Abstract
Canonical forms of duplex DNA are known to sample well defined regions of the α, β, γ, ε and ζ dihedral angles that define the conformation of the phosphodiester linkage in the backbone of oligonucleotides. While extensive studies of base composition and base sequence dependent effects on the sampling of the A, BI and BII canonical forms of duplex DNA have been presented, our understanding of the intrinsic contribution of the five dihedral degrees of freedom associated with the phosphodiester linkage to the conformational properties of duplex DNA is still limited. To better understand this contribution ab initio quantum mechanical (QM) calculations were performed on a model compound representative of the phosphodiester backbone to systematically sample the energetics about the α β γ ε and ζ dihedral angles relevant to the conformational properties of duplex DNA. Low energy regions of dihedral potential energy surfaces are shown to correlate with the regions of dihedral space sampled in experimental crystal structures of the canonical forms of DNA, validating the utility of the model compound and emphasizing the contribution of the intrinsic mechanical properties of the phosphodiester backbone to the conformational properties of duplex DNA. Those contributions include the relative stability of the A, BI and BII conformations of duplex DNA, where the gas phase energetics favor the BI form over the A and BII forms. In addition, subtle features of the potential energy surfaces mimic changes in the probability distributions of α, β, γ, ε and ζ dihedral angles in A, BI and BII forms of DNA as well as with conformations sampled in single-stranded DNA. These results show that the intrinsic mechanical properties of the phosphodiester backbone make a significant contribution to conformational properties of duplex DNA observed in the condensed phase and allow for the prediction that single stranded DNA primarly samples folded conformations thereby possibly lowering the entropic barrier to the formation of duplex DNA.
Keywords: DNA, quantum mechanics, conformational energies, single-stranded DNA, crystal survey
INTRODUCTION
Structural characteristics of the canonical forms of duplex DNA and RNA have been investigated for well over 50 years.1 These investigations initially involved X-ray diffraction studies of fibres from which overall features of the repeating units of DNA were elucidated.2,3 Information that, when combined with various chemical analysis, lead to the elucidation of the double helical form of DNA, a discovery that laid the foundation for modern molecular biology.4 Subsequent X-ray crystallographic5 and NMR studies of DNA and RNA verified the earlier observations concerning the repeating features of the canonical forms of these molecules and, importantly, revealed subtle variations in the helical structures that have have been shown to be related to base sequence and composition effects.6,7 For example, A tracts of DNA are known to favor bent forms of B DNA8 while regions high in GC content are predisposed to the A form of DNA.9,10
Significant work has gone into understanding the underlying properties of DNA that contribute to the relative stabilities of the A, BI and BII forms of DNA. In conditions of high water activity it has long been known that the BI form is favored over the A and BII forms based initially on fibre diffraction experiments and subsequently verified via X-ray crystallography and various solution techniques, including NMR spectroscopy. To achieve the A form of duplex DNA it is necessary to significantly lower the water activity of the environment of the DNA, via high salt, cosolvent (e.g. ethanol or trifluoroethanol) 11 12 or low relative humidity as used in fibre diffaction experiments.13 Concerning the BII form, it occurs at a higher probability when DNA is bound to certain proteins.14,15 However, it should be emphasized that both A and BII conformations are sampled to varying extents in duplex DNA under high water activity conditions. That sampling has been shown to be dependent on base composition and sequence, where the A form is favored by GC base pairs and CpA.TpG steps have been indicated to favor the BII state, based on a survey of crystal structures15 as well as experimental16 and theoretical studies.17
While the base composition and sequence determinants that favor the A and BII states have and continue to be studied, a full understanding of the energetic determinants of the relative stabilities of the A, BI and BII states is still lacking, though progress towards this goal has been made. Recent effort has been put towards understanding the factors impacting the A versus BI equilibrium. These efforts involved the application of MD simulations to study both the equilibrium18 19 and the transition20–25 between the A and B forms. All these studies have confirmed the essential role of solvent, consistent with the experimentally known impact of water activity on the equilibrium.26 However, the exact role of solvent is still not clear; a dominant role is certainly related to the minimization of intraphosphate repulsion,27 while important roles of both minor groove and major groove waters have been indicated.25,28 In the case of the BII form restructuring of the hydration shell is important for facilitation of the BI to BII transition29,30 and water activity has been implicated to contribute to their equilibrium.31,32 Concerning contributions from the intrinsic mechanical properties of DNA a role of base stacking to the equilibrium between the A and B forms has been proposed and quantum mechanical (QM) calculations have shown that cytosine at the nucleoside level intrinsically favors in the experimental regimen χ values.33 With the BII form detailed information on the intrinsic conformational properties is lacking though force field calculations indicate the transition between the BI and BII forms to involve torsional strain34 and QM calculations on a sugar analog with a 3’ methylphosphate indicate the BI form to be intrinsically energetically lower than the BII form.35
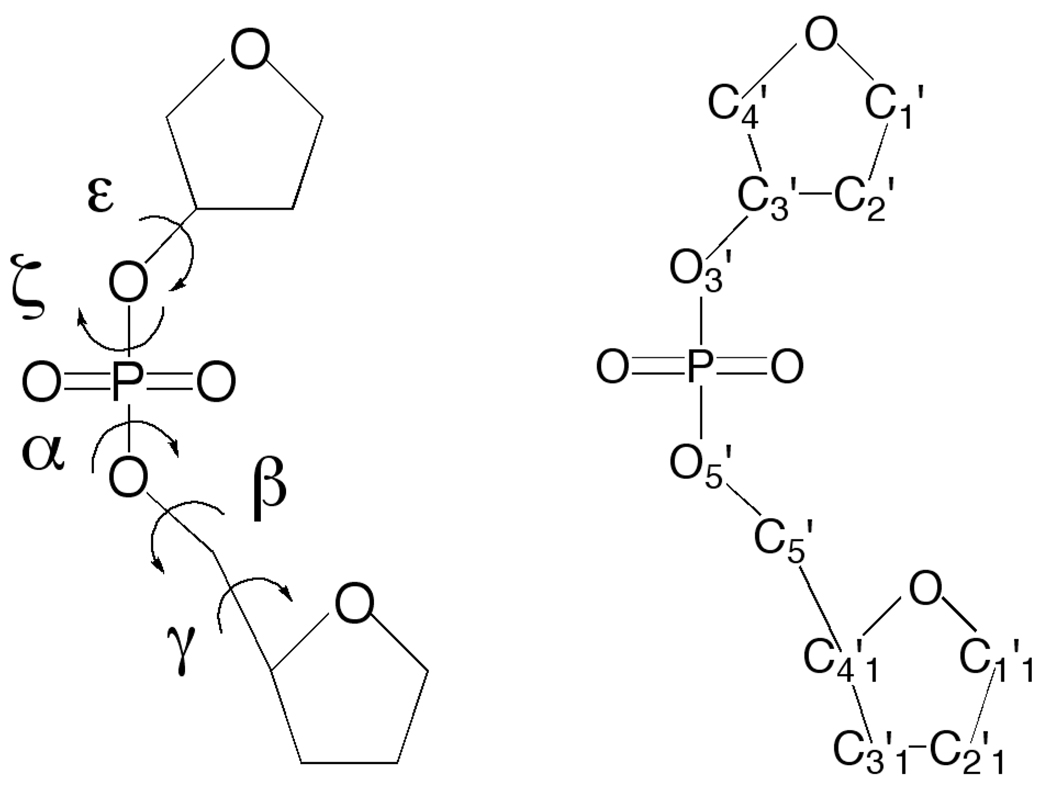
In the present study we extend previous QM studies of the energetics of DNA to investigate the intrinsic mechanical properties of the phosphodiester linkage to the conformational properties of the A, BI and BII forms of canonical DNA. Calculations were performed in the gas phase on a model compound (Figure 1) that contains a phosphodiester linkage capped by furanose rings. The compound includes all the essential features of the phosphodiester linkage allowing the regions of conformational space relevant to the A, BI and BII forms to be sampled explicitly, including the impact of sugar pucker. In particular, the compound was designed to omit i) a second phosphate moiety, thereby avoiding contributions from intrastrand phosphate repulsion, and ii) a nucleic acid base or base mimic, thereby avoiding contributions from base composition on the obtained energetics. In addition, all calculations are performed in the gas phase allowing the intrinsic mechanical properties of the linkage to be obtained without solvent. While QM methods do allow for solvation contributions to be taken into account via implicit solvent models, the hydration properties of DNA are complex36, making the results from continuum solvent model calculations questionable in the context of their impact an duplex DNA.
