Abstract
Background
Tonoplast intrinsic proteins (TIPs) are widely used as markers for vacuolar compartments in higher plants. Ten TIP isoforms are encoded by the Arabidopsis genome. For several isoforms, the tissue and cell specific pattern of expression are not known.
Results
We generated fluorescent protein fusions to the genomic sequences of all members of the Arabidopsis TIP family whose expression is predicted to occur in root tissues (TIP1;1 and 1;2; TIP2;1, 2;2 and 2;3; TIP4;1) and expressed these fusions, both individually and in selected pairwise combinations, in transgenic Arabidopsis. Analysis by confocal microscopy revealed that TIP distribution varied between different cell layers within the root axis, with extensive co-expression of some TIPs and more restricted expression patterns for other isoforms. TIP isoforms whose expression overlapped appeared to localise to the tonoplast of the central vacuole, vacuolar bulbs and smaller, uncharacterised structures.
Conclusion
We have produced a comprehensive atlas of TIP expression in Arabidopsis roots, which reveals novel expression patterns for not previously studied TIPs.
Background
Tonoplast intrinsic proteins (TIPs) are a subfamily of aquaporins, small integral membrane proteins belonging to the major intrinsic protein (MIPs) family. Aquaporins form channels that facilitate the movement of water, small uncharged solutes (glycerol, urea, boric acid, silicic acid, hydrogen peroxide) and gases (ammonia, carbon dioxide) across biological membranes. (For recent reviews see [1,2]). TIPs have been either detected, or predicted to localise, to the tonoplast [3].
The Arabidopsis genome encodes 10 TIP isoforms [4], further classified into five subgroups: three γ-TIP (TIP1), three δ-TIP (TIP2), the seed-specific α- and β-TIP (TIP3;1 and TIP3;2), one ε-TIP (TIP4;1) and one ζ-TIP (TIP5;1).
Several TIP isoforms have been studied in detail as regards their expression [3,5,6] and function [7,8]. TIPs have also been widely employed as intracellular markers for vacuolar biogenesis and identity. Immunofluorescence experiments in root tips and mature embryos of different plant species led to the identification of separate vacuolar compartments within the same cell [9-13]. These experiments indicated an association of γ-TIP (TIP1;1) with vegetative, lytic-type vacuoles and of α-TIP (TIP3;1) and δ-TIP (TIP2;1) with protein storage vacuoles. The detection of different TIP isoforms on separate tonoplasts provided evidence for multiple, functionally different vacuolar compartments within plant cells (reviewed in Frigerio et al, 2008). Recently we compared expression of TIP3;1 and TIP1;1 in Arabidopsis and found minimal overlap in the timing of their expression, with TIP3;1 being abundant in embryos of mature seeds and sharply declining during seed germination, to be replaced by TIP1;1 [14]. The latter was not present in root tips, thus raising some doubt as to the applicability of these particular isoforms as vacuolar markers in Arabidopsis [5,14]. As the investigation was limited to the three TIP isoforms against which peptide antibodies were raised for the immunofluorescence studies [10], the possibility remained that other TIP family members with similar immunoreactivity may be present in different vacuoles within Arabidopsis root tissues. Indeed, the tissue-specificity of expression of some TIP family members has not yet been investigated in detail.
In this report we have mapped the expression of every Arabidopsis TIP isoform that is predicted to be present in root tissues by transcriptomic analysis [15]. This excludes TIP3;1 and TIP3;2 (α and β-TIP), which have seed-specific expression patterns [14,16]; Gattolin and Frigerio, unpublished), and both TIP1;3 (γ-TIP3) and TIP5;1 (ζ-TIP), which are predicted by bioinformatic analysis to be expressed solely in floral organs and pollen [15-17].
Our results indicate that expression of some TIP isoforms under their native promoters is remarkably tissue and cell-specific. In general, when multiple isoforms are co-expressed in the same cell, they appear to localise mainly to the tonoplast of the central vacuole. Our identification of the sites of expression of every TIP isoform paves the way to understanding TIP specialisation and function in Arabidopsis root tissues.
Results
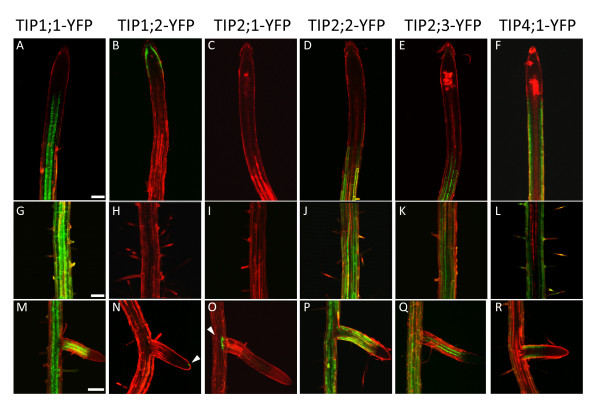
In addition to the fluorescent TIP reporters we generated previously for TIP1;1 (γ-TIP1; At2g36830) and TIP2;1 (δ-TIP1; At3g16240) [14], we cloned the genomic sequences of not previously studied isoforms: TIP1:2 (γ-TIP2; At3g26520), TIP2;2, TIP2;3 (δ-TIP2 and δ-TIP3; At4g17340 and At5g47450) and TIP4;1 (ε-TIP; At2g25810). We produced chimeric constructs in which either YFP or monomeric RFP were fused in frame to the C-terminus of each TIP genomic sequence (including their promoter regions, 5' UTR and introns), and generated transgenic plants which were analysed for TIP-XFP expression patterns by confocal laser scanning microscopy (CLSM). We first observed TIP-YFP expression at low magnification. 8-day old roots from seedlings expressing individual YFP-tagged TIPs were stained with propidium iodide and analysed by CLSM. With the possible exception of TIP1;2, no YFP-tagged TIP isoforms yielded a detectable signal in the root cap or meristem (Fig. 1, panels A to F). In general, TIP-YFP expression initiates at the elongation zone. While TIP1;1-YFP and TIP4;1-YFP are detectable from the base of the elongation zone (panels A and F), TIP2;2-YFP and TIP2;3-YFP expression is first observed at the zone of transition with the differentiation zone (panels D and E). The onset of fluorescence occurs in different cell types depending on the isoforms. TIP1;1-YFP is initially visible in endodermal cells, before extending to every cell type in the differentiation zone (Fig. 1, compare panels A and G). This pattern is faithfully replicated in lateral roots (Fig. 1M). TIP1;1 expression is strongest at the differentiation zone (Fig. 1G). TIP2;2-YFP becomes first detectable in the cortex and epidermis, but its expression extends to the pericycle as the root matures (Fig. 1, compare panels D and J). TIP2;3-YFP has a similarly widespread distribution in more mature root axes but its expression initiates in the pericycle, then extends to cortex and epidermal cells (Fig. 1, panels E and K). Again, the initial expression patterns of TIP2;2 and TIP2;3 are mirrored in the lateral roots (Fig 1, panels P and Q). In contrast to the previous isoforms, TIP4;1-YFP is only expressed in epidermal and (less strongly) in cortical cells of the differentiation zone (Fig. 1, panels F and L), with fluorescence decreasing in more mature parts of the root where lateral roots emerge (Fig. 1R).
