Abstract
In vivo growth of Alveolar Soft Part Sarcoma (ASPS) was achieved using subcutaneous xenografts in sex matched NOD.SCID\NCr mice. One tumor, currently at passage 6, has been maintained in vivo for 32 months and has maintained characteristics consistent with those of the original ASPS tumor including (1) tumor histology and staining with Periodic Acid Schiff/Diastase (2) the presence of the ASPL-TFE3 type 1 fusion transcript (3) nuclear staining with antibodies to the ASPL-TFE3 type 1 fusion protein, (4) maintenance of the t(X;17)(p11;q25) translocation characteristic of ASPS, (5) stable expression of signature ASPS gene transcripts and finally, the development and maintenance of a functional vascular network, a hallmark of ASPS. The ASPS xenograft tumor vasculature encompassing nests of ASPS cells is highly reactive to antibodies against the endothelial antigen CD34 and is readily accessible to intravenously administered FITC-dextran. The therapeutic vulnerability of this tumor model to anti-angiogenic therapy, targeting vascular endothelial growth factor (VEGF) and hypoxiainducible factor-1 alpha (HIF-1 α), was examined utilizing bevacizumab and topotecan alone and in combination. Together, the two drugs produced a 70% growth delay accompanied by a 0.7 net log cell kill which was superior to the antitumor effect produced by either drug alone. In summary, the current study describes a pre-clinical in vivo model for ASPS which will facilitate investigation into the biology of this slow growing soft tissue sarcoma and demonstrates the feasibility of employing an anti-angiogenic approach in the treatment of ASPS.
Keywords: Alveolar Soft Part Sarcoma (ASPS), In-Vivo Growth Model, Anti-Angiogenic Therapy of ASPS
INTRODUCTION
Alveolar Soft Part Sarcoma (ASPS) is an extremely rare soft tissue sarcoma affecting primarily young children and adolescents (1). This slow growing neoplasm is believed resistant to existing chemotherapeutic agents and radiation, limiting treatment primarily to surgical resection of the tumor. ASPS exhibits a unique histopathology which is the basis for clinical diagnosis. In addition to the alveolar architecture and the presence of cytoplasmic- rhomboid crystals and granules that stain with periodic acid-Schiff (PAS) reagent and are resistant to digestion with diastase (2), ASPS tumors possess a dense capillary vasculature. Investigation into the biology of ASPS as well as preclinical evaluation of potential ASPS therapeutics has been severely hampered by the lack of both in vitro and in vivo models of this soft tissue sarcoma. This can be attributed, in part, to its rarity, making it very difficult to obtain fresh ASPS tumors for study. Additionally, the slow growth rate as well as the histological makeup of the ASPS tumor, which is frequently populated with areas of necrotic cells (1), make in-vivo propagation and in-vitro culture of ASPS cells challenging. Nevertheless, in an attempt to develop a model which could be utilized to facilitate investigation into the biology of this soft tissue sarcoma and to identify potential ASPS therapeutics, we have utilized primary and metastatic ASPS tumors for growth in immunocompromised mice. In this report we describe the development of the first pre-clinical in vivo model for growth of ASPS in NOD.SCID\NCr mice and the therapeutic vulnerability of this highly vascular tumor to anti-angiogenic therapy.
MATERIALS AND METHODS
Tumorigenicity Studies: In Vivo Growth of ASPS
Fresh ASPS tumors from 12 separate surgical interventions on 9 patients were obtained following informed consent under NCI clinical research protocol 05-C-N138 with assistance of the Alliance Against Alveolar Soft Part Sarcoma (TAAASPS). Tumors were implanted into 6-8 week-old SCID and NOD.SCID\NCr mice. Several routes, including intrapulmonary (i.l.), intrasplenically (i.s.), intravenously (i.v.), and subcutaneously (s.c.) were evaluated for tumor growth. For the i.l., i.s. and i.v. routes, ASPS cells were prepared by mincing small tumor fragments in DME:F12 (1:1 v/v) (Mediatech, Herndon, VA.) containing 10 % fetal bovine serum (Hyclone Laboratories, Logan, UT.), 100 units/ml penicillin, 100μg/ml streptomycin , 2.5 μg/ml fungizone and 100 ng/ml DNase (Sigma-Aldrich, St. Louis, MO.). The mixture was transferred to a 15 ml conical centrifuge tube and undissociated tumor fragments were removed by settling at unit gravity for 1 minute. Under these conditions both individual ASPS cells and nests consisting of 15-25 ASPS cells are produced. The resulting cell suspension was utilized for injection. ASPS tumor fragments (1-2 mm) utilized for s.c. implantation were implanted directly. To measure the impact of vascular support on tumor growth, some tumor fragments were embedded in high-protein Matrigel® (BD BioSciences, Bedford, MA.) containing 100 ng/ml Vascular Endothelial Growth Factor (VEGF; R&D Systems, Minneapolis, MN). Twenty-four hours later these fragments were implanted s.c. into recipient NOD.SCID\NCr mice and tumor growth was monitored. Established tumors were maintained in sex-matched NOD.SCID\NCr mice by serial passage every 4-5 months when the tumors reached 15 mm in diameter.
