Abstract
Toll-like receptors (TLRs) recognize conserved molecular patterns in invading pathogens and trigger innate immune responses. TLR3 recognizes dsRNA, a molecular signature of most viruses via its ectodomain (ECD). The TLR3-ECD structure consists of a 23 turn coil bent into the shape of a horseshoe with specialized domains capping the N and C terminal ends of the coil. TLR3-ECDs bind as dimeric units to dsRNA oligonucleotides of at least 45 bp in length, the minimal length required for signal transduction. X-ray analysis has shown that each TLR3-ECD of a dimer binds dsRNA at two sites located at opposite ends of the TLR3 “horseshoe” on the one lateral face that lacks N-linked glycans. Intermolecular contacts between the C-terminal domains of two TLR3-ECDs stabilizes the dimer and positions the C-terminal residues within 20–25Å of each other, which is thought to be essential for transducing a signal across the plasma membrane in intact TLR3 molecules. Interestingly, in TLRs 1, 2 and 4, which bind lipid ligands using very different interactions from TLR3, the ligands nevertheless promote the formation of a dimer in which the same two lateral surfaces as in the TLR3-ECD:dsRNA complex face each other, bringing their C-termini in close proximity. Thus, a pattern is emerging in which pathogen-derived substances bind to TLR-ECDs, thereby promoting the formation of a dimer in which the glycan-free ligand-binding surfaces face each other and the two C-termini are brought in close proximity for signal transduction.
Keywords: Toll-like receptor, innate immunity, inflammation, leucine-rich repeat, dsRNA, Pattern recognition receptor
1. Introduction
Higher organisms rely on their innate immune system to block the growth and dissemination of pathogens at early stages of infection. The recognition of pathogens by the innate immune system is mediated by receptors that are encoded in the genome and selected during evolution to sense pathogen-associated molecular patterns (PAMPs) that cannot be altered to escape detection because they are essential for pathogen survival [11]. Bacterial membrane or cell wall components such as lipopolysaccharide, lipoteichoic acid, and peptidoglycan are well-known targets of pattern recognition receptors (PRRs) [17]. In addition, some forms of RNA and DNA from pathogens exhibit immutable features that distinguish them from nucleic acids of higher organisms. For example, dsRNA, is a common intermediate of viral replication and a potent indicator of infection. As such, it is recognized by several PRRs of the innate system, including Toll-like receptor 3 (TLR3) and a number of cytoplasmic sensors [28]. TLR3 is normally located in acidic endosomes where its luminal ectodomain (ECD) encounters dsRNA. The interaction of dsRNA with the TLR3-ECD leads to receptor dimerization and to the recruitment of the adapter molecule, TRIF, to the cytoplasmic domain of TLR3, known as a TIR (Toll-IL-1R-Resistance) domain due to its homology with the signaling domains of the IL-1 receptor and plant resistance proteins [26]. TRIF initiates signaling pathways that activate downstream transcription factors IRF3, AP-1 and NF-κB which in turn trigger the expression and secretion of type I interferons, inflammatory cytokines, and chemokines, as well as the maturation of dendritic cells, a key event in the generation of acquired immunity [13]. How TLR3 specifically recognizes dsRNA and initiates signaling is the subject of this review.
2. The Structure of the TLR3 Ectodomain
TLRs are type-I receptors with ligand-binding ECDs linked by a single transmembrane helix to cytoplasmic TIR signaling domains. The ligand binding domains of all TLRs consist of tandem copies (19–25 in the ten human TLRs) of the leucine-rich repeat (LRR) motif, capped by specialized N- and C- terminal sequences known as LRR-NT and LRR-CT domains, respectively. The consensus LRR sequence found in TLRs is xLxxLxLxxNxLxxLxxxxFxxLx, where leucine residues may be replaced by other hydrophobic amino acids. Based upon the structures of other LRR proteins of this type, it was predicted that all TLR-ECDs would resemble a long solenoid bent into the shape of a horseshoe, each turn of the solenoid corresponding to a single LRR sequence [3]. This horseshoe structure has now been observed in the crystal structures of several TLR-ECDs, the first reported being the TLR3-ECD structure [2,4,12,15,21].
