Abstract
Background
Although correlations of cervical cytology to human papillomavirus (HPV) load and histopathology are recognized, it is largely undetermined whether viral load-related risks of cervical intraepithelial neoplasia III (CIN3) differ by cytology.
Methods
Study subjects were 821 women enrolled in the ASCUS-LSIL Triage Study who were positive for HPV16 at entry. Women were followed semi-annually over 2 years. Baseline HPV16 load was measured by real-time PCR; expressed as log10 [HPV16 copies per-nanogram of cellular DNA].
Results
CIN3 was confirmed in 34.8% of 821 women during 2-year follow-up. The adjusted odds ratio (OR) associating 2-year cumulative risk of CIN3 with per log10–unit increase in HPV16 load was 1.46 (95%CI, 1.29-1.64). The ORs varied from 1.66 (95%CI, 1.16-2.37) for women with normal cytology at enrollment to 0.86 (95%CI, 0.61-1.20) for those with HSIL. Among women with normal cytology at enrollment, the area under the receiver operating characteristic curve for detecting CIN3 by viral load was 0.70 (95%CI, 0.61-0.78).
Conclusion
HPV16 DNA load was associated with CIN3 risk but the associations varied with cytology detected at the time when the viral load was measured. Clinical utility of testing for HPV16 load for CIN3 detection was minimal even in women with normal cytology.
Keywords: Human Papillomavirus, Viral load, Cervical Intraepithelial Neoplasia
INTRODUCTION
Rather than simply testing for the presence of human papillomavirus (HPV) infection, an assessment of HPV DNA load additionally reflects the amount of the virus infected. Risk associations between high-grade cervical intraepithelial neoplasia (CIN) and elevated viral load have been found in some [1-18] but not other studies [19-24]. Identification of factors that affect the viral load-related risk association may help to clarify these discrepant results and better understand the etiologic role of HPV DNA load.
Abnormalities of cervical cytology are thought to be manifestations of productive HPV infections. HPV DNA load has been shown significantly correlated to concurrent presence [14, 25-29] and future development of cytologic abnormalities [30, 31]. A study by Sherman et al. [32] has demonstrated that among women with CIN3, the amount of HPV DNA was substantially affected by the number of atypical squamous cells of undetermined significance (ASCUS) and low-grade squamous intraepithelial lesion (LSIL) cells present in exfoliative cervical samples. Given a close correlation between cervical cytology and histopathology, it is likely that the viral load-related risk of high-grade CIN may differ by cytomorphology detected in Pap smears.
Although fewer studies [4, 30, 33] have attempted to assess risk association in women with normal cytology and ASCUS/LSIL, separately, the findings are limited by either semi-quantitative measurement of additive viral loads from different HPV types or a lack of sufficient statistical power. Fully understanding of effects of cervical cytology on viral load-related risk of CIN would be clinically important, because testing for these viruses has now been recommended to be added into programs as an adjunct to Pap testing for screening and clinical management [34-37].
To address this, we evaluated in a longitudinal setting, cytology-stratified associations between baseline HPV16 DNA load and 2-year cumulative risk of CIN3 among women who participated in the ASCUS-LSIL Triage Study (ALTS) and performance of testing for HPV16 DNA load for predicting risk of underlying CIN3.
STUDY SUBJECTS AND METHODS
Study subject and design
The ASCUS-LSIL Triage Study was a randomized multicenter clinical trial designed to evaluate strategies for management of equivocal and mildly abnormal cervical cytology. A detailed description of the ALTS design and study population is presented elsewhere [38, 39]. Briefly, between January 1997 and December 1998, a total of 5,060 women were enrolled an average of 2 months after a referral Pap of ASCUS or LSIL was obtained and randomly assigned into one of three management arms (i.e., immediate colposcopy, HPV triage, and conservative management). These arms differed only in referral for colposcopy at enrollment: an entry colposcopy with colposcopic-directed biopsy of visible lesions was referred for all women in the immediate colposcopy arm, women who had an enrollment cytologic diagnosis of high-grade SIL (HSIL) in the conservative management arm, and women who had an enrollment HPV testing result of positive for oncogenic types or a cytologic diagnosis of HSIL in the HPV arm. All women, regardless of the study arm, were scheduled for follow-up with cytology and HPV testing at 6-month intervals for 2 years. During follow-up, women were re-referred for colposcopy and biopsy if HSIL was found. At exit, participants were required to undergo an exit procedure including cytology, HPV testing, and colposcopic examination with biopsy of any visible lesions. Women who had received a diagnosis of ≥CIN2 were promptly treated usually by Loop Electrosugical Excision Procedure (LEEP).
An ALTS participant was eligible for the present study, if her enrollment cervical sample was positive for HPV16 DNA by polymerase chain reaction (PCR)-based reverse-line blot. The ALTS protocol was approved by the institutional review board at National Cancer Institute and each of the four clinical centers involved in the trial. The protocol for this study was approved by the institutional review board at University of Washington. Of 846 eligible women, 7 were excluded because their enrollment samples were later confirmed to be negative for HPV16 DNA. We additionally excluded 12 women whose enrollment samples were unavailable for viral load testing and 6 women who had an unsatisfactory cytologic diagnosis at enrollment, leaving 821 in the analysis.
