Abstract
Neurodevelopmental disorders that disturb speech and language are highly heritable. Isolation of the underlying genetic risk factors has been hampered by complexity of the phenotype and potentially large number of contributing genes. One exception is the identification of rare heterozygous mutations of the FOXP2 gene in a monogenic syndrome characterised by impaired sequencing of articulatory gestures, disrupting speech (developmental verbal dyspraxia, DVD), as well as multiple deficits in expressive and receptive language. The protein encoded by FOXP2 belongs to a divergent subgroup of forkhead-box transcription factors, with a distinctive DNA-binding domain and motifs that mediate hetero- and homodimerisation. FOXP1, the most closely related member of this subgroup, can directly interact with FOXP2 and is co-expressed in neural structures relevant to speech and language disorders. Moreover, investigations of songbird orthologues indicate that combinatorial actions of the two proteins may play important roles in vocal learning, leading to the suggestion that human FOXP1 should be considered a strong candidate for involvement in DVD. Thus, in this study, we screened the entire coding region of FOXP1 (exons and flanking intronic sequence) for nucleotide changes in a panel of probands used earlier to detect novel mutations in FOXP2. A non-synonymous coding change was identified in a single proband, yielding a proline-to-alanine change (P215A). However, this was also found in a random control sample. Analyses of non-coding SNP changes did not find any correlation with affection status. We conclude that FOXP1 mutations are unlikely to represent a major cause of DVD.
Keywords: speech and language, developmental verbal dyspraxia, transcription factor, FOXP2, FOXP1, heterodimerisation
Introduction
Developmental speech and language disorders are highly heritable, but the identification of genetic risk factors through classical mapping or association studies is hampered by genotypic and phenotypic complexity.1 The implication of FOXP2 in a rare monogenic form of disorder2 provides novel entry points into the critical molecular pathways.3 A heterozygous missense mutation of FOXP2 co-segregates with speech and language disorder in the well-studied multigenerational KE family,2 disrupting the function of the encoded protein.4, 5 People carrying this mutation have problems sequencing mouth movements underlying speech (developmental verbal dyspraxia, DVD; MIM: 602081), along with impaired expressive and receptive language skills whether oral or written. Further cases of FOXP2 disruptions causing verbal dyspraxia include a heterozygous nonsense mutation co-segregating with impairment in a small pedigree6 and several gross chromosomal rearrangements.7, 8, 9 However, aetiological point mutations of FOXP2 likely account for only ∼2% of children with DVD;6 other key genetic risk factors remain to be discovered. Importantly, data from functional studies of FOXP2 can identify related genes acting in the same pathways to be considered as candidates for involvement in disorder.
FOXP2 encodes a member of the FOX group of transcription factors, featuring a characteristic forkhead-box DNA-binding domain. It belongs to a divergent subgroup (FOXP1–4) displaying a number of distinctive characteristics. Co-immunoprecipitation studies of murine orthologues showed that Foxp1, Foxp2 and Foxp4 form homo- and heterodimers, which are thought to be necessary for their efficient binding to target DNA.10 In contrast, most other forkhead proteins act as monomers.10 Furthermore, a FOXP3 mutation that disturbs dimerisation results in immunodysregulation, polyendocrinopathy, enteropathy, X-linked syndrome.11 Overall, for this subgroup of forkhead transcription factors, activity and/or specificity may be determined by the relative levels and combinations of different FOXP proteins at a given time point, developmental stage or tissue/cell type.
