEN-based chemosensitivity prediction is efficient development of GEMs for any single drug or combination with observed effects on cell lines in vitro. However, an important question is whether this technique sacrifices predictive performance compared to classical patient sample-trained GEMs that may reflect in vivo activities and/or synergistic effects of certain compounds. We thus compared whether COXEN-derived GEMs were equally effective as previous GEMs developed using a training microarray set from human patients with known clinical responses to the drug(s). For example, in the BL-Jap study, the original authors reported a 14-gene signature GEM, which we call the Takata-GEM, predictive of patients’ response to MVAC combination chemotherapy.(15) This Takata-GEM was developed from the initial set of 18 patients and evaluated on an additional set of 27 patients (total 45) from the same center. We compared the performance of the COXEN-based MVAC combination GEM, COXEN-GEM, to Takata-GEM for patients’ disease-free survival time stratification. Both GEMs were first evaluated both with the test subset of BL-Jap (N=27) and the whole BL-Jap cohort (N=45). Finally, both GEMs were evaluated for their predictive ability on a third independent set, BL-Den.(16) COXEN-GEM was found to be an effective predictor of patient outcome in the whole BL-Jap set (Figure 3A upper panel) as well as its test subset (Figure 3A lower panel). Takata-GEM was certainly able to stratify patients’ outcomes well when applied to the whole BL-Jap cohort (Figure 3B upper panel). However, when this model was used to predict the test subset independent from its GEM training, its statistical significance became considerably weaker (p-value=0.078; Figure 3B lower panel). When applied to the independent BL-Den set, the COXEN-GEM retained its prediction performance (Figure 3C) while the Takata-GEM was unable to stratify the patient outcome in this independent cohort (Figure 3D).
Figure 3. Comparison of predictive performance of COXEN-based GEMs to conventional patient data-derived GEMs.
Patient disease-free (DFS) or overall (OS) survival prediction was performed either by the COXEN-derived GEM stratification of MVAC (COXEN-GEM) used in Figure 2A or that using conventional human patient-based approach (Takata-GEM).(4) Kaplan-Meier survival analysis by (A) COXEN-GEM on the whole BL-Jap set (N=45), (B) COXEN-GEM on the test subset of BL-Jap (N=27), (C) Takata-GEM on the whole BL-Jap set (N=45), (D) Takata-GEM on the test subset of BL-Jap (N=27), (E) COXEN-GEM on the BL-Den set (N=14), and (F) Takata-GEM on the BL-Den set (N=14). P-values by log-rank test.
DISCUSSION
The conceptual framework and the high potential of in vitro drug activity-based GEMs for predicting patient response to chemotherapy has been laid out in previous studies.(6) In this study we demonstrated two major breakthroughs in GEM-based prediction for patients’ chemotherapeutic responses: 1) concordant prediction performance of unaltered in vitro-based GEMs on geographically and ethnically diverse cancer patient cohorts in three different cancer types, and 2) simultaneous use of multiple, parallel historical patient data sets for efficient GEM assessment. Together, we believe these provided a highly encouraging possibility in applying such in vitro-based GEMs to guide patients with more effective chemotherapies.
Some recent GEMs initially reported with their superior prediction performance failed to perform well by an independent group’s validation.(11) This may be due to the so-called selection bias when such multi-gene predictors were trained both with modeling and applying microarray datasets of numerous candidate genes. Our current study has avoided this pitfall by independently testing multiple geographically- and ethnically-diverse patient sets and by consistently predicting both clinical tumor response and patient survival outcome of 477 patients across three different cancer types. Therefore, we believe the prediction performance of the in vitro-based GEMs here will be highly likely realized in clinical practice even though the statistical power of some of these GEMs may appear to be a bit lower than those reported in other recent studies applied only to a single patient set at a time.(17)
We found that the GEM score was independent of conventional clinical parameters and was always a significant predictor of tumor response to pharmacotherapy even when all other clinical variables were considered together, suggesting that the GEMs are not simply surrogates of standard tumor characteristic variables. Also, GEMs generated by various different training sets that contained distinct stages of bladder cancer successfully stratified patient survival with only a slight decrease in statistical significance whereas the accuracy to distinguish the ability to responders from non-responders was maintained (Supplementary Figure S2). While the assumption of independence of relevant agents was necessary due to our GEM derivation from the single-agent drug activity data of NCI-60, our GEM scores correlated well with clinical responses and survival of diverse patient cohorts of the three cancer types treated with multi-agent pharmacotherapy in this study. This demonstrated that even though our combination GEMs could not capture synergistic drug effects, the main efficacy of combination regimens appeared to be reflected by simple additive effects of single drug GEMs. It is conceivable that future in vitro work on doublet or higher-order drug combinations may help in modifying the algorithm to effectively incorporate drug synergies.(18)
We, however, note that each GEM’s reported sensitivity, specificity, NPV, PPV values, or Kaplan-Meier curves here (derived from its ROC curve) showed its optimal performance on the applying particular data set. This does not yet represent validation of each drug GEM’s pre-set threshold to call a patient case + (responder) or − (non-responder), but rather shows proof-of-a-concept, illustrating that such GEM scores are informative in stratifying patients’ responses to the agent. In order to derive an exact cutoff criterion on a specific GEM assay for clinical use, a standard diagnosis assay platform and procedure should be developed for routine clinical practice, from which a fixed cutoff value can be defined for a target patient population, which, we believe, is quite feasible in the near future.