The inoculated mice were held in microisolator cages with ad libitum autoclaved feed and hyperchlorinated water. The facility was maintained on a 12 h light/dark cycle. The mice were monitored regularly for the appearance of tumor growth. Tumors occurring at the s.c. implant site were harvested and sectioned into small fragments (2-5 mm). These fragments were placed in RNAlater (Ambion, Austin. TX) for isolation of RNA to determine the ASPL-TFE3 type 1 and ASPL-TFE3 type 2 fusion transcripts as well as for gene expression analysis. Some fragments were fixed in 10% neutral buffered formalin for histological evaluations (H&E, Periodic Acid Schiff/ Diastase and immunohistochemical staining for both TFE3 and the ASPL-TFE3 type 1 and ASPL-TFE3 type 2 fusion proteins. Finally fragments were implanted into sex-matched NOD.SCID\NCr recipients. The detailed methodologies utilized for isolation of RNA, RT-PCR detection of the ASPL-TFE3 type 1 and ASPL-TFE3 type 2 fusion transcripts, Periodic Acid Schiff staining and immunohistochemical detection of TFE3 and the ASPL-TFE3 type 1 and ASPL-TFE3 type 2 fusion proteins have been previously described (3).
Microarray Data Acquisition and Analysis
RNA Isolation and Reverse Transcription
Total RNA was isolated from patient and xenograft tumor as described previously (3). RNA was reverse transcribed using Omniscript RT according to the manufacturers' instructions. A standard reaction comprised 2μg total RNA, 0.5mM each dNTP, 2μM random decamers (Ambion, Austin, TX) and 4 units of reverse transcriptase in 20μl total volume of 1x RT buffer. The reaction was allowed to proceed for 90 min at 37°C and the cDNA product diluted to 1μg/ml and stored at -80 °C.
Microarray Data Acquisition
Microarray data was collected at Expression Analysis, Inc. (Durham, NC) using the GeneChip® Human U133 plus 2.0 Array (Affymetrix), according to standardized operating procedures. A description of the microarray along with methodologies applied for hybridization, scanning and data analysis can be found in the supplementary data. Overall, analysis was performed on RNA from the initial ASPS patient tumor, 5 individual xenograft samples and universal RNA, where all samples were analyzed in duplicate. Affymetrix array data files (.CEL) have been deposited with the Gene Expression omnibus (http://www.ncbi.nlm.nih.gov/geo/).
Microarray Data Analysis
The Genesifter suite (VizX labs, Seattle, WA) was used for analysis of microarray data. In brief, compressed .CEL files containing array data were loaded into the Genesifter web portal (www.genesifter.net). Using the advanced upload function, probe-level data was then compiled, normalized and transformed using GC-RMA (4). Pairwise analysis was then performed comparing universal RNA arrays with ASPS patient arrays. The following criteria were applied to filter the differentially expressed transcript list: a fold change of >3 and a Wilcoxon rank sum test where p<0.05 with Benjamini-Hochberg estimation of false discovery rate (FDR). The list of differentially regulated transcripts was then exported into excel for gene ontogeny analysis. The percentage of overlapping transcripts between datasets was calculated using Digdb (www.digdb.com).
Chromosomal Analysis of ASPS Xenograft Tumors
ASPS xenograft tumor fragments, obtained at passage 3 from a female NOD.SCID\NCr mouse, were dissociated by mincing into small fragments and allowed to settle at unit gravity as described above. ASPS cells and nests in the supernatant were centrifuged for 10 minutes at 300xg, resuspended in complete medium and incubated at 37°C with shaking. Chromosomes were prepared for analysis as follows: Colcemid (Karyomax, Invitrogen) (0.1μg/ml) was added for 16 hours prior to harvest. Cells were centrifuged as described above, suspended in 75 mM KCl for 15 minutes at 37°C and fixed with methanol: acetic acid 3:1 (v/v). G banding patterns were obtained with trypsin treatment followed by Giemsa stain (5).
Fluorescence In Situ Hybridization (FISH)
Human chromosome paints were obtained as previously described by chromosome flow sorting (6), followed by degenerate oligonucleotide primed PCR amplification (7). For whole chromosome painting of chromosomes X and 17, probes were labeled with Rhodamine 110-dUTP (Vector) and Spectrum Orange-dUTP (Vysis), respectively. In-situ hybridizations of xenograft preparations with human probes were performed with the following reagents: 5μl of Rh110 labeled chromosome X probe, 5μl of chromosome 17 probe previously labeled with Spectrum Orange, 5μg of human Cot-1 DNA and 2μg of Salmon Sperm DNA, 1.7μg Sodium Acetate. The mixture was precipitated and dissolved in 14 μl of hybridization buffer (formamide 50%, dextran sulfate 10%, 2 x SSC). The probe was denatured at 80°C for 10 min and reannealed at 37°C for 90 min before hybridization. A metaphase slide preparation was aged at 42°C overnight. The slide was denatured in 70% formamide/2 x SSC, at 65 °C for 80s, and quenched in ice-cold 70% ethanol followed by dehydration in a room temperature 70%, 90%, and 100% ethanol series. Hybridization was carried out in a humidity chamber at 37°C overnight. Post-hybridization wash was in 50% formamide in 2xSSC at 42°C for 10 minutes, followed by 10 minute washes each in 2XSSC, 4XSSC with 0.1% Tween 20. Counterstaining was performed with DAPI (0.8ng/μl) for 10 min and the slides were mounted with Antifade. Analyses were performed under an Axioplan 2 (Zeiss) fluorescence microscope coupled with a CCD camera (Photometrics), and images were captured with FISH view 4.5 software (Applied Spectral Imaging Inc., Vista, CA).
