Abstract
Cells lacking telomerase cannot maintain their telomeres and undergo a telomere erosion phase leading to senescence and crisis in which most cells become nonviable. On rare occasions survivors emerge from these cultures that maintain their telomeres in alternative ways. The movement of five marked telomeres in Saccharomyces cerevisiae was followed in wild-type cells and through erosion, senescence/crisis and eventual survival in telomerase-negative (est2::HYG) yeast cells. It was found that during erosion, movements of telomeres in est2::HYG cells were indistinguishable from wild-type telomere movements. At senescence/crisis, however, most cells were in G2 arrest and the nucleus and telomeres traversed back and forth across the bud neck, presumably until cell death. Type I survivors, using subtelomeric Y′ amplification for telomere maintenance, continued to show this aberrant telomere movement. However, Type II survivors, maintaining telomeres by a sudden elongation of the telomere repeats, became indistinguishable from wild-type cells, consistent with growth properties of the two types of survivors. When telomere-associated proteins Sir2p, Sir3p and Rap1p were tagged, the same general trend was seen—Type I survivors retained the senescence/crisis state of protein localization, while Type II survivors were restored to wild type.
Electronic supplementary material
The online version of this article (doi:10.1007/s10577-007-1178-2) contains supplementary material, which is available to authorized users.
Key words: Rap1, Saccharomyces cerevisiae, Sir2, Sir3, telomere
Introduction
Telomeres are the physical ends of eukaryotic chromosomes. They usually consist of a GC-rich DNA track bound with telomere-associated proteins forming a nucleoprotein structure. Maintenance of this structure is important for genome stability and chromosome preservation. Furthermore, a minimal telomere length is crucial for the cell to maintain the ability to divide. Without a special dedicated mechanism for replication, the telomere tract erodes with each cell cycle as a result of the inability of the DNA replication machinery to fully replicate the ends of the chromosome and by processing of the ends by nucleases. This will finally result in critically short telomeres, leading to cell cycle arrest.
Telomeres are normally maintained by use of the reverse transcriptase, telomerase (Blackburn 1991). However, several cell types have been found that maintain their chromosome ends in the absence of telomerase. These cells use telomerase-independent pathways to maintain their telomeres, including the alternative lengthening of telomeres (ALT) pathway. ALT and ALT-like pathways have been found in several species including yeast (Lundblad & Blackburn 1993), human (Bryan et al. 1995), mouse (Blasco et al. 1997) and more recently Arabidopsis (Watson et al. 2005).
In the yeast Saccharomyces cerevisiae the deletion of the RNA component of telomerase, TLC1, and the reverse transcriptase component, EST2, as well as deletion of associated proteins Est1p or Est3p, or mutations in Cdc13p (Est4) result in telomere erosion causing cell senescence/crisis and cell cycle arrest (Lundblad & Szostak 1989, Lendvay et al. 1996). On rare occasions cells survive in the absence of telomerase. At least two survival pathways are found in S. cerevisiae. Both pathways are dependent on proteins involved in homologous recombination (Lundblad & Blackburn 1993, Teng & Zakian 1999). Type I survivors maintain their telomeres by amplification of a subtelomeric Y′-element that is spread to all ends in large tandem arrays (Figure 1). These elements are naturally found at some chromosome ends and are flanked by telomere repeats (Louis & Haber 1992). This pathway is RAD51-, RAD52-, RAD54-, RAD55- and RAD57-dependent (Lundblad & Blackburn 1993, Le et al. 1999, Chen et al. 2001). The second pathway, resulting in Type II survivors, exhibits a sudden extension of the telomere repeats to several kb (Figure 1). This pathway is RAD50-, RAD52- and RAD59-dependent (Le et al. 1999, Chen et al. 2001) as well as SGS1-dependent (Cohen & Sinclair 2001, Huang et al. 2001, Johnson et al. 2001).
Figure 1.
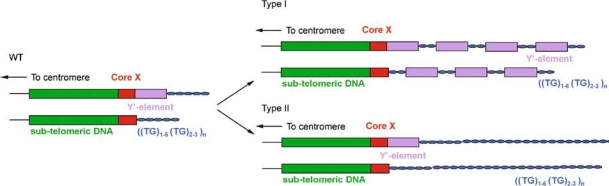
Cartoon of the model for telomere maintenance in yeast. There are two types of chromosome ends in WT cells: ends with only Core X and ends with Core X and Y′-elements. Both types of ends are followed by TG-rich telomeric sequences. In the presence of telomerase these telomeric sequences are maintained at approximately 350 bp. In the absence of telomerase the telomeric sequence will shorten with every round of replication and will finally become critically short, resulting in cell cycle arrest. However, on rare occasions survivors will appear in a culture that maintain their telomeres in a telomerase-independent manner. Type I survivors use a Y′ amplification mechanism to elongate their telomeres, at both ends with and without Y′-elements. Type II survivors show a sudden elongation of both the Y′ and non-Y′ ends with tracts of telomere repeats several kb in length.
Regulation of telomere length in yeast is the result of a counting mechanism using the number of Rap1 proteins bound to the telomere (Lustig et al. 1990, Sussel & Shore 1991) and works via its interacting factors Rif1p and Rif2p (Wotton & Shore 1997, Levy & Blackburn 2004). Detection of Rap1p showed foci at the nuclear periphery (Klein et al. 1992, Gotta et al. 1996, Laroche et al. 2000). In addition to binding to the telomere, Rap1p is involved in many other processes including the silencing of the mating-type loci HMR and HML (Kurtz & Shore 1991). Rap1p also blocks nonhomologous end joining (NHEJ) in yeast, which is important for telomere protection (Pardo & Marcand 2005). In the absence of telomerase, end-to-end fusions of chromosomes occurs (Hackett et al. 2001, Hemann et al. 2001, Liti & Louis 2003) and it is possible that this is a result of loss of Rap1p at the telomeres caused by the shortening of the telomere tracts.
Other proteins recruited to the telomeres by Rap1p and found in foci at the nuclear periphery are the silent information regulator proteins Sir2p and Sir3p together with Sir4p (Gotta et al. 1996, Laroche et al. 2000). These proteins are also involved in silencing at the mating-type loci HML and HMR (Ivy et al. 1986) and at telomeres (Gottschling et al. 1990, Pryde & Louis 1999). Furthermore, Sir2p is involved in silencing of the rDNA locus (Gotta et al. 1997, Smith & Boeke 1997). In ageing cells, Sir3p, together with Sir4p, translocates from the telomeres to the nucleolus (Kennedy et al. 1997). Because shortening of telomeres is related to ageing, the localization of Sir proteins might be of importance in early stages of telomere erosion in the absence of telomerase, in senescence/crisis or in survivors.
The two classes of survivors in S. Cerevisiae differ from each other and from wild type (WT) cells in growth characteristics and telomere structure. Here we aim to investigate how the telomeres and some telomere-associated proteins behave in live cells during telomere erosion, at senescence/crisis, and in survivors. We relate changes in their localization to the process of telomere loss and identify differences in Type I and Type II survivors.
Material and Methods
Yeast strains
All of the strains were isogenic derivatives of S. cerevisiae strain S288c and are listed in Table 1. Yeast strains were cultured at 30°C on appropriate selection media for markers used for protein tagging (KanMX4, HIS3, and LEU2), telomere tagging (TRP1) and gene disruption (HYG) (Sherman et al. 1986). Single colony selection of telomerase-negative strains was performed on YPD plates. Diploid strains were sporulated at 25°C on sporulation medium for 10–14 days before dissection of asci (Sherman et al. 1986).
Table 1.