Generating GEMs using in vitro-based approaches does also have some theoretical and practical limitations. For example, it cannot be used to develop GEMs for agents which do not have any effect on cell lines in vitro. While this requirement is strictly embedded in the design of the approach, the composition of the cell panel may be tailored to the expected cellular and molecular target of the tested agent. For example, while using the NCI-60 panel may not result in an effective generation of a drug response profile for an anti-angiogenic agent that targets endothelial cells, carrying out such an experiment on an endothelial cell panel may provide the necessary data which can be used for GEM development. Similarly, for agents that target the immune system, panels composed of the appropriate cells may permit GEM development. As we have shown above, the critical requirement for the cell panel is to provide effective dose response information for the agent in question rather than be required to be composed of the same histological tumor types as the human tumors whose response to therapy is assessed. Hence, using an endothelial cell panel for an anti-angiogenic agent and generating GEMs for the use of such an agent in bladder cancer, for example, seems to be justified.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by National Institutes of Health grants T32DK069264 to PDW, R01CA075115 to DT, and R01HL081690 to JKL. We thank Drs. Yusuke Nakamura and Toyomasa Katagiri at the Tokyo University, Dr. Torben F. Ørntoft at the Aarhus University Hospital, Denmark, and Dr. John McDonald at the Ovarian Cancer Institute of the Georgia Tech University who provided us with unpublished patient outcome information. We also thank Drs. Christopher Moskaluk, Christopher Thomas, and Alton Sartor for their insightful comments and suggestions.
Key to abbreviations
- GEM
gene expression model
- NCI-60
National Cancer Institute 60 cancer cell line panel
- BL-Jap
Japanese bladder cancer patient cohort treated by MVAC combination chemotherapy
- BL-Den
Denmark bladder cancer patient cohort treated by MVAC combination chemotherapy
- OV-NC
North Carolina/Florida ovarian cancer patient cohort treated by platinum-based chemotherapy
- OV-GA
Georgia ovarian cancer patient cohort treated by carboplatin and Taxol combination chemotherapy
- BR-TX
Texas/Peru breast cancer patient cohort treated by TFAC combination chemotherapy
- BR-Tai
North Carolina/Taiwan breast cancer patient cohort treated by FAC combination chemotherapy
- BR-Ger
German breast cancer patient cohort treated by GED combination chemotherapy
- GI50
growth inhibition 50
Footnotes
Competing interests’ statement: None
REFERENCES
- 1.Chang JC, Wooten EC, Tsimelzon A, Hilsenbeck SG, Gutierrez MC, Elledge R, et al. Gene expression profiling for the prediction of therapeutic response to docetaxel in patients with breast cancer. Lancet. 2003;362:362–9. doi: 10.1016/S0140-6736(03)14023-8. [DOI] [PubMed] [Google Scholar]
- 2.Ma XJ, Wang Z, Ryan PD, Isakoff SJ, Barmettler A, Fuller A, et al. A two-gene expression ratio predicts clinical outcome in breast cancer patients treated with tamoxifen. Cancer Cell. 2004;5:607–16. doi: 10.1016/j.ccr.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 3.Dressman HK, Berchuck A, Chan G, Zhai J, Bild A, Sayer R, et al. An integrated genomic-based approach to individualized treatment of patients with advanced-stage ovarian cancer. J Clin Oncol. 2007;25:517–25. doi: 10.1200/JCO.2006.06.3743. [DOI] [PubMed] [Google Scholar]
- 4.Takata R, Katagiri T, Kanehira M, Shuin T, Miki T, Namiki M, et al. Validation study of the prediction system for clinical response of M-VAC neoadjuvant chemotherapy. Cancer Sci. 2007;98:113–7. doi: 10.1111/j.1349-7006.2006.00366.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Potti A, Dressman HK, Bild A, Riedel RF, Chan G, Sayer R, et al. Genomic signatures to guide the use of chemotherapeutics. Nat Med. 2006;12:1294–300. doi: 10.1038/nm1491. [DOI] [PubMed] [Google Scholar]