Spectral Karyotype
Using a previously G-banded metaphase slide, the slide was washed in 4xSSC with 0.1% Tween 20 for 5 minutes. The slide was dehydrated at room temperature in an ethanol series: 70%, 90% and 100% for 2 minutes each and allowed to air dry. The slide was denatured in 70% formamide/2 x SSC, at 72°C for 80s, and quenched in ice-cold 70% ethanol followed by dehydration in a room temperature 70%, 90%, and 100% ethanol series. The metaphases were hybridized with the 24-color human SKY paint kit (ASI) according to manufacturer's protocol (8). Hybridization was carried out in a humidity chamber at 37°C for 36 hours. The post-hybridization rapid wash procedure was used with 0.5xSSC at 72°C for 4 minutes. Detection was carried out following ASI manufacturer's protocol (8). Spectral images of the hybridized metaphases were acquired using a SD301 SpectraCubeTM system (Applied Spectral Imaging Inc., Vista, CA) mounted on top of an epifluorescence microscope Axioplan 2 (Zeiss). Images were analyzed using Spectral Imaging 4.0 acquisition software (Applied Spectral Imaging Inc., Vista, CA). G banding was simulated by electronic inversion of DAPI-counterstaining.
Vascular Infusion of FITC-Dextran
FITC -Dextran (Sigma-Aldrich, St. Louis, MO.)(FW 2.0 × 106) was dissolved in Phosphate Buffered Saline at a concentration of 5mg/ml, clarified by centrifugation at 12,000 × g for 5 minutes and 0.2ml injected into the tail vein of female NOD.SCID\NCr mice bearing ASPS tumors implanted 4 months previously. Tumors were harvested at 5, 30, 120 and 240 minutes following injection of the fluorochrome, fixed in 10% buffered formalin and embedded in paraffin.
Images were captured using a Nikon Eclipse 80i microscope mounted with a Retiga 2000R monochrome CCD camera (Qimaging, Surrey, BC, Canada). DAPI and FITC fluorescent signals were visualized using Nikon UV-2E/C and B-2E/C filter cubes respectively along with an Xcite-120 fluorescent illuminator (EXFO, Mississauga, Ontario, Canada). Either a Nikon 20x (NA: 0.75) or 40x (NA: 0.95) Plan Apo objective was utilized to capture each field. All images were captured under constant illumination settings for both DAPI and FITC signals within Image-Pro Plus v6.1 (Media Cybernetics, Bethesda, MD).
Anti-Angiogenic Therapy of ASPS
The therapeutic potential of bevacizumab (NSC 704865), topotecan (NSC 609699) and the combination of bevacizumab and topotecan were evaluated in female NOD.SCID/NCr mice with s.c. ASPS tumors. The tumored mice were randomized into one of 5 treatment groups with n=9/group. These included the control group which received water intraperitoneally (i.p.) once every 3 days for 6 cycles (Q3Dx6); bevacizumab administered i.p. at 100μg/dose Q3Dx6; topotecan administered i.p. at 12mg/kg/dose once every 4 days for 4 cycles (Q4Dx4); topotecan administered i.p. at 1mg/kg once daily for 15 days (QDx15); or topotecan at 1mg/kg QDx15 in combination with bevacizumab at 100 ug/dose Q3Dx6. Tumor growth was monitored by once weekly caliper measurements and the tumor weight (mg) was calculated with the formula (9): where the tumor length is the longest dimension (mm) and the tumor width is the narrowest dimension (mm).
Toxicity was assessed by clinical observation and by monitoring individual body weights weekly. During the course of monitoring the efficacy study, tumor samples were collected for molecular and histological studies to assess anti-tumor effects.