S288c yeast strains used in this study
| Name | Genotype |
|---|---|
| M139 | MATα, ura3–52, leu2Δ, his3Δ, trp1Δ, CyhR |
| M148 | MATα, ura3–52, leu2Δ, his3Δ |
| M206 | MATα, ura3–52, lys2Δ, leu2Δ |
| M207 | MATα, ura3–52, lys2Δ, his3Δ |
| yAM12 | MATα, ura3–52, leu2Δ, his3Δ, rap1::RAP1-GFP-LEU2 |
| yAM13 | MATa/MATα, ura3–52/ura3–52, leu2Δ/LEU2, his3Δ/his3Δ, lys2Δ/LYS2, SIR2-CFP-KANMX4/SIR2-CFP-KANMX4, SIR3-YFP-HIS3/SIR3-YFP-HIS3 |
| YKS43 | MATα, ura3–52, leu2Δ, his3Δ, SIR2-CFP-KanMX, SIR3-YFP-HIS3 |
| yKS50 | MATα, ura3–52, ade2Δ, trp1Δ, CYH2R, his3::KANMX4, LacI-GFP-HIS3 |
| yKS51 | MATα, ura3Y52, lys2D, trp1D, CYH2R, his3::KANMX4, LacI-GFP-HIS3 |
| yKS16.3 | MATα, ura3–52, ade2Δ, trp1Δ, CYH2R, his3::KANMX4, LacI-GFP-HIS3, TEL VIIR:: Y′acO-TRP1, TEL XVIR ::Y′acO-TRP1 |
| yKS16.9 | MATα, ura3–52, ade2Δ, trp1Δ, CYH2R, his3::KANMX4, LacI-GFP-HIS3, TEL XL:: Y′acO-TRP1 |
| yKS17.2 | MATa, ura3–52, lys2Δ, trp1Δ, CYH2R, his3::KANMX4, LacI-GFP-HIS3, TEL XVR:: Y′acO-TRP1 |
| yKS17.12 | MATa, ura3–52, lys2Δ, trp1Δ, CYH2R, his3::KANMX4, LacI-GFP-HIS3, TEL VL:: Y′acO-TRP1, TEL XIIR::Y′acO-TRP1 |
| yKS58 | MATa, ade2Δ, ura3–52, trp1D, his3::KanMX, LacI-GFP-HIS3, TEL IVR ::Y′acO-TRP1, TEL VL:: Y′acO-TRP1, TEL VIIR:: Y′acO-TRP1, TEL XIIR:: Y′acO-TRP1, TEL XVR:: Y′acO-TRP1, CyhR |
| yKS86 | MATa/MATα, ura3–52/ura3–52, leu2Δ/leu2Δ, HIS3/his3Δ, LYS2/lys2Δ, RAP1/rap1::RAP1-GFP-LEU2 |
| yKS303 | Type I survivor of yKS58 |
Tagging of telomeres with a lacO array
To visualize individual telomeres in yeast, a lacO array containing 256 lacO repeats was used and integrated in Y′-elements. The S288c strain has subtelomeric Y′-elements in 17 of its 32 telomeres (SGD database). To target lacO arrays to Y′-elements a 450 bp internal fragment of a Y′-element was amplified via PCR using primer pair KSY′fwd and KSY′rev with XhoI linkers (for primers see Table 2) and inserted in the SalI site of pAFS52 (gift from A. Straight, Harvard University, Cambridge, MA, USA), a plasmid containing 258 repeats of the lacO sequence, resulting in pKS3.12 (Table 3). The resulting plasmid was digested using HpaI, unique in the Y′ sequence in pKS3.12, and integrated into genomic Y′-elements of yeast strains yKS50 and yKS51. These strains contained pAFS135, the LacI-GFP construct, integrated into and under control of the HIS3 promoter. The resulting transformants were analysed using a clamped homogeneous electric field (CHEF) gel (Louis 1998), blotted on membrane and hybridized using a DIG nick-labelled pAFS52 probe (Roche Diagnostics GmbH, Penzberg, Germany). To further characterize which chromosome arm was labelled with the lacO array, samples were digested using the rare cutting NotI (Link & Olson 1991, Louis & Borts 1995). Strains with multiple Y′ foci were created by serial crossing of transformants with different lacO-tagged telomeres until several tagged telomeres with distinct foci were found in a single cell line.
Table 2.
Primers used in this study
| Name | Sequence |
|---|---|
| KSY′wd | CCGCTCGAGTGTCTGGTTGAATACGCAAC |
| KSY′ev | CCGCTCGAGCGAGGATATCGTTTGTACTCGGCGTGTATGTCGTTACATCAGATGAACATCCCAAAACCCTCGGT |
| KSSIR2CFPfwd | CGACGGATCCCCGGGATATTAATTTGGCACTTTTAAATTATTAAATTGCCTTCTACATCGA |
| KSSIR2CFPrev | TGAATTCGAGCTCG |
| KSSIR2fwd | ATGCGGGAATAAGCACAGAT |
| KSSIR2rev | TTCAAAGGGTTCATACGCAAGCCTTTTCGATGGATGAAGAATTCAAAAATATGGACTGCATTGGT |
| KSSIR3YFPfwd | CGACGGATCCCCGGGATAGGCATATCTATGGCGGAAGTGAAAATGAATGTTGGTGGATCG |
| KSSIR3YFPrev | ATGAATTCGAGCTCG |
| KSSIR3fwd | AATGATCATCACGCGACTCA |
| KSSIR3rev | CCGTGGAAGTTGGTCAATAA |
| KSC/YFPrev | TTGGCCATGGAACAGGTAGTT |
| His3fwd | TTGTCTGCGAGGCAAGAATG |
| KanMX4fwd | CATCCTATGGAACTGCCTCGG |
| Est2fwd | CTCATCATCCGCAACTGCTGC |
| Est2rev | TGGACAACCGCTGACTGTCAT |
| HYGrev | CGGCGGGAGATGCAATAGG |
| HYGfwd | TCGCCCGCAGAAGCGCGGCC |
Table 3.
Plasmids used in this study
| Name | Description | Marker | Reference/source |
|---|---|---|---|
| pAFS52 | 256 Lac operator repeat sequences | TRP1 | Straight et al. (1996) |
| pKS3.12 | pAFS52 with 450 bp of Y′-element | TRP1 | This article |
| pAFS135 | GFP12-LacI12 in pRS303 | HIS3 | Straight et al. (1996) |
| pDH3 | CFP | KanMX6 | YRC Microscopya |
| pDH5 | YFP | HIS5 | YRC Microscopya |
| pTY24 | pDsRed.T1.N1 | KanMX6 | YRC Microscopya |
| pAH52 | RAP1-GFP | LEU2 | Hayashi et al. (1998) |
| pEL30 | Y′-element and TG1Y3 sequences cloned in Sal1 site of pGEM3ZF(−) | Watt et al. (1996) | |
| pAG32 | Hygromycin B | Goldstein & McCusker (1999) |
aYRC Microscopy, Department of Biochemistry, University of Washington, Seattle, WA, USA.
Protein tagging
Sir2 and Sir3 proteins were C-terminally tagged with CFP or YFP in yeast strains M207 and M148 using the plasmids pDH3 and pDH5 respectively (obtained from Yeast Resource Center Microscopy, University of Washington, Seattle, WA, USA, supported by a P41 grant from the National Center for Research Resources) and primer sets KSSIR2CFPfwd-KSSIR2CFPrev and KSSIR3YFPfwd-KSSIR3YFPrev (see Table 2 for primers). Direct transformation of PCR products was performed according to Gietz & Woods (2002). The incorporation of the fluorescent proteins was confirmed by PCR amplification of the junctions using the appropriate forward primer in combination with KSC/YFPrev and reverse primer with HIS3fwd (Table 2). Mating these strains resulted in yAM13. Haploid strain yKS43 was obtained from this strain.
The protein Rap1 was tagged by replacing the RAP1 gene by the GFP-RAP1-LEU2 construct from plasmid pAH52 (Hayashi et al. 1998) in yeast strain M148, resulting in yAM12. This strain was crossed with M206 resulting in diploid yeast strain yKS86.
Telomerase-negative yeast strains
To obtain telomerase-negative yeast strains, EST2, encoding the catalytic subunit of telomerase, was disrupted in diploids yAM13 and yKS86, and haploid yKS58 using the hygromycin B disruption cassette from plasmid pAG32 (Goldstein & McCusker 1999). Genomic DNA from an est2::HYG strain was used to amplify the disruption cassette using primers Est2fwd and Est2rev and transformants were selected using primer combinations Est2fwd-HYGrev and HYGfwd-Est2rev (Table 2). The diploids were dissected and appropriate progeny were selected for analysis. Colonies derived from single cells were isolated. A single cell isolate was derived from each colony to propagate the cultures further while cells from each colony were analysed on a Zeiss LSM510 confocal microscope. Cultures were followed for up to ten successive re-streaks (250 generations; 25 generations per re-streak), which would take them through telomere erosion, senescence/crisis and postsenescence survival.
