Abstract
Hyperuricemia is associated with the metabolic syndrome, gout, renal and cardiovascular disease (CVD). American Indians have high rates of CVD and 25 % of individuals in the Strong Heart Family Study (SHFS) have high serum uric acid levels. The aim of this study was to investigate the genetic determinants of serum uric acid variation in American Indian participants of the SHFS. A variance component decomposition approach (implemented in SOLAR) was used to conduct univariate genetic analyses in each of three study centers and the combined sample. Serum uric acid was adjusted for age, sex, age*sex, BMI, estimated glomerular filtration rate, alcohol intake, diabetic status and medications. Overall mean ± SD serum uric acid for all individuals was 5.14 ± 1.5 mg/dl. Serum uric acid was found to be significantly heritable (0.46 ± 0.03 in all centers, and 0.39 ± 0.07, 0.51 ± 0.05, 0.44 ± 0.06 in Arizona, Dakotas and Oklahoma, respectively). Multipoint linkage analysis showed significant evidence of linkage for serum uric acid on chromosome 11 in the Dakotas center (logarithm of odds score (LOD) = 3.02) and in the combined sample (LOD = 3.56) and on chromosome 1 (LOD = 3.51) in the combined sample. A strong positional candidate gene in the chromosome 11 region is solute carrier family22, member 12 (SLC22A12) that encodes a major uric acid transporter URAT1. These results show a significant genetic influence and a possible role for one or more genes on chromosomes 1 and 11 on the variation in serum uric acid in American Indian populations.
Keywords: SLC22A12 gene, URAT1, variance component decomposition approach, chromosome
Introduction
Increased serum uric acid or hyperuricemia is associated with gout, renal and cardiovascular disease (CVD) and the metabolic syndrome (Nakagawa et al. 2006). Although serum uric acid has been more closely associated with gout, its association with CVD has recently been recognized (Alderman 2002; Johnson et al. 2005; Chen et al., 2009). Serum uric acid has been independently associated with insulin resistance, hypertension, dyslipidemia and obesity (Hayden and Tyagi 2004; Forman et al., 2009), all disorders that are elements of the metabolic syndrome (Conen et al. 2004; Onat et al. 2006; Nan et al. 2008). Even in individuals with serum uric acid levels in the normal range, it was found that increasing serum uric acid was associated with an increased risk of metabolic syndrome (Yoo et al. 2005; Hikita et al. 2007). Higher serum uric acid levels have also been linked to increased risk of developing type 2 diabetes (Dehghan et al. 2008a). Endothelial dysfunction, another common finding in renal and CVD patients, may be induced by increased serum uric acid (Khosla et al. 2005). The possible mechanisms by which serum uric acid may have pathogenic role in CVD are i) promoting lipid peroxidation by oxygenation of low density lipoprotein cholesterol, ii) increasing production of free oxygen radicals and thus increasing the risk for atherosclerosis, iii) increasing platelet aggregation, and iv) inducing renal hypertension by intrarenal crystal deposition (Alderman 2002). However, it is not clear whether serum uric acid is an independent risk factor of CVD or is merely associated with other CVD risk factors or metabolic syndrome components. Irrespective of its importance as an independent risk factor, elevated serum uric acid is a matter of concern in individuals at risk for CVD (Hayden and Tyagi 2004).
Serum uric acid concentrations are highly variable in humans. Age, ethnicity, sex, high purine nutrient intake, alcohol consumption, defects in purine metabolism and genetic factors influence serum uric acid levels (Johnson et al. 2003). In addition, medications including losartan, allopurinol, diuretics and other hypertension medications impact uric acid metabolism (Sica and Schoolwerth 2002). Serum urate levels have been found to be significantly heritable in family-based studies (Rao et al. 1982; Rice et al. 1990; Tang et al. 2003; Yang et al. 2005; Nath et al. 2007; Voruganti et al. 2009). A few genome-wide studies of serum uric acid have been conducted in Caucasian, African American and Mexican American population with relatively little replication of chromosomal locations (Tang et al. 2003; Yang et al. 2005; Nath et al. 2007; Voruganti et al. 2009).
The Strong Heart Family Study (SHFS) is an extension of an ongoing, longitudinal study of CVD in American Indians, the Strong Heart Study (SHS). American Indians have high rates of CVD and CVD-related mortality (Howard et al. 1999; North et al. 2003). Serum uric acid values can vary substantially based on ethnicity and geography (Alderman 2002). Serum uric acid levels > 7 mg/dl in men and 6mg/dl in women are considered high (Rathmann et al., 1998; Krishnan et al., 2007). However, serum uric acid levels > 4mg/dl have been associated with complications of atherogenesis and stroke in individuals who are diabetic or at risk for CVD (Lehto et al., 1998; Hayden and Tyagi, 2004). Serum uric acid in this cohort also run high with more than 25 % and 76 % of individuals having serum uric acid levels more than 6 and 4mg/dl, respectively. These findings are a public health concern in a cohort that is at a higher risk for CVD. Given the high rates of CVD and serum uric acid levels, investigation of genetic factors that influence the variation in serum uric acid in this population is of utmost importance. Therefore the primary aim of this study was to identify quantitative trait loci (QTLs) influencing the variation in serum uric acid levels in American Indians.
Methods
Study population
The SHFS is family-based genetic study in the American-Indian community. It is an extension of the Strong Heart Study which is a population-based observational study of CVD and its risk factors in this population. More than 3600 members of multigenerational families were enrolled from all three centers located in Arizona, North and South Dakota and Oklahoma. The North and South Dakota centers have been grouped together as one center as the SHFS participants in the Dakotas are members of Sioux tribes whose reservations exist in both North and South Dakota. The Indian Health Service Institutional Review Board and the institutional review boards from the participating centers approved the SHFS protocol. All subjects gave informed consent. Study design and methods of the SHFS have been described before (North et al. 2003).
Phenotyping
During a clinical visit, information related to anthropometry, alcohol intake, medical history and medication use was obtained using a questionnaire. Weight was measured to the nearest 0.1 kilogram, using an ISO-9001 certified Scale-Tronix electronic scale with a capacity of 880 pounds (400 kilograms) (White Plains, NY). Standing height was measured twice, to the nearest centimeter, using a SECA wall-mounted stadiometer (Seca Corp., Hanover, MD). Body mass index (BMI) was computed as the ratio between weight in kilograms and height (in meters) squared. Blood was collected after an overnight fast and plasma and serum samples were stored at −80°C until analyzed. Fasting serum uric acid was oxidized in the presence of uricase to form hydrogen peroxide, which was measured photometrically (Domagk et al. 1968). Serum creatinine was estimated by modified kinetic Jaffe reaction (Beckman Synchron LX System, Beckman Coulter, Fullerton, CA). Estimated glomerular filtration rate (eGFR) was computed using the simplified modified diet and renal disease (MDRD) equation:
[eGFR (ml/min/1.73m2) = 186 × sCr (mg/dl)−1.154 × age (years)−0.203 × (1.212 if black) × (0.742 if female)] (Levey et al. 1999; Myers et al. 2006).
Genotyping
Genotyping was conducted with DNA isolated from fasting blood samples using organic solvents. All study participants were genotyped for about 400 microsatellite markers, using the ABI PRISM Linkage Mapping Set-MD10 Version 2.5 (Applied Biosystems, Foster City, CA) with markers spaced at an average interval of 10cM across the autosomal chromosomes. Genotyping procedures have been described previously (North et al. 2006). Pedigree and Mendelian errors were detected and corrected utilizing the software PREST (pedigree relationship statistical tests) (Sun et al. 2002) and SIMWALK2 (Sobel et al. 1996), respectively. Multipoint identity-by-descent (IBD) matrices for genome-wide linkage analyses were calculated using the linkage analysis package (LOKI) (Heath 1997). The chromosomal map used in these computations was based on marker locations reported by DeCode genetics (Kong et al. 2002).