Figure 1.
Expression patterns of TIP isoforms in Arabidopsis roots. 8-day old roots from the indicated transgenic lines were excised, stained with propidium iodide for 2 min and visualised by CLSM. The images show representative results for each construct. The signals from YFP (green) and propidium iodide fluorescence (red) are merged. Top panels: single optical sections of the root tips, Middle panels: single optical sections of root differentiation zones. Bottom panels: maximal projection of 16 optical z sections (4 μm step-size) through mature root axes and young lateral roots. Scale bars: 100 μm.
In the case of TIP1;2, expression seems to be exclusively limited to the root cap and the columella (Fig. 1B). A very limited YFP signal can also be detected in the same region of the young lateral root (Fig 1N, arrowhead) and in older lateral roots (Additional file 1A).
Perhaps the most remarkable expression pattern observed is that of TIP2;1, which in 8-day old roots is only detectable in a small region at the base of the lateral roots (Fig. 1O, arrowhead).
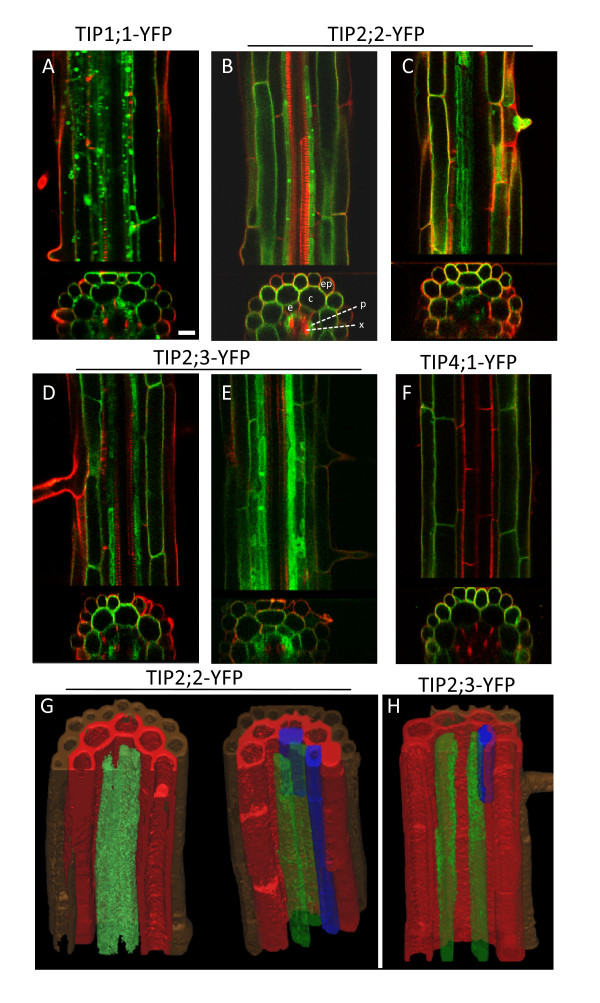
Having identified the general patterns of expression of the different isoforms at low magnification, we then studied the cell-specificity of TIP-YFP expression in more detail. We analysed propidium iodide -stained roots by CLSM by performing optical z-sections through differentiation zones at 63× magnification (Fig. 2).
Figure 2.
Cell-type specificity of TIP-YFP expression in the root axis. 8-day old roots from the indicated transgenic lines were excised, stained with propidium iodide for 2 min and visualised by CLSM. Stacks of 80 optical z sections (1 μm step-size) were collected from root axes at the differentiation zone. The images show representative results for each construct. A to F: for each panel, the top section shows a single xy optical section, and the bottom section shows the xz projection of the whole image stack, revealing the cross section of the root axis. The signals from YFP fluorescence (green) and propidium iodide fluorescence (red) are merged. G and H: the YFP fluorescence trace from representative image stacks for the indicated transgenic lines was reconstructed, segmented and rendered in 3D with Mimics 12.1. The different tissues are colour-coded as follows: brown, epidermis; red, cortex; blue, endodermis; green, pericycle. Ep, epidermis; c, cortex; e, endodermis; p, pericycle; x, xylem. Scale bar: 20 μm.
TIP1;1-YFP is clearly expressed in epidermis and cortex, but its expression is particularly strong in the endodermis and pericycle (Fig. 2A). Here TIP1;1-YFP highlights numerous bright circular structures in the lumen of the central vacuole. We hypothesise these are vacuolar 'bulbs', which have previously been described as tonoplast invaginations, which occur independently of the ectopic expression of XFP-tagged membrane proteins, and where TIP1;1-GFP is concentrated [18,19]. It is however difficult in some cases to observe a continuity between these structures and the central vacuole tonoplast.
At higher magnification, the overlapping patterns of expression of TIP2;2-YFP and TIP2;3-YFP are confirmed. Both are present in pericycle cells, particularly in the rows of pericycle cells that form the xylem poles [20]. Both TIP-YFPs tend to be absent from the endodermis (Fig. 2, panels B and D), although we could detect discontinuous endodermal expression at various positions along most root axes (Fig. 2, panels C and E; and highlighted in blue in panels G and H).
