Abstract
Pneumocystis organisms are opportunistic fungal pathogens that cause significant pneumonia in immune-compromised hosts. Recent evidence has suggested that Pneumocystis carinii exists as separate mating types, and expresses and regulates proteins that govern meiosis and progression of the life cycle. This study was undertaken to investigate the activity of three life cycle–regulatory proteins in Pneumocystis, including two proteins essential in mating signaling, and a putative meiotic regulator, to determine the conditions under which they are most active. This study used V5/HIS-tagged PCRan1p, PCSte20p, and PCCbk1, purified from Saccharomyces cerevisiae strain, INVSC, as well as an in vitro Escherichia coli protein expression system to determine the optimal expression conditions of each protein in the presence of varying pH, temperature, and metal ions. These studies demonstrate an atypical enzymatic activity in PCRan1p, whereby the kinase was most active in the environmental conditions between 10 and 25°C, compared with a dramatic reduction in activity above 30°C, temperatures typically found within mammalian hosts. Circular dichroism and fluorescence spectroscopy suggest that PCRan1p becomes partially unfolded at 25°C, leading to its most active conformation, whereas continued unfolding as temperature increases results in strongly suppressed activity. These studies suggest that, in vivo, while under conditions within the mammalian lung (typically 37°C), PCRan1p kinase activity is largely suppressed, allowing better conditions for the activation of meiosis, whereas in ex vivo environments, PCRan1p kinase activity increases to arrest progression of the life cycle until conditions become more favorable.
Keywords: Pneumocystis, meiosis, kinase, temperature
CLINICAL RELEVANCE
In the current investigation, we delineate key life cycle regulatory mechanisms in Pneumocystis (meiotic control), an important cause of severe pneumonia in immunocompromised patients.
Pneumocystis species represent opportunistic fungal pathogens that have become increasingly significant as a result of the ongoing acquired immunodeficiency syndrome pandemic. Pneumocystis pneumonia is typically associated with severely immune compromised individuals, such as patients with advanced acquired immunodeficiency syndrome, organ transplantation, chronic steroid treatment, or with other various hematological and solid malignancies (1, 2). The overall mortality rate varies from 10–50% (1, 2). In the immune-competent host, it is believed that primary Pneumocystis infection is contracted during infancy, whereby the resulting immune response confers lifelong immunity in most individuals (3, 4). However, the reservoir of this primary infection, as well as subsequent infections, remains a mystery (5).
Pneumocystis research continues to be limited, because the organism cannot sustain viability in long-term in vitro culture once removed from the host (6). As a result of this limitation, direct studies of the organism's life cycle have been hampered. To date, limited data have been gathered determining the manner in which Pneumocystis survives ex vivo, as well as how the organism is able to progress through its life cycle while replicating in the host. Microscopic structural studies of Pneumocystis carinii demonstrate progression of life cycle from the thick-walled cystic forms to the smaller pleomorphic trophic forms of the organism. In addition, further studies have confirmed the presence of a precyst form that acts as an intermediate between these other two morphologies (7–9).
Recent investigations into the molecular regulation of the Pneumocystis life cycle have been aided as a result of the Pneumocystis genome project. The sequencing results of this study have allowed us to use homology-based approaches to identify genes involved in the regulation of life cycle processes within P. carinii. In the current investigation, we sought to evaluate environmental regulation of three protein kinases—PCRan1p, PCSte20p, and PCCbk1p—previously identified to participate in Pneumocystis life cycle control (10–12).
PCRan1p has recently been shown by our laboratory to be crucial to the inhibition of meiotic entry in Pneumocystis (10). PCRan1p has been demonstrated to phosphorylate its cognate substrate, PCMei2p, blocking downstream signaling to activate the meiotic pathway (10). The loss of the homologous Pat1p in Schizosaccharomyces pombe is a lethal mutation, because the yeast can no longer prevent itself from entering meiosis, regardless of the genetic background (13–15). In contrast, PCSte20p has been shown, through heterologous expression in yeast, to be essential for the mating process (11). Additionally, PCSte20 expression is strongly induced by binding of the cell to fibronectin, vitronectin, or collagen. This suggests that the tropism of P. carinii allows the fungus to bind, activating the mating pathway, induced specifically by PCSte20 (11). The deletion of Ste20 in the yeast homolog results in the complete lack of pseudohyphal extensions and the ability of the yeast to mate, resulting in halting of the life cycle progression (16). Finally, PCCbk1p acts as a serine/threonine kinase in several fungal species, including Pneumocystis carinii, Candida albicans, and Sacharomyces cerevisiae. The role of Pneumocystis PCCbk1, as well as Cbk1 in yeast species, has been shown to be essential in normal mating projection formation, polarized cell growth, and normal cell separation (12, 17, 18). C. albicans strains that lack function of this homolog are rendered avirulent (19). Thus, PCRan1p, PCSte20p, and PCCbk1p each act as specific components regulating life cycle progression of the Pneumocystis organism.
Accordingly, we performed a series of biochemical characterization studies to determine the ambient environmental conditions that best suit the activity of each of these protein kinases, including pH, temperature, and metal ions. Interestingly, we found that, whereas the PCSte20p and PCCbk1p kinases were most active under conditions that best represent those present within the mammalian host lung (the preferred site of organism proliferation), PCRan1p was most active at subphysiological temperatures, regardless of the relative pH (conditions most commonly present ex vivo), suggesting a significant role for this protein outside of the host. Additional studies were performed to suggest a mechanism for this proposed suppression of PCRan1p activity. These results demonstrate a novel, temperature-sensitive profile of PCRan1 in Pneumocystis, and the first acquisition of results that suggest a putative role of a protein in the maintenance and long-term survival of the pathogen while it maintains stasis outside of the host.
MATERIALS AND METHODS
Media and Growth Conditions
P. carinii cells were obtained through the American Type Culture Collection (Manassas, VA) and propagated in corticosteroid-treated Long-Evans rats (Harlan, Inc., Indianapolis, IN). These rats were then killed and P. carinii–purified with differential filtration through 10-μm filters, as has been described previously (20, 21). For certain experiments, purified P. carinii was incubated in Dulbecco's modified Eagle's medium before protein was isolated with Y-PER (Pierce, Rockford, IL), as previously reported (11). For selection of transformants and maintenance of plasmids in yeast, cultures were incubated in pombe dropout media (QBioGene, Irvine, CA) lacking the correct dropout amino acid. Cultures were incubated overnight at 30°C. For the selection and maintenance of plasmids in Escherichia coli, cultures were performed in lysogeny broth containing 100 μg/ml carbenicillin overnight at 37°C.