Among the FOXP subfamily, FOXP1 is the most likely protein to influence language-related pathways in concert with FOXP2. The two proteins show a particularly high sequence homology and can cooperatively regulate downstream targets through heterodimerisation. Studies of FoxP1 in diverse vertebrates, including humans, mice and songbirds, indicate several key sites of neural expression, displaying a significant overlap with FoxP2 expression.12 In the developing mouse lung (another site of co-expression), Foxp1 and Foxp2 have been shown to bind the promoter of a shared target gene (T1α), acting together to modulate its expression.13 Moreover, although mice with heterozygous Foxp1 disruption develop normally, and those with homozygous Foxp2 loss show a subtly altered postnatal alveolarisation, combining both genotypes (Foxp1+/−, Foxp2−/−) yields much more severe lung defects, consistent with cooperative roles.13
Songbird studies further suggest that coordinated effects of FOXP2 and FOXP1 are relevant for spoken language. Like humans, songbirds display both innate vocalisations and vocal learning.14 In zebrafinches, only males learn and modify courtship song, doing so through the song system – neuronal networks spanning the cortical mantle, striatum and thalamus.12 A striatal nucleus, area X, is necessary for song development and vocalisation, and is only present in the male brain. FoxP2 and FoxP1 are well conserved in zebrafinches, and are expressed in several parts of the song system, including high levels in area X and surrounding striatum.12, 15 Knockdown of area-X FoxP2 expression during song development results in inaccurate and incomplete imitation of tutor songs,14 suggesting important postnatal roles in auditory-guided motor learning. However, FoxP2 does not show sexually dimorphic expression; there is no consistent differentiation between its expression in area X of males and that in the corresponding region in females.12 This suggests either that females have an unrealised potential for vocal learning or that this process is influenced by a sexually dimorphic co-regulator of FoxP2. Intriguingly, FoxP1 expression in area X of male zebrafinches displays a sexual dimorphism closely resembling the pattern observed for the song circuit itself.12 Thus, in songbirds, dimerisation with FoxP1 may confer sexually dimorphic activity on FoxP2 during vocal learning.
Integrating the above findings, it is plausible that development/function of language-related circuits in the human brain could be disturbed by imbalances in relative functional dosage of FOXP2 and FOXP1. Thus, as explicitly proposed by Teramitsu et al,12 FOXP1 represents a strong functional candidate for involvement in speech disorders. In this study, we directly tested this hypothesis by mutation screening of all coding FOXP1 exons and flanking splice sites in children with DVD.
Materials and methods
Children with DVD
A panel of 49 probands was assembled based on a primary clinical diagnosis of DVD (see Supplementary methods). Earlier analyses of this panel successfully identified novel mutations in the FOXP2 gene.6
Genomic organisation of FOXP1
The intron/exon structure of FOXP1 was determined by aligning GenBank entry AF146696 (FOXP1 mRNA) with human genomic sequence through the UCSC Genome Browser (hg17 assembly, NCBI Build 35).
Denaturing high-performance liquid chromatography screening
Primers were designed to amplify the 16 coding exons of FOXP1 (Supplementary Table S1), each with a fragment size optimal for denaturing high-performance liquid chromatography (DHPLC) (200–600 bp). After amplification using touchdown PCR, fragments were analysed through the Transgenomics WAVE DHPLC system followed by direct sequencing (Supplementary methods).
Results
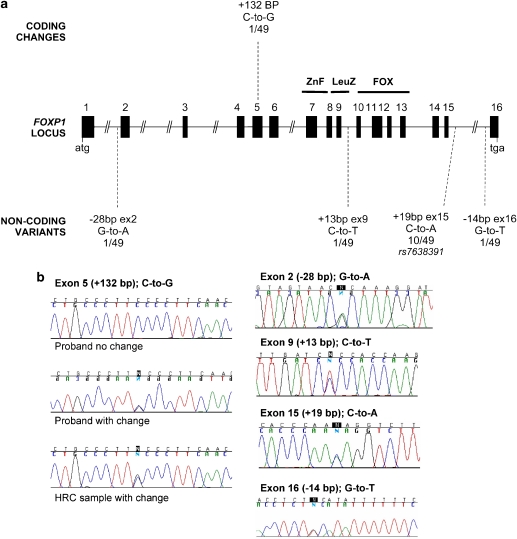
FOXP1 consists of 16 coding exons spanning ∼586 kb on 3p14.1 (Figure 1a). We screened these exons and flanking intronic sequence in 49 probands with clinically diagnosed DVD plus their siblings (59 individuals in total). DHPLC analysis, followed by sequencing of variants, revealed one exonic (coding) change and four intronic (non-coding) polymorphisms (Table 1). The coding change, identified in a singleton proband, was located in exon 5. This change constituted a heterozygous C-to-G transversion, yielding a proline-to-alanine substitution at position 215 of the encoded protein. Alignments of FOXP proteins from different species indicated that this was one of the few highly conserved residues in a region of low homology (Figure 2). To determine if this change was exclusive to DVD, 384 control chromosomes from Human Random Control (HRC) panels were screened using identical methods to the clinical samples. The same heterozygous C-to-G change was identified in a single chromosome within the controls (Figure 1b).