DNA was isolated after 225–250 generations to determine the survivor type. The DNA was digested using XhoI, run on an agarose gel, blotted on membrane and hybridized with digoxigenin (DIG)-labelled probe pEL30 containing Y′-element and telomere repeat sequences (Watt et al. 1996), which reveals the distinct terminal restriction fragment patterns of the two types of survivors (Huang et al. 2001).
Imaging
To image the localization of the LacI-GFP, Sir2-CFP, Rap1-GFP or Sir3-YFP, cells were taken from fresh YPD plates and immobilized on a microscope slide using 1% low-melting-point agarose (BMA, Rockland, ME, USA) in F1 medium (Walmsley et al. 1983). For the lacO/LacI-GFP est2::HYG experiment and for the Sir2-CFP/Sir3-YFP est2::HYG experiment, four colonies from a plate of 10 colonies were followed and imaged at each time point during 250 generations. Cells from the other colonies were viewed to confirm the results found in the colonies imaged. To control the imaging process during the experiment at eight stages, WT Sir2-CFP/Sir3-YFP cells were imaged. For Rap1-GFP est2::HYG cells were imaged during erosion, at senescence/crisis and in survivors at 50, 100 and 200 generations respectively. At every stage in these experiments Z-series were taken from single time points from at least 10 cells. To capture time series from WT cells, from cells in senescence/crisis and from survivors, cells were cultured at 30°C in a POC culture chamber (Laborbedarf, Reutlingen, Germany) on the microscope stage. The movements of the telomeres were followed in at least three cells at these stages. Single optical sections were captured at 2-, 5- or 10-second intervals for 100–1400 images (depending on signal intensity and movement of the telomeres) and the focus was manually adjusted when the signal went out of focus.
Images were taken using a Zeiss LSM 510 confocal laser scanning microscope on an inverted Zeiss Axiovert 100 microscope equipped with a 100× oil (1.4 NA) objective using a 458 nm laser line for CFP and 488 nm laser line for GFP or YFP. In most cases a 5× zoom was used, but at senescence/crisis the cells were enlarged and occasionally a smaller zoom setting was used.
Results
To visualize the movement of telomeres in mitotic cells in the presence and absence of telomerase, yeast strains were created with a lacO array in one of the subtelomeric Y′-elements. Transformants were checked for the presence of lacO arrays by Southern hybridization of pulsed-field gel-separated chromosomes and for LacI-GFP signal on the confocal microscope (not shown). Crosses were made between strains with inserts in different telomeres. These were dissected, checked, and crossed again. This was serially repeated and a strain was obtained with five marked telomeres, which could individually be identified as foci under the confocal microscope. This strain, yKS58, showed lacO arrays in the Y′-elements of telomeres IVR, VL, VIIR, XIIR and XVR (Figure 2A, lanes 1 and 3; Figure 2B, lane 6) and was a result of crosses between yKS16.3, yKS16.9, yKS17.2 and yKS17.12 (Figure 2B). Figure 2A shows the lacO tag in undigested chromosomes while in Figure 2B the chromosomes are digested with NotI to identify the lacO-tagged telomeres. As both telomeres XIIL and XIIR have Y′ elements, the NotI digest of XII would give a band of 65 kb for telomere XIIR or 105 kb for telomere XIIL (Link & Olson 1991). The results indicate that telomere XIIR is the most likely candidate.
Figure 2.
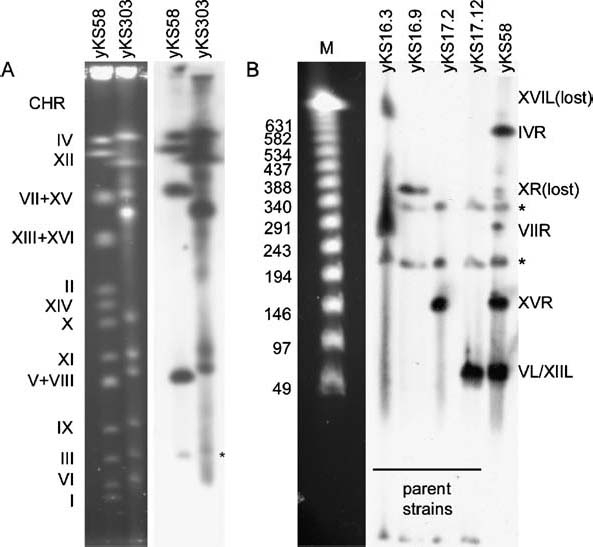
CHEF analysis of yKS58 and its Type I survivor yKS303 to identify the tagged Y′-elements. (A) Whole chromosomes were separated on a CHEF gel. Lanes 1 and 2, EtBr stained CHEF gel of yKS58 and its Type I survivor yKS303. The chromosome numbers at the left refer to the chromosome bands of yKS58. Lanes 3 and 4, the Southern blot of lanes 1 and 2 probed with DIG-labelled pAFS52 showing the locations of lacO repeats in yKS58 and its Type I survivor yKS303. (B) Blot of a NotI digest of the four parental strains yKS16.3, yKS16.9, yKS17.2 and yKS17.12 and the resulting strain yKS58, probed with DIG-labelled pAFS52 to identify the lacO tagged telomeres. The identified telomeres are named at the right. M = lambda ladder PFG marker (New England Biolabs). *Bands due to hybridization between probe fragments and sequences in the genetic background of the yeast strains, which are not a result of incorporation of the lacO arrays. These bands were found in all strains with this genetic background.
During construction of this strain, many Y′-elements lost lacO repeats during both mitotic and meiotic divisions, resulting in shorter lacO arrays with a weaker LacI-GFP signal or total loss of signal. In other cases lacO arrays were found at a Y′-element different from their parental location. For example, from Figure 2B it is clear that in yKS58 the lacO array of telomere XL from yKS 16.9 and telomere XVIL from yKS16.3 have been lost (only a faint band left of XL in yKS58) but that telomere IVR has gained a band that was not present in the strains used to obtain yKS58. This is confirmed in Figure 2A, where a clear lacO band can be found for chromosome IVR.
Movement of telomeres in EST2 cells
The strain with the five lacO-tagged telomeres was used to assess telomere movement and architecture in telomerase-positive and telomerase-negative cells. During G1-phase in yKS58, a telomerase-positive strain, the telomeres moved through the nucleus. In G2/M-phase the telomeres moved towards the bud neck (N = 6) and remained at the entrance of the bud neck, in one case for more than half an hour, before they started to migrate through it (Figure 3A; supplementary Video 1). The separation of a pair of chromatids took place in the mother cell, resulting in an increase in number of foci in the mother cell (Figure 3A, time point 3′20″), followed by movement of the telomeres one by one to the daughter cell. During mitosis the telomeres often gave the impression of being ‘lined up’ toward the bud neck as shown in Figure 3A, time point 0 and at 5′35″. Because telomeres were found to be very dynamic, this ‘lined up’ telomere configuration transiently appeared and disappeared during imaging.
Figure 3.