Clinical endpoint
In ALTS, cervical cytology and histology were initially diagnosed by clinical center pathologists and then reviewed by a panel of expert pathologists for quality control and safety monitoring. Histologic diagnosis was made on tissues obtained by biopsy, endocervical curettage, and/or LEEP. The most severe diagnosis was used, was there more than one tissue block examined at a single visit. We used a 2-year cumulative diagnosis of CIN3 as the clinical endpoint to overcome potential study-arm-related missing and/or delayed diagnoses of CIN3 at enrollment [40]. For women with more than one CIN3 diagnosis during the study period, only the first onset was counted. Cervical cytology and histology used in the present study were based on diagnoses by the panel of expert pathologists. The results did not alter appreciably when the diagnoses by the clinical center pathologists were used as clinical outcomes (data not shown).
Quantification of HPV16 DNA load
A multiplex real-time PCR was performed on all HPV16-positive enrollment samples for quantification of HPV16 DNA. The assay was set up in a reaction volume of 25 μl with the TaqMan Universal PCR Master Mix kit (Applied Biosystems, Foster City, CA). Sequences of primers and probe for HPV16 E7 gene were: forward primer, nucleotide position 700-720, 5’-CCGGACAGAGCCCATTACAAT; reverse primer, nucleotide position 782-762, 5’-ACGTGTGTGCTTTGTACGCAC; and florescence-labeled probe, nucleotide position 733-760, 5’ FAM-TGTTGCAAGTGTGACTCTACGCTTCGGT-TAMRA. The primers and probe for human β-actin gene were commercially available (Applied Biosystems, Foster City, CA). Amplification was carried out on Applied Biosystems 7900 HT Sequence Detection System with a cycling program of holding at 50°C for 2 minutes and then at 95°C for 10 minutes followed by a two-step cycle of 10 seconds at 95°C and 1 minute at 60°C for 40 cycles.
Two log-phase 5-point standard curves were implemented in each set of the assay, one for HPV16 and the other for cellular DNA. The number of HPV16 E7 copies was normalized according to the input amount of cellular DNA (β-actin). Each sample was assayed in triplicate and a mean of three measures was used for analysis. As assessed by one-way ANOVA with random effects, the estimated reliability of the mean of the triplicate measures was 0.981 with an intra-class correlation of 0.947 (95% CI, 0.941-0.954). The viral load was expressed as the mean of log10 [HPV16 E7 copy number per nanogram of cellular DNA].
HPV16 E7 DNA was undetectable by real-time PCR in 61 samples that were positive by initial PCR-based reverse-line blot. The negative result was not explained by the presence of potential inhibitors or a lack of sufficient sample inputs because the amount of cellular DNA between samples with and without detectable HPV16 E7 DNA was similar (data not shown). Considering that the “negativity” might result from a tiny amount of viral DNA, the load of one viral copy per nanogram of cellular DNA (equal to zero of log10-transformed value) was assigned to each of these samples. Similar results were obtained when these samples were excluded from the analysis. For simplicity, these results are not presented.
Statistical analysis
HPV16 DNA load was treated as a continuous variable. The normality of the distribution of the log10-transformed viral load was assessed by the Q-Q plot. The 2-year cumulative risk of CIN3 associated with baseline HPV16 DNA load was evaluated using unconditional logistic regression [41]. The effects of cervical cytology at enrollment on risk association were assessed by stratified analyses while controlling for age at enrollment, current use of hormonal contraceptives, lifetime number of sex partners, and study arm.
A receiver operating characteristic (ROC) analysis [42] was used to evaluate whether testing for baseline HPV16 DNA load would provide added value to identification of women with 2-year prevalent CIN3. The cytology-stratified ROC curves were constructed by computing the true positive rate against the false positive rate for various amounts of viral loads. We used R to implement the algorithms proposed by Delong et al. [43]. The accuracy of the test for discriminating women with from those without CIN3 was assessed by the area under the ROC curve that was computed using maximum likelihood estimates to fit a smooth curve to all data points. These areas represent the probabilities that randomly drawn pairs of women with and without CIN3 can be accurately classified by the test. An area of 1 represents a perfect test (100% sensitivity and 100% specificity) and an area of 0.5 represents a worthless test.
A linear trend of increasing baseline HPV16 DNA load with increasing severities of concurrent cervical cytology was tested by assigning scores to cytologic diagnoses and treating this scored factor as a continuous variable. A Student’s t-test or one way ANOVA analysis, whichever was appropriate, was used to compare HPV16 DNA load by demographics, sexual behavior, study arm, HPV16 variants, and co-infection with other types of HPV. All statistical tests were at the 5% two-sided significance level.
RESULTS
The mean value of log10 HPV16 E7 copy number per nanogram of cellular DNA was 2.78 (95% CI, 2.69-2.87) for 821 women who had an HPV16 infection initially detected at enrollment. As shown in figure 1, the points for the observed values against values from a normal distribution were clustered around a straight line, suggesting that the distribution of these log10-transformed values was approximately normal. Of the 821 women, 503 had referral cytology of ASCUS and the rest had referral cytology of LSIL. Abnormalities of cervical cytology were diagnosed at enrollment in 665 women (81.0%), including 240 (29.2%) with ASCUS, 271 (33.0%) with LSIL, and 154 (18.8%) with HSIL. The baseline HPV16 DNA load increased significantly with increasing severities of concurrent cytologic abnormalities in women with a referral diagnosis of either ASCUS or LSIL (table 1, p trend <0.001 for both). The trend remained similar when the analysis was restricted to women who did not have a diagnosis of 2-year prevalent CIN2-3 (data not shown).
