Abstract
The challenge in medical oncology has always been to identify compounds that will kill, or at least tame, cancer cells while leaving normal cells unscathed. Most chemotherapeutic agents in use today were selected primarily for their ability to kill rapidly dividing cancer cells grown in cell culture and in mice, with their selectivity determined empirically during subsequent animal and human testing. Unfortunately, most of the drugs developed in this way have relatively low therapeutic indices (low toxic dose relative to the therapeutic dose). Recent advances in genomics are leading to a more complete picture of the range of mutations, both driver and passenger, present in human cancers. Synthetic lethality provides a conceptual framework for using this information to arrive at drugs that will preferentially kill cancer cells relative to normal cells. It also provides a possible way to tackle 'undruggable' targets. Two genes are synthetically lethal if mutation of either gene alone is compatible with viability but simultaneous mutation of both genes leads to death. If one is a cancer-relevant gene, the task is to discover its synthetic lethal interactors, because targeting these would theoretically kill cancer cells mutant in the cancer-relevant gene while sparing cells with a normal copy of that gene. All cancer drugs in use today, including conventional cytotoxic agents and newer 'targeted' agents, target molecules that are present in both normal cells and cancer cells. Their therapeutic indices almost certainly relate to synthetic lethal interactions, even if those interactions are often poorly understood. Recent technical advances enable unbiased screens for synthetic lethal interactors to be undertaken in human cancer cells. These approaches will hopefully facilitate the discovery of safer, more efficacious anticancer drugs that exploit vulnerabilities that are unique to cancer cells by virtue of the mutations they have accrued during tumor progression.
Cancer drug discovery
It is not difficult to identify small organic molecules that will kill cancer cells. In fact, 0.1 to 1% of the molecules in a typical pharmaceutical compound library will kill cancer cells when tested at the concentrations used in high-throughput screens [1]. This leads to an embarrassment of riches because many pharmaceutical compound libraries contain millions of chemicals. The trick, however, is to find small organic molecules that will kill cancer cells while sparing normal cells. Unfortunately, the hits emerging from high-throughput screens for cytotoxic agents were historically prioritized using factors such as potency, ease of synthesis, drug-like characteristics, structural and mechanistic novelty, and intellectual property considerations [1]. Although these factors are potentially important, they do not necessarily address selectivity. Sadly, it is possible that small molecules capable of selectively killing cancer cells scored in the high-throughput cytotoxicity screens performed over the past 50 years, only to be discarded because they failed one or more of these other metrics. This thought is especially sobering when one considers the horrendous toxicity associated with most chemotherapeutic agents and their limited efficacy for most patients with advanced disease.
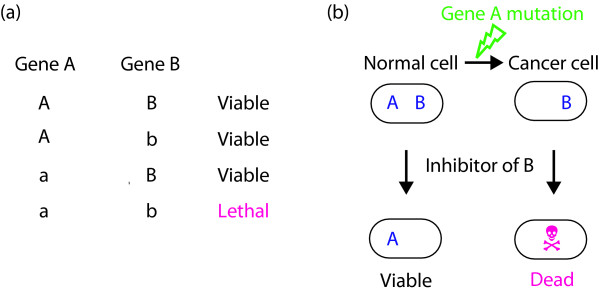
It is clear that cancer arises from the accumulation of genetic alterations in a susceptible cell. Fortunately, the mutations that are responsible for particular types of cancer are coming into view. This knowledge provides a foundation for discovering drugs that selectively kill cancer cells. In particular, it is almost certainly the case that some of the mutations within a given cancer cell will quantitatively or qualitatively alter the requirement of that cell for particular biochemical activities (or targets) [2]. This statement stems, in part, from studies of synthetic lethal interactions in model organisms, such as yeast and flies. Two genes are said to be 'synthetic lethal' if mutation in either gene alone is compatible with viability but simultaneous mutation of both genes leads to death [1,3-5] (Figure 1). Genome-wide studies in these model organisms suggest that synthetic lethal interactions are extremely common in biology [6-8]. Although synthetic lethal interactions are often thought of in terms of loss-of-function mutations, they can also be observed when one or both genes have sustained a gain-of-function mutation. This paradigm can be extended to include any situation in which the requirement for a particular gene in a cancer cell has been quantitatively or qualitatively altered by n non-allelic mutations, where n = 1 in the scenario outlined above. For example, mutations of two genes (such as simultaneous mutation of two tumor suppressor genes) might change the requirement for a third gene, and so on. Moreover, all the mutations in a cancer cell, whether contributing to the cancer phenotype (driver mutations) or not (passenger mutations), can potentially alter the cellular requirement for a particular target and hence contribute to selectivity [2,9].