In contrast to the previous isoforms labelling inner root cell layers, TIP4;1 expression is clearly restricted to the root epidermis and cortex, with no signal detectable in the inner layers (Fig. 2F).
Localisation of TIP1;1 and TIP1;2
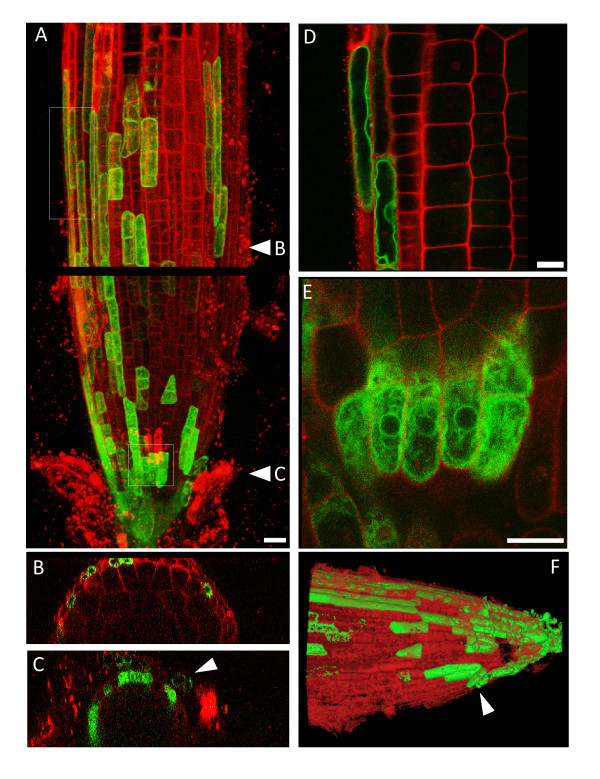
Having analysed the TIPs with the broadest expression patterns, we focussed on the two TIPs which seem to have a more limited expression range in roots. TIP1;2-YFP presented a patchy distribution in cells of the root cap (Fig 1B). To ascertain that this was not an artefact due to expression of our chimeric gene, we also generated a construct (YFP-TIP1;2) where YFP was fused downstream of the promoter and 5'UTR and in frame with the 5' of the TIP coding sequence. In transgenic plants, YFP-TIP1;2 presents a similar expression pattern to TIP1;2-YFP, thus ruling out YFP fusion artefacts (compare Fig. 3A with Fig. 1B). Expression is confined to the columella and the lateral root cap [21], with the labelled cells disappearing at the boundary with the elongation zone (Fig. 3A). Some of the labelled cells are in the process of detaching from the root (Fig. 3, panels C and F, arrowheads), suggesting that they may correspond to 'border-like' cells [22]. The distribution of YFP-TIP1;2 is therefore radically different to that observed for its paralogue, TIP1;1-YFP, which has the widest pattern of expression but is excluded from the root tip, including the root cap (Fig. 1, panels A to M).
Figure 3.
YFP-TIP1;2 is expressed in the root cap. 8-day old roots from the indicated transgenic lines were excised, stained with propidium iodide for 2 min and visualised by CLSM. Stacks of 80 optical z sections (1 μm step-size) were collected from root tips. The images show a representative result for this construct. The signals from YFP fluorescence (green) and propidium iodide fluorescence (red) are merged. A: maximal 3D projection of the root tip at the base of the elongation zone. The image shows two adjacent z-stacks of the same root, separated by a black line. B and C: xz projections of the image stack in panel a, revealing two cross-sections of the root axis, taken in the regions of the root indicated by the arrowheads in A. D and E: the regions indicated by dotted boxes in A were observed at high magnification. Single optical sections are shown. Note YFP-TIP1;2 in the ER of young root cap cells and in the tonoplast of root cap cells closer to the elongation zone. F: The fluorescent traces from YFP (green) and propidium iodide (red) from the image stack in panels A were reconstructed, segmented and rendered in 3D with Mimics 12.1. Scale bars: (a), 20 μm; (d) and (e), 10 μm.
At the subcellular level, YFP-TIP1;2 localises to the endoplasmic reticulum (ER) of young root cap cells (Fig 3e: note the characteristic reticular pattern and the nuclear envelope; see also Additional file 1B). The chimeric protein is mostly found on the tonoplast of elongated lateral root cap cells (Fig. 3D). This is likely to reflect different stages of TIP1;2 trafficking in cells of different ages, rather than impaired capacity to reach the tonoplast. This is further demonstrated by the fact that in the epidermis of cotyledonary cells, where TIP1;2 is uniformly expressed, the fusion protein appears to localise to the tonoplast (Additional file 1C-D).