Bacteria and Yeast Strains
Transformants containing the P. carinii PCRan1, PCSte20, or PCCbk1 were all maintained in S. cerevisiae strain INVSC− (Invitrogen, Carlsbad, CA). For the Rapid Translation System (RTS; Roche, Palo Alto, CA) in vitro transcription/translation experiments, plasmids were maintained in E. coli DH5α cells (Invitrogen).
Isolation of PCR Amplified P. carinii PCRan1, PCSte20, PCCbk1, and Pat1
P. carinii PCRan1, PCSte20, and PCCbk1 were PCR amplified with Taq polymerase (Invitrogen) using the following set of conditions: 1 cycle of 95°C for 5 minutes, 35 cycles of 95°C for 30 seconds, 55°C for 30 seconds, 72°C for 2.5 minutes, 1 cycle of 72°C for 7 minutes. The PCR setup contained 5 U Taq polymerase, 10 mM deoxyribonucleotide triphosphates, 2.5 mM MgCl2, 1× PCR amplification buffer, 25 pg forward and reverse primers, and H2O to 50 μl. The following primer sets were employed:
PCRan1 start: 5′-ATGTCAATGTTTCAAGAATCTAGGA-3′
PCRan1 stopV5: 5′-AAATCGTTTAGGTGAAATCGG-3′
PCSte20 start: 5′-ATGGCACATAATTTATCATCACAAT-3′
PCSte20 stopV5: 5′-TTTTCCAGACTTAAATGCCAAAAGA-3′
PCCbk1 start: 5′-ATGCAAAAAGTATCAGGTTCA-3′
PCCbk1 stopV5: 5′-TAAAATTCCTTTCTCCGTCATCAT-3′
Pat1 start: 5′-CACCATGATGCGCGAAAATCC-3′
Pat1 stopV5: 5′-GAGCGATTTTCGAGATGGATC-3′
All four reverse primers were engineered without the stop codon to permit subsequent V5/His-tag purification. The resulting amplicons were then gel purified using the MiniElute Gel Extraction kit (Qiagen, Valencia, CA) and cloned into the pYES2.1-TOPO (Invitrogen) vector containing the GAL1 promoter, as has been previously described (10). For the RTS in vitro transcription/translation experiments, P. carinii PCRan1 and S. pombe Pat1 were amplified using Pfx polymerase (Invitrogen) and the primers listed above under the same conditions. The resulting amplicons were gel purified and cloned into pET102-TOPO (Invitrogen) containing the T7 promoter, as has been previously described (10).
Purification of P. carinii PCRan1p–, PCSte20p–, and PCCbk1p–Expressed Protein from Yeast for Kinase Activity Assays
To perform the in vitro kinase activity assays, P. carinii PCRan1p, PCSte20p, and PCCbk1p were purified from cultures of S. cerevisiae INVSC− (Invitrogen) that expressed the plasmid of interest. For purification, 1.5 liters of culture broths of minimal media without uracil, containing 2% galactose, were grown to mid-log phase. Proteins were isolated, as described previously, using a Y-PER HIS6-purification column (Pierce) (10).
RTS In Vitro E. coli Protein Synthesis of P. carinii PCRan1 and S. pombe Pat1 Kinase
Additional kinase assays were performed using proteins synthesized from the RTS in vitro transcription/translation system. RTS reactions were performed according to the manufacturer's procedure (Roche). For protein synthesis, 500 ng of either P. carinii PCRan1 or S. pombe Pat1 containing the V5 tag in the pET102-TOPO vector were added to 12 μl E. coli lysate, 10 μl complete amino acids, 12 μl reaction mix, 1 μl methionine, 5 μl reconstitution buffer, and H2O, to a final volume of 50 μl, as is instructed in the manufacturer's protocol. The reaction was then placed in the Roche Proteomaster at 30°C for 8 hours. The resulting reactions were then purified by adding Tris-buffered saline (TBS) containing complete protease inhibitors (Roche) and V5 antibody conjugated to agarose beads (Sigma, St. Louis, MO), and incubating end over end at 4°C for 1 hour. The isolated products were then washed with TBS containing protease inhibitors, and vortexed, before being added to the kinase assay. To confirm that the RTS proteins were indeed synthesizing the proteins of interest, mass spectrometry was used to sequence major bands resolved by SDS-PAGE and Coomassie staining (Pierce). Mass spectrometry was performed at the Mayo Clinic Protein Sequencing Core Facility.
In Vitro Kinase Assays for P. carinii PCRan1p, PCCbk1p, and PCSte20p
To measure the overall biochemical activity of the proteins, 1 μg of each respective enzyme was added to a mixture that contained the common kinase substrate, eukaryotic translation initiation factor 4E binding protein 1 (PHAS-I) (2 μg; Stratagene, Cedar Creek, TX), kinase buffer (150 mM Tris, 20 mM MgCl2, 10 mM MnCl2, 15 mM β-mercaptoethanol), and 10 μCi [γ-32P] ATP (6,000 Ci/mmol). The mixtures were incubated for 1 hour at the conditions specified, and resolved on a 12% SDS-PAGE agarose gel, followed by autoradiography. In the reactions that contained RTS-generated proteins, the enzymes were added while still conjugated to anti-V5 beads.