Quantitative genetic analysis
To detect genetic influence on the variation in serum uric acid and localize quantitative trait loci (QTL) that affect serum uric acid, we employed a multipoint linkage analysis based on the variance components decomposition approach, implemented in the software program SOLAR and described in detail elsewhere (Blangero and Almasy 1997; Almasy and Blangero 1998). In short, it is an extension of the variance components approach in which variance due to a specific QTL is added to the basic model. It is based on estimating the effect of a specific QTL on the variation in phenotype, and can be modeled as a function of the IBD relationship at the marker locus between family members. Traditionally, a logarithm of the odds (LOD) score, which is computed directly from the likelihood ratio tests, is reported in linkage analyses (Almasy and Blangero 1998).
An inverse normalization was performed for all traits after removing outliers greater than four standard deviation from the mean. To conduct multipoint linkage analysis in the combined sample, we combined samples from individual centers and used the cumulative data for analysis, incorporating center as a covariate in the final model in addition to other covariates. The serum uric acid was adjusted for the effects of factors that are known to influence its variation using regression methods. The covariates age, sex, age*sex, BMI, eGFR, type 2 diabetes status, alcohol intake and medications were included in the final model. Relative pairs that were major contributors to this study were parent-offspring, siblings, grandparent-grandchild, avuncular, half siblings, first, second and third cousins. Details of these pairs are shown in Table 1.
Table 1.
Relative pairs utilized in this study
| Relationship | ALL | AZ | DK | OK |
|---|---|---|---|---|
| Parent-offspring | 2822 | 842 | 1026 | 954 |
| Siblings | 2657 | 846 | 941 | 870 |
| Grandparent-grandchild | 1068 | 361 | 375 | 332 |
| Avuncular | 6741 | 1961 | 2555 | 2225 |
| Half siblings | 943 | 304 | 323 | 316 |
| Grand avuncular | 3027 | 782 | 1236 | 1009 |
| Half avuncular | 1393 | 441 | 422 | 530 |
| First cousins | 9148 | 2801 | 3517 | 2830 |
| First cousins, once removed | 12562 | 3676 | 5027 | 3859 |
| Half first cousins | 1325 | 523 | 375 | 427 |
| First cousins, twice removed | 1485 | 354 | 389 | 742 |
| Half first cousins, once removed | 1607 | 799 | 266 | 542 |
| Second cousins | 7083 | 2184 | 3060 | 1839 |
| Second cousins, once removed | 2697 | 869 | 967 | 861 |
| Half second cousins | 801 | 421 | 133 | 247 |
| Third cousins | 915 | 180 | 562 | 173 |
| Others | 3075 | 689 | 1312 | 1074 |
| Total | 59349 | 18033 | 22486 | 18830 |
Results
3604 individuals (men = 1443, women = 2161) participated in this study. Of these, 1215 were from Arizona, 1186 from Dakotas and 1203 from Oklahoma. The distribution of age, BMI and serum uric acid by sex and center is given in Table 2. The overall mean ± SD serum uric acid for all individuals was 5.14 ± 1.5 mg/dl. According to the criteria for increased serum uric acid levels (men > 7 mg/dl and women > 6mg/dl) 12 %, 21 % and 17 % of individuals from Arizona, Dakotas and Oklahoma, respectively had hyperuricemia. As has been observed in previous studies (Mikkelsen et al. 1965; Freedman et al. 1995; Conen et al. 2004), men had higher levels of serum uric acid than women in all centers, despite being younger and having lower BMI. Approximately 20 % and 23 % of individuals had hypertension and type 2 diabetes, respectively. With respect to medication usage, 13 % of the individuals were taking aspirin and 15 % were taking angiotensin-converting enzyme (ACE) inhibitors. Approximately 7 % were taking diuretics. Diabetic drugs, insulin, metformin and sulfonyureas were taken by 6 %, 8 %, and 10 % of the participants, respectively. Each medication group was tested separately for significant effects on serum uric acid levels. The medication groups included aspirin, statins, fibrates, insulin, sulfonylureas, metformins, thiazolidinediones, ACE inhibitors, ARBs, beta blockers, calcium channel blockers, hydrocholrophiazides (diuretics), hypotensive agents, alpha blocking agents and vasodilating agents. Of these, aspirin, sulfonylureas, metformin, beta-blockers, angiotensin-converting enzyme (ACE) inhibitors, calcium channel blockers and hydrochlorothiazide (diuretics) were found to be significantly associated with serum uric acid. The main effects of all covariates included in the final model are given in Table 3. Individuals who were taking diabetic medications, metformin or sulfonylureas had lower serum uric acid levels as compared to those who did not. However, individuals on aspirin or other blood pressure lowering medications had higher serum uric acid levels than others (Table 4).
Table 2.
Distribution of age, BMI and serum uric acid (mg/dl) in Strong Heart Study participants by center
| Age | BMI | Serum uric acid | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Center | All | Women Mean (SD) |
Men | All | Women Mean (SD) |
Men | All | Women Mean (SD) |
Men |
| ALL | 39.96 (17.0) | 41.07 (17.2) | 38.30 (16.6) | 32.14 (7.6) | 32.67 (7.7) | 31.24 (7.5) | 5.14 (1.5) | 4.58 (1.9) | 5.98 (1.4) |
| AZ | 37.21 (16.0) | 38.16 (16.5) | 35.65 (15.0) | 35.13 (8.2) | 35.73 (8.2) | 34.12 (8.1) | 4.87 (1.5) | 4.35 (1.2) | 5.72 (1.4) |
| DK | 39.04 (17.11) | 40.37 (17.3) | 37.12 (16.7) | 30.17 (6.8) | 30.83 (6.9) | 29.22 (6.7) | 5.34 (1.5) | 4.81 (1.3) | 6.12 (1.5) |
| OK | 43.65 (17.3) | 44.92 (17.2) | 41.89 (17.3) | 31.15 (6.9) | 31.29 (6.9) | 30.96 (6.8) | 5.22 (1.5) | 4.60 (1.3) | 6.08 (1.4) |
Table 3.
Covariate effects on serum uric acid*
| All centers | Arizona | Dakotas | Oklahoma | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Covariate | P value | Effect | P value | Effect | P value | Effect | P value | Effect | |
| Age | 0.002 | 0.004 | 2.3 × 10−6 | −0.013 | NS | - | NS | - | |
| Sex (female) | 9.2 × 10−139 | −1.028 | 5.4 × 10−53 | −0.102 | 4.2 × 10−44 | −0.949 | 3.7 × 10−49 | −0.973 | |
| Age × sex | 2.0 × 10−6 | 0.008 | 0.012 | 0.008 | 0.029 | 0.005 | 5.2 × 10−7 | 0.014 | |
| Center | 0.0003 | 0.107 | NA | - | NA | - | NA | - | |
| BMI | 6.7 × 10−17 | 0.019 | 0.018 | 0.008 | 4.7 × 10−17 | 0.036 | 6.3 × 10−13 | 0.030 | |
| eGFR | 9.8 × 10−38 | −0.008 | 5.1 × 10−11 | −0.006 | 1.1 × 10−10 | −0.007 | 1.9 × 10−18 | −0.010 | |
| Diabetes status | 2.0 × 10−7 | −0.202 | 1.7 × 10−8 | −0.341 | NS | - | NS | - | |
| Alcohol intake | NS | - | 0.014 | −0.250 | NS | - | NS | - | |
| Medications | |||||||||
| Aspirin | 0.0002 | 0.171 | NS | - | 0.0116 | −0.207 | 0.004 | 0.212 | |
| Sulfonylureas | 1.5 × 10−7 | −0.278 | 5.9 × 10−8 | −0.430 | NS | - | NS | - | |
| Metformin | 6.6 × 10−6 | −0.265 | 5.4 × 10−7 | −0.430 | NS | - | NS | - | |
| ACE inhibitors | 0.0018 | 0.140 | NS | - | 0.027 | 0.186 | 0.0039 | 0.220 | |
| Beta blockers | 0.00002 | 0.344 | 0.0017 | 0.483 | 0.0225 | 0.315 | 0.053 | 0.258 | |
| Calcium channel blockers |
0.00018 | 0.256 | NS | - | 0.0008 | 0.438 | 0.023 | 0.253 | |
| Hydrochlorothiazide (diuretics) |
1.6 × 10−13 | 0.470 | 0.034 | 0.255 | 7.1 × 10−10 | 0.717 | 3.0 × 10−10 | 0.465 | |
All covariates were tested separately without having other covariates in the model
Covariates which were significant in at least one center were included in the final model
Table 4.