- 6.Lee JK, Havaleshko DM, Cho H, Weinstein JN, Kaldjian EP, Karpovich J, et al. A strategy for predicting the chemosensitivity of human cancers and its application to drug discovery. Proc Natl Acad Sci U S A. 2007;104:13086–91. doi: 10.1073/pnas.0610292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crunkhorn S. Forecasting drug responses. Nature Reviews Drug Discovery. 2007;6 doi:10.1038. [Google Scholar]
- 8.Wu Z, Siadaty MS, Riddick G, Frierson HF, Jr., Lee JK, Golden W, et al. A novel method for gene expression mapping of metastatic competence in human bladder cancer. Neoplasia. 2006;8:181–9. doi: 10.1593/neo.05727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hendrix ND, Wu R, Kuick R, Schwartz DR, Fearon ER, Cho KR. Fibroblast growth factor 9 has oncogenic activity and is a downstream target of Wnt signaling in ovarian endometrioid adenocarcinomas. Cancer Res. 2006;66:1354–62. doi: 10.1158/0008-5472.CAN-05-3694. [DOI] [PubMed] [Google Scholar]
- 10.Miller LD, Smeds J, George J, Vega VB, Vergara L, Ploner A, et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci U S A. 2005;102:13550–5. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coombes KR, Wang J, Baggerly KA. Microarrays: retracing steps. Nat Med. 2007;13:1276–7. doi: 10.1038/nm1107-1276b. author reply 7-8. [DOI] [PubMed] [Google Scholar]
- 12.Youden W. Index for rating diagnostic tests. Cancer. 1950;3:32–5. doi: 10.1002/1097-0142(1950)3:1<32::aid-cncr2820030106>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 13.Rouzier R, Pusztai L, Delaloge S, Gonzalez-Angulo AM, Andre F, Hess KR, et al. Nomograms to predict pathologic complete response and metastasis-free survival after preoperative chemotherapy for breast cancer. J Clin Oncol. 2005;23:8331–9. doi: 10.1200/JCO.2005.01.2898. [DOI] [PubMed] [Google Scholar]
- 14.Muller KE, Stewart PW. Linear Model Theory: Univariate, Multivariate, and Mixed Models. Wiley; New York: 2006. [Google Scholar]
- 15.Takata R, Katagiri T, Kanehira M, Tsunoda T, Shuin T, Miki T, et al. Predicting response to methotrexate, vinblastine, doxorubicin, and cisplatin neoadjuvant chemotherapy for bladder cancers through genome-wide gene expression profiling. Clin Cancer Res. 2005;11:2625–36. doi: 10.1158/1078-0432.CCR-04-1988. [DOI] [PubMed] [Google Scholar]
- 16.Als AB, Dyrskjot L, von der Maase H, Koed K, Mansilla F, Toldbod HE, et al. Emmprin and survivin predict response and survival following cisplatin-containing chemotherapy in patients with advanced bladder cancer. Clin Cancer Res. 2007;13:4407–14. doi: 10.1158/1078-0432.CCR-07-0109. [DOI] [PubMed] [Google Scholar]
- 17.Bonnefoi H, Potti A, Delorenzi M, Mauriac L, Campone M, Tubiana-Hulin M, et al. Validation of gene signatures that predict the response of breast cancer to neoadjuvant chemotherapy: a substudy of the EORTC 10994/BIG 00-01 clinical trial. Lancet Oncol. 2007;8:1071–8. doi: 10.1016/S1470-2045(07)70345-5. [DOI] [PubMed] [Google Scholar]
- 18.Havaleshko DM, Cho H, Conaway M, Owens CR, Hampton G, Lee JK, Theodorescu D. Prediction of drug combination chemosensitivity in human bladder cancer. Mol Cancer Ther. 2007;6:578–86. doi: 10.1158/1535-7163.MCT-06-0497. [DOI] [PubMed] [Google Scholar]
- 19.Moreno CS, Matyunina L, Dickerson EB, Schubert N, Bowen NJ, Logani S, et al. Evidence that p53-mediated cell-cycle-arrest inhibits chemotherapeutic treatment of ovarian carcinomas. PLoS ONE. 2007;2:e441. doi: 10.1371/journal.pone.0000441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hess KR, Anderson K, Symmans WF, Valero V, Ibrahim N, Mejia JA, et al. Pharmacogenomic predictor of sensitivity to preoperative chemotherapy with paclitaxel and fluorouracil, doxorubicin, and cyclophosphamide in breast cancer. J Clin Oncol. 2006;24:4236–44. doi: 10.1200/JCO.2006.05.6861. [DOI] [PubMed] [Google Scholar]
- 21.Bild AH, Potti A, Nevins JR. Linking oncogenic pathways with therapeutic opportunities. Nat Rev Cancer. 2006;6:735–41. doi: 10.1038/nrc1976. [DOI] [PubMed] [Google Scholar]
- 22.Thuerigen O, Schneeweiss A, Toedt G, Warnat P, Hahn M, Kramer H, et al. Gene expression signature predicting pathologic complete response with gemcitabine, epirubicin, and docetaxel in primary breast cancer. J Clin Oncol. 2006;24:1839–45. doi: 10.1200/JCO.2005.04.7019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.