TIP2;1 is localised in lateral root primordia
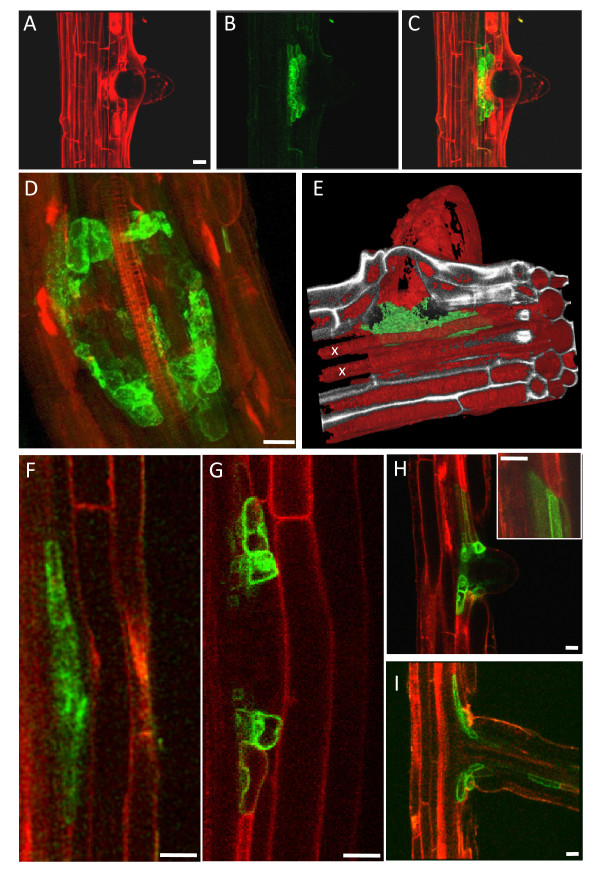
We have previously shown that TIP2;1 expression becomes detectable in old root regions nearing the hypocotyl, and is then widespread in hypocotyl and cotyledonary leaves [14]. We did not initially notice expression in young roots, but closer analysis revealed that in 8-day old roots TIP2;1-YFP has a very specialised expression pattern (Fig. 4). The YFP signal is detected in a ring-like cluster at the base of emerging lateral roots (Fig. 4, panels A-D). In very early lateral root primordia (LRP), TIP2;1 expression is detectable in 2-4 cells at the LRP. As the LRP grows further, the number of cells expressing TIP2;1-YFP increases but remains confined to a cluster underlying the base the lateral root (Fig. 4, panels F-I). In rare cases, when the lateral root is fully emerged, the expression of TIP2;1-YFP can extend to some cells within the lateral root axis (Fig 4I). Co-labelling with propidium iodide shows that the TIP2;1 expressing cells are located in close proximity to the xylem (fig. 4E), suggesting a pericycle localisation. Co-expression with TIP2;3-RFP, which we found to be enriched in the pericycle (Fig. 2, panels D, E, H) confirms that TIP2;1-YFP expression originates from pericycle cells (Fig. 4, panels F-I). This indicates that the initial expression of TIP2;1-YFP is likely to occur in the LRP founder cells. Remarkably, the expression of TIP2;1-YFP and TIP2;3-RFP appears to be mutually exclusive, with a clear boundary between cells expressing one or the other isoform (Fig. 4H, inset; see Additional file 2 for individual channels).
Figure 4.
TIP 2;1-YFP expression in lateral root primordia. A-E: 8-day old roots from TIP2;1-YFP transgenic seedlings were excised, stained with propidium iodide and visualised by CLSM. Stacks of 80 optical z sections (1 μm step-size) were collected from mature root axes. The images show representative results for this construct. Maximal projections of the z-stacks are shown, with the individual signals for YFP (A), propidium iodide (B) or the merged signals (C and D). E: The fluorescent traces from YFP (green) and propidium iodide (red) from the image stack in panels (A-C) was reconstructed, segmented and rendered in 3D with Mimics 12.1. Note that the TIP2;1-YFP-expressing cells are in close proximity to the xylem (labelled with x). F-I: Roots from 8-day old transgenic seedlings expressing TIP2;1-YFP (green) and TIP2;3-RFP (red) were imaged. Sequential stages of lateral root development are shown. Inset in H: note the boundary between pericycle cells expressing TIP2;3-RFP (top) and TIP2;1-YFP (bottom). Scale bars: 20 μm.
Overlapping TIP isoforms are mostly detectable at the central vacuole tonoplast
We have shown that the various TIP isoforms under study present diverse tissue specificity within roots. Several isoforms, however, are co-expressed in certain tissues, namely TIP1;1, TIP2;2, TIP2;3, and TIP4;1. In order to ascertain whether these isoforms were specific to distinct vacuolar compartments, we focused on the subcellular localisation of selected pairs of the above isoforms, tagged with different spectral variants of fluorescent proteins and co-expressed in transgenic Arabidopsis (Fig. 5).
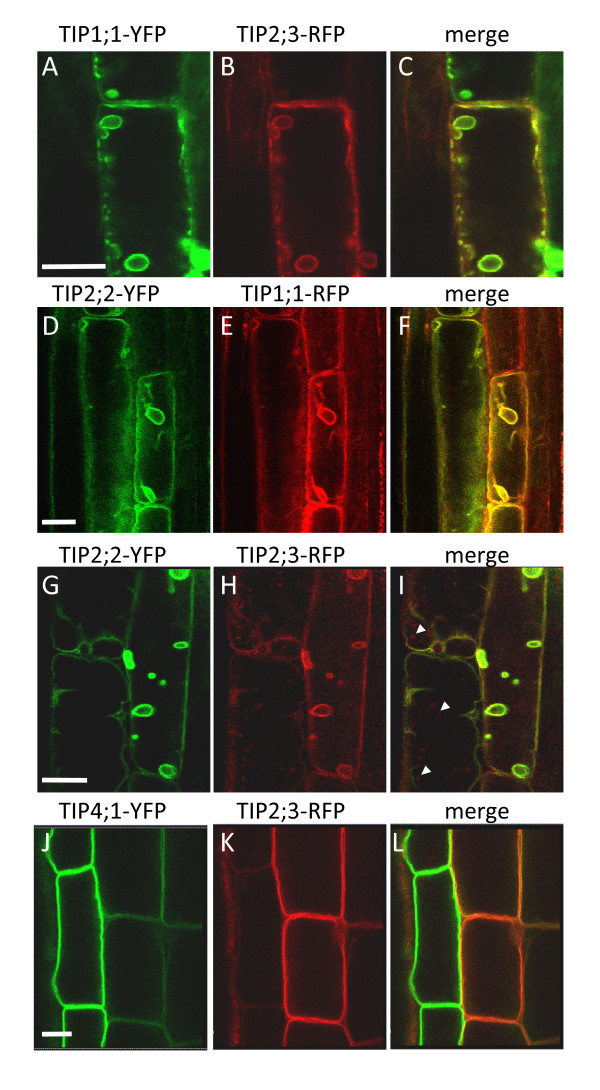
Figure 5.
Overlapping TIP isoforms are mainly detected at the tonoplast of the central vacuole. Transgenic seedlings co-expressing the indicated TIP-YFP and TIP-RFP constructs were grown for 8 days on MS medium--agar plates. Roots were excised and visualised by CLSM. (A, D, G, J): YFP fluorescence (green); (B, E, H, K): RFP fluorescence (red); (C, F, I, L): merged images. Arrowheads in panel I indicate structures labelled by TIP2;3-RFP but not TIP2;2-YFP. Scale bars:10 μm.