Ability of PCRan1p to Phosphorylate Its Native Substrate, PCMei2p
We further sought to determine whether PCRan1p would phosphorylate PCMei2p, its putative native target protein, in a temperature-dependant manner. To address this, full-length cDNAs of both PCRan1 and PcMei2 were amplified by PCR and sequenced. Both genes were subsequently subcloned into pGEX-4T-1 (GE Life Sciences, Piscataway, NJ) in frame with the N-terminal glutathione S-transferase tag and under control of the tac promoter. These two constructs were transformed into E. coli BL21(DE3)pLysS cells (Novagen, Darmstadt, Germany). After overnight growth of one colony in 10 ml media, the culture was added to 1.0-liter media and grown until an optical density of 600 nm of 0.6 at 22°C was attained. At an optical density of 600 nm, 0.1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) was added and the cultures again grown overnight at 18°C. The following day, cells were recovered by centrifugation and washed once with cold TBS. Cells were then resuspended in 20 ml lysis buffer (TBS containing 1% Triton X-100, 1 mM DTT, and 1 mM PMSF) and applied to a French press for lysis. Cells were then sonicated three times for 15 seconds on ice and collected by centrifugation at 20,000 × g for 30 minutes. Supernatant was removed and glutathione S-transferase–tagged PcRan1 and PcMei2 proteins were purified over glutathione-sepharose, according to manufacturer's directions (GE Life Sciences). After purification of the proteins, dialysis of the proteins was conducted three times in TBS with 1 mM dithiothreitol (DDT) and 10% glycerol for 8 hours at 4°C. Because PCMei2p is known to have low-grade autophosphorylation ability, this substrate was heat inactivated at 72°C for 30 minutes before use. Next, the PCRan1 kinase assays were performed as described above, using PCMei2 (3.0 μg) rather than PHAS-I as the target substrate, with reaction time of 1 hour and the various temperature conditions as specified.
Kinase Assays with In Vivo–Derived P. carinii PCRan1p
To identify the native activity of P. carinii PCRan1p kinase within the organism, P. carinii cells were purified and protein extracts obtained as described above. Subsequently, 100 μg of protein were precleared with a 1:250 dilution of PCRan1p preimmune sera, followed by a 1:250 dilution of a specific PCRan1p antibody generated previously (10). The proteins were then immunoprecipitated using protein A-Sepharose beads (Sigma). The resulting proteins were then assayed for kinase activity as described above.
Western Blotting of P. carinii PCRan1p and PCCbk1p
Western blots of P. carinii PCRan1p and PCCbk1p were performed by adding 5 μg of P. carinii protein lysate to a 12% SDS-PAGE gel and resolving the proteins for 45 minutes at 200 V. The proteins were then transferred to nitrocellulose at 100 V for 1 hour. Next, a 1:10,000 dilution of anti-PCRan1p or anti-PCCbk1p antibody was added and incubated for 2 hours. The membranes were washed, and a 1:5,000 dilution of goat anti-rabbit IgG horseradish peroxidase conjugate (Southern Biotech, Birmingham, AL) was added and incubated for an additional hour before being washed and developed with enhanced chemiluminescence reagent (GE Healthcare, Piscataway, NJ).
Circular Dichroism and Fluorescence Spectroscopy Analysis of P. carinii PCRan1
Analyses of the secondary and tertiary structure of P. carinii PCRan1 were next undertaken to determine the basis for the mechanism of the activity profiles that we observed in the kinase assays. Circular dichroism (CD) was used to resolve the secondary structure. To obtain enough protein for analysis, PCRan1p was synthesized by pooling six separate RTS reactions before the V5 purification step. The protein was then resuspended in 70 μl TBS plus protease inhibitors, yielding a final protein concentration of approximately 90 ng/μl. Protein secondary structure was monitored at 4°C by far ultraviolet (UV)-CD from 260 to 200 nm in a 0.1-mm cuvette. Measurements were taken every 1 nm with a scanning speed of 50 nm/min. Thermal denaturation experiments followed the ellipticity at 222 nm over a temperature range of 4–45°C. The temperature was increased by 30°C per hour in 2°C intervals. After the protein sample reached 45°C in the denaturation experiment, the protein refolding was immediately measured by following the ellipticity at 222 nm and decreasing the temperature from 45 to 4°C in 2°C intervals at a rate of 30°C per hour. A control reaction to determine the background generated by the V5 beads was performed by pooling six RTS reactions that lacked the addition of pET102-TOPO-PCRan1V5.
Furthermore, fluorescence spectroscopy was used to assess tertiary structure by monitoring intrinsic tryptophan fluorescence. For these experiments, the V5 beads containing the synthesized PCRan1p were resuspended in 100 μl TBS containing protease inhibitors (Roche). The sample was incubated at the appropriate temperature for 15 minutes, followed by excitation at 294 nm and an emission scan from 310 to 200 nm.
Assessment of P. carinii PCRan1 Kinase Activity after Thermal Unfolding and after Returning to More Favorable Temperatures
Finally, we sought to understand how reversible these thermal conformational changes are in PCRan1p by using an in vitro kinase assay. To accomplish this, PCRan1p was incubated at 25, 30, 37, or 45°C. After the incubation, the kinase reaction was returned to the favorable 25°C condition, and the assay performed at either 25°C to evaluate the extent of reversibility, or at 45°C as a control condition to define zero reversibility. This approach allowed us to evaluate whether the results seen in the thermal refolding assay could be corroborated by kinase activity. The results from multiple kinase assays were pooled, and the results quantified by scanning densitometry using Image J version 1.38x (Wayne Rasband, National Institutes of Health; http://rsb.info.nih.gov/nih-image/).
RESULTS
The Pneumocystis Meiotic Inhibitory PCRan1p Kinase Exhibits Temperature-Regulated Activity
To better understand how P. carinii PCRan1p may act to modulate meiosis over the course of the P. carinii life cycle, we undertook an initial series of experiments to determine the ambient environmental conditions at which the enzyme was most active. Our prior studies demonstrated that PCRan1p phosphorylates its native target substrate, PCMei2p (10). In the current study, we further assayed the ability of PCRan1p to phosphorylate a generic kinase substrate, PHAS-I, to directly contrast PCRan1p activity to other life cycle–regulatory kinases in Pneumocystis, including PCSte20p and PCCbk1p. Using this assay, we evaluated the effects of temperature (4–(45°C), pH (4–8), and the presence of various divalent metal cations for optimal activity (Figure 1).
Figure 1.