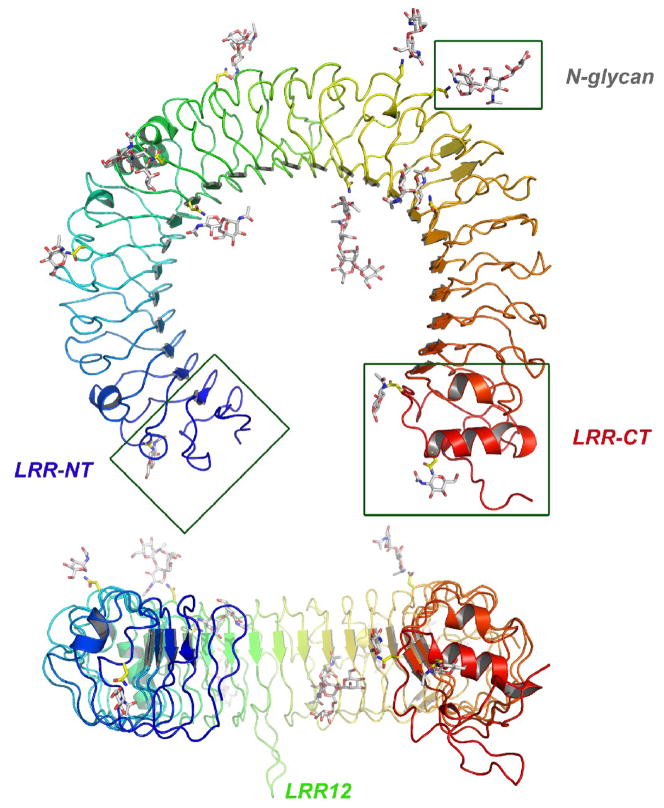
The TLR3-ECD horseshoe is composed of 23 LRRs and has an inner diameter of approximately 42 Å, an outer diameter of 90 Å, and a thickness of 35 Å. In common with all LRR structures, the consensus leucine side chains in the LRRs of the TLR3-ECD point towards the interior of the molecule and form a hydrophobic core, and the first eight residues of each LRR motif contribute a beta strand to an extended parallel beta-sheet that forms the concave, inner surface of the molecule (Fig. 1). The LRR-NT motif of the TLR3-ECD contains a hairpin loop stabilized by a single disulfide bond. The larger, LRR-CT domain is more globular in structure, and contains an internal α-helix and two disulfide bonds, which have been shown to be necessary for TLR3 function by mutational analysis [24]. The TLR3 LRR-CT has a similar fold to the C-terminal motifs of many other LRR proteins, for example all other TLRs, the variable lymphocyte receptors in agnathan vertebrates [7,14], glycoprotein 1B alpha [10] and Nogo receptor [8].
Fig. 1.
Structure of the TLR3-ECD. The TLR3-ECD consists of 23 LRRs that form a horseshoe-like solenoid with two capping motifs. The molecule has a surprisingly flat profile. There are 11 visible glycosylation sites.
Most LRR structures deviate from planarity, due to the inherent twist of the β-sheets that form their concave surfaces. By contrast, the TRL3-ECD horseshoe is flat, presumably because irregularities in the loops that connect the β strands in LRRs 8, 12, 14, 18 and 20 compensate for the β-sheet twist. These irregularities produce short alpha helices on the convex side of the horseshoe, and two large, conserved loops that protrude from the lateral and convex faces of LRRs 12 and 20, respectively. As we will discuss later, the flatness of the TLR3 horseshoe facilitates ligand binding and signaling.
The TLR3-ECD is highly glycosylated, with 15 predicted N-glycosylation sites, of which 11 are visible in the crystal structure. The glycan contributes 15 kDa to the total mass of 95 kDa in the TLR-ECD produced in insect cells, and this would be even greater in mammalian cells which introduce additional GlcNAc, GalNAc and sialyl rings to N-linked glycan moeities. The carbohydrates are distributed on the concave, convex, and on one lateral surface (at the N-terminal side of the β-sheet) of the TLR3-ECD. By contrast the opposite lateral surface completely lacks glycosylation and offers a large surface area for interaction with other molecules. The presence of glycan on the concave surface of TLR3 (and other TLRs), which would block access of ligand to this surface is surprising, since in most non-TLR LRR proteins, ligand binding occurs on the concave surface [9,10,16,25].
3. The interaction of soluble TLR3-ECD with dsRNA
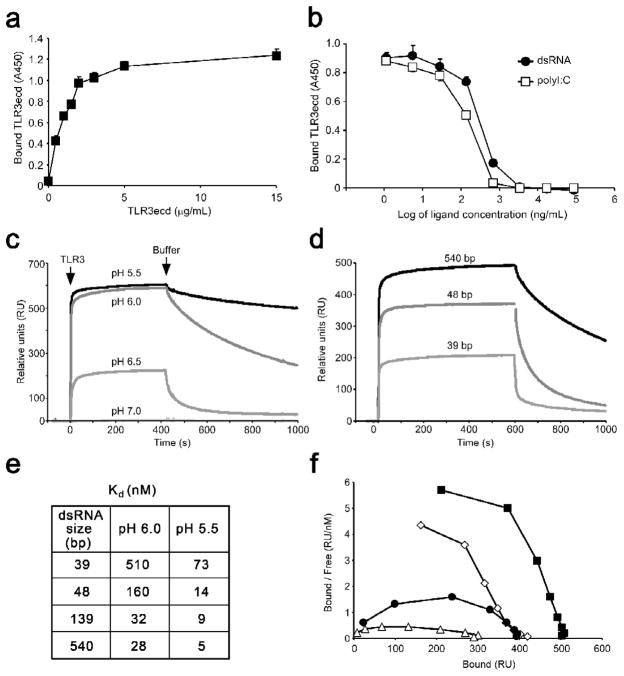
Studies using purified TLR3-ECD protein and homogenous dsRNA oligonucleotides of varying length have led to a detailed understanding of the interaction of TLR3 with its ligand. Binding of TLR3-ECD to immobilized dsRNA is saturable, reversible and can be inhibited by free dsRNA or the dsRNA analogue, p(I:C) (Fig. 2a,b), but not by dsDNA, ssRNA or ssDNA. Thus, TLR3 has a defined binding site that is specific for dsRNA but that does not distinguish between base sequences of dsRNA ligands [18]. TLR3 does not bind dsRNA at neutral pH [5,18], but strong binding does occur below pH 6.5 (Fig. 2c,e), which is the pH commonly found in the endocytic compartments that contain the TLR3-ECD. Moreover, the minimum length dsRNA oligonucleotide capable of binding to TLR3-ECD is unexpectedly long at 40–45 bp, corresponding to >110 Å for the A type helix adopted by dsRNA (Fig. 2d). Under optimal conditions the affinity of TLR3-ECD for dsRNA is reasonably high (~10 nM), but decreases sharply as the pH increases above pH 6, and the dsRNA length decreases below approximately 45 bp (Fig 2e). Interestingly, binding is positively cooperative (Fig 2f), which implies that the affinity of TLR3-ECD for dsRNA increases as the extent of binding increases.