Figure 1. Normal Q-Q plot of the log10-transformed HPV16 DNA load for women with baseline infection.
Observed values are plotted against values expected from a normal distribution. If the sample is from a normal distribution, the points are cluster around a straight line.
Table 1.
Baseline HPV16 DNA load by concurrent diagnoses of cervical cytology at enrollment: results stratified by referral cytology
| Referral cytology | Cervical cytology at enrollment | No. (%) of subjects | Mean (SD) of log10 HPV16 E7 copy number per nanogram of cellular DNA | P-value a |
|---|---|---|---|---|
| ASCUS (n = 503) | Within normal limits | 116 (23.1) | 1.66 (±1.39) | <0.001 |
| ASCUS | 157 (31.2) | 2.66 (±1.20) | ||
| LSIL | 143 (28.4) | 3.11 (±1.18) | ||
| HSIL | 87 (17.3) | 3.40 (±1.03) | ||
| LSIL (n = 318) | Within normal limits | 40 (12.6) | 2.06 (±0.86) | <0.001 |
| ASCUS | 83 (26.1) | 2.85 (±1.31) | ||
| LSIL | 128 (40.2) | 3.01 (±1.37) | ||
| HSIL | 67 (21.1) | 3.37 (±1.03) |
NOTE
Derived from a linear test
Among women with normal cytology at enrollment who did not have CIN2-3 during the study period, an increased baseline HPV16 DNA load was associated with <25 years of age (p=0.007), current use of hormonal contraceptives (p=0.006), and ≥6 lifetime number of sex partners (p=0.03). There were no appreciable differences in HPV16 DNA load by race, smoking status, number of Pap tests in the past 5 years, HPV16 variants, co-infection with other HPV types, and study arm (table 2).
Table 2.
Baseline HPV16 DNA load by characteristics of the women with normal cytology at enrollment who did not have CIN2-3 during the 2-year study period
| Variables | No. of subjects (n=138) | Mean (SD) of log10 HPV16 E7 copy number per nanogram of cellular DNA | P-value |
|---|---|---|---|
| Age at study entry | |||
| 18-24 | 86 | 1.94 (1.24) | 0.007 |
| ≥25 | 52 | 1.32 (1.30) | |
| Race/ethnicity a | |||
| White | 93 | 1.80 (1.28) | 0.27 |
| Non-white b | 44 | 1.54 (1.31) | |
| No. of life time male sex partners c | |||
| 0-5 | 70 | 1.46 (1.27) | 0.03 |
| ≥6 | 67 | 1.94 (1.29) | |
| Current use of hormonal contraceptives d | |||
| No | 77 | 1.44 (1.28) | 0.006 |
| Yes | 59 | 2.05 (1.27) | |
| Current smoking | |||
| No | 88 | 1.60 (1.35) | 0.19 |
| Yes | 50 | 1.90 (1.19) | |
| No. of Pap tests per year in the past 5 years | |||
| < 1 | 115 | 1.67 (1.33) | 0.45 |
| ≥ 1 | 23 | 1.89 (1.14) | |
| Study arms | |||
| Immediate colposcopy | 45 | 1.60 (1.41) | 0.48 |
| HPV triage | 33 | 1.57 (1.48) | |
| Conservative management | 60 | 1.86 (1.09) | |
| Co-infection with other HPV types | |||
| No | 52 | 1.72 (1.29) | 0.89 |
| Yes | 86 | 1.69 (1.32) | |
| HPV16 variants e | |||
| European | 90 | 2.07 (1.19) | 0.63 |
| Non-European | 25 | 1.95 (1.01) |
NOTE
Including African American, American Indian/Alaskan, or Asian/Pacific Islander women
Excluded was 1 woman whose race was ascertained
Excluded was 1 woman who did not provide information of number of lifetime sex partners
Excluded were 2 women who did not provide information on use of hormonal contraceptives
Excluded were 23 women whose samples were insufficient for variant characterization
CIN3 was histologically confirmed in 286 (34.8%) of 821 women during the 2-year study period: 30 (19.2%) of 156 with a diagnosis of normal cytology at enrollment, 86 (35.8%) of 240 with ASCUS, 76 (28.0%) of 271 with LSIL, and 94 (61.0%) of 154 with HSIL. The odds ratio associating 2-year cumulative risk of CIN3 with per 1 log10 increase in viral load was 1.46 (95% CI, 1.29-1.64) after adjusting for age at enrollment, current use of hormonal contraceptives, lifetime number of sex partners, and study arm (table 3). The increase in 2-year cumulative risk was statistically significant for women with normal cytology (OR adjusted = 1.66 (95% CI, 1.16-2.37), ASCUS (OR adjusted = 1.51, 95% CI, 1.17-1.94), or LSIL at enrollment (OR adjusted = 1.34, 95% CI, 1.07-1.69), although the size was less substantial in the latter group. There was no appreciable association of 2-year cumulative risk with baseline HPV16 DNA load among women with HSIL at enrollment.
Table 3.