Distribution of serum uric acid (mg/dl) based on the usage of medication*
| Medication | All centers | Arizona | Dakotas | Oklahoma | ||||
|---|---|---|---|---|---|---|---|---|
| No | Yes | No | Yes | No | Yes | No | Yes | |
| Aspirin | 5.10 (1.5) | 5.42 (1.7) | 4.85 (1.4) | 5.00 (1.6) | 5.29 (1.5) | 5.72 (1.7) | 5.17 (1.5) | 5.48 (1.7) |
| Sulfonylureas | 5.19 (1.5) | 4.72 (1.5) | 4.95 (1.4) | 4.37 (1.4) | 5.36 (1.5) | 5.09 (1.6) | 5.24 (1.5) | 5.02 (1.4) |
| Metformin | 5.17 (1.5) | 4.77 (1.5) | 4.94 (1.5) | 4.34 (1.2) | 5.34 (1.5) | 5.44 (1.7) | 5.23 (1.5) | 5.02 (1.4) |
| ACE inhibitors | 5.11 (1.5) | 5.32 (1.6) | 4.85 (1.5) | 4.96 (1.5) | 5.30 (1.5) | 5.69 (1.8) | 5.18 (1.5) | 5.43 (1.5) |
| Beta blockers | 5.12 (1.5) | 5.71 (1.9) | 4.84 (1.4) | 5.57 (1.7) | 5.32 (1.5) | 5.82 (1.8) | 5.19 (1.5) | 5.72 (2.0) |
| Calcium channel blockers |
5.12 (1.5) | 5.55 (1.7) | 4.85 (1.4) | 5.13 (1.6) | 5.30 (1.5) | 6.18 (1.7) | 5.20 (1.5) | 5.50 (1.6) |
| Hydrochlorothiazide (diuretics) |
5.08 (1.5) | 5.88 (1.9) | 4.84 (1.8) | 5.32 (2.0) | 5.27 (1.5) | 6.49 (1.9) | 5.14 (1.5) | 5.85 (1.9) |
Medications with significant effects on serum uric acid are shown in the table.
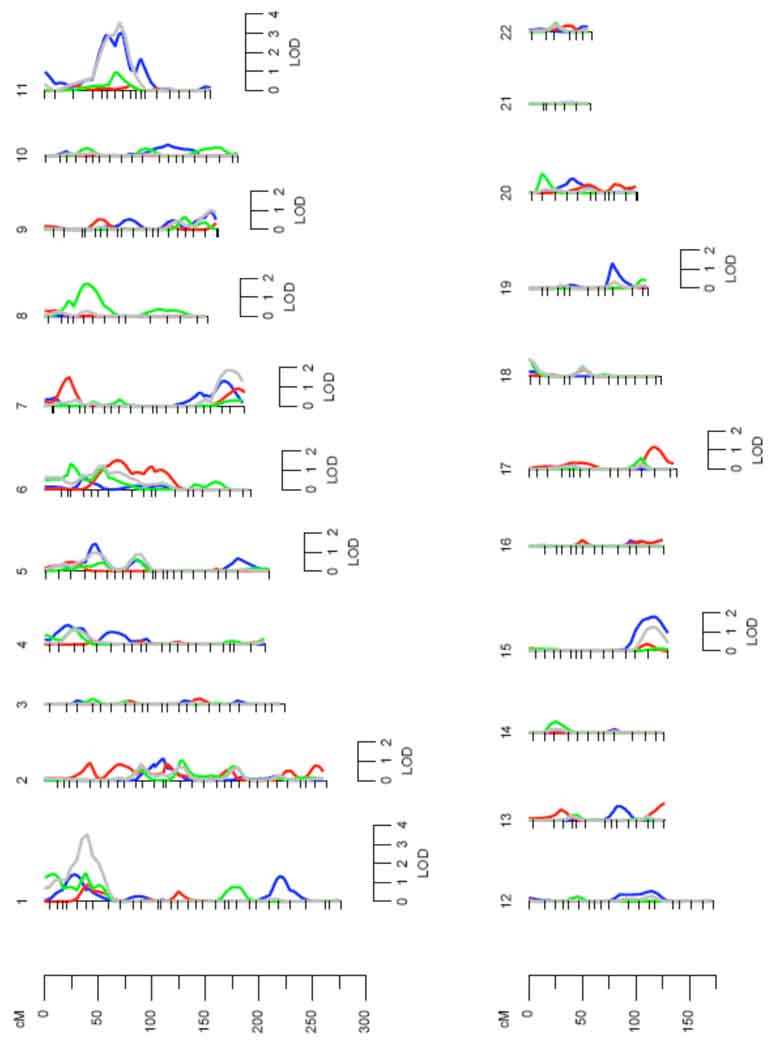
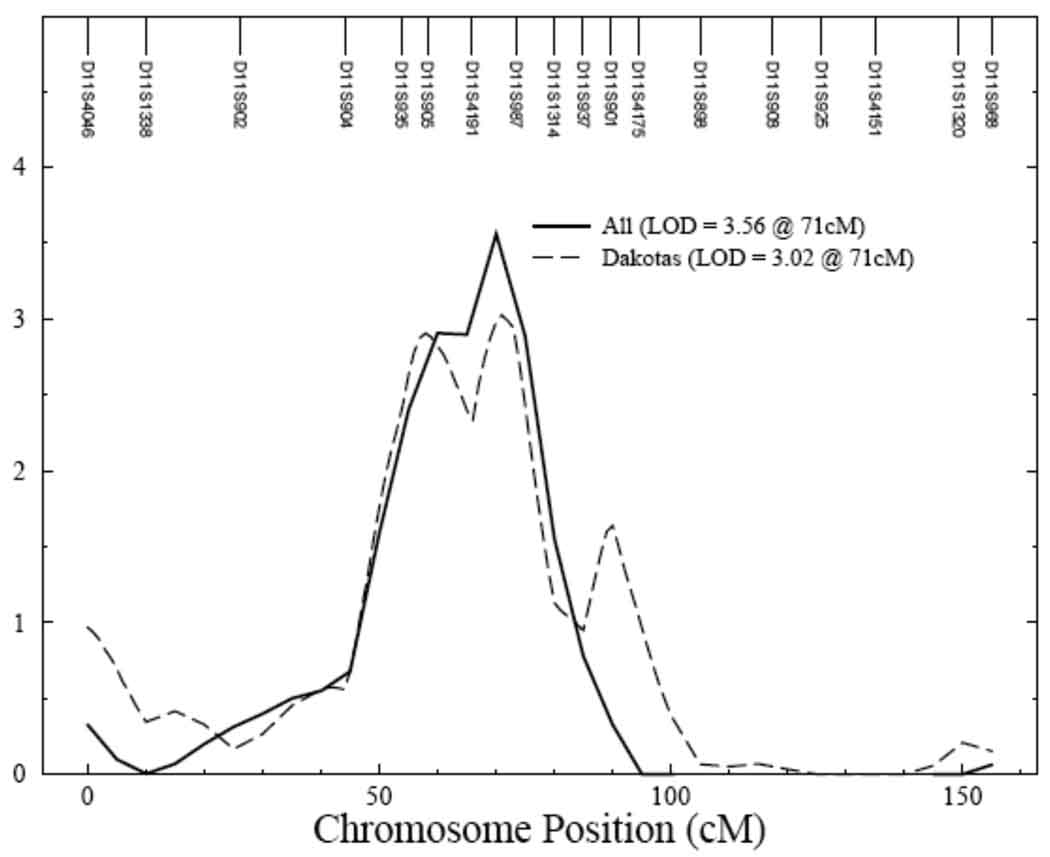
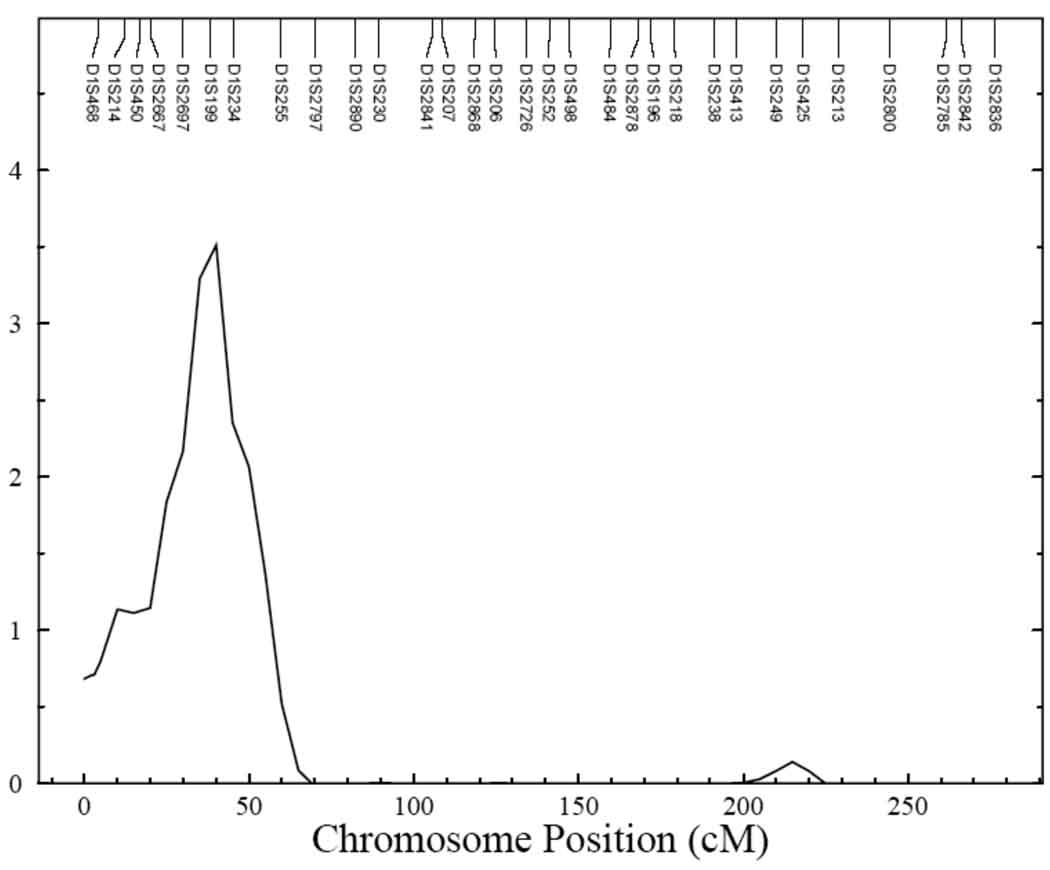
Univariate genetic analysis showed serum uric acid to be significantly heritable, with the heritability being 0.46 ± 0.03 (p = 6.5 × 10−63) in all centers and 0.39 ± 0.07 (p = 4.6 × 10−12), 0.51 ± 0.05 (p = 2.2 × 10−32), and 0.44 ± 0.06 (p = 1.5 × 10−18) in Arizona, Dakotas and Oklahoma, respectively. Multipoint linkage analysis for serum uric acid conducted with age, sex and their higher order terms and interactions as covariates showed evidence of linkage on chromosome 11 with LOD scores of 3.0 and 2.1 in the combined data from all centers and in the Dakota data, respectively (data not shown). Inclusion of additional, significant covariates, BMI, eGFR, type 2 diabetes, alcohol intake and medications in the final model improved the signal both in the Dakotas and in data from all centers, combined (Figure 1). Linkage signals for Dakotas (LOD score = 3.02) and all centers (LOD score = 3.56) on chromosome 11 at 71cM are shown in Figure 2. Both these QTLs had one LOD support intervals of 22cM (Table 5). An additional QTL for serum uric acid was obtained for all centers on chromosome 1, with a significant LOD score of 3.51 (Figure 3). For Arizona, the highest LOD score of 1.5 was obtained on chromosome 6 and for Oklahoma, a QTL on chromosome 8 was identified with a LOD score of 1.7.