Figure 1.
Synthetic lethality. (a) Table showing the effect of two mutants that are synthetically lethal. Lower case, mutant; upper case, wild-type. (b) The effect of mutations and inhibitors on a pair of synthetically lethal genes, A and B.
Exploiting synthetic lethal interactions to treat cancer cells is therefore very attractive insofar as it provides a conceptual framework for the development of drugs that will kill cancer cells (bearing the sensitizing mutation) while sparing normal cells (which do not; Figure 1). Moreover, it provides a framework for pharmacologically tackling targets that are not classically 'druggable'. For example, synthetic lethality theoretically provides an avenue for targeting cancer-causing loss-of-function mutations, such as mutations leading to the inactivation or loss of a particular tumor suppressor protein. The problem, however, is that synthetic lethal interactions, although common in biology, are difficult to predict a priori, especially given our current level of understanding of the molecular networks governing metazoan cells. Even once discovered, many synthetic lethal interactions are difficult to rationalize. For these reasons the study of synthetic lethal interactions has, until recently, been largely relegated to model organisms, such as bacteria and yeast, amenable to unbiased, genome-wide genetic screens. Unfortunately, many cancer-relevant genes, including tumor suppressor genes and oncogenes, are not conserved in these organisms.
Molecular pathway knowledge leads to synthetic lethal candidates
Nonetheless, a few synthetic lethal or 'synthetic sick' interactions (the latter refers to situations in which simultaneous mutation of two genes leads to a marked loss of fitness relative to mutation of either gene alone) involving cancer-relevant genes have been discovered using knowledge of particular molecular circuits. For example, many cancers have mutations that directly or indirectly inactivate the retinoblastoma tumor suppressor protein pRB, leading to hyperactivity of the E2F transcription factors. The E2F1 transcription factor can promote S-phase entry but can also induce apoptosis by p53-dependent and p53-independent pathways [10]. The timely neutralization of E2F1 activity in S-phase requires that it docks, via a peptidic sequence containing the core sequence Arg-x-Leu (RXL), with the substrate recognition pocket of Cyclin A [11-13]. Similar RXL motifs are present in additional proteins that physically interact with Cyclin A or Cyclin E, including other substrates and also p21-like cyclin-dependent kinase (CDK) inhibitors [14,15]. Several groups have shown that cancer cells, by virtue of high E2F1 activity, undergo apoptosis when treated with cell membrane-permeable versions of RXL-containing peptides whereas normal cells do not [16,17]. Unfortunately, it has not yet been possible to make non-peptidic, drug-like analogs of such RXL peptides. Loss of retinoblastoma protein (pRB), and consequent E2F1 deregulation, also seems to sensitize cells to drugs such as etoposide that lead to DNA damage after binding to topoisomerase II [18,19]. Loss of pRB increases S-phase entry and increases topoisomerase II levels. In addition, pRB may have a relatively direct role in the processing and repair of trapped topoisomerase-II-DNA complexes [20].
The c-Myc oncoprotein is a heterodimeric DNA-binding transcription factor. Unfortunately, such proteins, with the exception of the steroid hormone receptors, have not proven very tractable as drug targets. Quon and coworkers [21] showed, however, that overexpression of MYC sensitizes fibroblasts to agonists of the Trail death receptor DR5. They went on to show that c-Myc leads to increased DR5 expression and increased DR5-dependent signaling as a result of enhanced procaspase 8 autocatalytic activity [21]. Bishop and coworkers [22] found that MYC, among a panel of oncogenes tested, sensitized cells to undergo apoptosis when CDK1 is inhibited genetically or pharmacologically. Moreover, they showed that CDK1 inhibition leads to loss of the prosurvival protein survivin and that depletion of survivin selectively kills cells that overexpress MYC. Caron and colleagues [23] noted that CDK2 is often overexpressed in poor-risk neuroblastomas and that CDK2 is synthetic lethal with N-Myc amplification, which is a frequent genetic event in this disease. Genetic or pharmacological disruption of CDK2 led to p53-dependent apoptosis in N-Myc amplified neuroblastomas.