Figure 1.
Model compound T3PS used for calculation of the potential energy surface. Dihedrals sampled in the study are shown on the left panel and atom names are shown on the right panel.
METHODS
QM calculations were performed with Gaussian version 0337 and with QChem version 3.1.38 Geometry optimizations were performed to default tolerances, unless noted, using the 6–31+G(d) basis set 39 with electron correlation treated via 2nd order Møller-Plesset perturbation theory.40 During the initial minimizations constraints were applied to the α, β, γ, ε, and ζ dihedrals and a selected intra furanose ring dihedral (Table 1) to mimic the conformation of the A, BI or BII forms of DNA as described in the Results and Discussion. For all optimizations the initial geometries were generated using the program CHARMM41,42 with the all-atom nucleic acid force field.43,44. For the optimizations with the sugar pucker allowed to relax the initial structure was that optimized with the sugar pucker initially constrained via a single intra-ring torsion to enforce the C2’endo or C3’endo pucker followed by optimization in the absence of those constraints. This second optimization was performed using the tight optimization criteria in Gaussian. Single point energy calculations were performed using QChem with the double and triple zeta basis sets of Dunning and coworkers 45 using the resolution of identity MP2 method (RI-MP2).46,47 Estimates of relative energies at the complete basis set limit were performed using the extrapolation method of Helgaker 48,49 with the aug-cc-pVDZ and aug-cc-pVTZ energies obtained with the RI-MP2 treatment of electron correlation. Sugar pucker pseudorotation values were calculated following the method of Altona and Sundaralingam.50
Table 1.
Phosphodiester and sugar dihedrals defining the A and B forms of DNA as reported by Foloppeet al.33 and used as constraints in the present study.
| Dihedral Constraints | A DNA | BI DNA | BII DNA |
|---|---|---|---|
| α (O3’-P-O5’-C5’) | 291 | 298 | 298 |
| β (P-O5’-C5’-C4’) | 175 | 168 | 143 |
| γ (O5’-C5’-C4’-C3’) | 57 | 51 | 51 |
| ε (C4’-C3’-O3’-P) | 205 | 195.7 | 267.2 |
| ζ (C3’-O3’-P-O5’) | 287 | 270.3 | 159.2 |
| Sugar Constraints | |||
| C2’endo (south, B form) | C3’-C4’-O4’-C1’ = 0 | ||
| C3’endo (north, A form) | C4’-O4’-C1’-C2’ = 0 | ||
Dihedrals in degrees. Significant figures reported in the Table correspond to those applied in the calculations.
Surveys of the torsion angles in DNA were obtained from several sources. The A DNA and single-stranded DNA (ssDNA) data was obtained from the nucleic acid database (NDB, http://ndbserver.rutgers.edu).51 The search was performed using the NDB search engine selecting for X-ray crystallographic structures of A and ssDNA; in the case of A form DNA a 2.5 Å resolution cutoff was applied, yielding a total of 86 structures from which 1263 nucleotides were extracted. Following removal of undefined dihedrals 1108, 1115, 1263, 1104 and 1104 α, β, γ, ε and ζ dihedrals, respectively, were obtained. For the ssDNA search no cutoff based on resolution was applied to maximize the number of hits. A total of 135 structures were identified which included a total of 1428 nucleotides. Removal of undefined dihedrals yields 1213, 1298, 1402, 1216 and 1215 α, β, γ, ε and ζ dihedrals, respectively. Data on the BI and BII forms of DNA were obtained from the published study of Djuranovic and Hartmann.15 In addition to the above, survey results from the recent study by Svozil et al 10 were obtained to check that the selection criteria did not significantly bias the obtained distributions (Figure S1, supporting information). Those results, which more rigorously defined the A, BI and BII classes of DNA are similar to those from the present survey and from the Djuranovic and Hartmann study. All probability distributions were generated based on 5° bin widths.
RESULTS AND DISCUSSION
Model compound calculations have a long history with respect to improving our understanding of the conformational properties of biological macromolecules. The earliest studies relied on hard sphere models of the alanine dipeptide showing the low energies regions, as defined by the dihedral angles ϕ and ψ, are indicative of the range of the peptide backbone ϕ, and ψ, dihedrals sampled in proteins, thereby offering a better atomic-level understanding of the conformational space sampled by the protein backbone.52,53 Subsequently, numerous QM calculations were performed with this and related models from which more quantitative evaluations of the impact of the local energetics of the peptide backbone on conformational properties of proteins were obtained.54–56 With nucleic acids early work was also initiated using hard-sphere models 57 58 59 as well as force field 60 61 62,63 and semiempirical methods64 65–67. These studies yielded a range of insights into the conformational properties of nucleic acids, including oligonucleotides. However, the results from studies using such theoretical methods may be considered to be qualitative in nature. More recently, ab initio QM calculations have been undertaken on nucleic acid constitutents. A large number of these calculations have focused on the bases, including base pairing and stacking interactions; the later may be considered exceedingly challenging given the extensive overlap of orbitals of the bases such that this still represents an active area of research.68–70 Concerning the phosphodiester backbone, initial ab initio QM calculations involved dimethylphosphate (DMP). These have been followed by calculations on larger model compounds that include moieties representative of both the sugar and the phosphate as well as on nucleosides. These studies typically include relatively large basis sets (e.g. 6–31+G(d) or larger) and the explicit treatment of electron correlation with either MP2 or density functional methods. Notably these studies have shown that the low energy regions of dihedral energy surfaces for the model compounds correspond to regions sampled with high probabilities in surveys of the crystallographic structures.35,71 72 73 74 33,75–77 78 Analysis of these results yielded insights into the Analysis conformational properties of oligonucleotides as well as serving as part of the target data for the optimization of the CHARMM27 force field for nucleic acids.43,44 More recently, this approach was extended to the AMBER force field where extensive QM calculations on a model compound designed to treat both α and γ were used, in combination with MD simulations, to correct a deficiency in the original Cornell force field.79
Advances in computational power as well as in theoretical methods allow for higher-level QM calculations on larger model compounds that include more features of the macromolecule of interest. For example, in oligonucleotides a number of correlations exist between the rotatable degrees of freedom in the molecule. Examples include the relationship between ε and ζ and between χ and the sugar pseudorotation angle, among others (see Saenger for a detailed discussion of these correlations).1,10 To investigate such relationships using QM calculations it is necessary to have model compounds that include both of the dihedrals being studied. Examples of these include models to study the ε/ζ, 35 and α/γ relationships mentioned above.79 However the models used in those studies only contained a single sugar moiety, which may be limiting in understanding the full range of intrinsic mechanical properties in the phosphodiester backbone that influence the conformational properties of duplex DNA. To overcome that limitation we designed the model compound shown in Figure 1; the compound will be referred to as T3PS (tetrahydrofuran with 3’phosphate with a capping sugar, Figure 1). As stated above, T3PS was designed to allow all the features of a single phosphodiester moiety along with the capping sugars to be treated explicitly thereby allowing the contribution of the intrinsic mechanical properties of the phosphodiester linkage to the conformational properties of DNA duplexes to be investigated. Recently, work on a similar model, that also contained a phosphodiester moiety and two sugar moieties designed to investigate the α/γ conformational properties was published.80 That compound, referred to as SPSOM, in addition to the phosphodiester and capping sugars also contained 3’methoxy and 4’methoxymethyl (i.e. -CH2-O-CH3) moieties. Those functionalities were omitted in the present work to minimize the size of the model as well as avoid additional confounding effects from the 3’ and 4’ moieties that would limit the interpretation of the obtained energies to contributions solely from the sugars and phosphodiester linkage, a limitation noted by the authors of that study. However, while some differences exist in the two compounds, the values of the α and γ dihedrals at the minima reported in Table 2 of the SPSOM study correspond with the minima in the corresponding α and γ dihedral energy surfaces for T3PS (see Figure 2 and Figure 3), as expected given the similarity of the two model compounds.