Figure 1.
Univariate linkage analysis by each chromosome (all centers and individual centers). Chromosomal location (cM) is represented on the x-axis and the LOD score shown on the y-axis. All – grey; Dakotas – blue; Oklahoma – green; Arizona - red
Figure 2.
Evidence of significant linkage for serum uric acid on chromosome 11 in Dakotas and all centers. Chromosomal location (cM) is represented on the x-axis and LOD score is shown on the y-axis. All – Solid line; Dakotas – dashed line
Table 5.
Loci affecting serum uric acid levels
| Center | Highest LOD score |
Chromosome (location) |
Flanking markers | One LOD support interval |
|---|---|---|---|---|
| ALL | 3.51 | 1 (39cM) | D1S199 and D1S234 | 31–43cM |
| 3.56 | 11 (71cM) | D11S4191 and D11S987 | 55–77cM | |
| AZ | 1.5 | 6 (68cM) | D6S1610 and D6S257 | 60–81cM |
| DK | 3.02 | 11 (71cM) | D11S4191 and D11S987 | 55–77cM |
| OK | 1.7 | 8 (40cM) | D8S258 and D8S1771 | 35–44cM |
Figure 3.
Evidence of significant linkage for serum uric acid on chromosome 1 in all centers. Chromosomal location (cM) is represented on the x-axis and LOD score is shown on the y-axis
Discussion
This is the first genome-wide study of variation in serum uric acid performed in American Indians. We found significant genetic influence on the variation in serum uric acid levels and identified two novel QTLs on chromosomes 1 and 11. Increased serum uric acid is a risk factor for gout, renal disease and CVD and is known to aggregate in families (Stecher et al. 1949; Dixon 1960; Friedlander et al. 1988; Cameron and Simmonds 2005; Nakagawa et al. 2006). We found significant heritability for serum uric acid in all centers confirming a genetic influence on its variation. Furthermore, heritabilities obtained in this study are in the same range as those reported by previous family-based genetic studies of individuals of different ancestry (Rao et al. 1982; Tang et al. 2003; Yang et al. 2005; Nath et al. 2007; Voruganti et al. 2009).