In vitro kinase assay with HIS tag–purified PCRan1p from Saccharomyces cerevisiae strain INVSC, containing pYES2.1-TOPO plus PCRan1, tested for phosphorylation activity with a series of (A) temperatures, (B) pH, and (C) divalent metal cations, using eukaryotic translation initiation factor 4E binding protein 1 (PHAS-I) as the substrate. (D) In vitro kinase assay with glutathione S-transferase tag–purified PCRan1p and PCMei2p that were generated in Escherichia coli BL21(DE3)pLysS cells, and tested for similar PCRan1p temperature-dependant kinase activity as in A using the native substrate of PCRan1p, PCMei2p. Default conditions, unless otherwise stated in figure, are 25°C, 10 mM MnCl2 and 10 mM MgCl2, and pH 7.
We initially hypothesized that PCRan1p would be active at or near physiological temperatures and pH. Interestingly however, we observed that the temperature under which PCRan1p optimally phosphorylated PHAS-I was at the subphysiological temperature of 10°C (Figure 1A). In addition, the enzyme also seemed to function very efficiently at temperatures ranging between 4 and 25°C, typical of ambient environmental conditions. In contrast, when the temperature was increased above 25°C, the overall activity of the enzyme was dramatically reduced. At 30 and 37°C, temperatures typical of those found within mammalian lungs, phosphorylation of the target substrate was largely suppressed, with activity being nonexistent at 45°C. These findings strongly indicate that PCRan1p may function to prevent the advancement of the life cycle in intracystic bodies while ex vivo. Correspondingly, in vivo activity is reduced to a level that lends itself to the activation of meiosis under appropriate conditions.
Additionally, we assayed the ability of PCRan1p to phosphorylate its substrate in the presence of variable pH conditions. Again, we hypothesized that the enzymatic activity would be largely restricted to physiological pH. However, we observed that PCRan1p was able to phosphorylate the target substrate across a wide range of environmental pH (Figure 1B), suggesting that the enzyme was refractory to H+ concentration, and that high acidity or alkalinity had minimal effect on the overall conformation of the protein.
Lastly, we evaluated the ability of the PCRan1p enzyme to phosphorylate in the presence of a series of different divalent metal cations. Strikingly, PCRan1p was only able to phosphorylate PHAS-I in the presence of cobalt, manganese, and nickel ions (Figure 1C). These results were anticipated, as PCRan1p has been shown through homology studies to possess a manganese-requiring domain. However, these experiments did further verify that the kinase buffer in use did contain the essential components that permitted proper activity of the enzyme.
To verify that the effects of the temperature-dependent activity noticed in Figure 1A was not the result of a substrate-dependant interaction with PHAS-I, we performed an in vitro kinase assay using the native PCRan1p substrate, PCMei2p, at varying temperatures. By initiating this reaction, we were able to confirm that the kinase activity of PCRan1p does appear to be temperature dependent, regardless of substrate, suggesting that the effect noticed is likely the result of enzyme destabilization (Figure 1D).
In Contrast, Pneumocystis PCSTE20p and PCCbk1p Kinases Do Not Exhibit Similar Temperature Regulation
Based on the unique temperature-regulated kinase activity observed for PCRan1p, we next sought to contrast this with the environmental regulation of other life cycle–regulatory kinases previously identified in our laboratory. To accomplish this, we further analyzed two other serine/threonine kinases–PCSte20p and PCCbk1p–for enzymatic activity under varying environmental conditions of temperature and pH (Figure 2). As noted under the differential temperature conditions, PCRan1p lost most of its activity above 25°C. In contrast, PCSte20p and PCCbk1p remained equally as active from 4 to 37°C (Figure 2A). These findings underscore that PCRan1p exhibits unique temperature sensitivity compared with these other two life cycle–regulatory kinases of Pneumocystis.
Figure 2.
In vitro kinase assay with HIS tag–purified PCSte20p and PCCbk1p from S. cerevisiae INVSC, containing pYES2.1-TOPO plus PCSte20 or PCCbk1, tested for phosphorylation activity at a variety of (A) temperatures or (B) pH, using PHAS-I as a substrate. Default conditions in this figure same as previously listed.
Of further interest, although PCRan1p was able to phosphorylate the PHAS-I substrate over a wide range of pH, PCSte20p and PCCbk1p were only able to phosphorylate over the limited pH ranges of pH 5–7 and pH 6–8, respectively (Figure 2B). Both PCSte20p and PCCbk1p functioned most optimally at physiological pH conditions found within the lung, whereas PCRan1p retained its activity regardless of pH. Together, these findings suggest that the P. carinii organisms have the ability to differentially regulate the activity of key enzymes governing life cycle progression of the organisms while in the host compared with when the organism is expelled into the ambient environment.
The PCRan1p Homolog, Pat1p, Present in S. pombe, Demonstrates Similar Enzymatic Activity under Various Temperatures Conditions
We next sought to determine whether the unique temperature-based activity regulation observed for Pneumocystis PCRan1p was unique to this fungal species. To test this, we contrasted the kinase activity exhibited by Pat1p, the corresponding homolog present in S. pombe, the organism of closest phylogenic relatedness to Pneumocystis, under conditions of variable temperature. To accomplish this, PCRan1p and Pat1p were synthesized in vitro via an E. coli RTS in vitro transcription/translation expression system. The PCRan1 and Pat1 genes were cloned into vectors containing a T7 promoter and an in-frame V5 tail, and expressed through the RTS system and purified by immunoprecipitation against the V5 epitope tags. Western blotting was used to confirm the synthesis of the desired proteins, as well as to ensure the purity of the sample after immunoprecipitation (Figure 3A). Subsequent confirmation was further obtained by mass spectrometry protein sequencing of the expressed proteins (Figure E1 in the online supplement). In vitro kinase assays for the RTS-expressed PCRan1p and Pat1p confirmed that both RTS-expressed proteins were still active while conjugated to the V5 agarose beads, and able to actively phosphorylate their substrate (Figure 3B).
Figure 3.
(A) V5-tagged agarose beads used to immunoprecipitate PCRan1p or Pat1p, followed by Western blotting using the V5 antibody on Rapid Translation System (RTS)-generated protein made from pET102-TOPO plus PCRan1 or Pat1. (B) In vitro kinase assay on RTS-generated PCRan1p and Pat1p performed at 25°C for 1 hour using PHAS-I as a substrate. (C) In vitro kinase assay using RTS-generated PCRan1p and Pat1p assayed for enzymatic activity at a variety of different temperatures. V5 antibody was used to Western blot for overall protein levels of either PCRan1p or Pat1p.