Table 2.
Relative energies of the minima and selected high energy conformations as a function of the level of theory including extrapolation to the complete basis set.
| Alpha | RIMP2/cc-pVTZ | RIMP2/cc-pVTZ | RIMP2/aug-cc-pVTZ | CBS |
|---|---|---|---|---|
| A, minimum | 2.14 | 2.22 | 2.07 | 2.01 |
| A, highE | 5.12 | 4.90 | 4.89 | 4.89 |
| BI, minimum | 0.00 | 0.00 | 0.00 | 0.00 |
| BI, highE | 2.87 | 2.56 | 2.66 | 2.70 |
| BII, minimum | 2.57 | 2.98 | 2.77 | 2.69 |
| BII, highE | 5.14 | 6.05 | 5.29 | 4.96 |
| Beta | ||||
| A, minimum | 2.18 | 1.65 | 1.63 | 1.62 |
| A, highE | 8.03 | 11.39 | 11.33 | 11.30 |
| BI, minimum | 0.00 | 0.00 | 0.00 | 0.00 |
| BI, highE | 6.90 | 5.69 | 5.85 | 5.92 |
| BII, minimum | 2.03 | 2.45 | 2.11 | 1.96 |
| BII, highE | 3.47 | 3.28 | 3.42 | 3.48 |
| Gamma | ||||
| A, minimum | 2.15 | 2.26 | 2.07 | 1.99 |
| A, highE | 7.03 | 6.47 | 6.12 | 5.97 |
| BI, minimum | 0.00 | 0.00 | 0.00 | 0.00 |
| BI, highE | 5.94 | 5.44 | 4.99 | 4.79 |
| BII, minimum | 2.11 | 2.48 | 2.33 | 2.27 |
| BII, highE | 7.56 | 7.75 | 6.92 | 6.57 |
| Epsilon | ||||
| A, minimum | 1.78 | 2.20 | 1.90 | 1.77 |
| A, highE | 2.16 | 2.56 | 2.21 | 2.07 |
| BI, minimum | 0.00 | 0.00 | 0.00 | 0.00 |
| BI, highE | 1.16 | 1.67 | 1.38 | 1.25 |
| BII, minimum | 1.91 | 2.56 | 2.10 | 1.91 |
| BII, highE | 2.54 | 3.03 | 2.75 | 2.63 |
| Zeta | ||||
| A, minimum | 2.00 | 2.23 | 1.94 | 1.82 |
| A, highE | 4.73 | 5.82 | 4.81 | 4.39 |
| BI, minimum | 0.00 | 0.00 | 0.00 | 0.00 |
| BI, highE | 1.79 | 2.90 | 2.19 | 1.89 |
| BII, minimum | 1.02 | 0.93 | 0.82 | 0.77 |
| BII, highE | 2.85 | 3.22 | 2.95 | 2.83 |
All geometries at the MP2/6–31+G(d) level. Selected high energy conformations (highE) were 150, 135, 180, 270, and 150° for α, β, γ, ε, and ζ, respectively
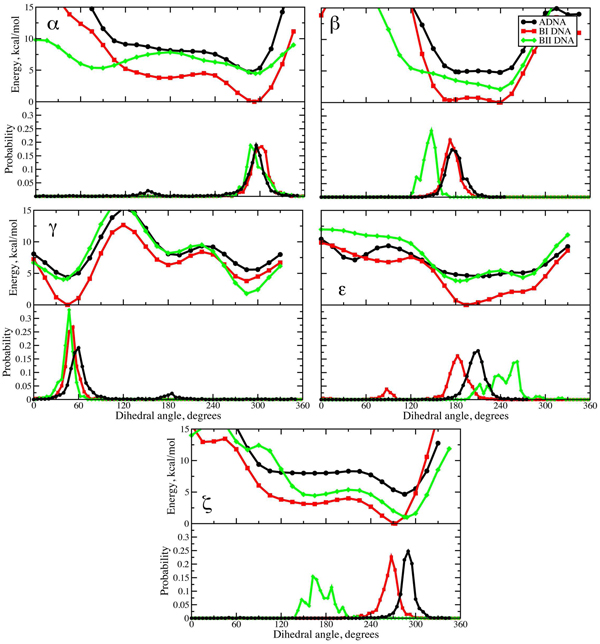
Figure 2.
Potential energy surfaces (upper panels) and crystallographic probability distributions for the α, β, γ, ε and ζ dihedrals. Energy surfaces obtained at the MP2/6–31+G(d) level with the sugar puckers fixed as described in the text and the surfaces in each panel are offset to the global minimum energy conformation in that panel (BI minimum in all cases). Probability distributions from a survey of the NDB for A form DNA and from Djuranovic and Hartmann 93 for the BI and BII forms of DNA.
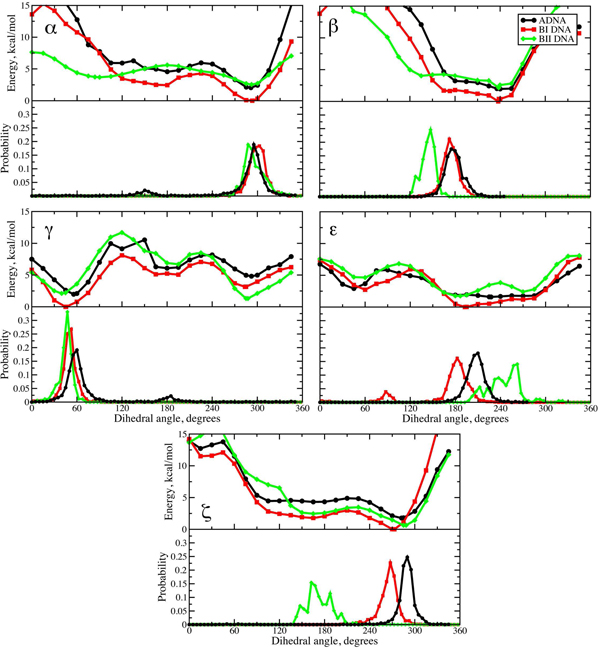
Figure 3.
Potential energy surfaces (upper panels) and crystallographic probability distributions for the α, β, γ, ε and ζ dihedrals. Energy surfaces obtained at the MP2/6-31+G(d)//MP2/cc-pVTZ level with Energy surfaces obtained at the MP2/6–31+G(d)//MP2/cc-pVTZ level with the sugar allowed to relax as described in the text and the surfaces in each panel are offset to the global minimum energy conformation in that panel (BI minimum in all cases). Probability distributions from a survey of the NDB for A form DNA and from Djuranovic and Hartmann 93 for the BI and BII forms of DNA.
Given the number of rotatable bonds in T3PS plus the two sugars the total number of conformations accessible to the molecule is large. Assuming a 15° grid and two states for each sugar the total number of conformations is approximately (2 × 2 × 25)5 or 106. While calculations on 106 conformers is accessible to force field methods this is still an intractable number when considering QM calculations at a sufficient level of theory. Accordingly, QM calculations focused on the regions of dihedral space relevant to the A, BI and BII forms of duplex DNA. This was performed by selecting values of the dihedrals associated with the three forms of DNA obtained from published data on surveys of the NDB,33 supplemented with a value of 143° for β in the BII form (Table 1).80 In addition, the C2’endo and C3’endo conformations of the furanose rings were restrained in the initial QM calculations using individual intra-ring dihedrals as listed in Table 1. Thus, in the dihedral energy scans to sample the potential energy surface of a selected dihedral, for example, α, for a given form of DNA the remaining 4 “non-target” phosphodiester dihedrals and the intra-ring furanose dihedrals were constrained to the values corresponding to the canonical values listed in Table 1. In the presence of those constraints the target dihedral, α, was then sampled from 0 to 345° or 360° in 15° increments with all remaining degrees of freedom in the molecule optimized at each increment. The resulting optimized structures were then subjected to additional minimization with the furanose rings allowed to fully relax while the four non-target dihedral constraints were retained. This approach allows for the regions of conformational space relevant to the canonical forms of duplex DNA to be sampled explicitly using the same model compound. Notably, use of the same model compound allows for the energies of the systems to be offset with respect to the “global” minimum for all the conformations investigated, thereby allowing for the intrinsic, mechanical conformational energy contributions to the A versus BI versus BII forms of DNA to be determined, as presented below. Finally, for all the minimum energy conformations for each dihedral and each form of DNA, full optimizations were performed allowing the impact of the constraints to be evaluated.