RESULTS
Establishment of the ASPS Xenograft Model: ASPS Histology, ASPL-TFE3 Fusion Transcript and Nuclear Staining with Antibodies to ASPL-TFE3
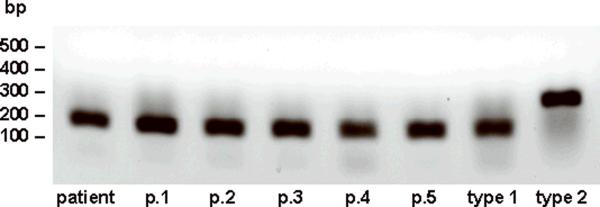
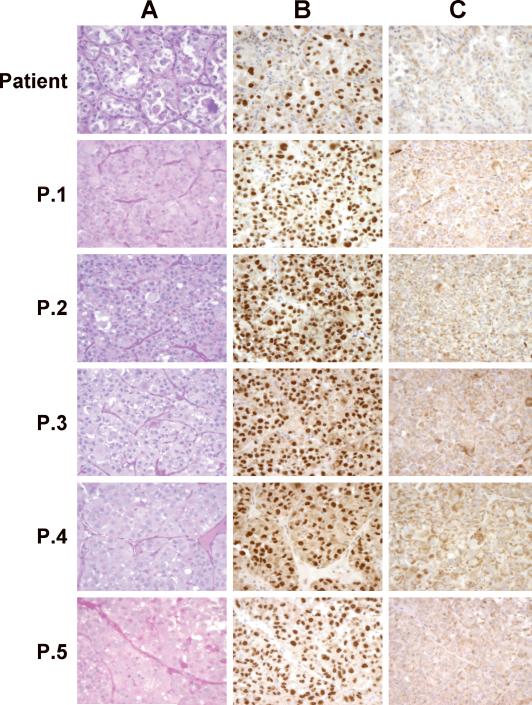
Attempts to grow fresh ASPS tumors from surgery in sex-matched SCID mice were not successful. Also, during the course of these studies no tumors were found growing following intrapulmonic, intrasplenic or intravenous implantation of ASPS tumor material derived from suspensions containing single cells and ASPS nests. In contrast, growth of fresh ASPS tumors did occur in sex-matched NOD.SCID\NCr mice, a non-obese diabetic derivative of the SCID mouse deficient in NK cells, at the subcutaneous site following implantation of ASPS tumor fragments. Implantation of ASPS tumor tissue from twelve surgical interventions on 9 ASPS cases have been performed to date. Successful growth of ASPS tumors occurred following implantation of tumor fragments from 4 patients (1 primary and 3 metastatic tumors: 2 lung tumors and 1 lymph node metastasis). ASPS tumor growth of fresh tumors was initially observed 7-12 months following s.c. implantation. At the present time, the tumor derived from the lymph node, the subject of the current report, is at passage 6 and has been maintained in NOD.SCID/NCr mice for 32 months (Figure 1). Two additional ASPS tumors, both derived from lung metastases of separate cases, are at passage 3. In contrast, the primary tumor as well as one of the lung tumors that grew following implantation from surgery did not grow following serial passage in mice. Successful growth, especially of fresh tumors, was dependent upon both implantation in Matrigel and inclusion of VEGF. ASPS xenograft tumors are routinely monitored at each passage for the presence of the appropriate ASPL-TFE3 fusion transcript, tumor histology and nuclear staining with antibodies to the two ASPL-TFE3 fusion proteins (3). As can be seen in Figure 2, the ASPL-TFE3 type 1 fusion transcript, present in the original patient tumor, is expressed at each passage. ASPS tumor histology, consisting of organoid nests of tumor cells surrounded by a dense endothelial network, is readily apparent at each passage as are the diagnostic PAS-diastase resistant crystals (Figure 3A). Nuclear expression of the ASPLTFE3 type 1 fusion protein (Figure 3B), consistent with the presence of the corresponding ASPL-TFE3 type 1 fusion transcript, was verified at each passage with an antibody to the ASPL-TFE3 type 1 fusion protein (3). No nuclear staining was observed with an antibody to the ASPL-TFE3 type 2 fusion protein (Figure 3C).
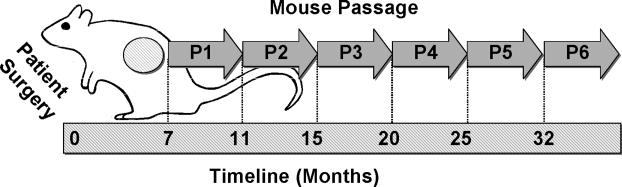
Figure 1.
Growth and Serial Passage of ASPS Xenograft Tumors in Immunocompromised NOD.SCID\NCr Mice. ASPS tumor fragments (1-2 mm) were implanted subcutaneously in NOD.SCID\NCr immunocompromised mice within 24 hours following patient surgery as described in Materials and Methods. Tumors were harvested at the indicated times and serially passaged.
Figure 2.
RT-PCR Determination of the ASPL-TFE3 Fusion Transcript.
RNA was isolated from ASPS xenograft tumors at the appropriate passage and the ASPL-TFE3 fusion transcript was determined by RT-PCR as described in Materials and Methods.
Figure 3.
ASPS Xenograft Histology and Nuclear Reactivity to ASPL-TFE3 type 1 and ASPL-TFE3 type 2 Antibodies.
ASPS xenograft tumors, maintained in female NOD.SCID\NCr mice, were harvested at the indicated passage (see timeline illustrated in Figure 1), fixed in 10% buffered formalin and embedded in paraffin. Sections (4μm) were stained with Periodic Acid Schiff (PAS)( Panel A) or with affinity-purified polyclonal antibodies to either the ASPLTFE3 type 1 fusion protein (Panel B) or to the ASPL-TFE3 type 2 fusion protein (Panel C). (See reference 3 for details of staining).