Many cancer cells show defects in the ability to sense and respond to DNA damage. This property, which can lead to a variety of genomic abnormalities, including point mutations, copy number changes and structural abnormalities such as translocations, seems to be a fertile area for synthetic lethal interactions. For example, many human tumors harbor mutations of the p53 tumor suppressor gene, which has an important role in the maintenance of genomic stability. Loss of p53 renders tumor cells dependent on signaling molecules such as ATM, CHEK2 and MK2 for survival in the face of chemotherapy-induced DNA damage [24,25]. This sensitivity seems to reflect loss of a p53-mediated checkpoint at the G1/S boundary, rendering cells more reliant on checkpoints operating later in the cell cycle. These findings were presaged by studies by Friend and coworkers [24], who noted that p53-/- mouse embryonic fibroblasts (MEFs) are more sensitive than their wild-type counterparts to combined treatment with ultraviolet radiation and pharmacological doses of caffeine, which acts as a checkpoint inhibitor.
In the most striking example to emerge from studies of this type, two groups [26,27] reasoned that cells defective for homologous recombination should be hypersensitive to loss of alternative, collateral DNA repair pathways, such as the base-excision repair pathway. Proteins encoded by the breast cancer genes BRCA1 and BRCA2 have important roles in the repair of double-strand breaks by homologous recombination, whereas poly(ADP-ribose) polymerase-1 (PARP1) is required for base-excision repair. Both groups showed that tumor cells lacking BRCA1 or BRCA2 are exquisitely sensitive to PARP1 inhibitors. Moreover, preliminary clinical data following treatment of BRCA1-defective and BRCA2-defective tumors with the PARP1 inhibitor olaparib are very encouraging [28]. A particularly exciting possibility is that sensitivity to PARP1 inhibition will extend beyond BRCA1/2 mutant tumors to other tumors that show defects in homologous recombi nation. In this regard, a recent study suggested that tumors lacking the tumor suppressor PTEN show such a defect [29], as do many basal-like breast cancers [30,31].
Screening for synthetic lethality - an unbiased approach
Synthetic lethal interactions, at least in hindsight, must explain the selectivity (however modest in most cases) of currently available anticancer drugs because these agents, including classical cytotoxic drugs and newer 'targeted' agents, invariably interact with targets that are shared between normal cells and cancer cells. For example, the ability to induce tumor regressions with tolerable doses of DNA-damaging cytotoxic agents might reflect underlying defects in DNA repair coupled with collateral pro-apoptotic signals delivered by oncoproteins such as E2F1 and c-Myc. A clearer understanding of these interactions might allow one to improve outcomes by pre-selecting patients who are most likely to benefit from existing agents.
To fully explore the number of synthetic lethal interactions in cancer cells will, however, require unbiased screening approaches for the reasons outlined above. One such approach has been to use chemical compound libraries, looking for compounds that preferentially kill cells with a particular cancer-causing mutation relative to isogenic cells lacking the cancer-causing mutation. In a series of studies, Stockwell and colleagues [32-34] used this approach to show that cells expressing oncogenic versions of Ras display enhanced sensitivity to compounds that bind to particular mitochondrial voltage-dependent anion channels and induce oxidative cell death. This sensitivity seems to be due, at least partly, to Ras-mediated increases in intracellular iron.
Inactivation of the von Hippel-Lindau tumor suppressor protein (pVHL) is a signature lesion in clear cell renal cancer, which is the most common form of kidney cancer, and leads to profound reprogramming of cellular metabolism. This reprogramming is partly due to increased activity of the hypoxia inducible (HIF) transcription factor [35]. Giaccia and colleagues [36] showed that renal carcinoma cells lacking pVHL are hypersensitive to a series of small molecules that promote autophagy. Interestingly, an earlier study showed that VHL-/- cells display increased sensitivity to mTOR (mammalian target of rapamycin) inhibition, which can also promote autophagy [37].
Lander and coworkers [38] confirmed that downregulation of E-cadherin in mammary epithelial cells induced an epithelial to mesenchymal transition and showed that this was associated with the acquisition of cancer stem-cell-like properties. They identified chemicals, including the potassium ionophore salinomycin, that were selectively toxic to cells after E-cadherin loss [38].
Chemical biology approaches are powerful, but the identification of protein targets for chemical 'hits' emerging from high-throughput screens remains laborious. The advent of short-interfering RNA (siRNA) and short-hairpin RNA (shRNA) methodologies now enables unbiased synthetic lethal screens to be conducted in mammalian cells in which defined genes are inactivated in conjunction with a cancer-relevant mutation of interest. Using this approach, Bartz et al. [39] identified genes that, when inhibited, selectively sensitized p53-defective cells to specific forms of chemotherapy. For example, they found that BRCA1 pathway components were synthetic lethal to p53 in cells treated with cis-platinum, whereas ribonucleo-tide reductase subunit M1 was synthetic lethal to p53 in cells treated with gemcitabine.