Initial QM calculations were undertaken at the MP2/6–31+G(d) level on T3PS to determine if the compounds energetic properties correspond to regions of dihedral space sampled in crystallographic studies of DNA. Presented in Figure 2 are the potential energy surfaces for α through ζ for the A, BI and BII canonical forms of DNA with the non-target dihedrals and sugars constrained to the values listed in Table 1. For each set of surfaces (i.e. all α surfaces) the energies are offset to the global energy minimum for that target dihedral, which is the BI minimum in all cases. The figure also contains distributions of the dihedrals from a NDB survey for A, BI and BII forms of DNA. Inspection of the plots in Figure 2 shows the correlation between the low energy regions of the QM potential energy surfaces with the high probability regions of the NDB survey data to be high. This is true for all five dihedrals. For most of the dihedrals, including α, β, γ andε, there is a very strong correspondence between the low energy regions of the potential energy surfaces and the NDB data, though some interesting differences do exist. The low energy region in the vicinity of 240° in β surface is not sampled in the survey data. This appears to be due to unfavorable interactions with the base of the preceding nucleotide in DNA being in the anti orientation as this region of β is sampled in Z-form DNA 35 and the relationship of a syn orientation of the base of the preceding nucleotide with β in the vicinity of 220–240° is evident in high resolution Z-form crystal structures.81,82 With, γ, the g- minima in the vicinity of 300° is not sampled in the survey; this appears to be due to constraints associated with the duplex structure as some sampling of this region of γ occurs in nucleosides and nucleotides83 as well as in single-stranded DNA (see below). In the case of, ζ, the correspondence of the A and BI form surfaces with the survey data is high, while the BII form samples a relatively high energy region of the QM surface in the crystal structures (ζ ~ 180°), indicating the influence of more remote regions of DNA in the sampling of ζ. While some differences are anticipated, the overall level of correspondence between the energy surfaces and the NDB survey results in Figure 2 emphasize the utility of T3PS for the study of the intrinsic conformational properties of DNA.
Another notable feature in Figure 2 are the relative energies of the A vs. BI vs. BII surfaces. In all cases the BI form represents the lowest energy state with the A and BII states being similar, with the exception of the β and ζ surfaces, where BII is lower than A. The relative ordering of the surfaces is consistent with the BI form being that most frequently observed in experimental studies, indicating that the intrinsic energetics of the phosphodiester backbone contribute to the relative stabilities of these three forms of DNA. This discussion is expanded below.
Concerning sugar puckering, the sugars generally stay in either the C2’endo or C3’endo states, consistent with the constraints used in the energy surfaces (Figure S2A and S2B, supporting information). The only exception occurs with sugar 2 in the β surface for the BI form (Figure S2B). In the range from 330 up through 75° the sugar switched to the C2’exo state, in which the C3’-C4’-O4’-C1’ dihedral is also 0°, consistent with the applied constraint. This switch in the sugar pucker is due to severe steric overlap of the two sugars in this region of the surface that leads to the high relative energies. Indeed, this steric clash was so severe at β values of 0 and 15° in several instances the QM optimization did not converge (not shown). However, since this region is of such high energy and is forbidden, as evidenced by the lack of sampling of this region in the NDB survey data, the switch in the sugar pucker is not relevant to interpretation of the results.
While the results in Figure 2 are generally in agreement with previously reported QM results comparing model compound with survey data, there are differences. For example, our previous study of β using tetrahydrofuran (THF) with a 5’ phosphate showed the low energy structures to occur at 240°,35 as in the present study, but there was a steady rise in energy upon going to lower values where sampling in the survey occurs. However, with T3PS the energies of both the A and BI forms stay low in this region. Concerning α and ζ the model compound DMP has been used to show the intrinsic energetic favorability of the highly sampled region at 300° (i.e. the g- state).84–88 consistent with present results. In addition, it is known that the g,g conformer is the global minimum of DMP while the g,t conformer is of higher energy consistent with the higher energy of the BII form with T3PS whose restraints (Table 2) approximate the g,t conformation. However, as the restraints defining both A and BI forms (Table 2) correspond approximately to the g,g conformation, calculations were undertaken to determine if DMP yields the relative energies of the A and BI forms. Optimization of the A and BI forms was performed by constraining the ζ dihedral in DMP to the value presented in Table 1 with the remainder of the molecule allowed to relax. Interestingly this yielded a relative energy of the A form being 0.7 kcal/mol more favorable than that of the BI form in contradiction to the results obtained with T3PS. This is due to the A form α dihedral being closer to the true g minimum value as compared to the BI form. Energy surfaces of α show this trend to be maintained through the entire region of dihedrals that are sampled in the crystal survey (i.e. 120 through 315°, not shown). Such a difference emphasizes the importance of including the full representation of the phosphodiester linkage as well as the capping furanoses in T3PS to allow for conformational energies relevant to the A, BI and BII canonical forms of DNA to be obtained.
While the results in Figure 2 indicate an important role for the intrinsic energetics of the phosphodiester backbone in contributing to the conformational properties of DNA additional calculations were undertaken to insure that the presence of constraints on the sugar puckers did not significantly impact the energy surfaces and to test the impact of basis set on the obtained results. These computations included allowing the sugars to fully relax while constraining the remaining dihedrals and then performing single point RIMP2/cc-pVTZ calculations on all the MP2/6–31+G(d) optimized structures (Figure 3). Presented in Figure 3 are the resulting MP2/6–31+G(d)//RIMP2/cc-pVTZ energy surfaces for the five targeted dihedrals for T3PS in the A, BI and BII forms of DNA. The plots include the conformers where the target dihedral was fully optimized and the energies are again all offset to the lowest energy structure for each target dihedral, corresponding to the BI form in all cases. As with the results in Figure 2, the low energy regions of the plots generally correspond to regions that are highly populated in the survey data and the result showing the BI conformation to systematically be of lower energy than either the A or BII conformations is maintained.
In general, allowing the sugar puckers to optimize yielded values of the pseudorotation angles that remained in the target north or south minima for the A or BI/BII forms, respectively, for the low energy regions of the energy surfaces (Figure S3A and S3B of the supporting information). However, in many cases the sugar pseudorotation values did deviate significantly from the idealized C3’endo (north) or C2’endo (south) conformations. For example, with the second sugar in the higher energy regions of the α and ζ surfaces (60 to 180° and beyond, Figure S3B) there is a tendency to sample pseudorotation values in the vicinity of 240°, values that are typically forbidden in duplex DNA structures. However, due to the lack of the base moieties in the model compound, the steric clash that normally occurs with the 5’ sugar substituent and leads to this region being forbidden cannot occur such that conformations in the vicinity of 240° will be allowed in T3PS. Similarly, in the case of β there are significant deviations on the sugar 2 (Figure S3B) pseudoroation values in the high energy regions (above 270° and below 90°). This is associated with the severe steric clash of that sugar with the remainder of the model compound, leading to the high potential energies away from the minimum energy region, as discussed above. As the north and south sugar puckers dominate in the regions of the energy surfaces observed in crystallographic structures, T3PS is yielding conformational properties consistent with those observed in crystallographic studies. Concerning the relative energies of the surfaces for the A, BI and BII conformations, comparison of Figure 2 and Figure 3 show that restriction of the puckers to either the C3’endo or C2’endo forms does contribute to the higher energies of the A and BII conformations, though they are still of higher energies than the BI form when the sugars are allowed to relax.