ASPS Xenograft Gene Stability
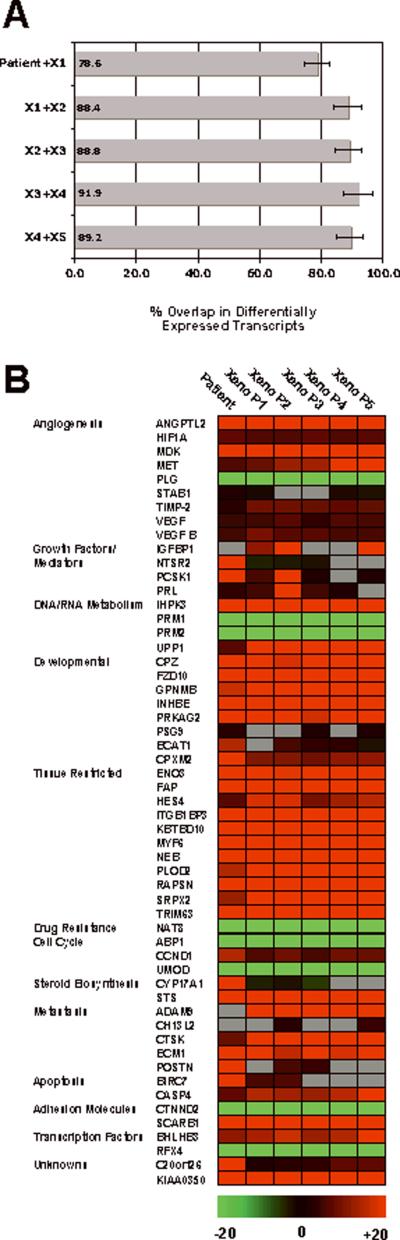
Transcriptomic analysis of individual xenograft passages was performed as a means of providing objective measures of biological consistency with time. Total RNA samples isolated from ASPS patient tumor, xenograft passages 1-5 and universal RNA were reverse transcribed and each sample (7 in total) hybridized to duplicate U133 plus 2.0 arrays (Affymetrix). Data files (.CEL) from these arrays (14 in total) were then uploaded into the Genesifter analysis package (VizX laboratories) using the advanced upload option. Six differentially expressed transcript lists were then generated through pairwise analysis of duplicate ASPS tumor or xenograft arrays with control universal RNA array data. Search criteria for the transcript lists were GC-RMA normalization, <0.05 adjusted p-value, Benjamini and Hochberg false discovery estimates with >+/- 3 fold changes in expression. As shown in Figure 4A, the transcript lists were then meshed using DigDb (www.DigDb.com) to determine the degree of overlap in transcript IDs between chronological neighbors. Results demonstrated that 78.6% of transcripts were conserved between the original patient material and xenograft P1 (Figure 4A). For the other xenograft passages rates of conservation held constant at approximately 90% suggesting that over time the tumor retained the ASPS transcriptomic signature (Figure 4A). Following this, for selected transcripts implicated in ASPS pathogenesis, expression differentials were extracted from the datasets and presented in the form of a heat map (Figure 4B). It is apparent that the transcriptomic profile of the ASPS xenograft tumor is similar not only to that of the original patient tumor (Figure 4B) but also to that recently described in a comprehensive analysis of seven ASPS patients with primary and metastatic disease (10). Of particular significance, for both in-vivo model validation of this highly vascular soft tissue sarcoma and in-vivo testing of potential ASPS therapeutics, was the consistent elevated expression of genes related to angiogenesis (ANGPTL2, HIF 1-α, MDK, MET, TIMP-2 , VEGF and VEGF B). For these and the majority of the remaining 55 genes, a high degree of consistency was observed from patient material out to the fifth xenograft passage (Figure 4B). The expression of several transcripts disappeared after the initial patient material (NTSR2, PCSK1, POSTN, CYP17A1), and these may reflect the influence of stromal cells that ultimately disappear from xenografts. Importantly, a subset of genes designated `tissue restricted' such as ITGB1BP3, MYF6 and TRIM63, currently under investigation as ASPS specific markers (10), showed consistent differentials in all samples as did the basic helix-loop helix domain containing, class 3B (BHLHB3), transcription factor. Overall, these data suggest that xenograft passages display a stable ASPS gene signature.
Figure 4.
Conservation and Stability of Selected Genes in ASPS Xenograft Tumors. Panel A. Transcript Conservation in ASPS Xenograft Tumors. Data was analyzed as described in Materials and Methods.
Panel B. ASPS Xenograft Tumor Heat Map. Data was analyzed as described in Materials and Methods.