Risk of 2-year prevalent CIN3 associated with per unit increase in log10-transformed baseline HPV16 DNA load: results stratified by baseline cytology
| Cervical cytology At enrollment | No. with/without CIN3 in the 2-year study period | Mean No. of visits between women with/without CIN3 | Mean (SD) of log10 HPV16 E7 copy number per nanogram of cellular DNA in women |
Adjusted OR a (95% CI) | |
|---|---|---|---|---|---|
| with CIN3 | without CIN3 | ||||
| Overall | 286/535 | 4.23/4.76 | 3.18 (±1.05) | 2.57 (±1.42) | 1.46 (1.29-1.64) |
| Within normal limits | 30/126 | 4.28/4.73 | 2.40 (±1.05) | 1.61 (±1.29) | 1.66 (1.16-2.37) |
| ASCUS | 86/154 | 4.05/4.77 | 3.08(±1.03) | 2.53 (±1.30) | 1.51 (1.17-1.94) |
| LSIL | 76/195 | 4.39/4.83 | 3.38 (±0.99) | 2.94 (±1.35) | 1.34 (1.07-1.69) |
| HSIL | 94/60 | 4.03/4.69 | 3.35 (±1.00) | 3.45 (±1.07) | 0.86 (0.61-1.20) |
NOTE
Odds ratio associating 2-year cumulative risk of CIN3 with per 1 log10 increase in viral load after adjusting for age at enrollment, current use of hormonal contraceptives, lifetime number of sex partners, and study arm
Approximately 23% of 821 women exited the trial prior to the scheduled last visit. The mean number of visits was 4.37, 4.31, 4.52, and 4.44 for women with baseline normal cytology, ASCUS, LSIL, and HSIL, respectively. The HPV16 DNA load was similar between women who completed the last visit and those who did not (2.73 versus 2.79 log10-tranfomed viral load, P = 0.54). Additional adjustment for the number of follow-up visits did not considerably change the overall risk estimate (OR adjusted = 1.47, 95% CI, 1.30-1.66) as well as cytology-stratified risk estimates (OR adjusted = 1.68, 95% CI, 1.17-2.43 for women with normal cytology, OR adjusted = 1.55, 95% CI, 1.19-2.00 for those with ASCUS, OR adjusted = 1.35, 95% CI, 1.07-1.71 for those with LSIL, and OR adjusted = 0.89, 95% CI, 0.62-1.27 for those HSIL). The risk estimates remained similar when the analysis was restricted to those who completed the scheduled last visit (data not shown).
Two hundred and thirty-two women were positive for HPV16 alone at enrollment. The mean (SD) of the log10-tranfomed viral load was 3.13 (±1.04) and 2.82 (±1.45) for women with and without CIN3, respectively, who were positive for HPV16 alone; 3.20 (±1.05) and 2.47 (±1.41) for women with and without CIN3, respectively, who were positive for multiple types of HPV. The increased risk of CIN3, although not statistically significant, remained associated with the elevated viral load in women positive for HPV16 alone (OR adjusted = 1.19, 95% CI, 0.96-1.47). The impact of cervical cytology on risk association was similar between women with and without co-infection with other HPV types, i.e., the association was evident in women with normal cytology or ASCUS (although not statistically significant in those positive for HPV16 alone) but not in women with HSIL (data not shown). Additional adjustment for co-infection with other types did not appreciably alter the risk estimates (data not shown).
Although the elevated HPV16 DNA load was significantly associated with risk of CIN3 for all except for those with HSIL, the clinical value of testing for viral load at a single point in time to discriminating women with and without CIN3 was minimal. As shown in figure 2, the area under the ROC curve for detecting 2-year prevalent CIN3 by testing for baseline HPV16 DNA load was 0.63 (95% CI, 0.59-0.67) for HPV16-positive women overall. Although the test performed slightly more accurately in women with a baseline diagnosis of normal cytology (the area under the ROC curve, 0.70; 95% CI, 0.61-0.78) relative to those with ASCUS (the area under the ROC curve, 0.62; 95% CI, 0.55-0.69) or LSIL (the area under the ROC curve, 0.60; 95% CI, 0.55-0.65), the difference was not statistically significant (p=0.25 and p=0.10, respectively). For women with HSIL at enrollment, the area under the curve was only 0.46 (95% CI, 0.38-0.54) i.e., no better than a random classification. The thresholds for a given sensitivity varied with baseline cervical cytology, e.g., corresponding to a sensitivity of 95%, the values of the log10-transforme viral loads were 0.65, 1.81, and 2.05 for women with normal cytology, ASCUS, and LSIL, respectively, which resulted in specificities of 34.70%, 22.73%, and 24.10%, accordingly.
Figure 2. Receiver operating characteristic curves of quantitative analysis of baseline HPV16 DNA load for detection of 2-year prevalent CIN3.