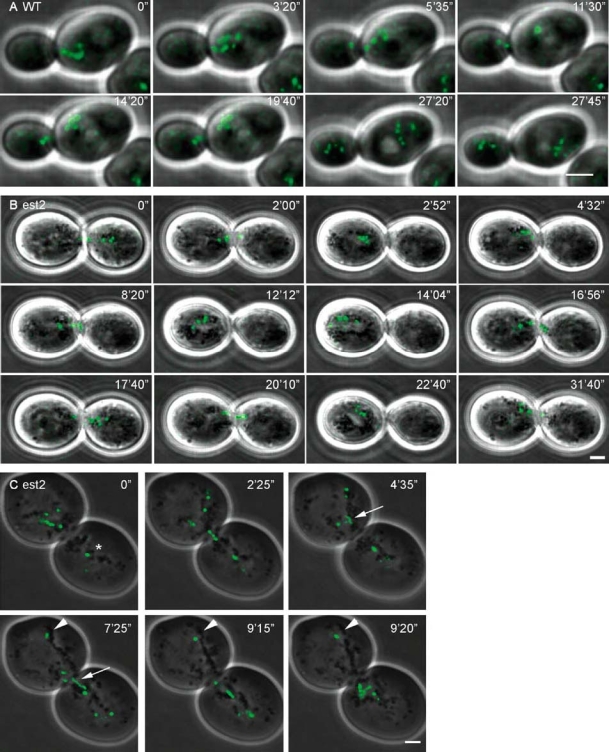
Time-lapse imaging of the movement of five lacO-tagged telomeres during mitosis. One optical section is shown per time point. (A) Telomerase-positive yKS58 yeast cell in mitosis. The telomeres move one by one from the mother cell into the daughter cell. (B) Senescence/crisis in telomerase-negative yeast cells derived from yKS58. During senescence/crisis the telomeres move between both cells without separation. Mother and daughter cell are equal in size and larger than WT cells. (C) Telomerase-negative yeast cell during senescence/crisis showing that most telomere foci move between cells but at least one telomere (arrowhead) and possibly a second telomere (star) stayed positioned away from the bud neck. Most foci move through the bud neck as a dot but in one case the signal is elongated (arrow). Bar = 2 µm.
Movement of telomeres in est2::HYG cells
Telomerase-negative strains obtained from yKS58 were followed through telomere erosion and went into senescence/crisis after 50–75 generations, as determined by their growth arrest. Rare survivors emerged and were followed to 225–250 generations before the type of survivor was determined using Southern blot analysis. For simplification we refer to cells at earlier time points as Type I lineage or Type II lineage. We are, however, aware that at senescence/crisis a culture is a mixture of cells that might generate Type I and Type II cells depending on the colony selected when colonies are re-streaked for single cells. Very slow growth in cells of the Type I lineage continued for up to 100 generations before the cell growth improved. However, even then the cell growth was still slower than the growth rate in WT and Type II survivors. Cultures of the Type II lineage often exhibited slow growth for only 25–50 generations before attaining a normal WT growth rate.
During telomere erosion the movement of the tagged telomeres in est2::HYG cells was as in WT cells, indicating no telomere defects during erosion. At senescence/crisis, in cells of both the Type I lineage and the Type II lineage, the tagged telomeres moved back and forth between the two dividing cells, which appeared to be at G2/M (Figure 3B; Supplementary Video 2). It was impossible to establish which cell was the mother cell as both cells were enlarged, compared with WT cells, and equal in size. In 19 from the 50 cells imaged, tagged telomeres moved continuously between the two cells in contrast to the movement of the tagged telomeres in the WT cells. This does not mean that in the other cells the nucleus did not move between the two cells, only that it did not happen during the period in which these cells were imaged.
From the 19 cells imaged showing movement of telomeres between the cells at senescence/crisis, the tagged telomeres moved as well-defined foci (see, for example, Figure 3C, time point 2′25″), but in two cases a tagged telomere spot elongated as it moved between the cells (Figure 3C, arrow). This might indicate end-to-end fusion between chromosomes. As only one optical section was taken per time point and not all the telomeres are in this focal plane, it is possible that this phenomenon happened more often than observed. This also is the case for the observation that in some cells one tagged telomere was positioned opposite to the bud in the plane of focus and did not move with the rest of the telomere foci between the two cells (Figure 3C, star and arrowhead).
After 250 generations the movement of the tagged telomeres in Type I survivors still resembled the movement of telomeres in cells at senescence/crisis (n = 6). However, in Type II survivors the movement of tagged telomeres resembles the movement of tagged telomeres in WT cells (n = 10). This correlates with the slow growth found in Type I survivors and apparently normal growth of Type II survivors on plates (see also McEachern & Haber 2006).
As Type I survivors maintain their telomeres by amplification of Y′-elements, we expected that some cells in the survivor population would have amplified Y′-elements carrying the lacO arrays to maintain telomere stability. In earlier experiments using est2::HYG strains with a single tagged telomere, cells were found that showed clear amplification of this telomere (data not shown). However, amplification of lacO sequences was rarely found after senescence/crisis and on only one occasion was a colony identified that had amplified a tagged lacO Y′-element, subsequently called yKS303. In these cells many foci can be found after survival/crisis but the signal from most of these was weaker than the signal from the original lacO-tagged telomeres (Figure 4A). Analysis of interphase nuclei of yKS303 showed up to 12 tagged telomeres (Figure 4B). However, from Figure 2A it is clear that the lacO array was not at all telomeres. Survivor yKS303 (Figure 2, lanes 2 and 4) showed major alterations in chromosome patterns when compared with yKS58 (lanes 1 and 3). The est2::HYG YKS303 (unstable Type I survivor) was rescued by crossing it with WT strain M139 and the chromosomes of nine independent diploids were analysed on a CHEF gel, blotted and probed with the lacO sequence containing plasmid pAFS52. Figure 4C shows again chromosomal rearrangements in the different strains obtained from the survivors. It also shows that in these strains different chromosomes were tagged with lacO arrays, indicating that the original survivor culture was a mixture of cells and that senescence/ crisis and/or survival in this culture was highly dynamic as these cells were all derived from a single cell. Eight colonies of the est2::HYG strain were cultured for another 300 generations to see whether the Y′lacO arrays continued to be amplified, but in all cases this resulted in loss of signal rather than amplification of lacO tagged subtelomeric sequences to other telomeres. It is not clear whether this loss of lacO arrays is a result of the instability of the lacO construct as was seen during the construction of yKS58 or a result of chromosomal instability initiated by the short telomeres, or a combination of both.
Figure 4.
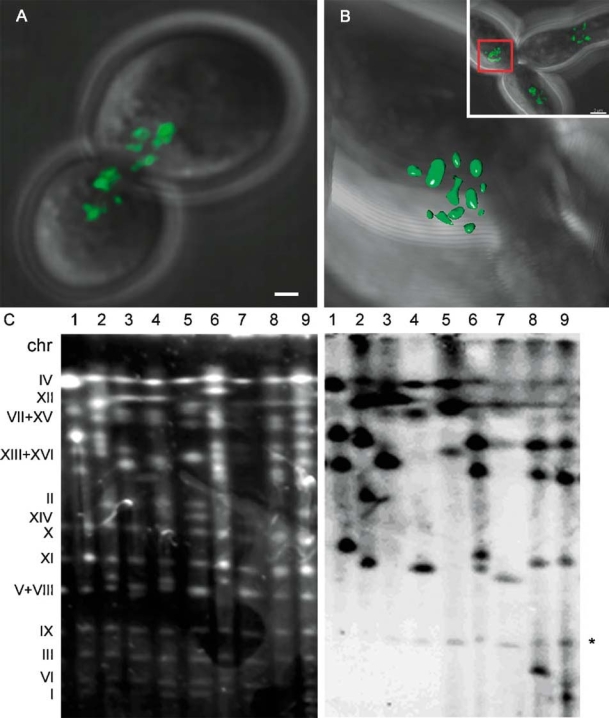
Type I survivor (yKS303) after amplification of tagged Y′ sequences to maintain telomere integrity. (A) At crisis many foci can be seen. One optical section is shown. (B) Imaris isosurface representation of a 3D reconstruction of the tagged telomeres after 200 generations in an interphase nucleus. Inset shows an overview from one optical section of this cell and its neighbours. Bar = 1 µm. (C) EtBr-stained CHEF gel and Southern blot of Type I survivor yKS303 rescued by crossing with telomerase-positive strain M139 and probed with DIGpAFS135 to identify the lacO construct, showing different bands in each rescued strain. The chromosome numbers at the left of the gel refer to the chromosome numbers in WT yeast cells. The gel results show major chromosomal rearrangement. *Bands due to hybridization between probe fragments and sequences in the genetic background of the yeast strains, which are not a result of incorporation of the lacO arrays. These bands were found in all strains with this genetic background.