Fig. 2.
The interaction of soluble TLR3-ECD with dsRNA. (a,b) ELISA showing the binding of TLR3-ECD to immobilized 540 bp dsRNA. (a) binding at increasing TLR3-ECD concentrations, (b) inhibition of binding by free 540 bp dsRNA or polyI:C. (c) Binding and dissociation of TLR3-ECD to immobilized 540 bp dsRNA at indicated pH values, using surface plasmon resonance (SPR). The TLR3-ECD was added at the arrow labeled TLR3, and was replaced by medium at the arrow Buffer. (d) TLR3-ECD binding at pH 6.0 to increasing lengths of immobilized dsRNA measured by SPR. (e) Dissociation constants of TLR3-ECD binding to varying lengths of dsRNA at pH 6.0 and 5.5, calculated from SPR data. (f) Scatchard plot of binding data at pH 6.0. Downward curvature indicates positive cooperativity. Data are taken from [18].
Gel filtration and equilibrium ultracentrifugation analyses at pH 6.0 and 5.5 showed that TLR3-ECD is monomeric in solution, but binds as dimeric units to dsRNA; two TLR3-ECD molecules bind to one 48 bp dsRNA oligonucleotide, four bind to 90 bp dsRNA, and six to 139 bp. Importantly, studies using different length dsRNA oligonucleotides to activate TLR3 in NF-κB reporter cells showed that the shortest oligonucleotide that activates TLR3 is 40–48 bp in length, which, in solution, binds no more than one TLR3 dimer. Thus, a complex consisting of two TLR3 molecules bound to a ~45 bp dsRNA oligonucleotide is the minimum TLR3 activation signal. Interestingly, in other cell lines which express lower amounts of TLR3, larger oligonucleotides were required for activation. This observation suggested that cells may be able to regulate the length or concentration of dsRNA that they recognize by altering either TLR3 expression levels, or the location of TLR3 within the cell.
4. Structure of the Signaling Complex
4.1. The overall structure
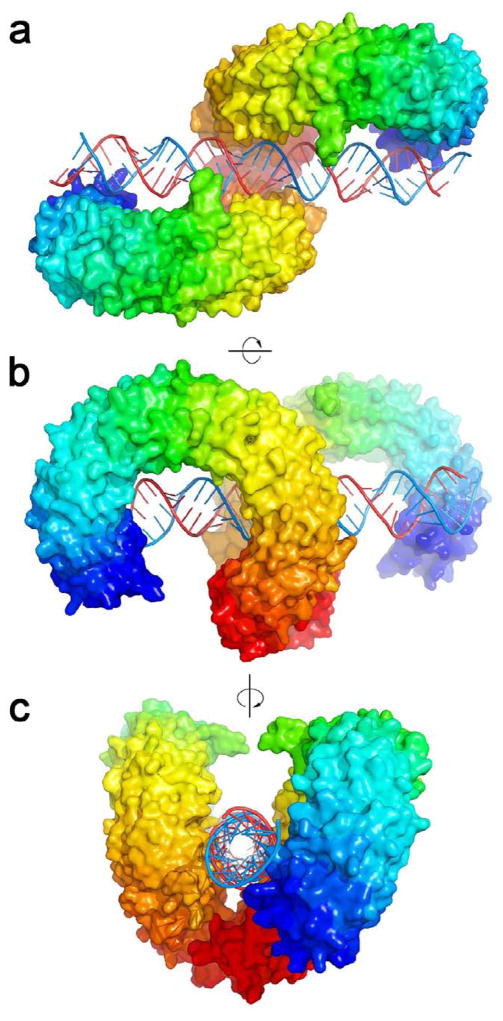
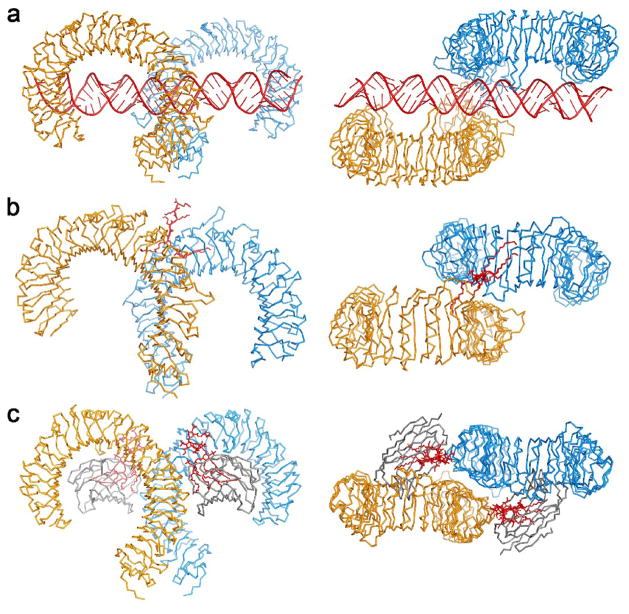
The crystal structure of mouse TLR3-ECD bound at pH5.5 to 46 bp dsRNA was recently published [19]. In the structure, the dsRNA molecule interacts with the glycan-free surfaces of two TLR3-ECDs, forming a complex in which the two TLR3-ECDs are related by two-fold symmetry (Fig. 3a). Viewed from the side (Fig. 3b) the two TLR3-ECDs resemble the letter “m” and the dsRNA crosses the full width of the “m”. At the C-terminal ends, the two TLR3-ECD molecules interact with each other homotypically to form the dimer (Fig. 3c). The dsRNA in the complex is a typical A-form RNA double-helix spanning over 120 Å. Superposing the α-carbons of the two TLR3-ECD molecules in the complex with the α-carbons of the apo-TLR3-ECD, indicated that the TLR3-ECD does not undergo a ligand-induced conformational change.