The temperature comparison of PCRan1p and Pat1p once again noted similar overall profiles, although differences were observed in overall magnitude. Overall, the profiles were still very similar, with the strongest overall phosphorylation occurring over 4–25°C, with a strong decrease in activity observed from 30 to 45°C (Figure 3C). It should be noted, however, that, in these reactions using the RTS-generated PCRan1p, the strongest overall phosphorylation occurred at 25°C, in contrast to 10°C, as was noted with the yeast-generated protein. The profile for Pat1p was slightly different from that observed for PCRan1p in that it only phosphorylated the PHAS-I target substrate from 10 to 30°C, with the strongest overall activity at 25°C. However, once again, the S. pombe Pat1p homolog also exhibited a dramatic decrease in phosphorylation above 25°C (Figure 3C). Thus, in both the case of Pneumocystis PCRan1p and with S. pombe Pat1p, temperature regulation favors optimal activity under conditions below the physiological range of approximately 37°C, conditions the organism would experience outside of the host lung.
The Activity of PCRan1p Derived from Pneumocystis Organisms Matches the In Vitro Kinase Assays, and Is Not the Result of Increased or Decreased PCRan1p Translation at Various Temperatures
Performing in vivo studies using live P. carinii organisms can be a daunting task, due to the limited viability of these organisms once they are purified from the host lung. Nonetheless, we next sought to directly verify the temperature sensitivity of PCRan1p by immunoprecipitating PCRan1p from freshly isolated P. carinii lysates, and then using immunoprecipitated PCRan1p to evaluate kinase activity under various temperature conditions (Figure 4). Using the freshly isolated PCRan1p derived from Pneumocystis organisms, we found a similar kinase activity profile to what had been previously demonstrated with both the yeast and in vitro–generated proteins. Native PCRan1p phosphorylated PHAS-I best between 4 and 25°C, with additional activity at 30°C (Figure 4A). However, the enzyme once again showed suppressed levels of activity at the physiological temperature of 37°C.
Figure 4.
(A) PCRan1p immunoprecipitated from Pneumocystis carinii cysts and trophic forms purified from infected rats, using a specific PCRan1p antibody and protein A sepharose, was then added to a kinase assay performed at a series of temperatures to determine enzymatic activity of in vivo–derived PCRan1p. Western blots of PCRan1p using a specific antibody were performed as a loading control. (B) P. carinii isolated from infected rats was incubated for 2 hours at a series of temperatures, followed by protein isolation and Western blotting using a specific PCRan1p antibody. PCCBK1 was used as a loading control to verify equal protein loading.
To further confirm that the enzymatic profile was not being confounded by fluctuating overall levels of PCRan1p at the different temperatures as a result of temperature-dependent translation, Western blot determination of total levels of PCRan1p was performed after incubating P. carinii organisms for 2 hours at each of the temperatures analyzed. Indeed, the overall levels of PCRan1p did not vary as a result of the temperature, demonstrating that the enzyme was equally expressed from 4 to 37°C (Figure 4B). As a control comparison, consistent with prior experiments from our laboratory, PCCbk1p was also equally expressed across these conditions. These findings further indicate that the PCRan1p kinase activity profile seen in the Pneumocystis organisms themselves is related to temperature-sensitive activity of the enzyme, rather than translational levels.
CD and Fluorescence Spectroscopy Verify the Temperature-Sensitive Profile of PCRan1p
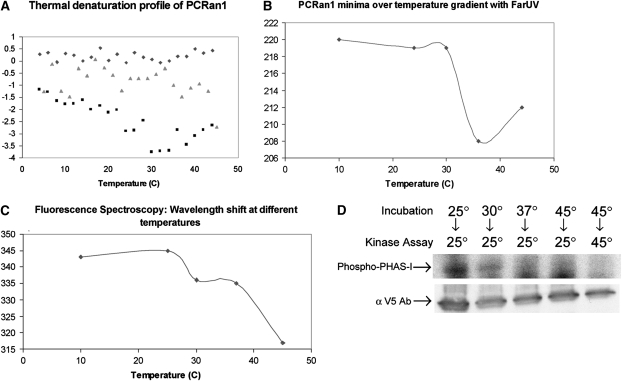
To decipher the mechanism for the depression of PCRan1p activity at or below physiological temperature, we undertook a structural protein chemistry approach. Using CD and fluorescence spectroscopy, we were next able to determine whether PCRan1p was indeed unfolding at higher overall temperatures, thus explaining the reduced activity observed under these conditions. CD was first used to determine the effect of temperature on the secondary structure of the protein (Figure 5A). Although the presence of the V5 beads used for protein purification increased the noise of the signal, a control reaction with only V5 beads did not have a protein signal. Thus, the ellipticity observed with the PCRan1p sample was attributed to the protein. The thermal denaturation profile of PCRan1p was striking. According to the denaturation profile, PCRan1p seems to begin altering its conformation around 15°C, with continuing shift occurring up to 30°C. This was interesting, because optimal activity of the enzyme in prior experiments was repeatedly shown to occur between 10 and 25°C. These findings suggest that PCRan1p optimally phosphorylates its substrate after some level of conformational change has already occurred. Additionally, we assessed the ability of PCRan1p to refold after the thermal denaturation of the protein (see Materials and Methods). As the temperature decreased, the refolding profile of PCRan1p did not follow the same pattern in reverse, further suggesting that this process is not reversible.
Figure 5.
(A) Circular dichroism of RTS-generated PCRan1p conjugated to V5 beads was used to determine a thermal denaturation and refolding profile by measuring ellipticity at 222 nm from 4 to 45°C. In addition, V5 beads were assayed alone to determine if noise was produced as a result of interference of the beads in the solution. Diamonds, V5 beads; squares, PCRan1 unfolding; triangles, PCRan1 refolding. (B) Far-ultraviolet (UV) spectra of RTS-generated PCRan1p conjugated to V5 beads were determined at five different temperatures ranging from 10 to 45°C, and the minima were plotted to demonstrate the trend line. (C) Fluorescence spectroscopy, with an excitation at 294 nm and emission measured from 310 to 400 nm, was performed to determine the conformational profile of RTS-generated PCRan1p conjugated to V5 beads. A V5 bead control was subtracted from all samples and then plotted at their respective maxima. (D) RTS-generated PCRan1p conjugated to V5 agarose beads was incubated at 25, 30, 37, or 45°C for 1 hour, followed by performing an in vitro kinase assay at 25 or 45°C for 1 hour to determine the reversibility of the unfolding reaction using PHAS-I as the substrate.