It is likely that multiple genes, and not a single gene influence the variation in serum uric acid (Wilk et al. 2000). Multiple QTLs have been reported in genome-wide scans of serum uric acid with little overlap in populations. This may be due to confounding by population stratification, or possibly due to genetic heterogeneity of this common, complex trait. Yang et al. (2005) reported a QTL on chromosome 15q (LOD = 3.3) for serum uric acid in a Caucasian population from the Framingham Heart Study. In another study, using serum uric acid as a part of the metabolic syndrome, a QTL was localized on chromosome 2q (Tang et al. 2003). Among Mexican Americans, a strong QTL for serum uric acid levels was found on chromosome 6 and a suggestive linkage on chromosome 3 (Nath et al. 2007). In a subsequent follow up study with a larger sample of individuals, the linkage on chromosome 3 reached genome-wide significance with a LOD score of 4.7 (Voruganti et al. 2009). Our study identified new QTLs for serum uric acid relevant to our population of American Indians.
Genome-wide association studies (GWAS) for gene discovery have been conducted mostly in individuals of European ancestry. These studies are biased towards common variants and the gene coverage varies depending on the platform used for genotyping. However, findings from GWAS have been more consistent. They identified common variants in solute carrier family2, member 9 (SLC2A9) that were associated with serum uric acid (Li et al. 2007; Vitart et al. 2008; Doring et al. 2008; Brandstattter et al. 2008; Stark et al. 2008; Wallace et al. 2008; Dehghan et al. 2008b; Matsuo et al., 2008). Interestingly, SLC2A9 encodes a transporter for urate as well as fructose, and is known to influence serum uric acid levels (Vitart et al. 2008). Within the confidence interval of our QTL for serum uric acid on chromosome 11 lies the gene for URAT1, solute carrier family22, member 12 (SLC22A12). URAT1 is an urate-anion exchanger and the main transporter responsible for reabsorption of uric acid from the glomerular filtrate in the apical membrane of the renal tubules (Enomoto et al. 2002). Mutations in SLC22A12 have been associated with hypouricemia in Japanese patients (Enomoto et al. 2002; Komoda et al. 2004; Takahashi et al., 2005; Wakida et al., 2005; Ichida et al. 2008; Lee et al., 2008) and Korean men (Jang et al. 2008). http://www.ncbi.nlm.nih.gov/entrez/dispomim.cgi?id=220150.
A strong linkage signal for serum uric acid was localized to chromosome 1p36. Prominent candidate genes in this region are the kidney chloride channel A and B (CLCNKA and CLCNKB) genes. The CLCNKA and CLCNKB channels are part of a family of mammalian chloride channels (Saito-Ohara et al. 1996). Mutations in these genes cause Bartter’s syndrome type 3 which is characterized by impaired salt reabsorption, salt wasting, hypokalemic metabolic alkalosis and hypercalciuria (Simon et al. 1997). Metabolic alkalosis can decrease the excretion of uric acid thus contributing to hyperuricemia in patients with this type of Bartter’s syndrome (Meyer et al. 1975). Thus these genes are strong positional candidate genes that may influence variation in serum uric acid in American Indians.
The rate of urate excretion and its renal handling play a major part in the variation of serum uric acid levels (Sica and Schoolwerth 2002). Therefore, medications that affect renal urate excretion have a significant influence on serum uric acid levels. Drugs such as losartan, have been shown to reduce serum uric acid levels by reducing its reabsorption from kidneys (Sica and Schoolwerth 2002). In the current study, among the medications that were associated with and adjusted against serum uric acid [aspirin, sulfonylureas, metformin, ACE inhibitors, beta blockers, calcium channel blockers and diuretics], diuretics was the most significant group. Diuretics, usually prescribed as the primary treatment for hypertension, are known to increase serum uric acid by increasing its reabsorption. They had the maximum effect on serum uric acid levels in all centers, except Arizona, when compared to other medications. Similarly beta blockers and ARBs, with the exception of losartan, are believed to increase serum levels of uric acid (Reyes 2003). Earlier genetic analyses explored diuretics and aspirin for effects on variation in serum uric acid (Wilk et al. 2000; Yang et al. 2005). However, the current study is the first one to investigate not only the effects of cardiovascular drugs such as ARBs, ACE inhibitors, beta blockers, calcium channel blockers and diuretics but also diabetic drugs such as sulfonylureas, metformin and insulin on the variation in serum uric acid levels. We observed that individuals taking metformin or sulfonyureas have comparatively lower levels of serum uric acid than those who don’t take these medications. Serum uric acid has been linked to insulin resistance (Clausen et al., 1998; Yoo et al., 2005) and has often been advocated to be a part of the metabolic syndrome (Nakagawa et al., 2005; Cirillo et al., 2006). It has been reported that drugs such as metformin that improve insulin sensitivity are also known to decrease serum uric acid levels (Tsouli et al., 2006). On the other hand, we observed that blood pressure lowering medications have an adverse effect on serum uric acid levels. Diuretics increase serum uric acid levels by either affecting its secretion or increased distal reabsorption (Langford et al., 1987). ACE inhibitors are known to have a mild reducing effect on serum uric acid levels helping blunt the effect of diuretics (Reyes, 2003) however this was not observed in individuals in our study. Even though drugs that lower serum uric acid have not shown substantial improvements in vascular mortality, increased serum uric acid values provide useful prognostic information in individuals at risk for CVD (Wannamethee 2005).
In summary, we identified two new loci on chromosomes 11q and 1q for variation in serum uric acid among American Indians, where strong positional candidate genes are located. Given the importance of serum uric acid as a likely biomarker and a potential independent risk factor for CVD, and the recent increase in CVD mortality in American Indians, these results assume considerable significance.
Acknowledgments
We thank the SHFS participants, Indian Health Services facilities, and participating tribal communities for their extraordinary cooperation and involvement, and without whose assistance, this project would not have been possible. The opinions expressed in this paper are those of the author(s) and do not necessarily reflect the views of the Indian Health Service. This work was also supported by grants U01-HL65520, U01-HL41642, U01-HL41652, U01-HL41654 and U01-HL65521 from the National Heart, Lung, and Blood Institute, Bethesda, MD. Development of SOLAR was supported by NIH grant MH59490. This investigation was conducted in part in facilities constructed with support from the Research Facilities Improvement Program under grant numbered C06 RR014578, C06 RR013556, C06 RR015456, and C06 RR017515.