Figure 1.
Mutation screening of FOXP1 in verbal dyspraxia probands. (a) Genomic organisation of FOXP1. FOXP1 is located on chromosome 3p14.1 and consists of 16 coding exons spanning 586 kb. Exons are represented by filled bars whose width is proportional to the length of the exon. Numbering scheme is based on alignment with the published mouse Foxp1 structure.20 Initiation codon is indicated by ‘atg' and stop codon by ‘tga'. The Zinc-Finger domain spans exons 7 and 8, the Leucine-Zipper domain spans exons 8 and 9 and the forkhead-box motif is encoded by exons 10–13. Locations of base changes summarised in Table 1 are indicated on figure by bars, with base change and frequency in probands. (b) Direct sequencing confirmed the presence of base changes in probands displaying aberrant DHPLC elution patterns (see also Table 1). The exon 5 polymorphism is shown for the proband and HRC samples that carried the C-to-G transversion, aligned with a proband that did not carry the change for comparison.
Table 1. Base changes identified through DHPLC mutation screening and direct sequencing.
| Exon | Change | DbSNP reference | Intronic/exonic | Positiona | No. of individuals (N=49) |
|---|---|---|---|---|---|
| 2 | G-to-A | — | Intronic | −28 bp | 1 |
| 5 | C-to-G | — | Exonic | +132 bp | 1 |
| 9 | C-to-T | — | Intronic | +13 bp | 1 |
| 15 | C-to-A | rs7638391 | Intronic | +19 bp | 10 |
| 16 | G-to-T | — | Intronic | −14 bp | 1 |
Position is given as upstream (−) or downstream (+) of exon/intron boundary.
Figure 2.
Proline-215 is conserved within the FOXP subfamily. Amino acids 206–260 encoded by exon 5 of FOXP1 were aligned (using CLUSTALW) with a range of sequences from FoxP1, FoxP2 and FoxP4 plus the closest drosophila homologue to the FOXP subgroup. Swiss-Prot/TrEMBL accession numbers are shown in parentheses. Conserved amino acids are shaded in black and similar amino acids are shaded in grey. Proline-215 is indicated by an asterisk and is completely conserved across all available FOXP sequences.
A heterozygous C-to-A transversion in the intron downstream of exon 15 (Figure 1, Table 1) was found in 10 of the 49 probands (an SNP frequency of ∼10%). We tested whether this non-coding SNP might be in linkage disequilibrium with an undiscovered functional variant. DHPLC screening of 146 HRC samples identified the SNP in 20 controls (an SNP frequency of ∼7%). As the SNP frequency is not significantly different between DVD cases and controls (χ2 P-value>0.25), it is unlikely to be associated with the disorder. The same heterozygous SNP was reported on dbSNP (http://www.ncbi.nlm.nih.gov/SNP/) at ∼12.5% in populations of European descent. Again, the frequency did not significantly differ from that of our DVD cases (χ2 P-value >0.5).