To further validate the energies presented in Figure 3, relative energies extrapolated to the CBS were obtained for the minima from each surface and for selected high energy conformations (Table 2). The same procedure used by Svozil et al. on the SPSOM model compound80 was applied. In addition, Svozil et al. applied a correction for the treatment of electron correlation to the CCSD(T) level. That correction only changed the relative energies by a maximum of 0.1 kcal/mol. Accordingly, such a correction was not included in the present study. Comparison of the RIMP2/cc-pVTZ energies extracted from Figure 3 and CBS energies in Table 2 show the agreement to generally be good, with the differences typically 0.3 kcal/mol or lower for the minimum energy conformations. With the selected high energy conformations the differences are larger. The largest difference of 3.3 kcal/mol occurs with β for high energy A form with several others high energy conformations differing by approximately 1 kcal/mol (i.e.β, BI;γ, A, BI and BII); however, these differences do not change the relative ordering amongst the different forms of DNA. Thus, the RIMP2/cc-pVTZ relative energies may be considered adequately converged and will act as the basis for further discussions of the data.
A final check of the use of restraints in the optimizations was performed by taking the minimum energy structures from Figure 3 and subjecting them to full optimizations in the absence of any restraints at the MP2/6–31+G(d) level followed by single point calculations at the RIMP2/cc-pVTZ level. Presented in Table 3 are the relative energies corresponding to the minima in Figure 3 with constraints on the non-target dihedrals (Cons in Table 3), the relative energies of the fully optimized conformations (Full Opt in Table 3) and their dihedral angles and pseudorotation values. In general, full optimization led to a lowering of the relative energies of the A and BII states. The largest decrease occurs with ζ while an exception to this trend is observed with the β and ε BII minimum energy structures, where the fully optimized relative energies are higher. With ζ the decrease in relative energies are associated with the β dihedral in the A form shifting to its global minimum (Figure 3) while with the BII form the ζ dihedr al shifts to its minimum conformation from the value of 159° observed in the crystal survey data (Table 1). In addition, with γ the BII conformer relaxes to the BI conformation. These results further validate the relative energies of the minima obtained in the presence of the non-target dihedral constraints as these conformations are also minima in the absence of constraints, with the only exception being the γ BII conformation.
Table 3.
Relative energies, dihedral angles and sugar pseudorotation values of the fully optimized minimum energy structures from each of the dihedral energy surfaces.
| RIMP2/cc-pVTZ | alpha | beta | gamma | epsilon | zeta | Puck1 | Puck2 | ||
|---|---|---|---|---|---|---|---|---|---|
| Cons | Full Opt | ||||||||
| Alpha | |||||||||
| A | 2.14 | 1.61 | 287.1 | 184.8 | 53.7 | 221.9 | 297.0 | 357.9 | 308.6 |
| BI | 0.00 | 0.00 | 288.5 | 181.6 | 46.6 | 201.1 | 281.6 | 149.4 | 167.8 |
| BII | 2.57 | 2.16 | 293.6 | 146.0 | 39.6 | 271.8 | 522.2 | 157.6 | 245.9 |
| Beta | |||||||||
| A | 2.18 | 1.98 | 288.8 | 236.4 | 56.6 | 191.0 | 282.5 | 355.8 | 303.2 |
| BI | 0.00 | 0.00 | 285.3 | 241.5 | 50.0 | 192.7 | 278.3 | 154.5 | 173.7 |
| BII | 2.03 | 2.26 | 289.5 | 242.5 | 49.1 | 267.1 | 167.5 | 157.1 | 171.9 |
| Gamma | |||||||||
| A | 2.15 | 1.61 | 287.1 | 184.8 | 53.7 | 221.9 | 297.0 | 357.9 | 308.6 |
| BI | 0.00 | 0.00 | 288.5 | 181.6 | 46.6 | 201.1 | 281.6 | 149.4 | 167.8 |
| BII | 2.11 | 0.00 | 288.5 | 181.6 | 46.6 | 201.1 | 281.6 | 149.4 | 167.8 |
| Epsilon | |||||||||
| A | 1.78 | 1.61 | 287.1 | 184.8 | 53.7 | 221.9 | 297.0 | 357.9 | 308.7 |
| BI | 0.00 | 0.00 | 288.5 | 181.6 | 46.6 | 201.1 | 281.6 | 149.4 | 167.8 |
| BII | 1.91 | 2.16 | 293.6 | 146.0 | 39.6 | 271.8 | 162.2 | 157.6 | 245.9 |
| Zeta | |||||||||
| A | 2.00 | 0.68 | 288.8 | 236.4 | 56.6 | 191.0 | 282.5 | 355.7 | 303.2 |
| BI | 0.00 | 0.00 | 288.5 | 181.6 | 46.6 | 201.1 | 281.6 | 149.4 | 167.8 |
| BII | 1.02 | 0.70 | 293.6 | 157.0 | 44.6 | 266.4 | 294.2 | 151.1 | 166.6 |
Energies in kcal/mol and dihedral angles in degrees. Conformations are those optimized at the MP2/6-31+G(d) level in the presence of the four non-target dihedral constraints (Cons)and in the absence of any constraints (Full Opt). Single point RIMP2/cc-pVTZ relative energies are presented. Values in bold indicate dihedrals in which significant changes occurred from the constrained values associated with the respective forms of DNA studied upon full optimization.
Several aspects of the surfaces in Figure 3 are notable. These include the relative energies of the different forms of DNA mentioned above, subtle features of the potential energy surfaces that are consistent with the survey data, the more significant difference observed between the crystallographic probability distributions and the energy profiles for the BII form and the potential impact of the observed intrinsic properties on the structure of single stranded DNA. The remainder of the manuscript will address these issues.
For all 5 dihedrals the BI surfaces is systematically lower than both the A and BII forms. This effect was previously noted for the A versus BI forms of DNA using smaller model compounds35 where the sugar pucker was maintained in the C3’endo or C2’endo forms using the single intra-ring dihedral listed in Table 1. However, the present results go beyond those observations by extending them to the BII form as well as with respect to T3PS including the full phosphodiester backbone. The inherent stability of the BI form of DNA over the A and BII forms has long been known based on various experimental methods.1,12,13,25 The present results confirm that the intrinsic mechanical properties of the phosphodiester backbone contribute to the favored sampling of the BI form. From Table 2 it may be seen that the BI minima are at least 1.6 kcal/mol below the A form and at least 0.8 kcal/mol below the BII form. Moreover, the survey data show the BII form of duplex DNA to sample higher energy regions of theβ, ε and ζ surfaces (Figure 3) rather than sampling only the minimum energies regions. By selecting representative high energy conformations from these sampling regions (e.g. the 150° conformations for ζ and the 270° conformation forε) the present results suggest that approximately kcal/mol is required per nucleotide to overcome the intrinsic energetics of the phosphodiester backbone to achieve the BII conformation; a value of approximately 1.6 kcal/mol would be required to assume the A form conformations. While these energy requirements are not extremely large, being 3–4 kT at room temperature, T, where k is the Boltzmann constant, they are consistent with the need for additional energetic input from other regions of the DNA or from the environment to overcome these higher relative energies. However, it should be noted that other A, BI or BII conformations could exist in each local energy basin such that the true intrinsic energy differences may vary from the aforementioned values.
As discussed in a number of published studies, energy contributions to overcome the intrinsic mechanical energy contribution may come from base stacking, base-phosphodiester backbone interactions, solvent effects or protein-DNA interactions or a combination thereof. For example, QM calculations of nucleosides show the A versus B forms to be isoenergetic with adenine, guanine and thymine, while cytosine favors the A form by approximately 2 kcal/mol,71 an amount of energy that would overcome the intrinsic phosphodiester backbone energy of A versus BI DNA discussed above, contributing to the intrinsic ability of cytosine bases to favor A form DNA. Solvent contributions, including ionic strength effects, are an obvious means to overcome the intrinsic energy differences, consistent with studies using the Poisson Boltzmann/Molecular Mechanical (PB/MM) or Generalized Born/Molecular Mechanical (GB/MM) methods to study the A/B form equilibrium.18,19 Possible sources for such contributions are direct interactions of the solvent with backbone or indirectly via differential solvation in the minor or major grooves. An interesting consideration is that while the present model compound does not include intra-strand phosphate-phosphate repulsion, which further disfavors the A form with respect to the B form due to the intrastrand phosphate distance being significantly shorter in the A form89, it still predicts that the A, as well as BII, forms are still intrinsically unfavorable as compared to the BI form. Thus, the presented results represent an additional step towards better understanding the contribution of different moieties of oligonucleotides to their overall conformational properties.