D'Andrea and colleagues [40] systematically inactivated 230 DNA damage genes in isogenic cells that did or did not harbor mutations in the Fanconi anemia pathway, which responds to stalled replication forks during S phase. They showed that tumor cells with defects in this pathway are hypersensitive to loss of ATM activity, again in keeping with the idea that loss of a particular DNA repair pathway can increase dependency on alternative repair mechanisms.
Our group, in collaboration with Dorre Grueneberg and Ed Harlow [41], conducted a pilot synthetic lethal screen with shRNAs targeting 88 different kinases and multiple isogenic cell line pairs that differed only with respect to VHL status. Loss of pVHL sensitized cells to loss of MET, CDK6 and MEK1 in three independent, isogenic cell line pairs. MET activation has also been described in some kidney cancers and there is evidence for crosstalk between HIF and MET [42-44].
In all of the above studies, cells were grown in multiwell plates and different perturbants (chemicals, siRNAs or shRNA vectors) were added to the individual wells (an approach known as arrayed screens). Some laboratories have pioneered an alternative approach in which cells are infected en masse with pools of shRNA vectors and the abundance of individual shRNA is monitored over time as a reflection of their effect on cellular fitness (pooled screens) [45-50]. Typically the abundance of each shRNA vector has been determined by PCR amplification across a DNA sequence unique to that vector (a so-called DNA barcode) followed by hybridization of the PCR product to a custom microarray containing oligonucleotides complementary to the various barcodes present in the library. It is possible that quantitative sequencing techniques will eventually replace the use of microarrays to monitor changes in shRNA vector abundance.
KRAS is one of the most frequently mutated human oncogenes. Cancer-relevant KRAS mutations lead to loss of K-Ras GTPase activity, leading to constitutive signaling. So far K-Ras has not proven tractable as a drug target. Elledge and coworkers [51] infected isogenic colorectal cancer lines that did or did not harbor an oncogenic KRAS mutation with approximately 74,000 retroviral shRNA vectors corresponding to about 32,000 unique human sequences. These vectors were divided into six subpools and the abundance of each hairpin was monitored using PCR and custom microarrays. Importantly, the PCR products from the two different cell lines were labeled with two different fluorescent dyes (Cy5 and Cy3) before hybridization to facilitate the identification of products that were selectively depleted in the KRAS mutated cell line, indicating a potential synthetic lethal interaction. Hits emerging from the primary screen were validated in a second cell line pair and in low-throughput cellular fitness assays. They found that KRAS mutant cells are hypersensitive to loss of the polo-like kinase PLK1, components of the anaphase-promoting complex/cyclosome, and the proteasome. Note that all of these proteins are required for normal cells as well (PLK1 has been used as a control for shRNA-induced killing in some studies [39,41]). Therefore, the difference between KRAS wild-type and mutant cells with respect to these targets is quantitative, not qualitative.
Limitations and challenges for synthetic lethal screens
The synthetic lethal screens described above used isogenic cell line pairs. Exclusive reliance on this cell line model, however, creates certain technical and theoretical limitations. First, isogenic cell line pairs do not exist for every gene of interest. When they do exist, they may be derived from a different species or cell type than the tumor(s) of interest (for example, mouse embryo fibroblasts compared with human epithelial cells) or represent a genotype that is unlikely to be encountered in human cancers (for example, when p53 is inactivated in p53+/+ tumor cells in which the p53-regulatory protein ARF has already been deleted [52]). It is also not uncommon that cells isogenic for a particular oncogene or tumor suppressor gene differ with respect to variables such as proliferation rate and cell-cycle distribution, which can potentially confound synthetic lethal screens. Finally, it is important to interrogate multiple isogenic cell line pairs for any given gene of interest to ensure that the synthetic lethal interactions detected are truly robust rather than peculiar to a particular line [41].