Due to the unique nature of the thermal denaturation profile demonstrated for PCRan1p, additional experiments were performed to confirm these initial observations. Accordingly, far-UV spectra were captured after incubating PCRan1p at the respective temperature for 10-minute intervals (Figure 5B). By plotting the wavelength where the minimum ellipticity was observed in the far-UV scan at each temperature, it was apparent that there was a shift in the minimum as temperature increased. Whereas the PCRan1p protein was relatively stable up to 25°C, shortly after increasing to 30°C a dramatic shift occurred in the minimum (Figure 5B), suggesting that this is where the majority of the conformational change occurred.
The far UV data did not, however, fully corroborate the partial unfolding seen during the CD thermal denaturation experiments. Therefore, to further investigate this phenomenon, fluorescence spectroscopy was undertaken to assess the tertiary structure and determine the level of intrinsic tryptophan fluorescence at the various temperatures (Figure 5C). The hydrophobic nature of tryptophan typically renders these residues into the hydrophobic interior of the protein. Therefore, as a protein unfolds, tryptophan residues become more exposed to the exterior, and the overall level of fluorescence changes accordingly. Using this approach, we determined the wavelength of maximum fluorescence emission. We observed an apparent shift in the overall conformation of the protein that occurred between 25 and 30°C. Between 30 and 45°C, a second transition was observed, again suggesting that there is a secondary event that alters the conformation of PCRan1p near physiological temperature (Figure 5C). Taken together, these data elucidate an interesting biochemical profile for PCRan1p, suggesting a protein that is most active after a partial unfolding event, and that produces much reduced levels of activity as a result of conformational shift at physiological temperatures. Thus, PCRan1p demonstrates unique temperature sensitivity within a native kinase enzyme, regulating life cycle transitions within Pneumocystis.
Lastly, we wanted to verify the nonreversible nature of the conformational event in PCRan1p by using an in vitro kinase assay. In these experiments, PCRan1p was incubated at 25, 30, 37, or 45°C. After the incubation, the kinase reaction was performed at either 25°C to evaluate the reversibility, or 45°C as a negative control. This experiment allowed us to evaluate the nature of the conformational change, and whether the results seen in the thermal refolding assay could be corroborated by the kinase data. We observed that once the sample was incubated above 25°C for 1 hour, it could partially, but not fully, regain its optimal activity. These results were in direct correlation with the temperature at which it was incubated. When incubated at 45°C, only about half of the activity could be returned by performing the kinase assay at 25°C (46.1 ± 3.3% reversible; P = 0.003 compared with kinase reactions performed at 45°C, showing zero reversibility). However, at 37°C, the kinase was able to regain 58.3% of its optimal activity, and at 30°C, the kinase regained 65.8% of its optimal activity (Figure 5D). This suggests that, for the most part, the protein unfolding can only be partially reversed simply by returning the enzyme to a more favorable temperature condition.
DISCUSSION
Pneumocystis and other organisms have clearly evolved key mechanisms to regulate their life cycle by reacting to critical environmental signals. Currently, the majority of the information known about the Pneumocystis life cycle, such as the transformation of the organism from cystic to trophic forms within the host lung, has been deduced from ultrastructural studies of the organism (7–9). Almost nothing is known about life cycle regulation when the organisms are outside the host (5). Recent molecular studies have begun to unravel some of the puzzle surrounding P. carinii propagation and life cycle control through the identification of a number of regulatory kinases and related molecules (11, 12, 20, 22–24). The current study was undertaken to investigate three such protein kinases shown through prior studies to be crucial in the execution of critical life cycle junctures (10–12). Specifically, these studies were designed to investigate how these molecules react to common environmental signals to glean important details about the regulation of the Pneumocystis life cycle. Interestingly, we have demonstrated that the activity of the key inhibitory PCRan1p kinase, responsible for suppressing entry into meiosis, is strongly controlled by environmental temperature.
Although PCSte20 and PCCbk1 have been shown in previous studies to be integral in the mating pathway in P. carinii, a process that is believed to occur while the organisms are attached to human alveolar epithelial cells (11), the function of PCRan1 has been shown to participate throughout the life cycle of the organism. The exertion of PCRan1p kinase activity prevents the premature entry of the organism into meiosis, presumably allowing Pneumocystis to initiate meiosis only when conditions are favorable (10). Although these favorable conditions have not been formerly determined, these studies have allowed us to infer the optimal conditions under which PCRan1p activity is favored to permit Pneumocystis to maintain a viable life cycle.
Our initial characterization demonstrated that both the PCSte20p and PCCbk1p kinases function best at or near physiological conditions. In contrast, PCRan1p was found to have restricted activity and to exert a reduced function above 30°C, while it was able to phosphorylate the substrate protein over a wide range of pH. These data strongly suggest that ambient temperature represents an important environmental signal used by the organism to prevent entry into meiosis under hostile conditions. Thus, PCRan1p phosphorylates its substrate at subphysiological temperatures, such as those found when the organism is outside the host lung, to block entry into meiosis while ex vivo. This unique temperature-sensitive activity profile is further shared by the Pat1p homolog present in S. pombe, the closest phylogenetic relative of Pneumocystis.
Prior mouse studies have shown that exposure of the animals to Pneumocystis occurs within several hours of birth. These studies indicate that Pneumocystis species are ubiquitous in the environment, and are frequently contacted through inhalation or fomites (25). However, although these studies have determined the common presence of Pneumocystis outside the host, there have been no data to suggest how it may survive in such environments. The current study suggests a role for PCRan1p in maintaining stasis while outside of the host. In typical infection, the trophic form consists of 90% of the P. carinii forms present in the infected lung (26). However, as the infection progresses, there is an increase in the total number of cystic forms. It can be inferred from our data that PCRan1p may be one of many proteins that become up-regulated in the cystic form to aid in the maintenance of the organism. The results of these studies suggest that PCRan1p is most active in an environment outside the host, whether the reservoir is soil, fomites, et cetera. It is highly likely that the maintenance of the thick-walled cyst form is necessary to survive outside the host, and is the form putatively transmitted to another susceptible recipient (2).