An interesting aspect of the present results are that features of the energy surfaces are consistent with subtle aspects of the crystal survey distributions (Figure 3). Both α and ζ sample in the region of 300°, consistent with the present energy surfaces as well as with previous reported results based on DMP.84–88 However, subtle differences in the survey probability distributions are evident. In the case of α the flatter energy well with the BII form coincides with the widest range of sampling among the three forms while with ζ the location of the local minima differ for A versus BI which is mirrored in the survey distributions. Withβ, sampling in the BII form at values < 150° is consistent with the potential energy of this form not rising as rapidly as occurs with the A and BI forms. Subtle changes in the location of the γ maxima observed in the survey distributions where BII < BI < A are mimicked by the location of the local g+ minima for the three forms of DNA. With ε the location of the maxima in the survey results for A versus BI are consistent with the location of the minima in the energy surfaces, the somewhat broader distribution in A coincides with the flatter energy surface for that form and the significant shift in the location of the maxima in the distribution of for BII coincides with the local minimum at 270° in the corresponding energy surface. While such correlations between the energy surfaces and the survey distributions may simply be coincidence, the number of similarities suggests that the intrinsic mechanical properties of the phosphodiester backbone are contributing to subtle aspects of the sampling of conformational space by DNA in duplexes.
The relationship of the sampling of the dihedrals β, ε and ζ in the BII form in the crystal survey data deserves additional discussion. With β and ε sampling in the survey does not coincide with the global minima for the BII energy surfaces, but it does occur in allowed regions of the energy surfaces, further emphasizing the contribution of the phosphodiester backbone in dictating the sampling of backbone conformations in the experimental regimen. With ζ higher energy regions of the BII surface are being sampled. The sampling of these higher energy regions as well as the BII conformation being intrinsically unfavorable with respect to the BI form emphasizes the importance of specific environmental contributions to the stabilization of this form. These may include specific reorganization of the hydration layer,29,30 water activity effects31,32 or specific protein-DNA interactions.14 The present results will facilitate understanding how the energy contributions from those phenomena contribute to sampling of the BII conformation.
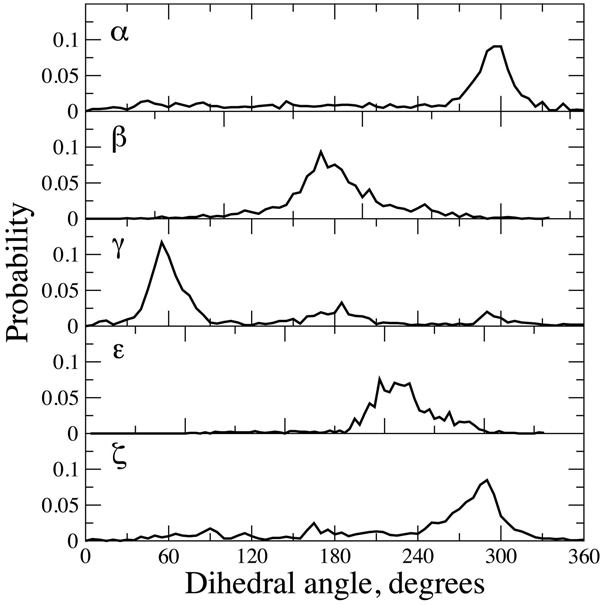
Correlation between the QM energy surfaces and the sampled regions of the backbone dihedrals in crystal structures may be of significance with respect to the structure of ssDNA. While solvent contributions have not been taken into account, the present QM results based on T3PS indicate that ssDNA will have a propensity to sample conformations that are similar to those occurring in the duplex form of DNA. In support of this suggestion a survey of all ssDNA structures in the NDB was performed, with the results presented in Figure 4. While all 5 dihedrals sample a large portion of the full 360° range it is evident that the most populated regions coincide with the low energy regions of the T3PS energy surfaces as well as with regions sampled in the duplex forms of DNA (Figure 3). In the case of γ significant sampling also occurs in the vicinity of the t and g- rotamers; with the sampling of those rotamers being significantly higher than occurring in the duplex DNA surveys. These also coincide with the three minima in the γ potential energy surfaces and sampling of the g- conformation as seen in nucleotides and nucleosides,83 as discussed above. These results are also consistent with a survey of single stranded DNA-protein complex crystal structures showing an average P-P distance of 6.8±0.8 Å,90 which is similar to the P-P distance of 7 Å in B form DNA1 and values of 6.3 91 and 5–7 Å 92 estimated for ssDNA in solution If ssDNA is sampling duplex-like phosphodiester backbone conformations such sampling would be anticipated to diminish the loss of configurational entropy in the oligonucleotide upon assuming the duplex form, thereby favoring is formation.
Figure 4.
Crystallographic probability distributions for α, β, γ, ε and ζ dihedrals from a survey of ssDNA in the Nucleic Acid Database.
CONCLUSION
QM calculations were performed on T3PS to systematically sample the conformational space of the molecule relevant to the A, BI and BII canonical forms of duplex DNA. Initial calculations in which the sugar moieties were constrained to be either C3’endo or C2’endo yielded surfaces in which the low energy regions are consistent with probability distributions of the respective dihedrals from a survey crystallographic structures, thereby validating both the selection of the model compound and the constraint method used to sample the selected regions of conformational space. Additional QM calculations included full optimization of the sugar moieties starting from the previously constrained puckers and energy calculations with more complete basis sets. The resulting energy surfaces showed the intrinsic mechanical properties of the phosphodiester backbone to favor the BI form over both the A and BII forms consistent with a range of experimental results in environments of high water activity. Notable is the intrinsic higher energy of the A form even in the absence of intra-strand phosphatephosphate repulsion. Clearly, significant contributions of the environment and/or the remainder of the DNA molecule are needed to assume the A or BII forms of duplex DNA. The contribution of the phosphodiester backbone to the structure of duplex DNA is further emphasized by the shape of the QM potential energy curves mimicking subtle details of sampling in dihedral probability distributions from the crystallographic survey data. Notably, the fact that the low energy regions of the potential energy surfaces are consistent those observed in DNA duplexes allows for the prediction that single-stranded DNA tends to assume the folded conformation, thereby lowering the entropic barrier to formation of duplex DNA. While the present results should not be interpreted as the phophodiester backbone dictating the structure of duplex DNA they do point to a significant contribution by that part of the molecule, yielding a quantitative estimate of the extent of that contribution. It is anticipated that similar QM studies on related model compounds will yield further insights into the structural properties of oligonucleotides.
Supplementary Material
ACKNOWLEDGMENT
Financial and computational support are acknowledged from the NIH (GM051501) and the DoD High Performance Supercomputing.