References
- 1.Alderman MH. Uric acid and cardiovascular risk. Curr Opin Pharmacol. 2002;2:126–130. doi: 10.1016/s1471-4892(02)00143-1. [DOI] [PubMed] [Google Scholar]
- 2.Almasy L, Blangero J. Multipoint quantitative trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brandstatter A, Keichl S, Kollerits B, Hunt SC, Heid IM, Coassin S, Willeit J, Adams TD, Illig T, Hopkins PN, Kronenberg F. The gender-specific association of the putative fructose transporter SLC2A9 variants with uric acid levels is modified by BMI. Diab Care. 2008;31:1662–1667. doi: 10.2337/dc08-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blangero J, Almasy L. Multipoint oligogenic linkage analysis of quantitative traits. Genet Epidemiol. 1997;14:959–964. doi: 10.1002/(SICI)1098-2272(1997)14:6<959::AID-GEPI66>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 5.Cameron JS, Simmonds HA. Hereditary hyperuricemia and renal disease. Semin Nephrol. 2005;25:9–18. doi: 10.1016/j.semnephrol.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 6.Chen JH, Chuang SY, Chen HJ, Yeh WT, Pan WH. Serum uric acid level as an independent risk factor for all-cause, cardiovascular, and ischemic stroke mortality: a Chinese cohort study. Arthritis Rheum. 2009;61:225–232. doi: 10.1002/art.24164. [DOI] [PubMed] [Google Scholar]
- 7.Cirillo P, Sato W, Reungui S, Heinig M, Gersch M, Sautin Y, Nakagawa T, Johnson Uric acid, the metabolic syndrome and renal disease. J Am Soc Nephrol. 2006;17:165–168. doi: 10.1681/ASN.2006080909. [DOI] [PubMed] [Google Scholar]
- 8.Clausen JO, Borch-Johnsen K, Ibsen H, Pedersen O. Analysis of the relationship between fasting serum uric acid and insulin sensitivity index in a population-based sample of 380 young healthy Caucasians. European J Epidemiol. 1998;138:63–69. doi: 10.1530/eje.0.1380063. [DOI] [PubMed] [Google Scholar]
- 9.Conen D, Wietlisbach V, Bovet P, Shamlaye C, Riesen W, Paccaud F, Burnier M. Prevalence of hyperuricemia and relation of serum uric acid with cardiovascular risk factors in a developing country. BMC Public Health. 2004;4:9. doi: 10.1186/1471-2458-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dehghan A, van Hoek M, Sijbrands EJG, Hofman A, Whiteman JCM. High serum uric acid as a novel risk factor for type 2 diabetes. Diabetes Care. 2008a;31:361–362. doi: 10.2337/dc07-1276. [DOI] [PubMed] [Google Scholar]
- 11.Dehghan A, Kottgen A, Yang Q, Hwang SJ, Kao WHL, Rivadeneira F, Boerwinkle E, Levy D, Hofman A, Astor BC, Benjamin EJ, van Dujin CM, Wittemant JC, Coresh J, Fox CS. Association of three genetic loci with uric acid concentrations and risk of gout: a genome-wide association study. Lancet. 2008b;372:1953–1961. doi: 10.1016/S0140-6736(08)61343-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dixon AS. Familial hyperuricemic nephropathy. Proc R Soc Med. 1960;53:967–968. [PMC free article] [PubMed] [Google Scholar]
- 13.Domagk GF, Schlicke HH. A colorimetric method using uricase and peroxidase for the determination of uric acid. Anal Biochem. 1968;22:219–224. doi: 10.1016/0003-2697(68)90309-6. [DOI] [PubMed] [Google Scholar]
- 14.Döring A, Gieger C, Mehta D, Gohlke H, Prokisch H, Coassin S, Fischer G, Henke K, Klopp N, Kronenberg F, Paulweber B, Pfeufer A, Rosskopf D, Völzke H, Illig T, Meitinger T, Wichmann HE, Meisinger C. SLC2A9 influences uric acid concentrations with pronounced sex-specific effects. Nat Genet. 2008;40:430–436. doi: 10.1038/ng.107. [DOI] [PubMed] [Google Scholar]
- 15.Enomoto A, Kimura H, Chairoungdua A, Shigeta Y, Jutabha P, Cha SH, Hosoyamada M, Takeda M, Sekine T, Igarashi T, Matsuo H, Kikuchi Y, Oda T, Ichida K, Hosoya T, Shimokata K, Niwa T, Kannai Y, Endou H. Molecular identification of a renal urate-anion exchanger that regulates blood urate levels. Nature. 2002;417:447–452. doi: 10.1038/nature742. [DOI] [PubMed] [Google Scholar]
- 16.Forman JP, Choi H, Curhan GC. Uric acid and insulin sensitivity and risk of incident hypertension. Arch Intern Med. 2009;169:155–162. doi: 10.1001/archinternmed.2008.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Freedman DS, Williamson DF, Gunter EW, Byers T. Relation of serum uric acid to mortality and ischemic heart disease. The NHANES I epidemiologic follow-up study. Am J Epidemiol. 1995;141:637–644. doi: 10.1093/oxfordjournals.aje.a117479. [DOI] [PubMed] [Google Scholar]
- 18.Friedlander Y, Kark JD, Stein Y. Family resemblance for serum uric acid in a Jerusalem sample of families. Hum Genet. 1988;79:58–63. doi: 10.1007/BF00291711. [DOI] [PubMed] [Google Scholar]
- 19.Hayden MR, Tyagi SC. Uric acid: A new look at an old risk marker for cardiovascular disease, metabolic syndrome, and type 2 diabetes mellitus: The urate redox shuttle. Nutr Metab. 2004:1–15. doi: 10.1186/1743-7075-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heath SC. Markov chain Monte Carlo segregation and linkage analysis for oligogenic models. Am J Hum Genet. 1997;61:748–760. doi: 10.1086/515506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hikita M, Ohno I, Mori Y, Ichida K, Yokose T, Hosoya T. Relationship between hyperuricemia and body fat distribution. Intern Med. 2007;46:1353–1358. doi: 10.2169/internalmedicine.46.0045. [DOI] [PubMed] [Google Scholar]
- 22.Howard BV, Lee ET, Cowan LD. Rising tide of cardiovascular disease in American Indians: the Strong Heart Study. Circulation. 1999;99:2389–2395. doi: 10.1161/01.cir.99.18.2389. [DOI] [PubMed] [Google Scholar]
- 23.Ichida K, Hosoyamada M, Kamatani N, Kamitsuji S, Hisatome I, Shibasaki T, Hosoya T. Age and origin of the G774A mutation in SLC22A12 causing renal hypouricemia in Japanese. Clin Genet. 2008;74:243–251. doi: 10.1111/j.1399-0004.2008.01021.x. [DOI] [PubMed] [Google Scholar]
- 24.Jang WC, Nam YH, Park SM, Ahn YC, Park SH, Choe JY, Shin IH, Kim SK. T6092C polymorphism of SLC22A12 gene is associated with serum uric acid concentrations in Korean male subjects. Clin Chim Acta. 2008;398:140–144. doi: 10.1016/j.cca.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 25.Johnson RJ, Kang DH, Feig D, Kivlighn S, Kanellis J, Watanabe S, Tuttle KR, Rodriguez-Iturbe B, Herrera-Acosta J, Mazzali M. Is there a pathogenetic role for uric acid in hypertension and cardiovascular and renal disease? Hypertension. 2003;41:1183–1190. doi: 10.1161/01.HYP.0000069700.62727.C5. [DOI] [PubMed] [Google Scholar]