Fig. 3.
Structure of the TLR3-ECD:dsRNA signaling complex. (a–c) 3 views of the TLR3-ECD dimer bound to a 46-mer dsRNA. (a,b) The glycan-free face of each TLR3 interacts with the dsRNA. There are two binding sites for each TLR3 molecule: C-terminal and N-terminal. (c) Looking down the axis of the dsRNA shows that dsRNA is linear and the C-termini of TLR3-ECD interact. The N-terminal region is colored purple, and the C-terminal, red.
4.2. TLR3-dsRNA interactions
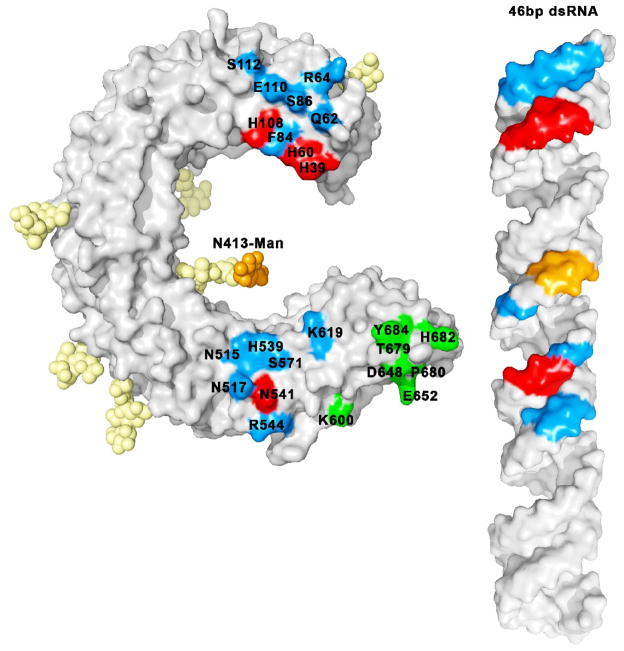
In the complex, both TLR3-ECDs interact with dsRNA at two sites separated by approximately 55Å, each on the glycan-free lateral surface (Fig. 3a,b). These sites interact only with the ribose-phosphate backbone of the dsRNA, which explains why ligand binding by TLR3 is independent of the base sequence. The first binding site is located toward the C-terminal end of the TLR3-ECD and includes residues in LRR19 and LRR20 (Fig. 4). The second binding site is located near the N-terminus, and includes residues in LRR-NT, LRR1, LRR2, and LRR3 (Fig. 4), also on the glycan-free surface. Interestingly a large N-glycosyl moiety linked to Asn-413 reaches out from the concave surface and makes contact with the dsRNA helix, but the significance of this interaction is unclear.
Fig. 4.
Residues involved in binding. Essential residues (which lead to loss of function when mutated to alanine) (in red) and interacting non-essential residues (which make contact with dsRNA in the crystal structure but retain function when mutated)(in blue) are mapped on the surface of the ECD with the corresponding colors on the dsRNA. Carbohydrate-ligand interactions are highlighted in orange. Some residues in the 600–684 range (in green) are involved in the homotypic protein-protein interactions between the two TLR-ECDs. The N-terminal ligand binding site is formed by labeled residues from the 39–112 range, and the C-terminal site by labeled residues from 515–619.
The total area buried by the dsRNA:TLR3-ECD interaction is 1103 Å2, only 4.4% of the TLR3 surface, and the great majority of interactions are hydrophilic, involving hydrogen bonds and salt bridges. Notably, several imidazole side chains of histidine residues appear to interact with phosphate groups on the dsRNA backbone. This could explain the pH-dependency of dsRNA binding, since these side chains would be protonated and therefore able to form ionic interactions with the RNA phosphates only below pH 6.5 (the pK of imidazole side chains).
The structure also suggests two reasons for the inability of TLR3 to interact with dsDNA. First, dsDNA and dsRNA assume different helical structures (B and A forms, respectively) and it is unlikely that the dsDNA B helix would be structurally compatible with the N and C terminal binding sites on the TLR3-ECD horseshoe. Secondly, there are a significant number of hydrogen bonds joining TLR3-ECD to 2′-hydroxyl groups of dsRNA, which are missing in DNA.