REFERENCES
- 1.Saenger W. Principles of Nucleic Acid Structure. New York: Springer-Verlag; 1984. [Google Scholar]
- 2.Franklin RE, Gosling RG. Nature. 1953:171–740. doi: 10.1038/171740a0. [DOI] [PubMed] [Google Scholar]
- 3.Arnott S, Hukins DWL. J. Mol. Biol. 1973:81–93. doi: 10.1016/0022-2836(73)90182-4. [DOI] [PubMed] [Google Scholar]
- 4.Watson JD, Crick FHC. Nature. 1953:171–737. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 5.Leslie AGW, Arnott S, Chandrasekaran R, Ratliff RL. J. Mol. Biol. 1980:143–49. doi: 10.1016/0022-2836(80)90124-2. [DOI] [PubMed] [Google Scholar]
- 6.Dickerson RE, Drew HR, Conner BN, Wing RM, Fratini AV, Kopka ML. Science. 1982:216–475. doi: 10.1126/science.7071593. [DOI] [PubMed] [Google Scholar]
- 7.Hartmann B, Lavery R. Quarterly Reviews of Biophysics. 1996:29–309. doi: 10.1017/s0033583500005874. [DOI] [PubMed] [Google Scholar]
- 8.Crothers DM, Shakked Z. Oxford Handbook of Nucleic Acid Structure. 1998. DNA bending by adenine-thymine tracts; p. 455. [Google Scholar]
- 9.Dickerson RE, Ng H-L. Proc Natl Acad Sci U S A. 2001:98–6986. doi: 10.1073/pnas.141238898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Svozil D, Kalina J, Omelka M, Schneider B. Nucl. Acid Res. 2008:36–3690. doi: 10.1093/nar/gkn260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ivanov VI, Minchenkova LE, Minyan EE, Frank-Kamenetskii MD, Schyolkina AK. J. Mol. Biol. 1974:87–817. doi: 10.1016/0022-2836(74)90086-2. [DOI] [PubMed] [Google Scholar]
- 12.Ivanov VI, Krylov DY. Methods in Enzymology. 1992:211–111. doi: 10.1016/0076-6879(92)11008-7. [DOI] [PubMed] [Google Scholar]
- 13.Franklin RE, Gosling RG. Acta Cryst. 1953:6–673. [Google Scholar]
- 14.Wecker K, Bonnet MC, Meurs EF, Delepierre M. Nucl. Acid Res. 2002;30:4452. doi: 10.1093/nar/gkf559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Djuranovic D, Hartmann B. Biopolymers. 2004;73:356. doi: 10.1002/bip.10528. [DOI] [PubMed] [Google Scholar]
- 16.Tisné C, Hantz E, Hartmann B, Delepierre M. Journal of Molecular Biology. 1998;279:127. doi: 10.1006/jmbi.1998.1757. [DOI] [PubMed] [Google Scholar]
- 17.Madhumalar A, Bansal M. J. Biomol. Struct. Dyn. 2005;23:13. doi: 10.1080/07391102.2005.10507043. [DOI] [PubMed] [Google Scholar]
- 18.Jayaram B, Sprous D, Young MA, Beveridge DL. J. Amer. Chem. Soc. 1998;120:10629. [Google Scholar]
- 19.Srinivasan J, Cheatham TE, III, Ceiplak P, Kollman PA, Case DA. J. Amer. Chem. Soc. 1998;120:9401. [Google Scholar]
- 20.Yang L, Pettitt BM. J. Phys. Chem. 1996;100:2550. [Google Scholar]
- 21.Cheatham TE, III, Kollman PA. Structure. 1997;5:1297. doi: 10.1016/s0969-2126(97)00282-7. [DOI] [PubMed] [Google Scholar]
- 22.Langley DR. J. Biomol. Struct. Dyn. 1998;16:487. doi: 10.1080/07391102.1998.10508265. [DOI] [PubMed] [Google Scholar]
- 23.Banavali NK, Roux B. J Am Chem Soc. 2005;127:6866. doi: 10.1021/ja050482k. [DOI] [PubMed] [Google Scholar]
- 24.Noy A, Pérez A, Laughton G, Orozco M. Nucl. Acid Res. 2007;35:3330. doi: 10.1093/nar/gkl1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knee KM, Dixit SB, Aitken CE, Ponomarev S, Beveridge D. Biophy. J. 2008;95:257. doi: 10.1529/biophysj.107.117606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manning GS. Quart. Rev. Biophy. 1978;11:179. doi: 10.1017/s0033583500002031. [DOI] [PubMed] [Google Scholar]
- 27.Saenger W, Hunter WN, Kennard O. Nature. 1986;324:385. doi: 10.1038/324385a0. [DOI] [PubMed] [Google Scholar]
- 28.Kopka ML, Yoon C, Goodsell DS, Pjura P, Dickerson RE. Proc Natl Acad Sci U S A. 1985;82:1376. doi: 10.1073/pnas.82.5.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flader W, Wellenzohn B, Winger RH, Hallbrucker A, Mayer E, Liedl KR. J. Phys. Chem. B. 2001;105:10379. doi: 10.1021/ja003639b. [DOI] [PubMed] [Google Scholar]
- 30.Pichler A, Rudisser S, Rauch C, Flader W, Wellenzohn B, Winger RH, Liedl KR, Hallbrucker A, Mayer E. J. Phys. Chem. B. 2002;106:3263. [Google Scholar]
- 31.Winger RH, Liedl KR, Pichler A, Hallbrucker A, Mayer E. J. Phys. Chem. B. 2000;104:11349. [Google Scholar]
- 32.Pichler A, Hallbrucker A, Winger RH, Liedl KR, Mayer E. J. Phys. Chem. B. 2000;104:11354. [Google Scholar]
- 33.Foloppe N, Hartmann B, Nilsson L, MacKerell AD., Jr Biophys. J. 2002;82:1554. doi: 10.1016/S0006-3495(02)75507-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hartmann B, Piazzola D, Lavery R. Nucl. Acid Res. 1993;21:561. doi: 10.1093/nar/21.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Foloppe N, MacKerell AD., Jr J. Phys. Chem. B. 1999;103:10955. [Google Scholar]
- 36.Berman HM. Curr. Opin. Struct. Biol. 1994;4:345. [Google Scholar]
- 37.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JA, Jr, Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA. Guassian03. Wallingford, CT: Gaussian, Inc; 2004. [Google Scholar]
- 38.Shao Y, Fusti-Molnar L, Jung Y, Kussmann J, Ochsenfeld C, Brown ST, Gilbert ATB, Slipchenko LV, Levchenko SV, O'Neill DP, RAD, Lochan RC, Wang T, Beran GJO, Besley NA, Herbert JM, Lin CY, Voorhis TV, Chien SH, Sodt A, Steele RP, Rassolov VA, Maslen PE, Korambath PP, Adamson RD, Austin B, Baker J, Byrd EFC, Dachsel H, Doerksen RJ, Dreuw A, Dunietz BD, Dutoi AD, Furlani TR, Gwaltney SR, Heyden A, Hirata S, Hsu C-P, Kedziora G, Khalliulin RZ, Klunzinger P, Lee AM, Lee MS, Liang W, Lotan I, Nair N, Peters B, Proynov EI, Pieniazek PA, Rhee YM, Ritchie J, Rosta E, Sherrill CD, Sherrill CD, Simmonett AC, Subotnik JE, HLW, Zhang W, Bell AT, Chakraborty AK, Chipman DM, Keil FJ, Warshel A, Hehre WJ, HFS, Kong J, Krylov AI, Gill PMW, Head-Gordon M, Gan Z, Zhao Y, Schultz NE, Truhlar D, Epifanovsky E, Oana. M. Q-Chem. Q-Chem 3.1 ed. Pittsburgh, PA: Q-Chem, Inc; 2007. [Google Scholar]
- 39.Krishnan R, Binkley JS, Seeger R, Pople JA. J. Chem. Phys. 1980;72:650. [Google Scholar]
- 40.Møller C, Plesset MS. Phys. Rev. 1934;46:618. [Google Scholar]
- 41.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. J. Comp. Chem. 1983;4:187. [Google Scholar]
- 42.MacKerell AD, Jr, Brooks B, Brooks CL, III, Nilsson L, Roux B, Won Y, Karplus M. CHARMM: The Energy Function and Its Paramerization with an Overview of the Program. In: Schleyer PvR, Allinger NL, Clark T, Gasteiger J, Kollman PA, Schaefer HF, III, Schreiner PR., editors. Encyclopedia of Computational Chemistry. Vol.1. Chichester: John Wiley & Sons; 1998. p. 271. [Google Scholar]