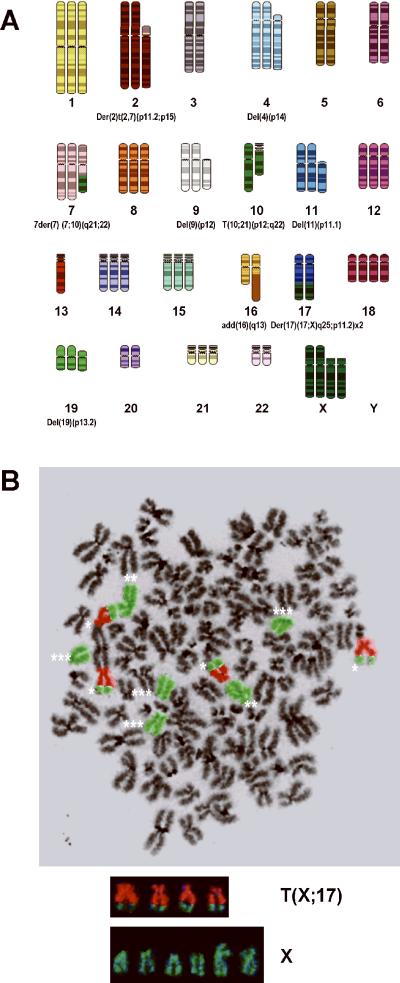
Xenograft Derived ASPS Cells Exhibit the t(X;17)(p11;q25) Translocation Characteristic of ASPS
Analysis of metaphase chromosomes of xenografted tumor cells revealed a near triploid modal chromosome number and the presence of numerous chromosomal rearrangements including the characteristic t(X;17)(q11;p25) translocation reported for this tumor (11,12). The SKY karyotype was converted in SKYGRAM format from an analyzed metaphase that was positive for the translocation (Figure 5A) (13). FISH analysis of metaphase cells using whole chromosome paints for X and 17 further confirmed the presence of the translocation (Figure 5B).
Figure 5.
ASPS Xenograft Tumors exhibit a Near-Triploid Karyotype and the t(X;17)(p11;q25) Chromosomal Translocation.
Panel A. ASPS Xenograft Spectral Karyotype (SKY). Numerous chromosomal aberrations are present in this near triploid karyotype including the characteristic t(X;17)(q11;p25) translocation.
Panel B. FISH Analysis of the ASPS Xenograft Tumor. Four copies of the t(X;17)(q11;p25) translocation (*), 2 normal X chromosomes (**) and 4 deleted X chromosomes (***) are present. X chromosome (green); chromosome 17 (orange).
Vascularity of the ASPS Xenograft Model
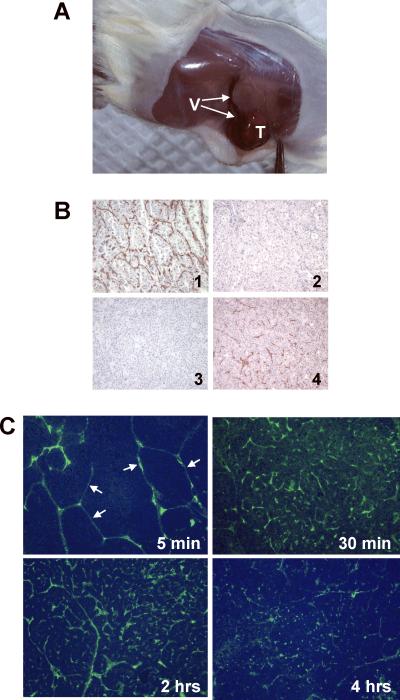
It is well documented that ASPS is a highly vascular soft tissue sarcoma. An in-vivo xenograft tumor model representative of the clinical disease should reflect this high degree of vascularity. As can be seen in Figure 6A, ASPS tumors grown subcutaneously in NOD.SCID\NCr mice are infiltrated by numerous blood vessels suggesting that growth of these ASPS xenograft tumors is accompanied by/dependent upon development of a complex tumor vasculature. We examined the reactivity of the capillary/endothelial network surrounding nests of ASPS cells to anti-human CD31 and anti-mouse CD34 antibodies, both in the original ASPS tumor and in ASPS tumors maintained as subcutaneous xenografts in NOD.SCID\NCr mice. As can be seen in Figure 6B, the capillary network of the original ASPS tumor is stained only by the anti-human CD34 antibody; no staining with the anti-mouse CD34 antibody is observed. The reactivity of the capillary network surrounding nests of ASPS cells to anti-human CD34 antibody observed in the original tumor is lost upon growth in mice (Figure 6B) and the capillary network becomes only reactive with anti-mouse CD34. To determine whether the ASPS capillary network was contiguous with and could be accessed from the general circulation, we infused FITC-dextran of high molecular weight into the tail veins of NOD.SCID\NCr mice bearing subcutaneous ASPS tumors implanted 4 months previously. As seen in Figure 6C, FITC-dextran readily locates to the ASPS xenograft tumor vascular network following intravenous infusion; fluorescent polymer is visible in and confined to the endothelial network surrounding nests of ASPS cells as soon as 5 minutes following infusion and, within 30 minutes, appears to achieve near maximum labeling of the capillary network surrounding the nests of ASPS cells. These results indicate that the vasculature surrounding the nests of ASPS cells is accessible from the general circulation.
Figure 6.
ASPS Xenograft Tumor Vasculature.
Panel A. ASPS Xenograft Tumors Exhibit a Complex Vascular Network. Numerous blood vessels (V) can be seen entering the subcutaneous ASPS tumor (T).