Hahn, Gilliland and coworkers [53,54] realized that if data for shRNA-mediated changes in cellular fitness were available for enough cancer cell lines representing two different classes (for example, K-Ras wild-type and K-Ras mutant) one could, in silico, look for shRNAs that differentially affected the viability of the two classes. Using this approach, they showed that inhibition of the protein kinases STK33 and TBK1 preferentially kills K-Ras mutant cells compared with K-Ras wild-type cells. In K-Ras mutant cells these kinases deliver critical pro-survival signals. STK33 indirectly targets the pro-apoptotic protein BAD1 for destruction, whereas TBK1 activates a pro-survival signal through the NF B transcription factor. This work should spur interest in these kinases as potential therapeutic targets and also suggests a paradigm for synthetic lethal screening of human cancer cells in the future.
siRNA screens, in contrast to chemical biology screens, are based on the downregulation of the abundance of a particular protein. The biological consequences of down-regulating a protein target need not phenocopy the effects of a small organic molecule bound to that target. For example, the phenotypes of downregulating an enzyme might reflect the loss of its catalytic activity, loss of a scaffolding function, or perhaps both. Moreover, the loss of catalytic activity as a result of protein elimination might allow forms of compensation that are prevented when the loss of catalytic activity is achieved with a drug. For example, the enzyme-drug complex might essentially act as a dominant negative. For these reasons, secondary screens that address these questions are required when the goal of a synthetic lethal screen is to identify new drug targets.
Equally importantly, targets emerging from in vitro synthetic lethal screens must eventually be validated in vivo to address the following questions: firstly, whether the synthetic lethal relationship within the tumor cell is maintained under conditions that more closely resemble those in patient tumors, and secondly, whether there are normal cells, perhaps derived from other cell lineages, that are also highly dependent on that target in vivo. These two questions obviously affect the potential efficacy and safety, respectively, of inhibiting that target, with the caveat that all preclinical models are imperfect replicas of human cancer.
Conclusions
In summary, synthetic lethality provides a conceptual framework for discovering drugs that selectively kill cancer cells while sparing normal tissues and for tackling 'undruggable' targets. Technological advances, coupled with the availability of large siRNA and shRNA libraries, now make unbiased synthetic lethal screens in mammalian cells feasible. Mapping synthetic lethal relationships in human cancer cells will hopefully enable us to use old drugs more wisely and to discover new drugs that are safer, and more efficacious, than existing agents.
Abbreviations
CDK: cyclin-dependent kinase; DR5: trail death receptor; PARP1: poly(ADP-ribose) polymerase-1; PLK1: polo-like kinase; pRB: retinoblastoma protein; pVHL: von Hippel-Lindau tumor suppressor protein; shRNA: short-hairpin RNA; siRNA: short-interfering RNA.
Competing interests
The author declares that he has no competing interests.
References
- Kaelin WG Jr. The concept of synthetic lethality in the context of anticancer therapy. Nat Rev Cancer. 2005;5:689–698. doi: 10.1038/nrc1691. [DOI] [PubMed] [Google Scholar]
- Kaelin WG Jr. Choosing anticancer drug targets in the postgenomic era. J Clin Invest. 1999;104:1503–1506. doi: 10.1172/JCI8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T. Genetics of natural populations. XIII. Recombination and variability in populations of Drosophila pseudoobscura. Genetics. 1946;31:269–290. doi: 10.1093/genetics/31.3.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchesi JC. Synthetic lethality and semi-lethality among functionally related mutants of Drosophila melanogaster. Genetics. 1968;59:37–44. doi: 10.1093/genetics/59.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L, Szankasi P, Roberts C, Murray A, Friend S. Integrating genetic approaches into the discovery of anti-cancer drugs. Science. 1997;278:1064–1068. doi: 10.1126/science.278.5340.1064. [DOI] [PubMed] [Google Scholar]