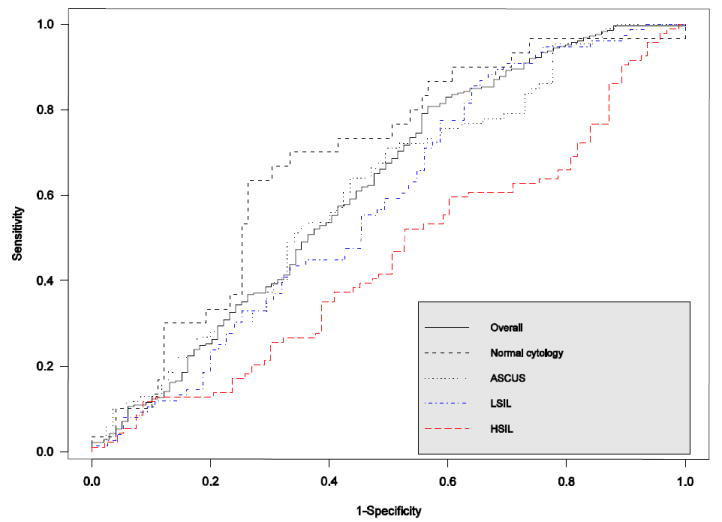
The area under the ROC curve was 0.63 (95% CI, 0.59-0.67) for the entire group of HPV16-positive women: 0.70 (95% CI, 0.61-0.78) for those with normal cytology at enrollment, 0.62 (95% CI, 0.55-0.69) for those with ASCUS, 0.60 (95% CI, 0.55-0.65) for those with LSIL, and 0.46 (95% CI, 0.38-0.54) for those with HSIL.
DISCUSSION
In this study of ALTS participants with HPV16 infection at enrollment, we found that the amount of baseline HPV16 DNA increased as a function of 2-year cumulative risk of CIN3. The viral load-related risk was not explained by factors previously shown to be related to risk for cervical neoplasia, including age, use of hormonal contraceptives, and lifetime number of sex partners or by different managements of the study arms. Because the measurement of HPV16 DNA load was performed without knowledge of any clinical or epidemiologic information and the diagnosis of cervical lesion was extensively reviewed by the panel of expert pathologists, potential bias in ascertainment of exposure and outcome was minimized. It is also unlikely that the risk association resulted from differential access to health care, because ALTS participants came from a screened population of women who were referred to the trial due to a mildly abnormal Pap smear; in this well-controlled trial setting, the majority of them were examined, followed, and diagnosed according to the standardized protocol.
The risk association observed in this large-scale longitudinal setting, perhaps the largest so far, agrees with previous reports that higher HPV DNA load was related to prevalence of [1, 7, 9-11] and progression to high-grade CIN [2-4, 12, 13]. This study further extends these reports by showing that HPV16 DNA load-associated risks of CIN3 varied with cervical cytology detected at the time when the viral load was measured, from a substantial risk increase for women with normal cytology to a lack of association for those with HSIL.
One explanation for the effect of cervical cytology on the viral load-related risk of CIN3 could be that the number of abnormal epithelial cells in cervical exfoliative samples increases as increasing severity of cervical cytology and that the increased number of these cells that reflect productive HPV infections makes the viral load contributed by CIN3 lesion be less apparent. As shown in a previous study of women with CIN3, HPV DNA load was highly correlated to the number of ASCUS and LSIL cells present in cervical samples [32]. Another possibility is that the effect of cervical cytology is mediated through a cytology-related lesion size. The size of cervical lesion is an important determinant of viral load detected in cervical samples [44]. Cervical lesion increases in size as increasing severities of cervical cytology; the increase is usually more substantial for the lesions surrounding the CIN3 [45]. These surrounding lesions were of importance in contribution to the viral load in cervical samples [32], because specimens obtained by scraping favor collection of cells shed from these extensive lesions even in the presence of CIN3. Consequently, the cytology-related change of risk association could be explained by differences in extent of the surrounding lesions. Further, because of aggressive follow-up, the majority of CIN3 lesions in ALTS were small. Thus, a fraction of HPV16 load contributed by cells shed from these lesions would be more likely to be masked by increasing severity of cytology.
In view of the risk association in women with baseline normal cytology, one would expect some benefits from testing for HPV16 DNA load to predicting risk of underlying CIN3. As indicated by the ROC analysis, however, the added value was too small to be clinically useful. Even among women with normal cytology, only about 70% of randomly drawn pairs of women with and without CIN3 would be accurately classified by the test. A rough guide for classifying the accuracy of a good diagnostic test requires an area of at least 0.85 in the traditional academic point system. Possible explanations for a lack of clinical usefulness of the test include wide variations in viral load within disease grades and a substantial overlap of viral load between women with and without CIN3 [4, 7, 32].
Nonetheless, the clinical implication of the present study is that for a given amount of HPV16 DNA, the interpretation for risk of underlying histopathology differs by cervical cytology. As a Pap smear with an adjunct of HPV testing is likely to continue to be the major approach for cervical cancer screening, physicians could one day apply this concept to their clinical management of women with HPV16 infections. It should be cautioned, however, that the results of HPV16 DNA load may not be generalizable to other types, because the associations between risk of CIN3 and viral load differ by HPV types [28, 46].