The individual TIP expression patterns in double transgenic lines mirrored those observed in the lines expressing individual isoforms (Additional file 3). The widespread TIP1;1-YFP and TIP2;3-RFP are co-expressed in epidermis, cortex and pericycle cells (Fig. 2). In these tissues, both proteins are detected on the tonoplast of the central vacuole (Fig. 5A-C). Both the tonoplast and the smaller, bulb-like vacuolar structures [18] are labelled. Likewise, TIP2;2-YFP and TIP1;1-RFP mostly label the same tonoplast in the cell layers where they are co-expressed (Fig. 5D-F). TIP2;2-YFP and TIP2;3-RFP, which almost overlap in root tissues (Fig. 2), are also co-localised on tonoplast and 'bulbs' (Fig. 5G-I). Occasionally, TIP2;3-RFP highlighted smaller vesicular structures that did not appear to contain TIP2;2-YFP (Fig 5I, arrowheads). The nature of these structures was not investigated further. Finally, TIP4;1-YFP, which is restricted to epidermis and cortex (Fig. 2F), co-localises with TIP2;3 in those tissues (Fig. 5J-L). Note that the relative abundance of these two isoforms mirrors the pattern observed in single isoform localisation, with TIP4;1 expression being strongest in the epidermis and weaker in the cortex (Fig. 2F), and TIP2;3 expression being stronger in cortex but weaker in epidermis (Fig. 2, panels D-E).
Taken together, these co-expression results indicate that each TIP isoform-fluorescent protein fusion we analysed is predominantly found at the central vacuole tonoplast in Arabidopsis root tissues.
Discussion
We have produced a complete expression map for all members of the TIP family that are present in Arabidopsis root tissues, including isoforms not previously studied. The use of XFP fusions to TIP genomic sequences allowed us to investigate both the tissue specificity and the subcellular localisation of these proteins.
In general, our fluorescent reporter - TIP localisation data correlate well with the relative TIP transcript levels, as observed by microarray analysis [15,23,24], with the exception of TIP1;2. TIP1;2 is indeed the isoform with the highest level of mRNA expression in the root cap [23], which matches our observations (Fig. 3). However, transcript levels for TIP1;2 have also been shown to be almost as high as TIP1;1 throughout the root axis [15,23,24]. We can only speculate at this stage that post-transcriptional control processes prevent TIP1;2-YFP protein from being detectable in these tissues.
TIP1;1 is the most widely expressed isoform along the root axis. TIP2;2 and 2;3 have very similar expression patterns, with their expression being low in the endodermis, but high in the xylem pole pericycle. It appears that TIP2;1 becomes strongly expressed in the pericycle when this undergoes differentiation to form the lateral root primordium (Fig. 4). This narrow range of localisation of TIP2;1 is intriguing. Transcripts of the maize aquaporin ZmTIP1 were also localised in the lateral root by in situ hybridisation, but every cell in the LRP seemed to contain the transcript [25]. TIP2;1 can therefore be considered an additional marker for the Arabidopsis LRP margins, alongside the auxin efflux carriers Pin4 and Pin6 [26] and the transcription factor CUC3 [27], which have a similar localisation. It will be interesting to study the specific role of TIP2;1 in these cells and determine why this stage of lateral root development demands such a precise TIP isoform activation.
As a general observation, we could not detect expression of any of the TIP-XFP fusions under study in the root tip meristem. This lack of expression was previously reported for TIP1;1 both by histochemical detection of GUS fusions [5] and YFP tagging [14,19]. It is of course possible that expression levels of our fusions are too low in this region to be detected by confocal microscopy. However, the fact that the more sensitive histochemical GUS staining also fails to detect expression of TIP1;1, which microarray data indicate is the most abundantly transcribed isoform in roots [16], strongly suggest that the protein is not expressed in the root meristem. This is in contrast with data from other species such as pea and barley, where TIPs have been located in isolated root tip cells [10,12] and in root tip sections by immunohistochemical methods [28]. Analysis in the Olbrich et al. study was performed on 3-day old seedlings. At the same age in Arabidopsis seedlings we could already detect all the TIP isoforms described in this study, with the exception of TIP2;1. However their expression pattern was already the same as observed at 8 days (data not shown). We therefore resolved to present results at 8 days, when the complete set of root TIPs is detectable.
This lack of observable expression in root tips makes it difficult to perform meaningful comparisons between the vacuolar complement of Arabidopsis root tip cells and that of other plant species.
TIP-YFP expression was also not detected in the root vasculature, regardless of the developmental stage. This mirrors observations in barley and pea root sections, where the stele was not labelled by TIP antisera [28]. While it is easy to rationalise the absence of a vacuole in the xylem cells, which underwent autolysis, and in mature sieve elements, which lack true vacuoles (reviewed in [29]) it was somewhat surprising not to find TIPs in the companion and parenchima cells. We think it unlikely that this lack of detection is caused by a loss of sensitivity by the confocal microscope detectors in the inner layers of the roots, because both propidium iodide staining and YFP signal are easily detected in the xylem and xylem pole pericycle, respectively (Fig. 2 and Additional file 3). In addition, we could easily detect 35S::TIP2;1-YFP in the vascular tissue using the same settings (Additional file 4). Accordingly, Boursiac et al [24] recently showed that constitutively expressed TIP1;1-GFP and TIP1;2-GFP clearly label the vascular tissue [24].