In an effort to better understand the mechanism of the suppression of PCRan1p activity at physiological temperatures in the range of 37°C, protein structural experiments were further undertaken. All three approaches employed indicated that the overall conformation of PCRan1p does begin to shift as the temperature increases, suggesting native temperature sensitivity of the protein. However, it became apparent through the CD experiments that the conformation of the protein appears to be in flux. By measuring the secondary structure of PCRan1p, we were able to discern a pattern whereby it appears that conformational shift in PCRan1p begins to occur somewhere around 15°C. Interestingly however, optimal activity of PCRan1p was not noted until 25°C. This suggests that the protein actually assumes its most active structure as PCRan1p undergoes a slight conformational change. However, complete denaturation is, of course, associated with conditions with loss of activity. Such a mechanism would be most useful in an organism in which its ability to propagate is tightly regulated by its immediate surroundings.
Notably, our previous work has demonstrated some function of PCRan1p at 37°C (10). This suggests that, although activity is dramatically reduced in vivo, the enzyme still maintains a basal capacity to phosphorylate and prevent the activation of meiosis by PCMei2p. Subsequently, although meiosis must be constitutively prevented in the intracystic bodies (which make up the entire known ex vivo form of Pneumocystis), the reduced activity of PCRan1p in vivo could allow for better modulation of activation based on host conditions and signaling pathways in the cell. Therefore, in the host, PCRan1p activity would become reduced, rendering it more susceptible to inactivation by additional means. However, when Pneumocystis is expelled from the host lung, the relative structure of PCRan1p would then favor a conformation with which it could strongly phosphorylate its target, and constitutively prevent meiosis from occurring until more favorable conditions are found, rendering the organism dormant.
Taken together, our studies strongly support that Pneumocystis PCRan1p appears to be a natively temperature-sensitive protein. Although temperature-sensitive proteins are a normal part of the metabolism of laboratory strains (including temperature-sensitive conditional lethal mutants), the existence and description of a temperature-sensitive kinase in nature, particularly one that may be thus exploited to govern life cycle control, is both exceedingly novel and interesting. Such phenomena have been noted before in other species, such as cholera toxin A from Vibrio cholerae (27). However, although cholera toxin A does become unfolded natively at 37°C, eukaryotic cofactors have been found to mitigate this instability (27). Such a phenomenon suggests the coevolution of these proteins to the needs of the organism for survival under their specific life cycle conditions, particularly involving a life cycle requiring proliferation within the mammalian lung and transmission of dormant forms though the external environment.
Overall control of the Pneumocystis life cycle remains largely unknown. Recent data from our laboratory have documented the presence of a conserved meiotic pathway in this fungus (10), representing the first molecular evidence supporting meiosis and a corresponding sexual life cycle phase of the organism. Our current studies of PCRan1p further reveal a protein with interesting biochemical qualities, including the ability to phosphorylate its substrate over a wide range of pH, and activity that is strongly regulated by environmental temperature. These data strongly indicate that these temperature and pH controls are essential in the correct progression of the life cycle and overall survival of Pneumocystis based on its ambient conditions.
Further studies will be required to understand how the temperature-based regulatory effects of PCRan1p interact on the whole with the meiotic pathway of P. carinii. In addition, other molecules necessary for the progression of meiosis, such as homologs to Mei3 in S. pombe and meiotic RNA (28, 29), must be discovered to fully understand Pneumocystis meiotic regulation. Further investigations also must be completed to determine whether regulation in P. carinii is fully dependent on the presence or absence of specific thermal conditions, or if secondary environmental signals also play a role. However, the description of this novel regulatory pathway represents an important advance in our understanding of the control pathways used by the organisms to ensure life cycle regulation and propagation.
A better understanding of life cycle regulation and environmental cues that regulate Pneumocystis proliferation may eventually provide insights into the in vitro cultivation of this intractable organism, as well as providing additional targets for therapeutic exploitation. Although these observations on the thermal regulation of PCRan1p by itself will not likely make Pneumocystis cultivation a reality, such information tied together with further studies of the molecular regulation of life cycle progression, may eventually make this possible.
Acknowledgments
The authors thank Deanne Hebrink for her assistance with the animal studies and propagation and purification of Pneumocystis. In addition, they thank Dr. James Maher, III, for his thoughtful perspectives to the advancement of this project.
This work was supported by National Institutes of Health grant RO1-HL55934 (A.H.L.).