4.3. Homotypic interactions
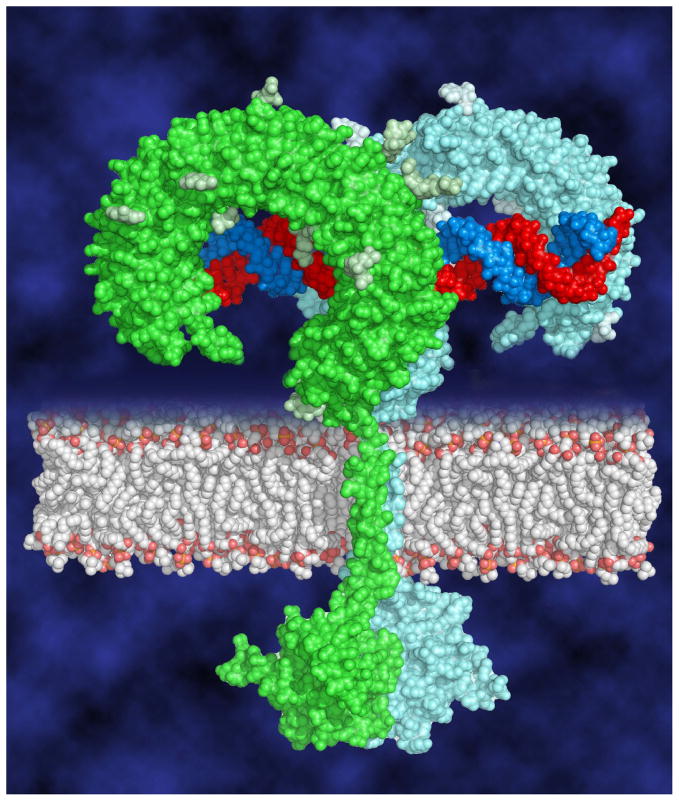
In the complex, the interaction between the two LRR-CT domains is the only force that directly drives two TLR3-ECD molecules to dimerize (Fig. 3c). This homotypic binding site contains amino acid side chains from both LRR-CTs in the dimer (Fig. 4), related by two-fold symmetry, and the intermolecular contacts include mainly hydrogen bonds and electrostatic interactions. Importantly, this interaction brings the two C-terminal residues within approximately 25Å of each other. In an intact TLR3:dsRNA complex in a cell, the close proximity of the two LRR-CT domains on the luminal side of an endosome would presumably bring the two TIR domains together on the cytoplasmic side, in a position capable of recruiting TRIF and initiating signaling (Fig. 5).
Fig. 5.
Model of the full-length signaling complex. Two TLR3 ECDs associate on the dsRNA ligand bringing the cytoplasmic TIR domains together to initiate downstream signaling.
5. Mutation analysis
5.1 Identification of essential residues
Mutation studies from several laboratories [1,6,22,24] have identified amino acid residues in the three TLR3-ECD binding site regions that are essential for function. Before the crystal structure of the TLR3-ECD:dsRNA complex was solved, two loss of function mutations, H539E and N541A located a binding region at LRR20 on the glycan-free surface of the TLR3-ECD [1,24]. It is now known that these amino acids lie in the C-terminal ligand binding site of TLR3, and that the Asn-541 amide side chain likely interacts with a 2′ hydroxyl on the dsRNA backbone. Removal of the amide group by mutation to alanine blocks activity, indicating that this interaction is necessary for forming an active TLR3:dsRNA complex. The imidazole side chain of His-539 interacts with a backbone phosphate group on the dsRNA, but this interaction is not necessary for complex formation since mutation to alanine does not significantly impair activity. However, the loss of activity in the H539E mutation indicates that repulsive forces from the negative charge of the glutamic acid destabilizes the complex and confirms that the His-539 side chain is in close proximity to a phosphate group. Mutation of other residues that make contact with the dsRNA in this region, including Arg-544, Asn-515, and Asn-517, have less pronounced effects on activity [1,24], suggesting that they contribute less stabilization energy to the complex than does Asn-541. Nevertheless, all amino acid residues that make contact with dsRNA at this site, except for Ser-571 (Fig 4), are totally conserved in 18 vertebrate orthologs, dating back to bony fish [19].
At the N-terminal binding site, three consecutive phosphates on the dsRNA backbone form salt bridges with the imidazole groups of His-39, His-60 and His-108, the only conserved amino acid residues at this site. Replacing His-39 and His-60 with Ala (or Glu) abrogates TLR3 activity, indicating that the imidazole groups of these residues are essential for function [6,19,22]. His-108 and two other residues that interact with the dsRNA backbone, Gln-62 and Arg-64, retain activity when mutated to Ala [22], and are therefore not essential for binding. Because His-39 and His-108 are the only histidine residues required for TLR3 function, it is likely that these residues are responsible for the pH dependence of dsRNA binding (Fig 2c). Interestingly, replacing His-39 with Arg produces an active molecule, providing further evidence that a positive charge at residue 39 is required for binding [22].