Localization of Sir2 and Sir3 proteins
After analysing the movement of the telomeres during erosion, at senescence/crisis and in survivors we were interested in the localization of the telomere-associated proteins during this process. To image Sir2p and Sir3p these proteins were tagged with CFP and YFP respectively, and their localization was analysed (Figure 5). It can be seen that in EST2 cells most Sir3-YFP foci co-localized with or were adjacent to Sir2-CFP foci (Figure 5D). The largest Sir2-CFP spot failed to co-localize with Sir3-YFP and was probably Sir2-CFP bound to rDNA in the nucleolus. During mitosis most of the Sir2 and Sir3 signal remained localized in foci, which were often adjacent to each other (Figure 5E).
Figure 5.
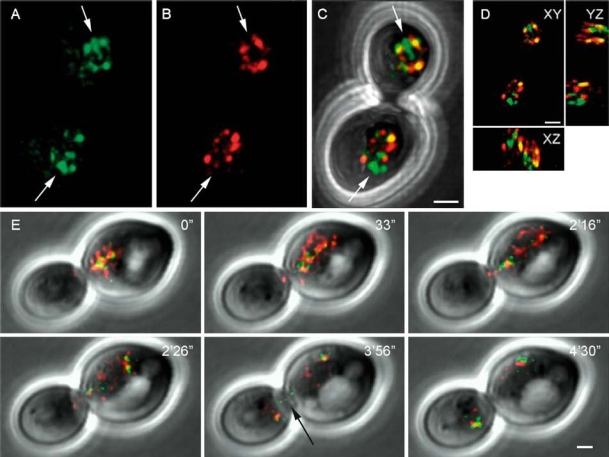
In vivo detection of Sir2-CFP (green) and Sir3-YFP (red) in haploid cells from strain yKS43. Maximum-intensity projection of Sir2-CFP (A) and Sir3-YFP (B) signal separate, merged (C), and as xy, yz, xz representation (D) in a mitotic cell. (E) Time-lapse imaging of single optical sections showing Sir2-CFP and Sir3-YFP signal during mitosis. Arrow indicates Sir2-CFP signal in the putative nucleolus. Bar = 1 µm.
During erosion and at senescence/crisis, Sir2-CFP signal was found most prominently in one or two bright spots, presumably bound to the nucleolar rDNA, and a weak signal in the nucleoplasm (Figure 6A, 75 and 100 generations; Figure 6B, 75 and 100 generations). Most of the Sir3-YFP was found in the nucleoplasm and the intensity of the signal increased over time. One Type I and one Type II lineages were further analysed and the results are shown in Figures 7A and 7B. During these experiments EST2 cells were imaged at eight time intervals as an internal control (Supplementary Figure S1). These results show that, although there was fluctuation in the signal intensity, the results fall within the error of the experiments, indicating that results from different imaging days can be compared. Figures 7A and 7B show that both Sir2-CFP and Sir3-YFP significantly increased in signal per nucleus during senescence/crisis (75 generations).
Figure 6.
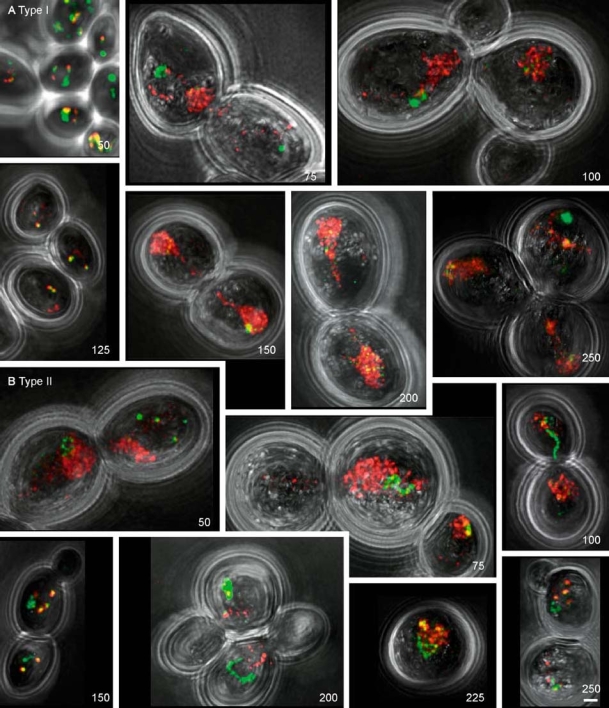
Maximum-intensity projections of Sir2-CFP (green) and Sir3-YFP (red) localization during telomere erosion, during senescence/crisis, and in survivors. (A) yeast cells of the Type I lineage from 50 to 250 generations. Number of generations is in the lower right corner. After 75 generations the localization of Sir2-CFP and Sir3-YFP is disrupted. The cells after 150 and 200 generations have almost finished the nuclear division; only a small stretch of Sir3-YFP-positive nucleoplasm connects the two nuclei. (B) Yeast cells of the Type II lineage from 50 to 250 generations. Bar = 1 µm.
Figure 7.
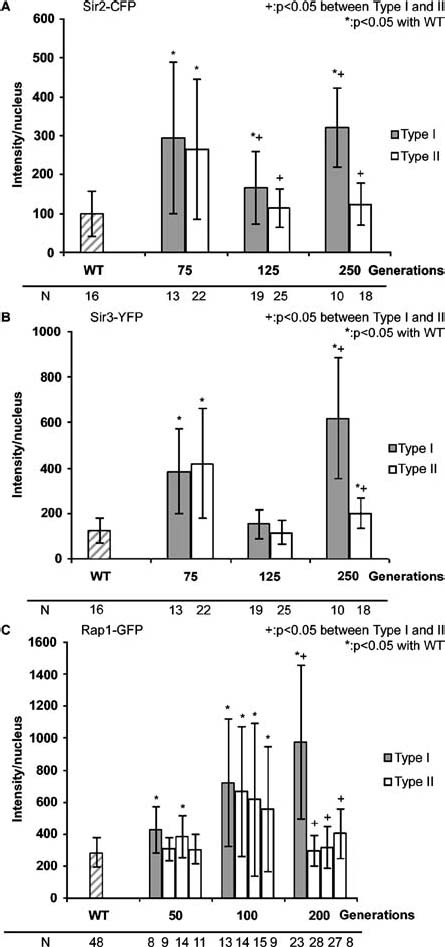
The intensity per nucleus and the number of cells analysed of Sir2-CFP (A), Sir3-YFP (B) and Rap1-GFP (C) in WT cells, during senescence/crisis and in survivors of cells of the Type I and Type II lineages. Bars represent standard deviation. Bars indicated with a star represent stages with a significant difference (p < 0.05) compared with the WT data. Bars indicated with a + represent stages with a significant difference (p < 0.05) between Type I and Type II data.
In cells of both the Type I and the Type II lineages, restoration of the telomeric localization of Sir2-CFP and Sir3-YFP was found after 125–150 generations (Figure 6A, 125 generations; Figure 6B 150 generations). This was reflected by a decrease of intensity for both proteins in both lineages (Figures 7A and 7B). Cells of the Type I lineage went back into senescence/crisis phenotype, and after 225–250 generations Type I survivors exhibited a phenotype of cells in senescence/crisis. Figure 7A and B shows that both the Sir2-CFP signal and the Sir3-YFP signal had further increased in these cells. Type II survivors (Figure 6, 200–250 generations) showed Sir2-CFP and Sir3-YFP signal in foci like in WT cells. The intensity of the signal was just significantly higher for Sir3-YFP than for WT cells (p < 0.05) (Figure 7B), but for Sir2-CFP no significant difference with WT cells was found (Figure 7A). However, we have to keep in mind that each lineage is derived from a single cell and therefore the results might be unique and specific for these two lineages.
When correcting for the change in nuclear size during erosion and senescence/crisis, we found some significant increase (p < 0.05) of signal compared with WT cells (Supplementary Figures S2A and S2B) but there was no clear difference between the Type I and Type II lineages. However, in both types of survivors the Sir3-YFP intensity was significantly higher than the intensity in WT cells, whereas the Sir2-CFP results showed no change at this stage when compared with WT cells.
Cells can be found in the Type I lineage that seem to have lost the bright putative nucleolar Sir2-CFP signal (Figure 6A, 150 and 200 generations). However, a large number of small foci were often found in the nucleus. In the nuclei that still showed putative nucleolar Sir2-CFP signal there was no co-localization with Sir3-YFP. The same was true for the Sir3-YFP localization in cells of the Type II lineage (Figure 6B).