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2008-0098OC on March 13, 2009
Conflict of Interest Statement: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Limper AH, Offord KP, Smith TF, Martin WJ II. Pneumocystis carinii pneumonia: differences in lung parasite number and inflammation in patients with and without AIDS. Am Rev Respir Dis 1989;140:1204–1209. [DOI] [PubMed] [Google Scholar]
- 2.Thomas CF Jr, Limper AH. Current insights into the biology and pathogenesis of Pneumocystis pneumonia. Nat Rev Microbiol 2007;5:298–308. [DOI] [PubMed] [Google Scholar]
- 3.Wakefield AE, Lindley AR, Ambrose HE, Denis CM, Miller RF. Limited asymptomatic carriage of Pneumocystis jiroveci in human immunodeficiency virus–infected patients. J Infect Dis 2003;187:901–908. [DOI] [PubMed] [Google Scholar]
- 4.Larsen HH, von Linstow ML, Lundgren B, Hogh B, Westh H, Lundgren JD. Primary Pneumocystis infection in infants hospitalized with acute respiratory tract infection. Emerg Infect Dis 2007;13:66–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morris A, Beard CB, Huang L. Update on the epidemiology and transmission of Pneumocystis carinii. Microbes Infect 2002;4:95–103. [DOI] [PubMed] [Google Scholar]
- 6.Cushion MT, Beck JM. Summary of Pneumocystis research presented at the 7th international workshop on opportunistic protists. J Eukaryot Microbiol 2001:101S–105S. [DOI] [PubMed]
- 7.Matsumoto Y, Yoshida Y. Sporogony in Pneumocystis carinii: synaptonemal complexes and meiotic nuclear divisions observed in precysts. J Protozool 1984;31:420–428. [DOI] [PubMed] [Google Scholar]
- 8.Haidaris PJ, Wright TW, Gigliotti F, Fallon MA, Whitbeck AA, Haidaris CG. In situ hybridization analysis of developmental stages of Pneumocystis carinii that are transcriptionally active for a major surface glycoprotein gene. Mol Microbiol 1993;7:647–656. [DOI] [PubMed] [Google Scholar]
- 9.Itatani CA, Marshall GJ. Ultrastructural morphology and staining characteristics of Pneumocystis carinii in situ and from bronchoalveolar lavage. J Parasitol 1988;74:700–712. [PubMed] [Google Scholar]
- 10.Burgess JW, Kottom TJ, Limper AH. Pneumocystis carinii exhibits a conserved meiotic control pathway. Infect Immun 2008;76:417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kottom TJ, Kohler JR, Thomas CF Jr, Fink GR, Limper AH. Lung epithelial cells and extracellular matrix components induce expression of Pneumocystis carinii Ste20, a gene complementing the mating and pseudohyphal growth defects of Ste20 mutant yeast. Infect Immun 2003;71:6463–6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kottom TJ, Limper AH. Pneumocystis carinii cell wall biosynthesis kinase gene Cbk1 is an environmentally responsive gene that complements cell wall defects of Cbk-deficient yeast. Infect Immun 2004;72:4628–4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beach D, Rodgers L, Gould J. Ran1+ controls the transition from mitotic division to meiosis in fission yeast. Curr Genet 1985;10:297–311. [DOI] [PubMed] [Google Scholar]
- 14.Iino Y, Yamamoto M. Negative control for the initiation of meiosis in Schizosaccharomyces pombe. Proc Natl Acad Sci USA 1985;82:2447–2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watanabe Y, Shinozaki-Yabana S, Chikashige Y, Hiraoka Y, Yamamoto M. Phosphorylation of RNA-binding protein controls cell cycle switch from mitotic to meiotic in fission yeast. Nature 1997;386:187–190. [DOI] [PubMed] [Google Scholar]
- 16.Leberer E, Ziegelbauer K, Schmidt A, Harcus D, Dignard D, Ash J, Johnson L, Thomas DY. Virulence and hyphal formation of Candida albicans require the Ste20p-like protein kinase CaCla4p. Curr Biol 1997;7:539–546. [DOI] [PubMed] [Google Scholar]
- 17.Bidlingmaier S, Weiss EL, Seidel C, Drubin DG, Snyder M. The Cbk1p pathway is important for polarized cell growth and cell separation in Saccharomyces cerevisiae. Mol Cell Biol 2001;21:2449–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Colman-Lerner A, Chin TE, Brent R. Yeast Cbk1 and Mob2 activate daughter-specific genetic programs to induce asymmetric cell fates. Cell 2001;107:739–750. [DOI] [PubMed] [Google Scholar]
- 19.McNemar MD, Fonzi WA. Conserved serine/threonine kinase encoded by Cbk1 regulates expression of several hypha-associated transcripts and genes encoding cell wall proteins in Candida albicans. J Bacteriol 2002;184:2058–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kottom TJ, Thomas CF Jr, Mubarak KK, Leof EB, Limper AH. Pneumocystis carinii uses a functional Cdc13 B-type cyclin complex during its life cycle. Am J Respir Cell Mol Biol 2000;22:722–731. [DOI] [PubMed] [Google Scholar]
- 21.Hahn PY, Evans SE, Kottom TJ, Standing JE, Pagano RE, Limper AH. Pneumocystis carinii cell wall beta-glucan induces release of macrophage inflammatory protein-2 from alveolar epithelial cells via a lactosylceramide-mediated mechanism. J Biol Chem 2003;278:2043–2050. [DOI] [PubMed] [Google Scholar]
- 22.Kottom TJ, Thomas CF Jr, Limper AH. Characterization of Pneumocystis carinii Phr1, a pH-regulated gene important for cell wall integrity. J Bacteriol 2001;183:6740–6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomas CF, Anders RA, Gustafson MP, Leof EB, Limper AH. Pneumocystis carinii contains a functional cell-division–cycle Cdc2 homologue. Am J Respir Cell Mol Biol 1998;18:297–306. [DOI] [PubMed] [Google Scholar]
- 24.Vohra PK, Puri V, Kottom TJ, Limper AH, Thomas CF Jr. Pneumocystis carinii Ste11, an Hmg-BOX protein, is phosphorylated by the mitogen activated protein kinase Pcm. Gene 2003;312:173–179. [DOI] [PubMed] [Google Scholar]
- 25.Icenhour CR, Rebholz SL, Collins MS, Cushion MT. Early acquisition of Pneumocystis carinii in neonatal rats using targeted PCR and oral swabs. J Eukaryot Microbiol 2001;135S–136S. [DOI] [PubMed]
- 26.Wyder MA, Rasch EM, Kaneshiro ES. Quantitation of absolute Pneumocystis carinii nuclear DNA content: trophic and cystic forms isolated from infected rat lungs are haploid organisms. J Eukaryot Microbiol 1998;45:233–239. [DOI] [PubMed] [Google Scholar]
- 27.Pande AH, Scaglione P, Taylor M, Nemec KN, Tuthill S, Moe D, Holmes RK, Tatulian SA, Teter K. Conformational instability of the cholera toxin A1 polypeptide. J Mol Biol 2007;374:1114–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watanabe Y, Yamamoto MS. Pombe mei2+ encodes an RNA-binding protein essential for premeiotic DNA synthesis and meiosis I, which cooperates with a novel RNA species meiRNA. Cell 1994;78:487–498. [DOI] [PubMed] [Google Scholar]
- 29.McLeod M, Beach D. A specific inhibitor of the Ran1+ protein kinase regulates entry into meiosis in Schizosaccharomyces pombe. Nature 1988;332:509–514. [DOI] [PubMed] [Google Scholar]