- Sharom JR, Bellows DS, Tyers M. From large networks to small molecules. Curr Opin Chem Biol. 2004;8:81–90. doi: 10.1016/j.cbpa.2003.12.007. [DOI] [PubMed] [Google Scholar]
- Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, Pagé N, Robinson M, Raghibizadeh S, Hogue CW, Bussey H, Andrews B, Tyers M, Boone C. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- Tong AH, Lesage G, Bader GD, Ding H, Xu H, Xin X, Young J, Berriz GF, Brost RL, Chang M, Chen Y, Cheng X, Chua G, Friesen H, Goldberg DS, Haynes J, Humphries C, He G, Hussein S, Ke L, Krogan N, Li Z, Levinson JN, Lu H, Ménard P, Munyana C, Parsons AB, Ryan O, Tonikian R, Roberts T. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- Reddy A, Kaelin WG. Using cancer genetics to guide the selection of anticancer drug targets. Curr Opin Pharmacol. 2002;2:366–373. doi: 10.1016/S1471-4892(02)00178-9. [DOI] [PubMed] [Google Scholar]
- Ginsberg D. E2F1 pathways to apoptosis. FEBS Lett. 2002;529:122–125. doi: 10.1016/S0014-5793(02)03270-2. [DOI] [PubMed] [Google Scholar]
- Krek W, G Xu, Livingston DM. Cyclin A-kinase regulation of E2F1 DNA binding function underlies suppression of an S phase checkpoint. Cell. 1995;83:1149–1158. doi: 10.1016/0092-8674(95)90141-8. [DOI] [PubMed] [Google Scholar]
- Krek W, Ewen M, Shirodkar S, Arany Z, Kaelin WG, Livingston DM. Negative regulation of the growth-promoting transcription factor E2F-1 by a stably bound cyclin a-dependent protein kinase. Cell. 1994;78:161–172. doi: 10.1016/0092-8674(94)90582-7. [DOI] [PubMed] [Google Scholar]
- Xu M, Sheppard KA, Peng C-Y, Yee AS, Piwnica-Worms H. Cyclin A/cdk2 binds directly to E2F1 and inhibits the DNA-binding activity of E2F1/DP1 by phosphorylation. Mol Cell Biol. 1994;14:8420–8431. doi: 10.1128/mcb.14.12.8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PD, Sellers WR, Sharma SK, Wu AD, Nalin CM, Kaelin WG. Identification of a cyclin-cdk2 recognition motif present in substrates and p21-like cdk inhibitors. Mol Cell Biol. 1996;16:6623–6633. doi: 10.1128/mcb.16.12.6623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loog M, Morgan DO. Cyclin specificity in the phosphorylation of cyclin-dependent kinase substrates. Nature. 2005;434:104–108. doi: 10.1038/nature03329. [DOI] [PubMed] [Google Scholar]
- Chen Y, Sharma S, Ramsey T, Jiang L, Martin M, Baker K, Adams P, Bair K, Kaelin WJ. Selective killing of transformed cells by cyclin/cyclin-dependent kinase 2 antagonists. Proc Natl Acad Sci USA. 1999;96:4325–4329. doi: 10.1073/pnas.96.8.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza N, Fong S, Marsters J, Koeppen H, Schwall R, Wickramasinghe D. Selective cyclin-dependent kinase 2/cyclin A antagonists that differ from ATP site inhibitors block tumor growth. Cancer Res. 2003;63:1020–1024. [PubMed] [Google Scholar]
- Nip J, Strom D, Fee B, Zambetti G, Cleveland J, Hiebert S. E2F-1 cooperates with topoisomerase II inhibition and DNA damage to selectively augment p53-independent apoptosis. Mol Cell Biol. 1997;17:1049–1056. doi: 10.1128/mcb.17.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee D, Schnieders B, Fu J, Ashikari D, Zhao S-C, Bertino J. Role of E2F-1 in chemosensitivity. Cancer Res. 1998;58:4292–4296. [PubMed] [Google Scholar]
- Xiao H, Goodrich DW. The retinoblastoma tumor suppressor protein is required for efficient processing and repair of trapped topoisomerase II-DNA-cleavable complexes. Oncogene. 2005;24:8105–8113. doi: 10.1038/sj.onc.1208958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Engels IH, Knee DA, Nasoff M, Deveraux QL, Quon KC. Synthetic lethal targeting of MYC by activation of the DR5 death receptor pathway. Cancer Cell. 2004;5:501–512. doi: 10.1016/S1535-6108(04)00113-8. [DOI] [PubMed] [Google Scholar]
- Goga A, Yang D, Tward A, Morgan D, Bishop J. Inhibition of CDK1 as a potential therapy for tumors over-expressing MYC. Nat Med. 2007;13:820–827. doi: 10.1038/nm1606. [DOI] [PubMed] [Google Scholar]