Since single TLR3-ECD molecules do not bind stably to dsRNA, it is likely that the amino acid residues in the LRR-CT region that are involved in dimer formation are required for the formation of a stable, active dsRNA:TLR3 complex. Although no mutations in this region have been reported in the literature, preliminary results indicate that several residues at this site are required for function (Wang, Y., unpublished data). An important conclusion from the mutation studies is that all three sites of interaction, the N- and C-terminal dsRNA binding sites and the C-terminal dimerization site, are required for formation of a functional TLR3:dsRNA complex. Thus TLR3-ECD binds dsRNA through the cooperation of three widely spaced, relatively weak binding sites, no two of which provide sufficient energy for stable complex formation.
5.2 Importance of glycosylation
TLR3-ECD contains fifteen N-linked glycosylation sites, all of which have been mutated individually or in pairs [1,5,27]. Most mutations have no significant effect upon TLR3 activity or expression. However mutation of Asn-247 (to Ala or Asp) consistently results in partial or total loss of activity, without affecting expression. Since the glycan linked to this site does not play a role in stabilizing the complex in the crystal structure, it is likely that this glycan is involved in a cellular function, such as trafficking to endosomes or protein folding. The glycan linked to Asn-413 is the only carbohydrate that makes contact with the dsRNA ligand in the crystal structure. One group [27] found that mutation of Asn-413 causes a significant decrease in responsiveness to dsRNA while a second [1] found no decrease in function. Since activation studies are at best qualitative in nature, it will be important to resolve this issue and to determine whether mutation of Asn-413 results in a decrease in binding of dsRNA to TLR3.
5.3 SNPs - Natural Mutations
Four single nucleotide polymorphisms (SNPs) result in amino acid substitutions in the TLR3-ECD, including N284I, Y307D, L412F and P554S. Tyr-307 is a surface residue and the Y307D mutation is fully active [23]. Asn-284 and Leu-412 are LRR consensus residues that are located in the hydrophobic core of TLR3-ECD. Mutation of these residues results in total (N284I) or partial (L412F) loss of activity, presumably by destabilizing the LRR architecture [23]. Interestingly, P554S is an autosomal dominant mutation that strongly correlates with herpes simplex encephalitis, a rare infection caused by the herpes simplex virus (HSV) [29]. Pro-554 is located immediately after the loop protruding from LRR20 and is distant from the dsRNA binding site. It is therefore unlikely that this mutation would affect the binding of dsRNA to the TLR3-ECD, although this has not yet been tested directly. Transfection studies showed that the P554S mutant is both inactive and acts as a dominant negative. Western analysis of the P554S mutated protein in fibroblasts indicated that the mutant protein is cleaved to a lower molecular weight form compared to wild-type hTLR3. The authors proposed that the P554S mutation renders TLR3 susceptible to proteolysis, which in turn generates and inactive, dominant negative form of the receptor.
6. Comparison with other TLR signaling complexes
Crystallographic investigations have been carried out on two other TLR/PAMP complexes.
6.1. TLR1/TLR2 complexed with a triacylated lipopeptide
In this crystal structure of the complex formed between TLR1 and TLR2 bound to the triacylated hexapeptide Pam3CSK4, the binding is mediated by the three lipid chains of the lipopeptide [12](Fig. 6b). The two ester-bound chains of the ligand insert into a pocket in TLR2 while the amide bound lipid chain inserts into a hydrophobic channel in TLR1. In addition, the two TLR-ECDs interact by the formation of an extensive network of hydrogen bonds and hydrophobic interactions. The binding of the lipopeptide and the interactions between the two ectodomain surfaces result in bringing the two C termini of the ectodomains near to each other which would permit dimerization of the cytoplasmic TIR domains.
Fig. 6.
Crystal structures of TLR signaling complexes. (a) side and top views of the TLR3:dsRNA homodimer [19] (b) TLR1/TLR2:Pam3CSK4 heterodimer [12] and (c) TLR4/MD2:LPS homodimer [21]. The ligands are shown in red.
It is not clear whether the two C-terminal capping domains can interact with each other since both C-termini have been replaced by the C-terminal domains of hagfish VLRs. However, it appears that the hydrogen bonds and hydrophobic interactions between the two surfaces would result in rather a rigid structure in which the two C-termini would be too distant for the kind of interaction observed in the TLR3-ECD:dsRNA complex.
6.2. TLR4, MD-2 and LPS
The signaling by TLR4 in the presence of lipopolysaccharide requires the intervention of at least two accessory proteins, MD-2, which binds to the TLR4 and CD14 which delivers the LPS to the MD-2. Ohto et al. in 2007 have determined the structure of MD-2 bound to an antagonist, lipid IVa which has four lipid strands compared to the endotoxic Lipid A which has six [20]. The MD-2 structure consists of two sheets, one with three beta strands and the other with six. Between the sheets is a very large hydrophobic cavity into which the four lipid strands of Lipid IVa are buried. It was observed that the MD-2 cavity likely could not accommodate more than four lipid chains (e.g. lipid A) without conformational changes. The additional lipid A acyl chains might be displaced from the MD-2 pocket and might induce the reported oligomerization of TLR4.
The crystal structure of mouse TLR4-ECD with bound MD-2 containing eritoran, a lipid antagonist with four lipid chains was also reported [15]. The MD-2 molecule binds to the top surface of the TLR4-ECD, contacting residues over a broad area from LRR2 to LRR10. The structures of the mTLR4-ECD and several chimaeric fragments of human TLR4 capped with hagfish VLR domains were also determined. The TLR4-ECD structure is much less uniform than the TLR3-ECD structure and it can be divided into three domains, N-terminal, central and C-terminal, with different degrees of twist and curvature.