Several limitations of the study should be addressed. Because women in ALTS were those with referral cytology of ASCUS or LSIL, a diagnosis of normal cytology, ASCUS/LSIL, and HSIL at enrollment where the amount of HPV16 DNA was measured can also be interpreted as cytologic regression, persistent mild abnormality, and cytologic progression, respectively. Thus, the quantity of HPV16 DNA detected in a particular cytologic category may not be generalizable to that in the same category of general populations. Also, while the effects of cervical cytology on HPV16 DNA load-related risk of CIN3 might be somewhat attenuated by a previous presence of ASCUS or LSIL, an extent of misclassification of disease status that may bias the effects of cytology down towards was less substantial in this, as compared to general, population, because in ALTS, cervical cytology and histology were initially diagnosed by the clinical center pathologists and then meticulously reviewed by a panel of export pathologists. Secondly, although a rigorous tracking system was implemented in ALTS, approximately 23% of HPV16-positive women exited the trial prior to the scheduled last visit. Biases in assessment of the 2-year cumulative risk could have been introduced had a loss to follow-up been differentially related to the viral load. Arguing against this is the fact that the amount of baseline HPV16 DNA was highly comparable between women who completed the last visit and those who did not and additional adjustment for the number of follow-up visits did not change the risk estimates substantially. Finally, we recognize that women included in this study were those who were positive for HPV16 at enrollment without distinction between new and previously existing infections. The viral load may fluctuate during the course of natural infection. But, the comparison of groups is still valid [47].
In summary, higher HPV16 DNA load was associated with risk of CIN3 but the association varied with cervical cytology detected at the time when the viral load was measured, the finding that underscores the importance of cervical cytology in interpreting HPV16 DNA load-related risk of underlying histopathology. However, clinical utility of testing for HPV16 DNA load at a single point in time for detection of CIN3 was minimal even in women with normal cytology.
Acknowledgments
This study was part of the project ancillary to the ALTS clinical trial but does not represent the ALTS Group. The authors would like to thank the ALTS Group for providing the biological specimens and HPV typing results and Dr Kathrin Jansen for providing the HPV16 DNA standard.
Financial support: The work was supported by Public Health Service grant CA 84396 to L.F.X.
Footnotes
Potential conflicts of interest: L.A.K. receives research funds from Merck Research Laboratories. All other authors have no commercial or other associations that might pose a conflict of interest
Presented in part: 24th International Papillomavirus Conference and Clinical Workshop, Beijing, China, November 3-10, 2007 (abstract PS 26-1)
References
- 1.Swan DC, Tucker RA, Tortolero-Luna G, et al. Human papillomavirus (HPV) DNA copy number is dependent on grade of cervical disease and HPV type. J Clin Microbiol. 1999;37:1030–4. doi: 10.1128/jcm.37.4.1030-1034.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ylitalo N, Sorensen P, Josefsson AM, et al. Consistent high viral load of human papillomavirus 16 and risk of cervical carcinoma in situ: a nested case-control study. Lancet. 2000;355:2194–8. doi: 10.1016/S0140-6736(00)02402-8. [DOI] [PubMed] [Google Scholar]
- 3.Josefsson AM, Magnusson PK, Ylitalo N, et al. Viral load of human papilloma virus 16 as a determinant for development of cervical carcinoma in situ: a nested case-control study. Lancet. 2000;355:2189–93. doi: 10.1016/S0140-6736(00)02401-6. [DOI] [PubMed] [Google Scholar]
- 4.van Duin M, Snijders PJ, Schrijnemakers HF, et al. Human papillomavirus 16 load in normal and abnormal cervical scrapes: an indicator of CIN II/III and viral clearance. Int J Cancer. 2002;98:590–5. doi: 10.1002/ijc.10232. [DOI] [PubMed] [Google Scholar]
- 5.Moberg M, Gustavsson I, Gyllensten U. Type-specific associations of human papillomavirus load with risk of developing cervical carcinoma in situ. Int J Cancer. 2004;112:854–9. doi: 10.1002/ijc.20480. [DOI] [PubMed] [Google Scholar]
- 6.Monnier-Benoit S, Dalstein V, Riethmuller D, Lalaoui N, Mougin C, Pretet JL. Dynamics of HPV16 DNA load reflect the natural history of cervical HPV-associated lesions. J Clin Virol. 2006;35:270–7. doi: 10.1016/j.jcv.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 7.Fiander AN, Hart KW, Hibbitts SJ, et al. Variation in human papillomavirus type-16 viral load within different histological grades of cervical neoplasia. J Med Virol. 2007;79:1366–9. doi: 10.1002/jmv.20875. [DOI] [PubMed] [Google Scholar]