Recently it has been shown that Arabidopsis knockout mutants lacking TIP1;1, TIP1;2, or both isoforms, do not have any major defects [7,19]. This is in contrast with drastic defects observed in Arabidopsis upon downregulation of TIP1;1 by RNAi [30]. A possible explanation for the latter result is off-target silencing in the RNAi lines [7]. Our data provide a rationale for the lack of a macroscopic phenotype in the double TIP1 knockouts observed by Schussler et al. We have shown that, in roots, expression of TIP1;1 and TIP1;2 does not appear to overlap, with TIP1;1 being expressed in epidermis, cortex, endodermis and perycycle starting from the elongation zone, and TIP1;2 being restricted to the root cap. As TIP1;1 and 1;2 show different tissue specificities, it seems unlikely that they are reciprocally redundant. In addition, we have shown that other TIP isoforms, namely TIP2;2, TIP2;3 and TIP4;1 would still be present in the tissues lacking TIP1;1 (Fig. 2). It is therefore possible that these remaining isoforms compensate for the lack of TIP1;1 in the knockout. On the other hand, the effect of the absence of TIP1;2 from the root cap may be subtle and may have gone undetected under the experimental conditions adopted for the whole-plant analysis of the double mutants. A lack of phenotype in the aerial parts of the single knockout plants may be explained by the fact that both TIP1;1 and TIP1;2 are expressed in leaves [14] and Additional file 2) and may well be acting redundantly there. As for the double knockout, redundancy may be afforded by TIP2;1 [14] and TIP2;2 [16], which are also expressed in leaves.
Conclusion
We have identified novel patterns of expression of TIP isoforms in Arabidopsis roots. This information may provide a useful starting point for a more targeted approach to dissect the function of individual TIP isoforms in root tissues. It also provides the foundation for further analysis of the intracellular targeting of different TIPs.
Methods
Recombinant DNA and generation of transgenic plants
The constructs encoding native TIP1;1-YFP and native TIP2;1YFP have been described previously [14].
A full list of primers designed to amplify the genomic sequences of the root-expressed TIPs is shown in Additional file 5. Each TIP genomic sequence, including either the complete promoter region (up to the UTR of the gene immediately upstream in the chromosome) or 1.5 Kb of the promoter (if longer than 1.5 Kb), plus 5' UTR and introns, was amplified from total genomic DNA from Arabidopsis thaliana Columbia ecotype. Primers included restriction sites KpnI at the 5' and XhoI at the 3' of the target sequences. Amplified fragments were cloned into the KpnI and XhoI sites of pGREEN0029, upstream of a XhoI/SacI fragment containing the YFP coding sequence and the OCS 3' terminator fragment. A similar strategy was adopted to fuse TIP sequences to RFP, but in this case the forward primers included both KpnI and SacI restriction sites, generating a TIP-RFP cassette that could be mobilized with SacI. To obtain pairwise TIP-YFP/TIP-RFP combinations, selected TIP-RFP cassettes were excised with SacI and ligated into TIP-YFP vectors linearised with SacI, giving rise to constructs harbouring both reporter genes in a tandem. All the chimeric constructs were introduced into strain C58 of Agrobacterium tumefaciens harbouring the pSoup vector [31]. Arabidopsis plants were then transformed using the floral dip method [32].
Confocal analysis and image processing
About 30 seeds from at least 4 independent transgenic lines per construct were germinated onto agar plates containing half-strength Murashige and Skoog (MS) Basal Medium (Sigma-Aldrich) and grown for 8 days at 22°C, in a 16:8 light:dark regime. Roots were excised, mounted in half-strength liquid MS medium and immediately observed with a Leica TSC SP5 confocal laser scanning microscope, using either a 10× (NA 0.3) air or a 63× (NA 1.4) oil immersion objective. In some cases roots were preincubated for 2 min in 10 μg/ml propidium iodide, diluted in half-strength MS medium. YFP was excited at 514 nm and detected in the 525 to 550 nm range. RFP was excited at 561 nm and detected in the 553 to 638 nm range. Propidium iodide was excited at 561 nm and detected in the 650 to 720 nm range. Simultaneous detection of YFP and RFP or YFP and propidium iodide was performed by combining the settings indicated above in the sequential scanning facility of the microscope, as instructed by the manufacturer.
3D reconstruction of z-stacks of optical sections was performed with the Leica LAS-AF Lite free software (Leica Microsystems, Germany). Segmentation analysis and 3D rendering were performed with Mimics 12.1 (Materialise N.V., Leuwen, Belgium).
Abbreviations
CLSM: confocal laser scanning microscopy; ER: endoplasmic reticulum; PI: propidium iodide; TIP: tonoplast intrinsic protein.
Authors' contributions
SG generated the majority of the constructs and transgenic plants and performed the bulk of the confocal analysis. MS produced the TIP1;2-RFP and 35S:TIP2;1 constructs and transgenic lines and performed confocal analysis. PH produced the native TIP1;1-YFP and TIP2;1 YFP constructs and transgenic plants and performed confocal analysis. RK performed the 3D image analysis in MIMICS. LF designed the experimental programme, gave technical and intellectual guidance and wrote the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Expression and subcellular localisation of TIP1;2. 8-day old seedlings expressing YFP-TIP1;2 were visualised by CLSM. A: 10× magnification of a lateral root. The signals from YFP fluorescence (green) and propidium iodide fluorescence (red) are merged. B: single root cap cell with TIP1;2-YFP showing typical ER labelling. C-D: epidermal cells in cotyledons where TIP1;2-YFP shows typical tonoplast labelling (green). Red: chlorophyll autofluorescence (excitation 514 nm, detection 600-650 nm). Scale bars: A, 100 μm; B and D, 5 μm; C, 20 μm.
Mutually exclusive expression of TIP2;1 and TIP2;3 in lateral root primordia. Roots from 8-day old transgenic seedlings expressing TIP2;1-YFP (green) and TIP2;3-RFP (red) were visualised by CLSM. Scale bar, 20 μm.
Co-expression of selected TIP-XFP pairs. Transgenic seedlings co-expressing the indicated TIP-YFP and TIP-RFP constructs were grown for 8 days on MS medium-agar plates. Roots were excised and visualised by CLSM. Stacks of 80 optical z sections (1 μm step-size) were collected from root axes at the differentiation zone. The images show representative results for each construct. Each panel shows the xz projection of the whole image stack, revealing the cross section of the root axis.