Recently, the 3.1 Å resolution structure of the dimer formed by TLR4, MD-2 and the Ra chemotype of lipopolysaccharide from E.coli was determined [21] (Fig. 6c). The LPS has six lipid chains, five of which bind inside the MD-2 cavity, with the sixth chain on the outside making hydrophobic contacts with the other TLR4-ECD. In addition, there are hydrogen bonds between an LPS phosphate group and a hydroxyl group with this ECD. Finally, there are several hydrogen bonds that link the MD-2 to the opposite ECD. The net effect of all these interactions is bringing the two ECDs together in a manner similar to that seen in the TLR3-ECD:dsRNA complex and in the TLR1:lipopeptide:TLR2 complex, with the C termini within reasonable proximity to facilitate dimerization of the cytoplasmic TIR domains (Fig. 6).
The signaling complex structures of the TLRs with their respective ligands illustrate the tremendous diversity displayed in recognizing these very different PAMPS. However, in all three cases the complex contains two TLRs brought together in such a way that the cytoplasmic TIR domains will be close enough to dimerize, thus initiating the downstream signaling cascade (Fig. 6). It is worth noting that the three ligands do not make use of the concave beta-sheet surface for binding that is customarily used by the majority of the other leucine-rich repeat proteins.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Bell JK, Askins J, Hall PR, Davies DR, Segal DM. The dsRNA binding site of human Toll-like receptor 3. Proc Natl Acad Sci U S A. 2006;103:8792. doi: 10.1073/pnas.0603245103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bell JK, Botos I, Hall PR, Askins J, Shiloach J, Segal DM, Davies DR. The molecular structure of the Toll-like receptor 3 ligand-binding domain. Proc Natl Acad Sci U S A. 2005;102:10976. doi: 10.1073/pnas.0505077102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell JK, Mullen GE, Leifer CA, Mazzoni A, Davies DR, Segal DM. Leucine-rich repeats and pathogen recognition in Toll-like receptors. Trends Immunol. 2003;24:528. doi: 10.1016/s1471-4906(03)00242-4. [DOI] [PubMed] [Google Scholar]
- 4.Choe J, Kelker MS, Wilson IA. Crystal structure of human toll-like receptor 3 (TLR3) ectodomain. Science. 2005;309:581. doi: 10.1126/science.1115253. [DOI] [PubMed] [Google Scholar]
- 5.de Bouteiller O, Merck E, Hasan UA, Hubac S, Benguigui B, Trinchieri G, Bates EE, Caux C. Recognition of double-stranded RNA by human toll-like receptor 3 and downstream receptor signaling requires multimerization and an acidic pH. J Biol Chem. 2005;280:38133. doi: 10.1074/jbc.M507163200. [DOI] [PubMed] [Google Scholar]
- 6.Fukuda K, Watanabe T, Tokisue T, Tsujita T, Nishikawa S, Hasegawa T, Seya T, Matsumoto M. Modulation of double-stranded RNA recognition by the N-terminal histidine-rich region of the human toll-like receptor 3. J Biol Chem. 2008;283:22787. doi: 10.1074/jbc.M802284200. [DOI] [PubMed] [Google Scholar]
- 7.Han BW, Herrin BR, Cooper MD, Wilson IA. Antigen recognition by variable lymphocyte receptors. Science. 2008;321:1834. doi: 10.1126/science.1162484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He XL, Bazan JF, McDermott G, Park JB, Wang K, Tessier-Lavigne M, He Z, Garcia KC. Structure of the Nogo receptor ectodomain: a recognition module implicated in myelin inhibition. Neuron. 2003;38:177. doi: 10.1016/s0896-6273(03)00232-0. [DOI] [PubMed] [Google Scholar]
- 9.Hillig RC, Renault L, Vetter IR, Drell T, Wittinghofer A, Becker J. The crystal structure of rna1p: a new fold for a GTPase-activating protein. Mol Cell. 1999;3:781. doi: 10.1016/s1097-2765(01)80010-1. [DOI] [PubMed] [Google Scholar]
- 10.Huizinga EG, Tsuji S, Romijn RA, Schiphorst ME, de Groot PG, Sixma JJ, Gros P. Structures of glycoprotein Ibalpha and its complex with von Willebrand factor A1 domain. Science. 2002;297:1176. doi: 10.1126/science.107355. [DOI] [PubMed] [Google Scholar]
- 11.Janeway CA, Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 12.Jin MS, Kim SE, Heo JY, Lee ME, Kim HM, Paik SG, Lee H, Lee JO. Crystal Structure of the TLR1-TLR2 Heterodimer Induced by Binding of a Tri-Acylated Lipopeptide. Cell. 2007;130:1071. doi: 10.1016/j.cell.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 13.Kawai T, Akira S. TLR signaling. Cell Death Differ. 2006;13:816. doi: 10.1038/sj.cdd.4401850. [DOI] [PubMed] [Google Scholar]