- 8.Cricca M, Morselli-Labate AM, Venturoli S, et al. Viral DNA load, physical status and E2/E6 ratio as markers to grade HPV16 positive women for high-grade cervical lesions. Gynecol Oncol. 2007 doi: 10.1016/j.ygyno.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 9.Bavin PJ, Giles JA, Deery A, et al. Use of semi-quantitative PCR for human papillomavirus DNA type 16 to identify women with high grade cervical disease in a population presenting with a mildly dyskaryotic smear report. Br J Cancer. 1993;67:602–5. doi: 10.1038/bjc.1993.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cuzick J, Terry G, Ho L, Hollingworth T, Anderson M. Type-specific human papillomavirus DNA in abnormal smears as a predictor of high-grade cervical intraepithelial neoplasia. Br J Cancer. 1994;69:167–71. doi: 10.1038/bjc.1994.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cuzick J, Terry G, Ho L, Hollingworth T, Anderson M. Human papillomavirus type 16 in cervical smears as predictor of high-grade cervical intraepithelial neoplasia [corrected] Lancet. 1992;339:959–60. doi: 10.1016/0140-6736(92)91532-d. [DOI] [PubMed] [Google Scholar]
- 12.Dalstein V, Riethmuller D, Pretet JL, et al. Persistence and load of high-risk HPV are predictors for development of high-grade cervical lesions: a longitudinal French cohort study. Int J Cancer. 2003;106:396–403. doi: 10.1002/ijc.11222. [DOI] [PubMed] [Google Scholar]
- 13.Moberg M, Gustavsson I, Wilander E, Gyllensten U. High viral loads of human papillomavirus predict risk of invasive cervical carcinoma. Br J Cancer. 2005;92:891–4. doi: 10.1038/sj.bjc.6602436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lai HC, Peng MY, Nieh S, et al. Differential viral loads of human papillomavirus 16 and 58 infections in the spectrum of cervical carcinogenesis. Int J Gynecol Cancer. 2006;16:730–5. doi: 10.1111/j.1525-1438.2006.00390.x. [DOI] [PubMed] [Google Scholar]
- 15.Santos AL, Derchain SF, Martins MR, Sarian LO, Martinez EZ, Syrjanen KJ. Human papillomavirus viral load in predicting high-grade CIN in women with cervical smears showing only atypical squamous cells or low-grade squamous intraepithelial lesion. Sao Paulo Med J. 2003;121:238–43. doi: 10.1590/S1516-31802003000600004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Snijders PJ, Hogewoning CJ, Hesselink AT, et al. Determination of viral load thresholds in cervical scrapings to rule out CIN 3 in HPV 16, 18, 31 and 33-positive women with normal cytology. Int J Cancer. 2006;119:1102–7. doi: 10.1002/ijc.21956. [DOI] [PubMed] [Google Scholar]
- 17.Lo KW, Yeung SW, Cheung TH, Siu NS, Kahn T, Wong YF. Quantitative analysis of human papillomavirus type 16 in cervical neoplasm: a study in Chinese population. J Clin Virol. 2005;34:76–80. doi: 10.1016/j.jcv.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 18.Ho CM, Chien TY, Huang SH, Lee BH, Chang SF. Integrated human papillomavirus types 52 and 58 are infrequently found in cervical cancer, and high viral loads predict risk of cervical cancer. Gynecol Oncol. 2006;102:54–60. doi: 10.1016/j.ygyno.2005.11.035. [DOI] [PubMed] [Google Scholar]
- 19.Lorincz AT, Castle PE, Sherman ME, et al. Viral load of human papillomavirus and risk of CIN3 or cervical cancer. Lancet. 2002;360:228–9. doi: 10.1016/S0140-6736(02)09463-1. [DOI] [PubMed] [Google Scholar]
- 20.Cheung JL, Lo KW, Cheung TH, Tang JW, Chan PK. Viral load, E2 gene disruption status, and lineage of human papillomavirus type 16 infection in cervical neoplasia. J Infect Dis. 2006;194:1706–12. doi: 10.1086/509622. [DOI] [PubMed] [Google Scholar]
- 21.Andersson S, Safari H, Mints M, Lewensohn-Fuchs I, Gyllensten U, Johansson B. Type distribution, viral load and integration status of high-risk human papillomaviruses in pre-stages of cervical cancer (CIN) Br J Cancer. 2005;92:2195–200. doi: 10.1038/sj.bjc.6602648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Onan MA, Taskiran C, Bozdayi G, et al. Assessment of human papilloma viral load of archival cervical intraepithelial neoplasia by real-time polymerase chain reaction in a Turkish population. Eur J Gynaecol Oncol. 2005;26:632–5. [PubMed] [Google Scholar]
- 23.Wensveen CW, Kagie MJ, Nagelkerke NJ, Veldhuizen RW, Trimbos JB. Can viral load, semi-quantitatively evaluated, of human papillomavirus predict cytological or histological outcome in women with atypical squamous or glandular cells of undetermined significance cytology? Eur J Gynaecol Oncol. 2005;26:393–7. [PubMed] [Google Scholar]
- 24.Castle PE, Schiffman M, Wheeler CM. Hybrid capture 2 viral load and the 2-year cumulative risk of cervical intraepithelial neoplasia grade 3 or cancer. Am J Obstet Gynecol. 2004;191:1590–7. doi: 10.1016/j.ajog.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 25.Wang-Johanning F, Lu DW, Wang Y, Johnson MR, Johanning GL. Quantitation of human papillomavirus 16 E6 and E7 DNA and RNA in residual material from ThinPrep Papanicolaou tests using real-time polymerase chain reaction analysis. Cancer. 2002;94:2199–210. doi: 10.1002/cncr.10439. [DOI] [PubMed] [Google Scholar]
- 26.Gravitt PE, Burk RD, Lorincz A, et al. A comparison between real-time polymerase chain reaction and hybrid capture 2 for human papillomavirus DNA quantitation. Cancer Epidemiol Biomarkers Prev. 2003;12:477–84. [PubMed] [Google Scholar]