- Molenaar JJ, Ebus ME, Geerts D, Koster J, Lamers F, Valentijn LJ, Westerhout EM, Versteeg R, Caron HN. Inactivation of CDK2 is synthetically lethal to MYCN over-expressing cancer cells. Proc Natl Acad Sci USA. 2009;106:12968–12973. doi: 10.1073/pnas.0901418106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell S, DeFrank J, Connell P, Eogan M, Preffer F, Dombkowski D, Tang W, Friend S. Differential sensitivity of p53(-) and p53(+) cells to caffeine-induced radiosensitization and override of G2 delay. Cancer Res. 1995;55:1643–1648. [PubMed] [Google Scholar]
- Jiang H, Reinhardt HC, Bartkova J, Tommiska J, Blomqvist C, Nevanlinna H, Bartek J, Yaffe MB, Hemann MT. The combined status of ATM and p53 link tumor development with therapeutic response. Genes Dev. 2009;23:1895–1909. doi: 10.1101/gad.1815309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, Martin NM, Jackson SP, Smith GC, Ashworth A. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, Mortimer P, Swaisland H, Lau A, O'Connor MJ, Ashworth A, Carmichael J, Kaye SB, Schellens JH, de Bono JS. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- Shen WH, Balajee AS, Wang J, Wu H, Eng C, Pandolfi PP, Yin Y. Essential role for nuclear PTEN in maintaining chromosomal integrity. Cell. 2007;128:157–170. doi: 10.1016/j.cell.2006.11.042. [DOI] [PubMed] [Google Scholar]
- McCabe N, Turner NC, Lord CJ, Kluzek K, Bialkowska A, Swift S, Giavara S, O'Connor MJ, Tutt AN, Zdzienicka MZ, Smith GC, Ashworth A. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res. 2006;66:8109–8115. doi: 10.1158/0008-5472.CAN-06-0140. [DOI] [PubMed] [Google Scholar]
- Turner N, Tutt A, Ashworth A. Hallmarks of 'BRCAness' in sporadic cancers. Nat Rev Cancer. 2004;4:814–819. doi: 10.1038/nrc1457. [DOI] [PubMed] [Google Scholar]
- Dolma S, Lessnick SL, Hahn WC, Stockwell BR. Identification of genotype-selective antitumor agents using synthetic lethal chemical screening in engineered human tumor cells. Cancer Cell. 2003;3:285–296. doi: 10.1016/S1535-6108(03)00050-3. [DOI] [PubMed] [Google Scholar]
- Yang WS, Stockwell BR. Synthetic lethal screening identifies compounds activating iron-dependent, nonapoptotic cell death in oncogenic-RAS-harboring cancer cells. Chem Biol. 2008;15:234–245. doi: 10.1016/j.chembiol.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagoda N, von Rechenberg M, Zaganjor E, Bauer AJ, Yang WS, Fridman DJ, Wolpaw AJ, Smukste I, Peltier JM, Boniface JJ, Smith R, Lessnick SL, Sahasrabudhe S, Stockwell BR. RAS-RAF-MEK-dependent oxidative cell death involving voltage-dependent anion channels. Nature. 2007;447:864–868. doi: 10.1038/nature05859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin WG Jr. The von Hippel-Lindau tumour suppressor protein: O2 sensing and cancer. Nat Rev Cancer. 2008;8:865–873. doi: 10.1038/nrc2502. [DOI] [PubMed] [Google Scholar]
- Turcotte S, Chan DA, Sutphin PD, Hay MP, Denny WA, Giaccia AJ. A molecule targeting VHL-deficient renal cell carcinoma that induces autophagy. Cancer Cell. 2008;14:90–102. doi: 10.1016/j.ccr.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas GV, Tran C, Mellinghoff IK, Welsbie DS, Chan E, Fueger B, Czernin J, Sawyers CL. Hypoxia-inducible factor determines sensitivity to inhibitors of mTOR in kidney cancer. Nat Med. 2006;12:122–127. doi: 10.1038/nm1337. [DOI] [PubMed] [Google Scholar]
- Gupta P, Onder T, Jiang G, Tao K, Kuperwasser C, Weinberg R, Lander E. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;138:645–659. doi: 10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartz SR, Zhang Z, Burchard J, Imakura M, Martin M, Palmieri A, Needham R, Guo J, Gordon M, Chung N, Warrener P, Jackson AL, Carleton M, Oatley M, Locco L, Santini F, Smith T, Kunapuli P, Ferrer M, Strulovici B, Friend SH, Linsley PS. Small interfering RNA screens reveal enhanced cisplatin cytotoxicity in tumor cells having both BRCA network and TP53 disruptions. Mol Cell Biol. 2006;26:9377–9386. doi: 10.1128/MCB.01229-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy RD, Chen CC, Stuckert P, Archila EM, De la Vega MA, Moreau LA, Shimamura A, D'Andrea AD. Fanconi anemia pathway-deficient tumor cells are hypersensitive to inhibition of ataxia telangiectasia mutated. J Clin Invest. 2007;117:1440–1449. doi: 10.1172/JCI31245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommi-Reddy A, Almeciga I, Sawyer J, Geisen C, Li W, Harlow E, Kaelin WG Jr, Grueneberg DA. Kinase requirements in human cells: III. Altered kinase requirements in VHL-/- cancer cells detected in a pilot synthetic lethal screen. Proc Natl Acad Sci USA. 2008;105:16484–16489. doi: 10.1073/pnas.0806574105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchietti S, Michieli P, Galluzzo M, Mazzone M, Giordano S, Comoglio PM. Hypoxia promotes invasive growth by transcriptional activation of the met protooncogene. Cancer Cell. 2003;3:347–361. doi: 10.1016/S1535-6108(03)00085-0. [DOI] [PubMed] [Google Scholar]