- 43.Foloppe N, MacKerell AD., Jr J. Comp. Chem. 2000;21:86. [Google Scholar]
- 44.MacKerell AD, Jr, Banavali NK. J. Comp. Chem. 2000;21:105. [Google Scholar]
- 45.Woon DE, Dunning TH., Jr J. Chem. Phys. 1993;98:1358. [Google Scholar]
- 46.Bernholdt D, Harrison RJ. Chem. Phys. Let. 1996;250:477. [Google Scholar]
- 47.Weigend F, Kohn A, Hattig C. J. Chem. Phys. 2002;116:3175. [Google Scholar]
- 48.Helgaker T, Klopper W. J Chem Phys. 1997;106:9639. [Google Scholar]
- 49.Halkier A, Helgaker T, Jörgensen P, Klopper W, Olsen J. Chemical Physics Letters. 1999;302:437. [Google Scholar]
- 50.Altona C, Sundaralingam M. J. Am. Chem. Soc. 1973;95:2333. doi: 10.1021/ja00788a038. [DOI] [PubMed] [Google Scholar]
- 51.Berman HM, Olson WK, Beveridge DL, Westbrook J, Gelbin A, Demeny T, Hsieh S-H, Srinivasan AR, Schneider B. Biophys. J. 1992;63:751. doi: 10.1016/S0006-3495(92)81649-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ramachandran GN, Ramakrishnan C, Sasisekharan V. J. Mol. Biol. 1963;7:95. doi: 10.1016/s0022-2836(63)80023-6. [DOI] [PubMed] [Google Scholar]
- 53.Ramachandran GN, V S. Adv. Protein. Chem. 1968;23:283. doi: 10.1016/s0065-3233(08)60402-7. [DOI] [PubMed] [Google Scholar]
- 54.Brooks CL, III, Case DA. Chem. Rev. 1993;93:2487. [Google Scholar]
- 55.Beachy MD, Chasman D, Murphy RB, Halgren TA, Friesner RA. Journal of the American Chemical Society. 1997;119:5908. [Google Scholar]
- 56.MacKerell AD, Jr, Feig M, Brooks CL., III J Am Chem Soc. 2004;126:698. doi: 10.1021/ja036959e. [DOI] [PubMed] [Google Scholar]
- 57.Haschemeryer AEV, Rich A. J. Mol. Biol. 1967;27:369. doi: 10.1016/0022-2836(67)90026-5. [DOI] [PubMed] [Google Scholar]
- 58.Sundaralingam M. Biopolymers. 1969;7:821. [Google Scholar]
- 59.Lakshminarayanan AV, Sasisekharan V. Biochim. Biophys. Acta. 1970;204:49. doi: 10.1016/0005-2787(70)90489-2. [DOI] [PubMed] [Google Scholar]
- 60.Sasisekharan V, Lakshminarayanan AV. Biopolymers. 1969;8:505. doi: 10.1002/bip.360100707. [DOI] [PubMed] [Google Scholar]
- 61.Lakshminarayanan AV, Sasisekharan V. Biopolymers. 1969;8:475. doi: 10.1002/bip.360100707. [DOI] [PubMed] [Google Scholar]
- 62.Olson WK, Flory PJ. Biolpolymers. 1972;11:1. doi: 10.1002/bip.1972.360110102. [DOI] [PubMed] [Google Scholar]
- 63.Olson WK, Flory PJ. Biopolymers. 1972;11:25. doi: 10.1002/bip.1972.360110103. [DOI] [PubMed] [Google Scholar]
- 64.Saran A, Perahia D, Pullman B. Theoret. chim. Acta. 1973;30:31. [Google Scholar]
- 65.Berthod H, Pullman B. Biochim. Biophys. Acta. 1971;232:595. doi: 10.1016/0005-2787(71)90750-7. [DOI] [PubMed] [Google Scholar]
- 66.Berthod H, Pullman B. Biochim. Biophys. Acta. 1971;246:359. doi: 10.1016/0005-2787(71)90772-6. [DOI] [PubMed] [Google Scholar]
- 67.Berthod H, Pullman B. FEBS Letters. 1973;30:231. doi: 10.1016/0014-5793(73)80658-1. [DOI] [PubMed] [Google Scholar]
- 68.Leininger ML, Nielsen IMB, Colvin ME, Janssen CL. J. Phys. Chem. A. 2002;106:3850. [Google Scholar]
- 69.Hobza P, Sponer J. J Am Chem Soc. 2002;124:11802. doi: 10.1021/ja026759n. [DOI] [PubMed] [Google Scholar]
- 70.Dabkowska I, Gonzalez HV, Jurecka P, Hobza P. J. Phys. Chem. A. 2005;109:1131. doi: 10.1021/jp046738a. [DOI] [PubMed] [Google Scholar]
- 71.Foloppe N, MacKerell AD., Jr Biophys. J. 1999;76:3206. doi: 10.1016/S0006-3495(99)77472-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Leulliot N, Ghomi M, Scalmani G, Berthier GJ. J. Phys. Chem. A. 1999;103:8716. [Google Scholar]
- 73.Shishkin OV, Pelmenshikov A, Hovorun DM, Leszczynski J. J. Mol. Struct. 2000;526:329. [Google Scholar]
- 74.Hocquet A, Luelliot N, Ghomi M. J. Phys. Chem. B. 2000;104:4560. [Google Scholar]
- 75.Hocquet A, Ghomi M. Phys. Chem. Chem. Phys. 2000;2:5351. [Google Scholar]
- 76.Foloppe N, Nilsson L, MacKerell AD., Jr Biopolymers (Nucleic Acid Sciences) 2002;61:61. doi: 10.1002/1097-0282(2001)61:1<61::AID-BIP10047>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 77.Banavali NK, MacKerell AD., Jr J. Am. Chem. Soc. 2001;128:6747. doi: 10.1021/ja010295w. [DOI] [PubMed] [Google Scholar]
- 78.Shishkin OV, Gorb L, Zhikol OA, Leszczynski J. J. Biomol. Struct. Dyn. 2004;21:537. doi: 10.1080/07391102.2004.10506947. [DOI] [PubMed] [Google Scholar]
- 79.Pérez A, Marchán I, Svozil D, Sponer J, Cheatham ITE, Laughton CA, Orozco M. Biophy. J. 2007;92:3817. doi: 10.1529/biophysj.106.097782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Svozil D, Sponer JE, Marchan I, Pérez A, Cheatham TE, Forti F, Luque FJ, Orozco M, Sponer J. J. Phys. Chem. B. 2008;112:8188. doi: 10.1021/jp801245h. [DOI] [PubMed] [Google Scholar]
- 81.Gessner RV, Frederick CA, Quigley GJ, Rich A, Wang AH. J.Biol.Chem. 1989;264:7921. doi: 10.2210/pdb1dcg/pdb. [DOI] [PubMed] [Google Scholar]
- 82.Egli M, Williams LD, Gao Q, Rich A. Biochem. 1991;30:11388. doi: 10.1021/bi00112a005. [DOI] [PubMed] [Google Scholar]
- 83.Gelbin A, Schneider B, Clowney L, Hsieh S-H, Olsen WK, Berman HM. J. Amer. Chem. Soc. 1996;118:519. [Google Scholar]
- 84.Alagona G, Ghio C, Kollman PA. J. Am. Chem. Soc. 1985;107:2229. [Google Scholar]
- 85.Jayaram B, Ravishanker G, Beveridge DL. J. Phys. Chem. 1988;92:1032. [Google Scholar]
- 86.FloriáE1n J, Baumruk V, Strajbl M, Bednárová L, Stepánek J. J. Phys. Chem. 1996;100:1559. [Google Scholar]
- 87.MacKerell AD., Jr J. Chim. Phys. 1997;94:1436. [Google Scholar]
- 88.Florián J, Strajbl M, Warshel A. J. Amer. Chem. Soc. 1998;120:7959. [Google Scholar]
- 89.Saenger W. Annu. Rev. Biophys. Biophys. Chem. 1987;16:93. doi: 10.1146/annurev.bb.16.060187.000521. [DOI] [PubMed] [Google Scholar]
- 90.Murphy MC, Rasnik I, Cheng W, Lohman TM, Ha T. Biophy. J. 2004:86. doi: 10.1016/S0006-3495(04)74308-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Smith SB, Cui Y, Bustamante C. Science. 1996;271:795. doi: 10.1126/science.271.5250.795. [DOI] [PubMed] [Google Scholar]
- 92.Mills JB. J. Mol. Biol. 1999;285:245. doi: 10.1006/jmbi.1998.2287. [DOI] [PubMed] [Google Scholar]
- 93.Djuranovic D, Hartmann B. J. Biomol. Struct. Dyn. 2003;20:771. doi: 10.1080/07391102.2003.10506894. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.