Panel B. CD31 and CD34 Staining of the ASPS Endothelial Network. ASPS xenograft tumors were harvested, fixed in 10% buffered formalin, embedded in paraffin and stained (3) with antibodies to human and mouse CD34. Panel 1: Patient Tumor (anti-human CD34). Panel 2: Patient Tumor (anti-mouse CD34). Panel 3: ASPS Xenograft Tumor (anti-human CD 34). Panel 4: ASPS Xenograft Tumor (anti-mouse CD 34).
Panel C. FITC-Dextran Localizes to the Endothelium Surrounding Nests of ASPS Cells Following Intravenous Administration. ASPS tumors were harvested at 5, 30, 120 and 240 minutes following intravenous administration of the fluorochrome and processed as described in Material and Methods. The fluorochrome readily locates to the endothelial network surrounding nests of ASPS tumor cells (5 minutes; arrows) and slowly diffuses into the tumor.
Anti-Angiogenic Therapy of ASPS
The upregulation of several genes involved in angiogenesis in both primary and metastatic ASPS tumors (10) and in the ASPS xenograft model presently described (Figure 4B) prompted an experimental therapeutic study designed to target genes involved in angiogenesis. The observation that hypoxia inducible factor 1 (HIF-1α), the master regulator of hypoxia responses, is elevated in primary and metastatic ASPS both at the RNA and protein expression levels (10) and that this gene controls the expression of several additional up-regulated angiogenesis-related ASPS genes (Angiopoietin-2, VEGF , c-Met. MDK) in the ASPS transcriptome (10) prompted a therapeutic study centered about targeting HIF-1α and VEGF. Bevacizumab (Avastin) (14), a humanized anti-vascular endothelial growth factor A (VEGF-A) monoclonal antibody, was employed alone an in combination with Topotecan, a topoisomerase 1 poison with antiangiogenic properties (15, 16). Interestingly, a single case report (17) detailed a possible response to bevacizumab in an ASPS patient previously treated with multiple cytotoxic drugs.
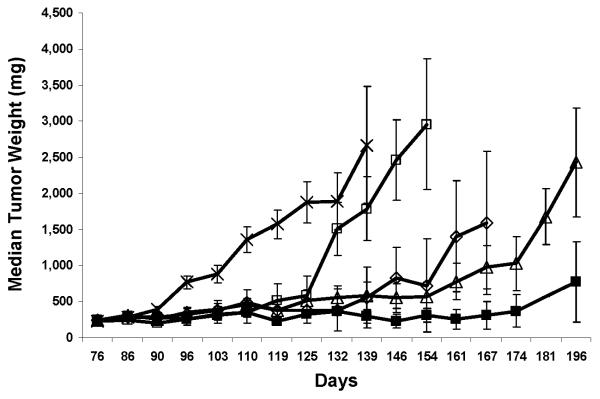
Topotecan has been shown to be a potent inhibitor of HIF-1α (18, 19) and daily, but not intermittent administration of the drug inhibits angiogenesis and both HIF-1α protein accumulation and HIF-1α dependent gene expression (20). The ASPS tumors were significantly impacted by treatment with bevacizumab and topotecan alone as well as the combination of bevacizumab and topotecan (Figure 7). As shown in Table 1 none of the treatments were associated with body weight losses of greater than 15%. Thus, the measured antitumor activities are presumed to be the result of specific rather than nonspecific effects since the body weight losses were within acceptable limits (less than 20%). As seen in Table 1, all treatments produced optimal % T/C values of less than 40%, the standard value associated with an antitumor effect (9). Because the majority of the control animals were lost due to large tumor masses by day 110, it is not possible to calculate %T/C values beyond day 110. If the % growth delay, time to 600mg and net log cell kill values are compared then the combination of topotecan plus bevacizumab appears superior to either agent alone (Figure 7). In fact, the combination treatment group had a 70% growth delay, a 0.7 net log cell kill and a time to 600mg of 159 days which indicate a greater antitumor effect than the results seen for the groups receiving only one treatment arm. Statistical analyses demonstrate all 4 treatment groups have significantly smaller tumors than the control group (P < 0.05) at all time points after 90 days. A comparison of the bevacizumab alone to the bevacizumab plus topotecan also demonstrates a significantly smaller tumor burden in the mice receiving the combination treatment compared to the bevacizumab alone. In contrast, a comparison of the topotecan alone to the combination group does not demonstrate a statistically significant difference in tumor burden between these 2 groups. However, the data shown in Table 1 demonstrate the combination treatment group has an apparent prolongation in tumor growth delay with a smaller tumor burden from day 119 till the end of the experimental observation period.
Figure 7.
Antiangiogenic Therapy of ASPS Xenograft Tumors.