- Hara S, Nakashiro KI, Klosek SK, Ishikawa T, Shintani S, Hamakawa H. Hypoxia enhances c-Met/HGF receptor expression and signaling by activating HIF-1alpha in human salivary gland cancer cells. Oral Oncol. 2006;42:593–598. doi: 10.1016/j.oraloncology.2005.10.016. [DOI] [PubMed] [Google Scholar]
- Hayashi M, Sakata M, Takeda T, Tahara M, Yamamoto T, Okamoto Y, Minekawa R, Isobe A, Ohmichi M, Tasaka K, Murata Y. Up-regulation of c-met protooncogene product expression through hypoxia-inducible factor-1alpha is involved in trophoblast invasion under low-oxygen tension. Endocrinology. 2005;146:4682–4689. doi: 10.1210/en.2005-0416. [DOI] [PubMed] [Google Scholar]
- Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, Carpenter AE, Foo SY, Stewart SA, Stockwell BR, Hacohen N, Hahn WC, Lander ES, Sabatini DM, Root DE. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- Luo B, Cheung HW, Subramanian A, Sharifnia T, Okamoto M, Yang X, Hinkle G, Boehm JS, Beroukhim R, Weir BA, Mermel C, Barbie DA, Awad T, Zhou X, Nguyen T, Piqani B, Li C, Golub TR, Meyerson M, Hacohen N, Hahn WC, Lander ES, Sabatini DM, Root DE. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci USA. 2008;105:20380–20385. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlabach MR, Luo J, Solimini NL, Hu G, Xu Q, Li MZ, Zhao Z, Smogorzewska A, Sowa ME, Ang XL, Westbrook TF, Liang AC, Chang K, Hackett JA, Harper JW, Hannon GJ, Elledge SJ. Cancer proliferation gene discovery through functional genomics. Science. 2008;319:620–624. doi: 10.1126/science.1149200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva JM, Marran K, Parker JS, Silva J, Golding M, Schlabach MR, Elledge SJ, Hannon GJ, Chang K. Profiling essential genes in human mammary cells by multiplex RNAi screening. Science. 2008;319:617–620. doi: 10.1126/science.1149185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddison PJ, Silva JM, Conklin DS, Schlabach M, Li M, Aruleba S, Balija V, O'Shaughnessy A, Gnoj L, Scobie K, Chang K, Westbrook T, Cleary M, Sachidanandam R, McCombie WR, Elledge SJ, Hannon GJ. A resource for large-scale RNA-interference-based screens in mammals. Nature. 2004;428:427–431. doi: 10.1038/nature02370. [DOI] [PubMed] [Google Scholar]
- Berns K, Hijmans EM, Mullenders J, Brummelkamp TR, Velds A, Heimerikx M, Kerkhoven RM, Madiredjo M, Nijkamp W, Weigelt B, Agami R, Ge W, Cavet G, Linsley PS, Beijersbergen RL, Bernards R. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature. 2004;428:431–437. doi: 10.1038/nature02371. [DOI] [PubMed] [Google Scholar]
- Luo J, Emanuele MJ, Li D, Creighton CJ, Schlabach MR, Westbrook TF, Wong KK, Elledge SJ. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009;137:835–848. doi: 10.1016/j.cell.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown J, Sedivy J, Kinzler K, Vogelstein B. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- Scholl C, Fröhling S, Dunn IF, Schinzel AC, Barbie DA, Kim SY, Silver SJ, Tamayo P, Wadlow RC, Ramaswamy S, Döhner K, Bullinger L, Sandy P, Boehm JS, Root DE, Jacks T, Hahn WC, Gilliland DG. Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells. Cell. 2009;137:821–834. doi: 10.1016/j.cell.2009.03.017. [DOI] [PubMed] [Google Scholar]
- Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, Schinze AC, Sandy P, Meylan E, Scholl C, Frohling S, Chan EM, Sos ML, Michel K, Mermel C, Silver JS, Weir BA, Reiling JH, Q S, Gupta PB, Wadlow RC, Le H, Hoersch S, Wittner BS, Ramaswamy S, Livingston DM, Sabatini DM, Meyerson M, Thomas RK, Lander ES. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. in press . [DOI] [PMC free article] [PubMed]