The brighter labelling of the nucleoplasm by Sir3-YFP in cells of the Type I lineage made the whole nucleus visible and the nuclear movement between the two cells could be followed. This confirmed the results found in the experiment with the multiple lacO-tagged telomeres that the nucleus moved between the two cells during senescence/crisis and in Type I survivors. It also showed that when most of the nucleus was divided between the two cells there was often still a faint Sir3-YFP-positive labelled connection between them (Figure 6A, 150 and 200 generations).
Rap1 protein
To image the movements of Rap1p, RAP1 was replaced with a RAP1-GFP construct (Hayashi et al. 1998), resulting in yeast strain yKS86. During normal cell growth in haploid cells obtained from yKS86 up to eight foci could be seen at the nuclear periphery as has previously been reported (Klein et al. 1992, Gotta et al. 1996, Laroche et al. 2000). In haploid {tiest2::HYG} strains obtained from yKS86 after the disruption of the EST2 gene with the hygromycin B cassette, the Rap1-GFP localization for the first 50 generations was as in WT (Figure 7C; Figures 8A, 8B 50 generations and 8C 50 generations). After 100 generations the cells were in crisis and the nucleoplasm was brightly labelled. After 200 generations Type I survivors continued to show the senescence/crisis phenotype, whereas Type II survivors exhibited brighter foci and a weaker labelling in the nucleoplasm like WT cells (Figure 8C, 200 generations). During mitosis a normal pattern of telomere movement was observed in these Type II survivor cells (Figure 8D). The intensity of fluorescence signal per nucleus in both Type I (n = 1) and Type II (n = 3) lineages had significantly increased during senescence/crisis (100 generations) with no difference between the two lineages at this stage (Figure 7C). In Type I survivors the intensity had further increased, whereas in Type II survivors the intensity was as in WT cells. However, when corrected for the change in nuclear size during the different stages, the concentration of Rap1-GFP did not show a real increase—if anything the signal was significantly less in cells of the Type II lineage than in WT cells (Supplementary Figure S2C).
Figure 8.
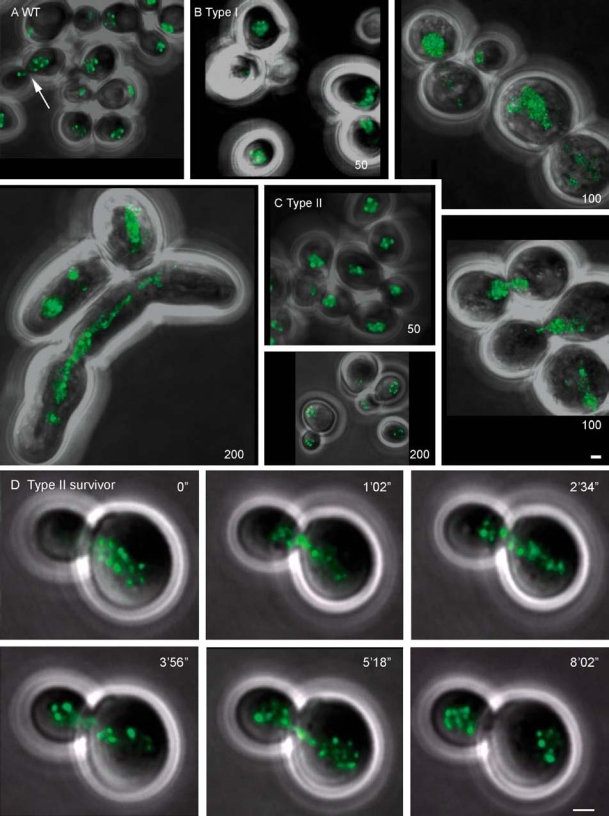
Maximum-intensity projection of the localization of Rap1-GFP in WT cells, during senescence/crisis and in survivors. (A) WT cells (yAM12) in different stages of the cell cycle. Arrow shows a cell in mitosis. (B) Yeast cells of the Type I lineage after 50, 100 and 200 generations. After 200 generations these cells still show a senescence/crisis phenotype. (C) Yeast cells of the Type II lineage after 50, 100 and 200 generations. (D) Localization of Rap1-GFP in a Type II survivor during mitosis. One optical section shown per time point. Bar = 1 µm.
Monitoring of the general nuclear movement between two cells confirmed that the nucleus moved between these two cells during crisis/senescence in cells of both the Type I and Type II lineages as well as in Type I survivors (see Supplementary Video 3). In 29 out of 59 cells analysed, foci could be seen at the nuclear periphery at this stage. If the nucleus was divided between the two cells (n = 12) a connection could often be found between both nuclei (n = 6) and in two cases a nucleus was stretched between more than two cells (Figure 8B, 200 generations).
Discussion
To study the dynamics of telomeres in vivo, a yeast strain was created containing repetitive lacO arrays in five sub-telomeric Y′- sequences. After binding of LacI-GFP, these sequences could be visualized in a fluorescent microscope as five individual foci. Different methods have indicated that there are four to nine telomere foci in a yeast nucleus. These methods include in situ hybridization using a Y′-probe (Gotta et al. 1996, Heun et al. 2001) and immunolabelling using antibodies against Rap1p and Sir proteins (Gotta et al. 1996, Laroche et al. 2000). This indicates that the five foci in this strain represent more than 50% of the telomeric foci that can be found in the yeast nucleus. The tagged telomeres showed dynamic movements during interphase, as has previously been reported for individual telomeres (Heun et al. 2001). When the bud starts to appear, the nucleus moves towards the bud neck and can stay close to this position for more than half an hour, as previously described (Shaw et al. 1998). During mitosis the telomeres were occasionally found lined up in the mother cell or in both mother and daughter cells. This is in agreement with previous results (Straight et al. 1997, Laroche et al. 2000) where the spindle microtubules form one long spindle with telomeres attached to them in a linear fashion. This all indicates that the lacO tagged telomeres behave like normal telomeres.
In telomerase-negative strains, cell growth slows down after 50 to 75 generations and the cells enter senescence/crisis. In the est2::HYG cells the telomeres become critically short, which might result in end-to-end fusions, recombination events and G2/M arrest that result in a senescence/crisis phenotype (Lundblad & Blackburn 1993, Lundblad & Szostak 1989, Hackett et al. 2001). At this stage the telomere foci do not separate but move between both cells. This nuclear movement between mother and daughter cells has recently also been described by Shimada & Gasser (2007) during checkpoint-induced G2/M delay in orc2-depleted S. cerevisiae. This indicates that this specific nuclear movement might be a result of the G2/M arrest, in our case caused by the critical short telomeres. However, we point out that Nautiyal et al. (2002) showed that the gene expression pattern as a result of loss of telomerase activity share features with G2/M arrest response but is not identical to it and there is a specific telomerase deletion response.
End-to-end fusions could explain the elongated telomere signal seen in some of the nuclei as well as the nuclear tracks found in Sir3-YFP and Rap1-GFP tagged cells when most of the nucleus has been divided between the two cells. However, in many cells there are still forces, presumably from the spindle microtubules, that try to separate the chromosomes and as a result they are pulled from one cell to the other. This indicates that these forces are not equally distributed between the two cells at any given moment because, if they were, the fused telomeres would end up in the bud neck. At later stages some nuclei can be found distributed between more than two cells (Figure 6A 250 generations and Figure 6B 200 generations), presumably the spindle is no longer functioning properly. More evidence for this is the fact that many nuclei have lost their bright putative nucleolar Sir2-CFP labelling, reflecting an uneven division of rDNA between the dividing cells. This dynamic and aberrant movement may underlie the increased gross chromosomal rearrangements seen at senescence/crisis and in survivors (see also Hackett et al. 2001).