- 26.Johnson RJ, Segal MS, Srinivas T, Ejaz A, Mu W, Roncal C, Sanchez-Lozada L, Gersch M, Rodriguez-Iturbe B, Kang DH, Acosta JH. Essential hypertension, progressive renal disease and uric acid: a pathogenic link? J Am Soc Nephrol. 2005;16:1909–1919. doi: 10.1681/ASN.2005010063. [DOI] [PubMed] [Google Scholar]
- 27.Khosla UM, Zharikov S, Finch JL, Nakagawa T, Roncal C, Mu W, Krotova K, Block ER, Prabhakar S, Johnson RJ. Hyperuricemia induces endothelial dysfunction. Kid Intl. 2005;67:1739–1742. doi: 10.1111/j.1523-1755.2005.00273.x. [DOI] [PubMed] [Google Scholar]
- 28.Komoda F, Sekine T, Inatomi J, Enomoto A, Endou H, Ota T, Matsuyama T, Ogata T, Ikeda M, Awazu M, Muroya K, Kamimaki I, Igarashi T. The W258X mutation in SLC22A12 is the predominant cause of Japanese renal hypouricemia. Pediatr Nephrol. 2004;19:728–733. doi: 10.1007/s00467-004-1424-1. [DOI] [PubMed] [Google Scholar]
- 29.Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardson B, Sigurdardottir S, Barnard J, Hallbeck B, Masson G, Shlien A, Palsson ST, Frigge ML, Thorgeirsson TE, Gulcher JR, Stefansson K. A high-resolution recombination map of the human genome. Nat Genet. 2002;31:241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- 30.Krishnan E, Kwoh CK, Schumacher HR, Kuller L. Hyperuricemia and incidence of hypertension among men without metabolic syndrome. Hypertension. 2007;49:298–303. doi: 10.1161/01.HYP.0000254480.64564.b6. [DOI] [PubMed] [Google Scholar]
- 31.Langford HG, Blaufox MD, Borhani NO, Curb JD, Molteni A, Schneider KA, Pressel S. Is thiazide-produced uric acid elevation harmful? Analysis of data from the Hypertension Detection and Follow-up Program. Arch Intern Med. 1987;147:645–649. doi: 10.1001/archinte.147.4.645. [DOI] [PubMed] [Google Scholar]
- 32.Lee JH, Choi HJ, Lee BH, Kang HK, Chin HJ, Yoon HJ, Ha IS, Kim S, Choi Y, Cheong HI. Prevalence of hypouricaemia and SLC22A12 mutations in healthy Korean subjects. Nephrology (Carlton) 2008;13:661–666. doi: 10.1111/j.1440-1797.2008.01029.x. [DOI] [PubMed] [Google Scholar]
- 33.Lehto S, Niskanen L, Ronnemaa T, Laakso Serum uric acid is a strong predictor of stroke in patients with non-insulin-dependent diabetes mellitus. Stroke. 1998;29:635–639. doi: 10.1161/01.str.29.3.635. [DOI] [PubMed] [Google Scholar]
- 34.Levey AS, Bosch JP, Lewis JB, Greene T, Rogers N, Roth D. A more accurate method to estimate glomerular filtration rate from serum creatinine: a new prediction equation. Ann Intern Med. 1999;130:461–470. doi: 10.7326/0003-4819-130-6-199903160-00002. [DOI] [PubMed] [Google Scholar]
- 35.Li S, Sanna S, Maschio A, Busonero F, Usala G, Mulas A, Lai S, Dei M, Orru M, Albai G, Bandinedlli S, Schlessinger D, Lakatta E, Scuteri A, Najjar SS, Guralnik J, Naitza S, Crisponi L, Cao A, Abecasis G, Ferrucci L, Uda M, Chen WM, Nagaraja R. The GLUT9 gene is associated with serum uric acid levels in Sardinia and Chianti cohorts. PloS Genet. 2007;3:2156–2162. doi: 10.1371/journal.pgen.0030194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsuo H, Chiba T, Nagamori S, Nakayama A, Domoto H, Phetdee K, Wiriyasermkul P, Kikuchi Y, Oda T, Nishiyama J, Nakamura T, Morimoto Y, Kamakura K, Sakurai Y, Nonoyama S, Kanai Y, Shinomiya N. Mutations in glucose transporter 9 gene SLC2A9 cause renal hypouricemia. Am J Hum Genet. 2008;83:744–751. doi: 10.1016/j.ajhg.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mikkelsen WM, Dodge HJ, Valkenburg H. The distribution of serum uric acid values in a population unselected as to gout or hyperuricemia. Am J Med. 1965;39:242–251. doi: 10.1016/0002-9343(65)90048-3. [DOI] [PubMed] [Google Scholar]
- 38.Myers WJ, 3rd, Gill JR, Jr, Bartter FC. Gout as a complication of Bartter’s syndrome. A possible role for alkalosis in the decreased clearance of uric acid. An Intern Med. 1975:56–59. doi: 10.7326/0003-4819-83-1-56. [DOI] [PubMed] [Google Scholar]
- 39.Myers GL, Miller WG, Coresh J, Fleming J, Greenberg N, Greene T, Hostetter T, Levey AS, Panteghini M, Welch M, Eckfeldt JH for the National Kidney Disease Education Program Laboratory Working Group. Recommendations for improving serum creatinine measurement: A report from the laboratory working group of the National Kidney Disease Education Program. Clin Chem. 2006;52:5–18. doi: 10.1373/clinchem.2005.0525144. [DOI] [PubMed] [Google Scholar]
- 40.Nakagawa T, Tuttle KR, Short RA, Johnson RJ. Hypothesis: fructose-induced hyperuricemia as a causal mechanism for the epidemic of the metabolic syndrome. Nat Clin Pract Nephrol. 2005;1:80–86. doi: 10.1038/ncpneph0019. [DOI] [PubMed] [Google Scholar]
- 41.Nakagawa T, Kang D-H, Feig D, Sanchez-Lozada LG, Srinivas TR, Ejaz AA, Segal M, Johnson RJ. Unearthing uric acid: An ancient factor with recently found significance in renal and CVD. Kid Int. 2006;69:1722–1725. doi: 10.1038/sj.ki.5000391. 2006. [DOI] [PubMed] [Google Scholar]
- 42.Nan H, Qiao Q, Soderberg S, Gao W, Zimmet P, Shaw J, Alberti G, Dong Y, Uusitalo U, Pauvaday V, Chitson P, Tuomilehto J. Serum uric acid and components of the metabolic syndrome in non-diabetic populations in Mauritian Indians and Creoles and in Chinese in Qingdao, China. Metab Syndr Relat Disord. 2008;6:47–57. doi: 10.1089/met.2007.0028. [DOI] [PubMed] [Google Scholar]
- 43.Nath SD, Voruganti VS, Arar NH, Thameem F, Lopez-Alvarenga JC, Bauer R, MacCluer JW, Blangero J, Comuzzie AG, Abboud HE. Genome Scan for Determinants of Serum Uric Acid Variability. J Am Soc Nephrol. 2007;18:3156–3163. doi: 10.1681/ASN.2007040426. [DOI] [PubMed] [Google Scholar]
- 44.North KE, Howard BV, Welty TK, Best LG, Lee ET, Yeh JL, Fabsitz RR, Roman MJ, MacCluer JW. Genetic and environmental contributions to cardiovascular disease risk in American-Indians. Am J Epidemiol. 2003;157:303–314. doi: 10.1093/aje/kwf208. [DOI] [PubMed] [Google Scholar]
- 45.North KE, Göring HHH, Cole SA, Diego VP, Almasy L, Laston S, Cantu T, Howard BV, Lee ET, Best LG, Fabsitz RR, MacCluer JW. Linkage analysis of LDL cholesterol in American Indian populations: the Strong Heart Family Study. J Lipid Res. 2006;47:59–66. doi: 10.1194/jlr.M500395-JLR200. [DOI] [PubMed] [Google Scholar]
- 46.Onat A, Uyarel H, Hergenc G, Karabulut A, Albayrak S, San I, Yazici M, Keles I. Serum uric acid is a determinant of metabolic syndrome in a population-based study. Am J Hypertension. 2006;19:1055–1062. doi: 10.1016/j.amjhyper.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 47.Rao DC, Laskarzewski PM, Morrison JA, Kelly K, Glueck CJ. The clinical Lipid Research Clinic Family Study: familial determinants of plasma uric acid. Hum Genet. 1982:257–261. doi: 10.1007/BF00303013. [DOI] [PubMed] [Google Scholar]