- 14.Kim HM, Oh SC, Lim KJ, Kasamatsu J, Heo JY, Park BS, Lee H, Yoo OJ, Kasahara M, Lee JO. Structural diversity of the hagfish variable lymphocyte receptors. J Biol Chem. 2007;282:6726. doi: 10.1074/jbc.M608471200. [DOI] [PubMed] [Google Scholar]
- 15.Kim HM, Park BS, Kim JI, Kim SE, Lee J, Oh SC, Enkhbayar P, Matsushima N, Lee H, Yoo OJ, Lee JO. Crystal structure of the TLR4-MD-2 complex with bound endotoxin antagonist Eritoran. Cell. 2007;130:906. doi: 10.1016/j.cell.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 16.Kobe B, Deisenhofer J. The leucine-rich repeat: a versatile binding motif. Trends Biochem Sci. 1994;19:415. doi: 10.1016/0968-0004(94)90090-6. [DOI] [PubMed] [Google Scholar]
- 17.Kumar H, Kawai T, Akira S. Pathogen recognition in the innate immune response. Biochem J. 2009;420:1. doi: 10.1042/BJ20090272. [DOI] [PubMed] [Google Scholar]
- 18.Leonard JN, Ghirlando R, Askins J, Bell JK, Margulies DH, Davies DR, Segal DM. The TLR3 signaling complex forms by cooperative receptor dimerization. Proc Natl Acad Sci U S A. 2008;105:258. doi: 10.1073/pnas.0710779105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu L, Botos I, Wang Y, Leonard JN, Shiloach J, Segal DM, Davies DR. Structural basis of toll-like receptor 3 signaling with double-stranded RNA. Science. 2008;320:379. doi: 10.1126/science.1155406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohto U, Fukase K, Miyake K, Satow Y. Crystal structures of human MD-2 and its complex with antiendotoxic lipid IVa. Science. 2007;316:1632. doi: 10.1126/science.1139111. [DOI] [PubMed] [Google Scholar]
- 21.Park BS, Song DH, Lee H, Lee JO. The structural basis of lipopolysaccharide recognition by the TLR4–MD-2 complex. Nature. 2009 doi: 10.1038/nature07830. advance online publication. [DOI] [PubMed] [Google Scholar]
- 22.Pirher N, Ivicak K, Pohar J, Bencina M, Jerala R. A second binding site for double-stranded RNA in TLR3 and consequences for interferon activation. Nat Struct Mol Biol. 2008;15:761. doi: 10.1038/nsmb.1453. [DOI] [PubMed] [Google Scholar]
- 23.Ranjith-Kumar CT, Miller W, Sun J, Xiong J, Santos J, Yarbrough I, Lamb RJ, Mills J, Duffy KE, Hoose S, Cunningham M, Holzenburg A, Mbow ML, Sarisky RT, Kao CC. Effects of single nucleotide polymorphisms on Toll-like receptor 3 activity and expression in cultured cells. J Biol Chem. 2007;282:17696. doi: 10.1074/jbc.M700209200. [DOI] [PubMed] [Google Scholar]
- 24.Ranjith-Kumar CT, Miller W, Xiong J, Russell WK, Lamb R, Santos J, Duffy KE, Cleveland L, Park M, Bhardwaj K, Wu Z, Russell DH, Sarisky RT, Mbow ML, Kao CC. Biochemical and functional analyses of the human Toll-like receptor 3 ectodomain. J Biol Chem. 2007;282:7668. doi: 10.1074/jbc.M610946200. [DOI] [PubMed] [Google Scholar]
- 25.Schubert WD, Urbanke C, Ziehm T, Beier V, Machner MP, Domann E, Wehland J, Chakraborty T, Heinz DW. Structure of internalin, a major invasion protein of Listeria monocytogenes, in complex with its human receptor E-cadherin. Cell. 2002;111:825. doi: 10.1016/s0092-8674(02)01136-4. [DOI] [PubMed] [Google Scholar]
- 26.Seya T, Matsumoto M, Ebihara T, Oshiumi H. Functional evolution of the TICAM-1 pathway for extrinsic RNA sensing. Immunol Rev. 2009;227:44. doi: 10.1111/j.1600-065X.2008.00723.x. [DOI] [PubMed] [Google Scholar]
- 27.Sun J, Duffy KE, Ranjith-Kumar CT, Xiong J, Lamb RJ, Santos J, Masarapu H, Cunningham M, Holzenburg A, Sarisky RT, Mbow ML, Kao C. Structural and functional analyses of the human Toll-like receptor 3. Role of glycosylation. J Biol Chem. 2006;281:11144. doi: 10.1074/jbc.M510442200. [DOI] [PubMed] [Google Scholar]
- 28.Takeuchi O, Akira S. Innate immunity to virus infection. Immunol Rev. 2009;227:75. doi: 10.1111/j.1600-065X.2008.00737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang SY, Jouanguy E, Ugolini S, Smahi A, Elain G, Romero P, Segal D, Sancho-Shimizu V, Lorenzo L, Puel A, Picard C, Chapgier A, Plancoulaine S, Titeux M, Cognet C, von BH, Ku CL, Casrouge A, Zhang XX, Barreiro L, Leonard J, Hamilton C, Lebon P, Heron B, Vallee L, Quintana-Murci L, Hovnanian A, Rozenberg F, Vivier E, Geissmann F, Tardieu M, Abel L, Casanova JL. TLR3 deficiency in patients with herpes simplex encephalitis. Science. 2007;317:1522. doi: 10.1126/science.1139522. [DOI] [PubMed] [Google Scholar]