The Sir2-CFP and Sir3-YFP signals exhibit partial co-localization in WT cells. Although several models predict that a Sir2-Sir3-Sir4 protein complex will bind to telomeres via Rap1p interaction, we found foci with only Sir2-CFP or only Sir3-YFP as well as foci adjacent to each other but not overlapping. In our system the CFP signal is always weaker than the YFP signal so it is possible that some weaker Sir2-CFP foci are missed. However, these results might indicate that some telomeres are only labelled with Sir2-CFP and some other telomeres with Sir3-YFP. Whether this is the result of tagging the proteins is not clear. Mating type silencing is intact, the Sir2-CFP and Sir3-YFP foci localize towards the nuclear periphery and the number of foci seems to be in line with earlier published results (Gotta et al. 1996, Laroche et al. 2000), but tagging the proteins with CFP and YFP might have disrupted the co-localization of these proteins. Alternatively, the numbers of Sir2 and Sir3 molecules per telomere might differ and the ratio between these proteins on telomeres might fluctuate. Evidence for this was found by Lieb et al. (2001) in microarray experiments.
During senescence/crisis no co-localization of the Sir2-CFP putative nucleolar signal with Sir3-YFP was found. This suggests that the process of telomere shortening does not automatically initiate an early ageing phenotype when Sir3p co-localizes in the nucleolus with Sir2p (Kennedy et al. 1997). An attempt to follow the movement of Sir2-CFP and Sir3-YFP in a strain containing mRFP-tagged Sik1p, a nucleolar protein (Huh et al. 2003), failed as most of the foci were lost.
Type I survivors maintain their ends by amplification of Y′ elements. However, the imaging results and the quantification suggest that most cells of a Type I survivor culture still show a high degree of cell cycle arrest. This resulted in a phenotype that resembles the phenotype of a nucleus in crisis/senescence, although there is growth. The Sir2-CFP and Sir3-YFP labelling showed a short period of restoration of foci before cells went back towards a senescence/crisis phenotype. This shows that amplification of Y′-elements does not result in stable telomere maintenance (see also McEachern & Haber 2006).
In one case a survivor was found where the lacO-tagged telomere was involved in amplification. This strain contained as many as 12 foci in interphase nuclei. This is more than has been seen by Y′ in situ hybridization or using tagged telomere-associated proteins, suggesting that in this strain the clustering of telomeres might have been disrupted. Alternatively, failure of proper segregation of chromosomes during mitosis might have lead to aneuploidy or even duplication of the genome.
Type II survivors maintain their telomeres by addition of several kb of telomeric repeats. This system seems more efficient, and no unprotected ends or end-to-end fusions cause any further problems in mitosis at the stages analysed. Only a slight increase in Sir3-YFP and no increase in intensity of Sir2-CFP or Rap1-GFP was found in these survivors to relate to the increase in telomere tract of several kb in Type II survivors, indicating that neither Rap1-GFP nor the tagged Sir proteins cover the whole telomere sequence. Phenotypically these cells look like WT cells in terms of telomere movement and protein localization
We have shown that the telomeres in S. cerevisiae move between the mother and daughter cells during senescence/crisis and in Type I survivors. In these cells Sir3-YFP and Rap1-GFP were prominently found in the nucleoplasm. In Type II survivors the movement of the telomeres, the localization of Sir2-CFP, Sir3-YFP and Rap1-GFP and the intensity of these proteins resembled the movement and localization found in the WT yeast cells. No obvious difference could be found in the telomere movement and protein localization to identify at an early stage of senescence/crisis which culture will result in Type I or Type II survivors. The movement and localization are, however, correlated with the growth and stability phenotypes of the two survivor types
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
The number of nuclei analysed for Sir2-CFP and Sir3-YFP during the different stages of Type I and Type II survivor development (DOC 26.5 KB).
The intensity of Sir2-CFP and Sir3-YFP of WT cells imaged during different stages of the experiment as an internal control for the experiment. Bars are standard deviations. The numbers of nuclei analysed per sample are below the graph (TIF 4.43 MB).
The intensity of Sir2-CFP (A), Sir3-YFP (B) and Rap1-GFP (C) in nuclei corrected for the nuclear size during the different stages of Type I and Type II survivor development. Bars are standard deviations (TIF 6.12 MB).
Movie showing the movement of lacO tagged telomeres during mitosis in an EST2 yeast strain (WT) (AVI 645 KB).
Movie showing the movement of lacO tagged telomeres during senescence/crisis (AVI 7.03 MB).
Movie showing the movement of the nucleus of a Type I survivor tagged with Rap1-GFP (AVI 14.4 MB).
Acknowledgements
This work was supported by a Wellcome Trust Grant. The Yeast Resource Center is supported by a P41 grant (P41 RR11823), from the National Center for Research Resources at the US National Institutes of Health.
Abbreviations
- ALT
alternative lengthening of telomeres
- CHEF
clamped homogeneous electric field gel
- CFP
cyan fluorescent protein
- CHR
chromosome
- DIG
digoxigenin
- EST2
ever shorter telomeres 2
- GFP
green fluorescent protein
- HIS3
histidine 3
- HML
mating type cassette — left
- HMR
mating type cassette -right
- HYG
hygromycin B
- KanMX4
kanamycin resistance cassette 4
- LacO
Lac operon
- LacI
Lac repressor
- LEU2
leucine 2
- MIP
maximum intensity projection
- RADx
radiation sensitive x
- RAP1
repressor activator protein 1
- rDNA
ribosomal DNA
- RIF1/2
RAP1-interacting factor 1/2
- SGD
Saccharomyces genome database
- SIR2/3/4
silent information regulator 2/3/4
- SGS1
slow growth suppressor 1
- TLC1
telomerase component 1
- TRP1
tryptophan 1
- WT
wild-type yeast strain for the EST2 gene
- YFP
yellow fluorescent protein
- YPD
yeast extract/peptone/dextrose medium
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s10577-007-1178-2) contains supplementary material, which is available to authorized users.
References
- Blackburn EH (1991) Structure and function of telomeres. Nature350: 569–73. [DOI] [PubMed]
- Blasco MA, Lee HW, Rizen M, Hanahan D, DePinho R, Greider CW (1997) Mouse models for the study of telomerase. Ciba Found Symp211: 160–70. [DOI] [PubMed]
- Bryan TM, Englezou A, Gupta J, Bacchetti S, Reddel RR (1995) Telomere elongation in immortal human cells without detectable telomerase activity. Embo J14: 4240–248. [DOI] [PMC free article] [PubMed]
- Chen Q, Ijpma A, Greider CW (2001) Two survivor pathways that allow growth in the absence of telomerase are generated by distinct telomere recombination events. Mol Cell Biol21: 1819–827. [DOI] [PMC free article] [PubMed]
- Cohen H, Sinclair DA (2001) Recombination-mediated lengthening of terminal telomeric repeats requires the Sgs1 DNA helicase. Proc Natl Acad Sci USA98: 3174–179. [DOI] [PMC free article] [PubMed]
- Gietz RD, Woods RA (2002) Transformation of yeast by the Liac/SS carrier DNA/PEG method. Methods Enzymol350: 87–6. [DOI] [PubMed]
- Goldstein AL, McCusker JH (1999) Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast15: 1541–553. [DOI] [PubMed]
- Gotta M, Laroche T, Formenton A, Maillet L, Scherthan H, Gasser SM (1996) The clustering of telomeres and co-localization with Rap1, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae. J Cell Biol134: 1349–363. [DOI] [PMC free article] [PubMed]
- Gotta M, Strahl-Bolsinger S, Renauld H, et al. (1997) Localization of Sir2p: the nucleolus as a compartment for silent information regulators. EMBO J16: 3243–255. [DOI] [PMC free article] [PubMed]