- 48.Rathmann W, Funkhouser E, Dyer AR, Roseman JM. Relations of hyperuricemia with the various components of the insulin resistance syndrome in young black and white adults: the CARDIA study. Coronary Artery Risk Development in Young Adults. Ann Epidemiol. 1998;8:250–261. doi: 10.1016/s1047-2797(97)00204-4. [DOI] [PubMed] [Google Scholar]
- 49.Reyes AJ. Cardiovascular drugs and serum uric acid. Cardiovasc Drugs Ther. 2003;17:397–414. doi: 10.1023/b:card.0000015855.02485.e3. [DOI] [PubMed] [Google Scholar]
- 50.Rice T, Vogler GP, Perry TS, Laskarzewski PM, Province MA, Rao DC. Heterogenity in the familial aggregation of fasting uric acid levels in five North American populations: the Lipid Research Clinics Family Study. Am J Med Genet. 1990:219–225. doi: 10.1002/ajmg.1320360216. [DOI] [PubMed] [Google Scholar]
- 51.Saito-Ohara F, Uchida S, Takeuchi Y, Sasaki S, Hayashi A, Maraumo F, Ikeuchi T. Assignment of the genes encoding the human chloride channels, CLCNKA and CLCNKB, to 1p36 and of CLCN3 to 4q32-q33 by in situ hybridization. Genomics. 1996;36:372–374. doi: 10.1006/geno.1996.0479. [DOI] [PubMed] [Google Scholar]
- 52.Sica DA, Schoolwerth AC. Uric acid and losartan. Curr Opin Nephrol Hypertens. 2002;11:475–482. doi: 10.1097/00041552-200209000-00001. [DOI] [PubMed] [Google Scholar]
- 53.Simon DB, Bindra RS, Mansfield TA, Nelson-Williams C, Mendonca E, Stone R, Schurman S, Nayir A, Alpay H, Bakkaloglu A, Rodriguez-Soriano J, Morales JM, Sanjad SA, Taylor CM, Pilz D, Brem A, Trachtman H, Griswold W, Richard GA, John E, Lifton RP. Mutations in the chloride channel gene, CLCNKB, cause Bartter's syndrome type III. Nat Genet. 1997;17:171–178. doi: 10.1038/ng1097-171. [DOI] [PubMed] [Google Scholar]
- 54.Sobel E, Lange K. Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker sharing statistics. Am J Hum Genet. 1996;58:1323–1337. [PMC free article] [PubMed] [Google Scholar]
- 55.Stark K, Reinhard W, Neureuther K, Wiedmann S, Sedlacek K, Baessler A, Fischer M, Weber S, Kaess B, Erdmann J, Schunkert H, Hengstenberg C. Association of common polymorphisms in GLUT9 gene with gout and not with coronary artery disease in a large case-control study. PloS One. 2008;3:1–9. doi: 10.1371/journal.pone.0001948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stecher RM, Hersh AH, Solomon WM. The heredity of gout and its relationship to familial hyperuricemia. Ann Intern Med. 1949;31:595–614. doi: 10.7326/0003-4819-31-4-595. [DOI] [PubMed] [Google Scholar]
- 57.Sun L, Wilder K, McPeek MS. Enhanced pedigree error detection. Hum Hered. 2002;54:99–110. doi: 10.1159/000067666. [DOI] [PubMed] [Google Scholar]
- 58.Tang W, Miller MB, Rich SS, North KE, Panlow JS, Borecki I, Myers RH, Hopkins PN, Leppert M, Arnett DK. Linkage analysis of a composite factor for the multiple metabolic syndrome. The National Heart, Lung and Blood Institute Family Heart Study. Diabetes. 2003;52:2840–2847. doi: 10.2337/diabetes.52.11.2840. [DOI] [PubMed] [Google Scholar]
- 59.Takahashi T, Tsuchida S, Oyamada T, Ohno T, Miyashita M, Saito S, Komatsu K, Takashina K, Takada G. Recurrent URAT1 gene mutations and prevalence of renal hypouricemia in Japanese. Pediatr Nephrol. 2005;20:576–578. doi: 10.1007/s00467-005-1830-z. [DOI] [PubMed] [Google Scholar]
- 60.Tsouli SG, Liberopoulos EN, Mikhailidis DP, Athyros VG, Elisa MS. Elevated serum uric acid levels in metabolic syndrome: an active component or an innocent bystander? Metabolism Clinical and Experimental. 2006;55:1293–1301. doi: 10.1016/j.metabol.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 61.Vitart V, Rudan I, Hayward C, Gray NK, Floyd J, Palmer CN, Knott SA, Kolcic I, Polasek O, Graessler J, Wilson JF, Marinaki A, Riches PL, Shu X, Janicijevic B, Smolej-Narancic N, Gorgoni B, Morgan J, Campbell S, Biloglav Z, Barac-Lauc L, Pericic M, Klaric IM, Zgaga L, Skaric-Juric T, Wild SH, Richardson WA, Hohenstein P, Kimber CH, Tenesa A, Donnelly LA, Fairbanks LD, Aringer M, McKeigue PM, Ralston SH, Morris AD, Rudan P, Hastie ND, Campbell H, Wright AF. SLC2A9 is a newly identified urate transporter influencing serum urate concentration, urate excretion and gout. Nat Genet. 2008;40:437–442. doi: 10.1038/ng.106. [DOI] [PubMed] [Google Scholar]
- 62.Voruganti VS, Nath SD, Cole SA, Thameem F, Jowett JB, Bauer R, MacCluer JW, Blangero J, Comuzzie AG, Abboud HE, Arar NH. Genetics of Variation in Serum Uric Acid and Cardiovascular Risk Factors in Mexican-Americans. J Clin Endocrinol Metab. 2009;94:632–638. doi: 10.1210/jc.2008-0682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wallace C, Newhouse SJ, Braund P, Zhang F, Tobin M, Falchi M, Ahmadi K, Dobson RJ, Marçano AC, Hajat C, Burton P, Deloukas P, Brown M, Connell JM, Dominiczak A, Lathrop GM, Webster J, Farrall M, Spector T, Samani NJ, Caulfield MJ, Munroe PB. Genome-wide association study identifies genes for biomarkers of cardiovascular disease: serum urate and dyslipidemia. Am J Hum Genet. 2008;82:139–149. doi: 10.1016/j.ajhg.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wakida N, Tuyen DG, Adachi M, Miyoshi T, Nonoguchi H, Oka T, Ueda O, Tazawa M, Kurihara S, Yoneta Y, Shimada H, Oda T, Kikuchi Y, Matsuo H, Hosoyamada M, Endou H, Otagiri M, Tomita K, Kitamura K. Mutations in human urate transporter 1 gene in presecretory reabsorption defect type of familial renal hypouricemia. J Clin Endocrinol Metab. 2005;90:2169–2174. doi: 10.1210/jc.2004-1111. [DOI] [PubMed] [Google Scholar]
- 65.Wannamethee SG. Serum uric acid and risk of coronary heart disease. Curr Pharm Des. 2005;11:4125–4132. doi: 10.2174/138161205774913200. [DOI] [PubMed] [Google Scholar]
- 66.Wilk JB, Djousse L, Borecki I, Atwood LD, Hunt SC, Rich SS, Eckfeldt JH, Arnett Dk, Rao DC, Myers RH. Segregation analysis of serum uric acid in the NHLBI Family Heart Study. Hum Genet. 2000;106:355–359. doi: 10.1007/s004390000243. [DOI] [PubMed] [Google Scholar]
- 67.Yang Q, Guo CY, Cupples A, Levy D, Wilson PWF, Fox CS. Genome-wide search for genes affecting serum uric acid levels: the Framingham Heart Study. Metab Clin Exp. 2005:1435–1441. doi: 10.1016/j.metabol.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 68.Yoo TW, Sang KC, Shin HS, Kim BJ, Kim NS, Kang JH, Lee MH, Park JR, Kim H, Rhee EJ, Lee WY, Kim SW, Ryu SH, Keum DG. Relationship between serum uric acid concentration and insulin resistance and metabolic syndrome. Circ J. 2005;69:928–933. doi: 10.1253/circj.69.928. [DOI] [PubMed] [Google Scholar]