Discussion
A range of FOXP2 mutations has been identified in DVD, clearly showing the aetiological importance of this gene.2, 6, 7, 8, 9 Given the high heritability of speech/language impairments it is highly likely that other genes contribute. Here, we investigated the FOXP1 functional candidate by mutation screening in a panel of cases that had earlier enabled us to identify FOXP2 coding changes.6
A heterozygous FOXP1 coding change was identified in a proband with no affected siblings. The resulting substitution (P215A) replaced a proline with an alanine in the region encoded by exon 5. The region lies outside known functional domains and its structure is unknown. However, alignments of FOXP orthologues from diverse species (human to Drosophila melanogaster) indicate the high conservation of this proline (Figure 2). Mutations affecting proline residues commonly affect protein structure/stability.16 Owing to their rigid conformation, prolines frequently induce bends or kinks. Although a proline may sometimes enhance structural stability (eg, as the first residue of an α-helix), it often disrupts the secondary structure (eg, when located internally within an α-helix or a β-sheet).17 Thus, P215 could be important for maintaining local structure. Nonetheless, in the absence of a thorough characterisation of the structure of full-length FOXP1, it is difficult to determine the structural impact of the P215A substitution.
It is worth noting that a heterozygous P215A change was also identified in one of 384 control chromosomes from HRC panels (compared with a frequency of 1-in-98 chromosomes in the DVD panel). The possibility remains that this change produces a functional effect, particularly as developmental language disorders are observed in up to 7% of school-age children.18 Given that information is unavailable regarding phenotypes of HRC individuals, we cannot exclude the possibility that the control individual carrying the P215A change was affected with a mild speech/language disorder.
Despite evidence from human, mouse and songbirds suggesting the importance of FoxP1–FoxP2 neural interactions in pathways mediating speech and language, our study found no indication of clear correlations between FOXP1 variants and DVD risk. Nevertheless, given emerging roles for this gene in nervous system development,19 it continues to represent a candidate for involvement in neurodevelopmental disorders. Further investigations of FoxP1/FoxP2 in model systems promise greater insights into their coordinated effects on brain function.
Acknowledgments
We thank Nuala Sykes and Thomas Scerri for assistance in setting up the DHPLC mutation analysis and Thomas Bajorek for assistance during screening. The project was funded by the Wellcome Trust and Autism Speaks. SCV was supported by a Christopher Welch Biological Sciences Scholarship from the University of Oxford. SEF is a Royal Society Research Fellow.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Fisher SE, Lai CS, Monaco AP. Deciphering the genetic basis of speech and language disorders. Annu Rev Neurosci. 2003;26:57–80. doi: 10.1146/annurev.neuro.26.041002.131144. [DOI] [PubMed] [Google Scholar]
- Lai CS, Fisher SE, Hurst JA, Vargha-Khadem F, Monaco AP. A forkhead-domain gene is mutated in a severe speech and language disorder. Nature. 2001;413:519–523. doi: 10.1038/35097076. [DOI] [PubMed] [Google Scholar]
- Vernes SC, Spiteri E, Nicod J, et al. High-throughput analysis of promoter occupancy reveals direct neural targets of FOXP2, a gene mutated in speech and language disorders. Am J Hum Genet. 2007;81:1232–1250. doi: 10.1086/522238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groszer M, Keays DA, Deacon RM, et al. Impaired synaptic plasticity and motor learning in mice with a point mutation implicated in human speech deficits. Curr Biol. 2008;18:354–362. doi: 10.1016/j.cub.2008.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernes SC, Nicod J, Elahi FM, et al. Functional genetic analysis of mutations implicated in a human speech and language disorder. Hum Mol Genet. 2006;15:3154–3167. doi: 10.1093/hmg/ddl392. [DOI] [PubMed] [Google Scholar]