Constitutively expressed TIP2;1-YFP is detectable in every root tissue. Roots from 8-day old transgenic seedlings expressing 35S::TIP2;1-YFP (green) were excised, stained with propidium iodide (red) for 2 min and visualised by CLSM. A: stacks of 80 optical z sections (1 μm step-size) were collected from root axes at the differentiation zone. The images show representative results for this construct. The signals from YFP fluorescence (green) and propidium iodide fluorescence (red) are merged. B-D: single optical section through the vascular tissue, indicating that constitutive expression of TIP2;1 is easily detectable in these cell types. B: YFP, C, propidium iodide, D, merged images. Scale bar, 10 μm.
Primers used in this study. The diagram indicates the target sequences for the indicated primers in the final constructs. Restriction sites are shown in bold.
Contributor Information
Stefano Gattolin, Email: s.gattolin@warwick.ac.uk.
Mathias Sorieul, Email: m.r.p.sorieul@warwick.ac.uk.
Paul R Hunter, Email: paul_hunter@talk21.com.
Roman H Khonsari, Email: roman.khonsari@kcl.ac.uk.
Lorenzo Frigerio, Email: l.frigerio@warwick.ac.uk.
Acknowledgements
We are grateful to Robert Spooner and Alessandro Vitale for critical reading of the manuscript. This work was orted in part by the European Union (LSH-2002-1.2.5-2 "Recombinant Pharmaceuticals from Plant for Human Health -Pharma-Planta") and by the Leverhulme Trust (grant F/00215/AP). A grant from the Fondation 'Les Gueules Cassées' funded the acquisition of the Mimics software.
References
- Kaldenhoff R, Fischer M. Functional aquaporin diversity in plants. Biochim Biophys Acta. 2006;1758:1134–1141. doi: 10.1016/j.bbamem.2006.03.012. [DOI] [PubMed] [Google Scholar]
- Maurel C. Plant aquaporins: Novel functions and regulation properties. FEBS Letters. 2007;581:2227–2236. doi: 10.1016/j.febslet.2007.03.021. [DOI] [PubMed] [Google Scholar]
- Hofte H, Hubbard L, Reizer J, Ludevid D, Herman EM, Chrispeels MJ. Vegetative and Seed-Specific Forms of Tonoplast Intrinsic Protein in the Vacuolar Membrane of Arabidopsis thaliana. Plant Physiol. 1992;99:561–570. doi: 10.1104/pp.99.2.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson U, Karlsson M, Johansson I, Gustavsson S, Sjovall S, Fraysse L, Weig AR, Kjellbom P. The complete set of genes encoding major intrinsic proteins in Arabidopsis provides a framework for a new nomenclature for major intrinsic proteins in plants. Plant Physiol. 2001;126:1358–1369. doi: 10.1104/pp.126.4.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludevid D, Hofte H, Himelblau E, Chrispeels MJ. The Expression Pattern of the Tonoplast Intrinsic Protein gamma-TIP in Arabidopsis thaliana Is Correlated with Cell Enlargement. Plant Physiol. 1992;100:1633–1639. doi: 10.1104/pp.100.4.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels MJ, Chaumont F, Mirkov TE, Chrispeels MJ. Characterization of a new vacuolar membrane aquaporin sensitive to mercury at a unique site. Plant Cell. 1996;8:587–599. doi: 10.1105/tpc.8.4.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schussler MD, Alexandersson E, Bienert GP, Kichey T, Laursen KH, Johanson U, Kjellbom P, Schjoerring JK, Jahn TP. The effects of the loss of TIP1;1 and TIP1;2 aquaporins in Arabidopsis thaliana. Plant J. 2008;56:756–767. doi: 10.1111/j.1365-313X.2008.03632.x. [DOI] [PubMed] [Google Scholar]
- Loque D, Ludewig U, Yuan L, von Wiren N. Tonoplast Intrinsic Proteins AtTIP2;1 and AtTIP2;3 Facilitate NH3 Transport into the Vacuole. Plant Physiol. 2005;137:671–680. doi: 10.1104/pp.104.051268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jauh GY, Fischer AM, Grimes HD, Ryan CA Jr, Rogers JC. delta-Tonoplast intrinsic protein defines unique plant vacuole functions. Proc Natl Acad Sci USA. 1998;95:12995–12999. doi: 10.1073/pnas.95.22.12995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jauh G-Y, Phillips TE, Rogers JC. Tonoplast intrinsic protein isoforms as markers for vacuolar functions. Plant Cell. 1999;11:1867–1882. doi: 10.1105/tpc.11.10.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie J, Rogers SW, Deery M, Dupree P, Rogers JC. A unique family of proteins associated with internalized membranes in protein storage vacuoles of the Brassicaceae. Plant J. 2005;41:429–441. doi: 10.1111/j.1365-313X.2004.02303.x. [DOI] [PubMed] [Google Scholar]
- Paris N, Stanley CM, Jones RL, Rogers JC. Plant cells contain two functionally distinct vacuolar compartments. Cell. 1996;85:563–572. doi: 10.1016/S0092-8674(00)81256-8. [DOI] [PubMed] [Google Scholar]
- Poxleitner M, Rogers SW, Lacey Samuels A, Browse J, Rogers JC. A role for caleosin in degradation of oil-body storage lipid during seed germination. Plant J. 2006;47:917–933. doi: 10.1111/j.1365-313X.2006.02845.x. [DOI] [PubMed] [Google Scholar]
- Hunter PR, Craddock CP, Di Benedetto S, Roberts LM, Frigerio L. Fluorescent Reporter Proteins for the Tonoplast and the Vacuolar Lumen Identify a Single Vacuolar Compartment in Arabidopsis Cells. Plant Physiol. 2007;145:1371–1382. doi: 10.1104/pp.107.103945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU. A gene expression map of Arabidopsis thaliana development. Nat Genet. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- Frigerio L, Hinz G, Robinson DG. Multiple vacuoles in plant cells: rule or exception? Traffic. 2008;9:1564–1570. doi: 10.1111/j.1600-0854.2008.00776.x. [DOI] [PubMed] [Google Scholar]
- Soto G, Alleva K, Mazzella MA, Amodeo G, Muschietti JP. AtTIP1;3 and AtTIP5;1, the only highly expressed Arabidopsis pollen-specific aquaporins, transport water and urea. FEBS Lett. 2008;582:4077–4082. doi: 10.1016/j.febslet.2008.11.002. [DOI] [PubMed] [Google Scholar]