- 27.Carcopino X, Henry M, Benmoura D, et al. Determination of HPV type 16 and 18 viral load in cervical smears of women referred to colposcopy. J Med Virol. 2006;78:1131–40. doi: 10.1002/jmv.20673. [DOI] [PubMed] [Google Scholar]
- 28.Kovacic MB, Castle PE, Herrero R, et al. Relationships of human papillomavirus type, qualitative viral load, and age with cytologic abnormality. Cancer Res. 2006;66:10112–9. doi: 10.1158/0008-5472.CAN-06-1812. [DOI] [PubMed] [Google Scholar]
- 29.Flores R, Papenfuss M, Klimecki WT, Giuliano AR. Cross-sectional analysis of oncogenic HPV viral load and cervical intraepithelial neoplasia. Int J Cancer. 2006;118:1187–93. doi: 10.1002/ijc.21477. [DOI] [PubMed] [Google Scholar]
- 30.Schlecht NF, Trevisan A, Duarte-Franco E, et al. Viral load as a predictor of the risk of cervical intraepithelial neoplasia. Int J Cancer. 2003;103:519–24. doi: 10.1002/ijc.10846. [DOI] [PubMed] [Google Scholar]
- 31.Ho CM, Cheng WF, Chu TY, et al. Human papillomaviral load changes in low-grade squamous intraepithelial lesions of the uterine cervix. Br J Cancer. 2006;95:1384–9. doi: 10.1038/sj.bjc.6603430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sherman ME, Wang SS, Wheeler CM, et al. Determinants of human papillomavirus load among women with histological cervical intraepithelial neoplasia 3: dominant impact of surrounding low-grade lesions. Cancer Epidemiol Biomarkers Prev. 2003;12:1038–44. [PubMed] [Google Scholar]
- 33.Castle PE, Schiffman M, Scott DR, et al. Semiquantitative human papillomavirus type 16 viral load and the prospective risk of cervical precancer and cancer. Cancer Epidemiol Biomarkers Prev. 2005;14:1311–4. doi: 10.1158/1055-9965.EPI-04-0799. [DOI] [PubMed] [Google Scholar]
- 34.Wright TC, Jr, Schiffman M. Adding a test for human papillomavirus DNA to cervical-cancer screening. N Engl J Med. 2003;348:489–90. doi: 10.1056/NEJMp020178. [DOI] [PubMed] [Google Scholar]
- 35.Wright TC, Jr, Schiffman M, Solomon D, et al. Interim guidance for the use of human papillomavirus DNA testing as an adjunct to cervical cytology for screening. Obstet Gynecol. 2004;103:304–9. doi: 10.1097/01.AOG.0000109426.82624.f8. [DOI] [PubMed] [Google Scholar]
- 36.Saslow D, Runowicz CD, Solomon D, et al. American Cancer Society guideline for the early detection of cervical neoplasia and cancer. CA Cancer J Clin. 2002;52:342–62. doi: 10.3322/canjclin.52.6.342. [DOI] [PubMed] [Google Scholar]
- 37.Cuzick J, Szarewski A, Cubie H, et al. Management of women who test positive for high-risk types of human papillomavirus: the HART study. Lancet. 2003;362:1871–6. doi: 10.1016/S0140-6736(03)14955-0. [DOI] [PubMed] [Google Scholar]
- 38.Schiffman M, Adrianza ME. ASCUS-LSIL Triage Study. Design, methods and characteristics of trial participants. Acta Cytol. 2000;44:726–42. doi: 10.1159/000328554. [DOI] [PubMed] [Google Scholar]
- 39.Results of a randomized trial on the management of cytology interpretations of atypical squamous cells of undetermined significance. Am J Obstet Gynecol. 2003;188:1383–92. doi: 10.1067/mob.2003.457. [DOI] [PubMed] [Google Scholar]
- 40.A randomized trial on the management of low-grade squamous intraepithelial lesion cytology interpretations. Am J Obstet Gynecol. 2003;188:1393–400. doi: 10.1067/mob.2003.462. [DOI] [PubMed] [Google Scholar]
- 41.Hosmer DW, Jr, Wang CY, Lin IC, Lemeshow S. A computer program for stepwise logistic regression using maximum likelihood estimation. Comput Programs Biomed. 1978;8:121–34. doi: 10.1016/0010-468x(78)90047-8. [DOI] [PubMed] [Google Scholar]
- 42.Metz CE, Herman BA, Shen JH. Maximum likelihood estimation of receiver operating characteristic (ROC) curves from continuously-distributed data. Stat Med. 1998;17:1033–53. doi: 10.1002/(sici)1097-0258(19980515)17:9<1033::aid-sim784>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 43.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–45. [PubMed] [Google Scholar]
- 44.Sun CA, Lai HC, Chang CC, Neih S, Yu CP, Chu TY. The significance of human papillomavirus viral load in prediction of histologic severity and size of squamous intraepithelial lesions of uterine cervix. Gynecol Oncol. 2001;83:95–9. doi: 10.1006/gyno.2001.6336. [DOI] [PubMed] [Google Scholar]
- 45.Sherman ME, Wang SS, Tarone R, Rich L, Schiffman M. Histopathologic extent of cervical intraepithelial neoplasia 3 lesions in the atypical squamous cells of undetermined significance low-grade squamous intraepithelial lesion triage study: implications for subject safety and lead-time bias. Cancer Epidemiol Biomarkers Prev. 2003;12:372–9. [PubMed] [Google Scholar]
- 46.Gravitt PE, Kovacic MB, Herrero R, et al. High load for most high risk human papillomavirus genotypes is associated with prevalent cervical cancer precursors but only HPV16 load predicts the development of incident disease. Int J Cancer. 2007;121:2787–93. doi: 10.1002/ijc.23012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brookmeyer R, Gail MH. Biases in prevalent cohorts. Biometrics. 1987;43:739–49. [PubMed] [Google Scholar]