- MacDermot KD, Bonora E, Sykes N, et al. Identification of FOXP2 truncation as a novel cause of developmental speech and language deficits. Am J Hum Genet. 2005;76:1074–1080. doi: 10.1086/430841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuk L, Kalervo A, Lipsanen-Nyman M, et al. Absence of a paternally inherited FOXP2 gene in developmental verbal dyspraxia. Am J Hum Genet. 2006;79:965–972. doi: 10.1086/508902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennon PA, Cooper ML, Peiffer DA, et al. Deletion of 7q31.1 supports involvement of FOXP2 in language impairment: clinical report and review. Am J Med Genet A. 2007;143:791–798. doi: 10.1002/ajmg.a.31632. [DOI] [PubMed] [Google Scholar]
- Shriberg LD, Ballard KJ, Tomblin JB, Duffy JR, Odell KH, Williams CA. Speech, prosody, and voice characteristics of a mother and daughter with a 7;13 translocation affecting FOXP2. J Speech Lang Hear Res. 2006;49:500–525. doi: 10.1044/1092-4388(2006/038). [DOI] [PubMed] [Google Scholar]
- Li S, Weidenfeld J, Morrisey EE. Transcriptional and DNA binding activity of the Foxp1/2/4 family is modulated by heterotypic and homotypic protein interactions. Mol Cell Biol. 2004;24:809–822. doi: 10.1128/MCB.24.2.809-822.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatila TA, Blaeser F, Ho N, et al. JM2, encoding a fork head-related protein, is mutated in X-linked autoimmunity-allergic disregulation syndrome. J Clin Invest. 2000;106:R75–R81. doi: 10.1172/JCI11679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teramitsu I, Kudo LC, London SE, Geschwind DH, White SA. Parallel FoxP1 and FoxP2 expression in songbird and human brain predicts functional interaction. J Neurosci. 2004;24:3152–3163. doi: 10.1523/JNEUROSCI.5589-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu W, Lu MM, Zhang Y, Tucker PW, Zhou D, Morrisey EE. Foxp2 and Foxp1 cooperatively regulate lung and esophagus development. Development. 2007;134:1991–2000. doi: 10.1242/dev.02846. [DOI] [PubMed] [Google Scholar]
- Haesler S, Rochefort C, Georgi B, Licznerski P, Osten P, Scharff C. Incomplete and inaccurate vocal imitation after knockdown of FoxP2 in songbird basal ganglia nucleus Area X. PLoS Biol. 2007;5:e321. doi: 10.1371/journal.pbio.0050321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haesler S, Wada K, Nshdejan A, et al. FoxP2 expression in avian vocal learners and non-learners. J Neurosci. 2004;24:3164–3175. doi: 10.1523/JNEUROSCI.4369-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajaj K, Madhusudhan MS, Adkar BV, et al. Stereochemical criteria for prediction of the effects of proline mutations on protein stability. PLoS Comput Biol. 2007;3:e241. doi: 10.1371/journal.pcbi.0030241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhoof G, Goossens F, De Meester I, Hendriks D, Scharpe S. Proline motifs in peptides and their biological processing. FASEB J. 1995;9:736–744. [PubMed] [Google Scholar]
- Tomblin JB, Records NL, Buckwalter P, Zhang X, Smith E, O'Brien M. Prevalence of specific language impairment in kindergarten children. J Speech Lang Hear Res. 1997;40:1245–1260. doi: 10.1044/jslhr.4006.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasen JS, De Camilli A, Wang B, Tucker PW, Jessell TM. Hox repertoires for motor neuron diversity and connectivity gated by a single accessory factor, FoxP1. Cell. 2008;134:304–316. doi: 10.1016/j.cell.2008.06.019. [DOI] [PubMed] [Google Scholar]
- Wang B, Lin D, Li C, Tucker P. Multiple domains define the expression and regulatory properties of Foxp1 forkhead transcriptional repressors. J Biol Chem. 2003;278:24259–24268. doi: 10.1074/jbc.M207174200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.