- Saito C, Ueda T, Abe H, Wada Y, Kuroiwa T, Hisada A, Furuya M, Nakano A. A complex and mobile structure forms a distinct subregion within the continuous vacuolar membrane in young cotyledons of Arabidopsis. Plant J. 2002;29:245–255. doi: 10.1046/j.0960-7412.2001.01189.x. [DOI] [PubMed] [Google Scholar]
- Beebo A, Thomas D, Der C, Sanchez L, Leborgne-Castel N, Marty F, Schoefs B, Bouhidel K. Life with and without AtTIP1;1, an Arabidopsis aquaporin preferentially localized in the apposing tonoplasts of adjacent vacuoles. Plant Mol Biol. 2009;70:193–209. doi: 10.1007/s11103-009-9465-2. [DOI] [PubMed] [Google Scholar]
- Parizot B, Laplaze L, Ricaud L, Boucheron-Dubuisson E, Bayle V, Bonke M, De Smet I, Poethig SR, Helariutta Y, Haseloff J. Diarch Symmetry of the Vascular Bundle in Arabidopsis Root Encompasses the Pericycle and Is Reflected in Distich Lateral Root Initiation. Plant Physiol. 2008;146:140–148. doi: 10.1104/pp.107.107870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan L, Janmaat K, Willemsen V, Linstead P, Poethig S, Roberts K, Scheres B. Cellular organisation of the Arabidopsis thaliana root. Development. 1993;119:71–84. doi: 10.1242/dev.119.1.71. [DOI] [PubMed] [Google Scholar]
- Vicre M, Santaella C, Blanchet S, Gateau A, Driouich A. Root border-like cells of Arabidopsis. Microscopical characterization and role in the interaction with rhizobacteria. Plant Physiol. 2005;138:998–1008. doi: 10.1104/pp.104.051813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN. A gene expression map of the Arabidopsis root. Science. 2003;302:1956–1960. doi: 10.1126/science.1090022. [DOI] [PubMed] [Google Scholar]
- Boursiac Y, Chen S, Luu DT, Sorieul M, Dries N van den, Maurel C. Early effects of salinity on water transport in Arabidopsis roots. Molecular and cellular features of aquaporin expression. Plant Physiol. 2005;139:790–805. doi: 10.1104/pp.105.065029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumont F, Barrieu F, Herman EM, Chrispeels MJ. Characterization of a maize tonoplast aquaporin expressed in zones of cell division and elongation. Plant Physiol. 1998;117:1143–1152. doi: 10.1104/pp.117.4.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkova E, Michniewicz M, Sauer M, Teichmann T, Seifertova D, Jurgens G, Friml J. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell. 2003;115:591–602. doi: 10.1016/S0092-8674(03)00924-3. [DOI] [PubMed] [Google Scholar]
- Vroemen CW, Mordhorst AP, Albrecht C, Kwaaitaal MA, de Vries SC. The CUP-SHAPED COTYLEDON3 gene is required for boundary and shoot meristem formation in Arabidopsis. Plant Cell. 2003;15:1563–1577. doi: 10.1105/tpc.012203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olbrich A, Hillmer S, Hinz G, Oliviusson P, Robinson DG. Newly formed vacuoles in root meristems of barley and pea seedlings have characteristics of both protein storage and lytic vacuoles. Plant Physiol. 2007;145:1383–1394. doi: 10.1104/pp.107.108985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De D. Plant Cell Vacuoles. Collingwood, Australia: CSIRO Publishing; 2000. [Google Scholar]
- Ma S, Quist TM, Ulanov A, Joly R, Bohnert HJ. Loss of TIP1;1 aquaporin in Arabidopsis leads to cell and plant death. Plant J. 2004;40:845–859. doi: 10.1111/j.1365-313X.2004.02265.x. [DOI] [PubMed] [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM. pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol. 2000;42:819–832. doi: 10.1023/A:1006496308160. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression and subcellular localisation of TIP1;2. 8-day old seedlings expressing YFP-TIP1;2 were visualised by CLSM. A: 10× magnification of a lateral root. The signals from YFP fluorescence (green) and propidium iodide fluorescence (red) are merged. B: single root cap cell with TIP1;2-YFP showing typical ER labelling. C-D: epidermal cells in cotyledons where TIP1;2-YFP shows typical tonoplast labelling (green). Red: chlorophyll autofluorescence (excitation 514 nm, detection 600-650 nm). Scale bars: A, 100 μm; B and D, 5 μm; C, 20 μm.
Mutually exclusive expression of TIP2;1 and TIP2;3 in lateral root primordia. Roots from 8-day old transgenic seedlings expressing TIP2;1-YFP (green) and TIP2;3-RFP (red) were visualised by CLSM. Scale bar, 20 μm.
Co-expression of selected TIP-XFP pairs. Transgenic seedlings co-expressing the indicated TIP-YFP and TIP-RFP constructs were grown for 8 days on MS medium-agar plates. Roots were excised and visualised by CLSM. Stacks of 80 optical z sections (1 μm step-size) were collected from root axes at the differentiation zone. The images show representative results for each construct. Each panel shows the xz projection of the whole image stack, revealing the cross section of the root axis.
Constitutively expressed TIP2;1-YFP is detectable in every root tissue. Roots from 8-day old transgenic seedlings expressing 35S::TIP2;1-YFP (green) were excised, stained with propidium iodide (red) for 2 min and visualised by CLSM. A: stacks of 80 optical z sections (1 μm step-size) were collected from root axes at the differentiation zone. The images show representative results for this construct. The signals from YFP fluorescence (green) and propidium iodide fluorescence (red) are merged. B-D: single optical section through the vascular tissue, indicating that constitutive expression of TIP2;1 is easily detectable in these cell types. B: YFP, C, propidium iodide, D, merged images. Scale bar, 10 μm.
Primers used in this study. The diagram indicates the target sequences for the indicated primers in the final constructs. Restriction sites are shown in bold.