- Gottschling DE, Aparicio OM, Billington BL, Zakian VA (1990) Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell63: 751–62. [DOI] [PubMed]
- Hackett JA, Feldser DM, Grieder CW (2001) Telomere dysfunction increases mutation rate and genomic instability. Cell106: 275–86. [DOI] [PubMed]
- Hayashi A, Ogawa H, Kohno K, Gasser SM, Hiraoka Y (1998) Meiotic behaviours of chromosomes and microtubules in budding yeast: relocalization of centromeres and telomeres during meiotic prophase. Genes Cells3: 587–01. [DOI] [PubMed]
- Hemann MT, Strong M, Hao L-Y, Greider CW (2001) The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell107: 67–7. [DOI] [PubMed]
- Heun P, Laroche T, Shimada K, Furrer P, Gasser SM (2001) Chromosome dynamics in the yeast interphase nucleus. Science294: 2181–186. [DOI] [PubMed]
- Huang P, Pryde FE, Lester D, et al. (2001) SGS1 is required for telomere elongation in the absence of telomerase. Curr Biol11: 125–29. [DOI] [PubMed]
- Huh WK, Falvo JV, Gerke LC, et al. (2003) Global analysis of protein localization in budding yeast. Nature425: 686–91. [DOI] [PubMed]
- Ivy JM, Klar AJS, Hicks JB (1986) Cloning and characterization of four SIR genes of Saccharomyces cerevisiae. Mol Cell Biol6: 688–02. [DOI] [PMC free article] [PubMed]
- Johnson FB, Marciniak RA, McVey M, Stewart SA, Hahn WC, Guarente L (2001) The Saccharomyces cerevisiae WRN homolog Sgs1p participates in telomere maintenance in cells lacking telomerase. EMBO J20: 905–13. [DOI] [PMC free article] [PubMed]
- Kennedy BK, Gotta M, Sinclair DA, et al. (1997) Redistribution of silencing proteins from telomeres to the nucleolus is associated with extension of life span in S. cerevisiae. Cell89: 381–91. [DOI] [PubMed]
- Klein F, Laroche T, Cardenas M, Hofmann J, Schweizer D, Gasser S (1992) Localization of RAP1 and topoisomerase II in nuclei and meiotic chromosomes of yeast. J Cell Biol117: 935–48. [DOI] [PMC free article] [PubMed]
- Kurtz S, Shore D (1991) RAP1 protein activates and silences transcription of mating-type genes in yeast. Genes Dev5: 616–28. [DOI] [PubMed]
- Laroche T, Martin SG, Tsai-Pflugfelder M, Gasser SM (2000) The dynamics of yeast telomeres and silencing proteins through the cell cycle. J Struct Biol129: 159–74. [DOI] [PubMed]
- Le S, Moore JK, Haber JE, Greider CW (1999) RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics152: 143–52. [DOI] [PMC free article] [PubMed]
- Lendvay TS, Morris DK, Sah J, Balasubramanian B, Lundblad V (1996) Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics144: 1399–412. [DOI] [PMC free article] [PubMed]
- Levy DL, Blackburn EH (2004) Counting of Rif1p and Rif2p on Saccharomyces cerevisiae telomeres regulates telomere length. Mol Cell Biol24: 10857–0867. [DOI] [PMC free article] [PubMed]
- Lieb JD, Liu X, Botstein D, Brown PO (2001) Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat Genet28: 327–34. [DOI] [PubMed]
- Link AJ, Olson MV (1991) Physical map of the Saccharomyces cerevisiae genome at 110-kilobase resolution. Genetics127: 681–98. [DOI] [PMC free article] [PubMed]
- Liti G, Louis EJ (2003) NEJ1 Prevents NHEJ-dependent telomere fusions in yeast without telomerase. Mol Cell11: 1373–378. [DOI] [PubMed]
- Louis EJ (1998) Whole chromosome analysis. In: Tuite MF, Brown AP, eds. Methods in Microbiology: Yeast Gene Analysis. London: Academic Press, pp. 15–31.
- Louis EJ, Borts RH (1995) A complete set of marked telomeres in Saccharomyces cerevisiae for physical mapping and cloning. Genetics139: 125–36. [DOI] [PMC free article] [PubMed]
- Louis EJ, Haber JE (1992) The structure and evolution of subtelomeric Y′ repeats in Saccharomyces cerevisiae. Genetics131: 559–74. [DOI] [PMC free article] [PubMed]
- Lundblad V, Blackburn EH (1993) An alternative pathway for yeast telomere maintenance rescues est1- senescence. Cell73: 347–60. [DOI] [PubMed]
- Lundblad V, Szostak JW (1989) A mutant with a defect in telomere elongation leads to senescence in yeast. Cell57: 633–43. [DOI] [PubMed]
- Lustig AJ, Kurtz S, Shore D (1990) Involvement of the silencer and UAS binding protein RAP1 in regulation of telomere length. Science250: 549–53. [DOI] [PubMed]
- McEachern MJ, Haber JE (2006) Break-induced replication and recombinational telomere elongation in yeast. Annu Rev Biochem75: 111–35. [DOI] [PubMed]
- Nautiyal S, DeRisi JL, Blackburn EH (2002) The genome-wide expression response to telomerase deletion in Saccharomyces cerevisiae. PNAS99: 9316–321. [DOI] [PMC free article] [PubMed]
- Pardo B, Marcand S (2005) Rap1 prevents telomere fusions by nonhomologous end joining. Embo J24: 3117–127. [DOI] [PMC free article] [PubMed]
- Pryde FE, Louis EJ (1999) Limitations of silencing at native yeast telomeres. EMBO J18: 2538–550. [DOI] [PMC free article] [PubMed]
- Shaw SL, Maddox P, Skibbens RV, Yeh E, Salmon ED, Bloom K (1998) Nuclear and spindle dynamics in budding yeast. Mol Biol Cell9: 1627–631. [DOI] [PMC free article] [PubMed]
- Sherman F, Fink GR, Hicks JB (1986) Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Shimada K, Gasser SM (2007) The origin recognition complex functions in sister-chromatid cohesion in Saccharomyces cerevisiae. Cell128: 85–9. [DOI] [PubMed]
- Smith JS, Boeke JD (1997) An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev11: 241–54. [DOI] [PubMed]
- Straight AF, Belmont AS, Robinett CC, Murray AW (1996) GFP tagging of budding yeast chromosomes reveals that protein–protein interactions can mediate sister chromatid cohesion. Curr Biol6: 1599–608. [DOI] [PubMed]
- Straight AF, Marshall WF, Sedat JW, Murray AW (1997) Mitosis in living budding yeast: anaphase A but no metaphase plate. Science277: 574–78. [DOI] [PubMed]
- Sussel L, Shore D (1991) Separation of transcriptional activation and silencing functions of the RAP1-encoded repressor/activator protein 1: isolation of viable mutants affecting both silencing and telomere length. Proc Natl Acad Sci USA88: 7749–753. [DOI] [PMC free article] [PubMed]
- Teng S-H, Zakian VA (1999) Telomere–telomere recombination is an efficient bypass pathway for telomere maintenance in Sacchatomyces cerevisiae. Mol Cell Biol19: 8083–093. [DOI] [PMC free article] [PubMed]
- Walmsley RM, Gardner DC, Oliver SG (1983) Stability of a cloned gene in yeast grown in chemostat culture. Mol Gen Genet192: 361–65. [DOI] [PubMed]
- Watson JM, Bulankova P, Riha K, Shippen DE, Vyskot B (2005) Telomerase-independent cell survival in Arabidopsis thaliana. Plant J43: 662–74. [DOI] [PubMed]
- Watt PM, Hickson ID, Borts RH, Louis EJ (1996) SGS1, a homologue of the Bloom’s and Werner’s syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae. Genetics144: 935–45. [DOI] [PMC free article] [PubMed]
- Wotton D, Shore D (1997) A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev11: 748–60. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Below is the link to the electronic supplementary material.
The number of nuclei analysed for Sir2-CFP and Sir3-YFP during the different stages of Type I and Type II survivor development (DOC 26.5 KB).
The intensity of Sir2-CFP and Sir3-YFP of WT cells imaged during different stages of the experiment as an internal control for the experiment. Bars are standard deviations. The numbers of nuclei analysed per sample are below the graph (TIF 4.43 MB).
The intensity of Sir2-CFP (A), Sir3-YFP (B) and Rap1-GFP (C) in nuclei corrected for the nuclear size during the different stages of Type I and Type II survivor development. Bars are standard deviations (TIF 6.12 MB).
Movie showing the movement of lacO tagged telomeres during mitosis in an EST2 yeast strain (WT) (AVI 645 KB).
Movie showing the movement of lacO tagged telomeres during senescence/crisis (AVI 7.03 MB).
Movie showing the movement of the nucleus of a Type I survivor tagged with Rap1-GFP (AVI 14.4 MB).