Median tumor weights of mice treated with bevacizumab, topotecan or a combination of both drugs. Female NOD.SCID mice bearing subcutaneous ASPS tumors were randomized into one of 5 groups (n=9/group): ---x--- vehicle control Q3D x 6; ---□ ---100 ug bevacizumab Q3D x 6; ---◊--- 12 mg topotecan/kg Q4D x 4; ---Δ---1mg topotecan/kg QD x 15; ---■--- 100ug bevacizumab Q3D x 6 and 1 mg topotecan/kg QD x 15. All treatments were administered intraperitoneally starting on day 76 when tumors staged approximately 230mg. Two additional treatment cycles were initiated on days 96 and 116. Mice were euthanized when tumors reached a size of 2500mg or at the termination of the experiment.
Table 1.
Inhibition of ASPS Tumor Growth by Cytotoxic and Anti-Angiogenic Therapy
| Max Mean % Weight loss | Optimal %T/C | median days to 600 mg | %growth delay | net log cell kill | |
|---|---|---|---|---|---|
| vehicle control Q3D×6 | 7.7 | NA | 94 | NA | NA |
| 100 μg bevacizumab Q3D×6 | 5.4 | 27 | >119 | 27 | 0.1 |
| 12 mg topotecan/kg Q4D×4 | 9.7 | 36 | >114 | 21 | 0.1 |
| 1 mg topotecan/kg QD×15 | 14.6 | 34 | 122 | 30 | 0.2 |
| 100 μg bevacizumab Q3D×6 + 1 mg topotecan/kg QD×15 | 7.9 | 26 | 159 | 70 | 0.7 |
DISCUSSION
Alveolar Soft Part Sarcoma (ASPS), first described over 56 years ago (1), remains a little understood soft tissue sarcoma. Investigation into many aspects of ASPS including the basic biology of the disease, its resistance to standard treatment modalities, elucidation of tissue of origin and, importantly, identification of potential drug targets has been severely compromised due to a lack of pre-clinical in-vitro and in-vivo models of the disease. Tumor procurement for the development of these models has been difficult due to the rarity of the disease which has limited access to ASPS tumors for study. Lack of an ASPS model system and the resulting paucity of investigations on the disease has, most likely, contributed to the lack of therapeutic options thereby limiting treatment for ASPS mainly to surgical excision of the tumor. In an effort to facilitate investigation into this poorly understood rare soft tissue sarcoma, we sought to establish a pre-clinical in-vivo xenograft model of the disease in immunocompromised mice. The results here detail these efforts and describe the establishment of the first model system for this disease. The ASPS tumor, maintained as a xenograft in immunocompromised NOD.SCID\NCr mice, exhibits characteristics consistent with those of the patient tumor. These include relative tumor growth rate, tumor histology, presence of the appropriate ASPL-TFE3 fusion transcript and ASPL-TFE3 fusion protein, maintenance of the t(X;17)(p11;q25) non-reciprocal chromosomal translocation, stable expression of ASPS genes from the ASPS transcriptome (10) and, importantly, the presence of a complex, functional vasculature characteristic of ASPS. Comparison between the original patient tumor and the xenograft model suggests marked similarities in each of the above areas which, importantly, are consistently maintained in serial passage in immunocompromised mice. First, as is observed clinically, growth of the ASPS tumor as a xenograft is extremely slow. Subcutaneous implantation in sex-matched NOD.SCID\NCr mice is accompanied by a 2 month lag phase during which little, if any, tumor growth is observed. This lag phase is followed by a multi-month period of tumor growth resulting in tumors reaching 2000-3000mg in size (Figure 7). Such a growth pattern is often seen clinically in ASPS cases where long periods of latency are observed in the disease followed by tumor growth. Second, it is apparent from these studies that both the morphology of the xenograft-derived ASPS tumors as well as their staining with PAS/Diastase is comparable to that observed in the original tumor obtained from the patient (Figure 3A). The xenograft tumors are composed of organoid nests of ASPS cells encompassed by a vascular sinus comparable to that observed in the original ASPS tumor from the patient (Figure 3A). Third, the original patient-derived ASPS tumor and the ASPS xenograft tumor exhibit the same ASPL-TFE3 type 1 fusion transcript which is conserved at each passage (Figure 2). Detection of the ASPL-TFE3 type 1 fusion transcript is accompanied by nuclear staining with an antibody to the ASPL-TFE3 type 1 fusion protein (3), thereby confirming expression of the corresponding ASPL-TFE3 type 1 fusion protein (Figure 3B). Fourth, ASPS cells derived from mice bearing xenografts maintain the t(X;17)(p11;q25) chromosomal translocation characteristic of ASPS (11, 12) and exhibit a near-triploid karyotype (Figure 5). Fifth, the gene expression profile of the ASPS xenograft tumor is similar to that of the original patient tumor. Extensive gene analysis of primary and metastatic ASPS tumors has recently been completed (10) using freshly isolated patient tumor RNA. These studies resulted in the generation of a transcriptomic profile of the disease (10), which was validated using real-time PCR and immunohistochemistry, amongst others, several genes involved in angiogenesis (ANGPTL2, HIF-1α, MDK, VEGF and TIMP-2) and metastasis (c-Met, ADAM9, ECM1 and POSTN) were seen to have markedly elevated expression. Four muscle specific transcripts (ITGB1BP3/MIBP), MYF5, MYF6 and TRIM63) were identified as potential ASPS markers. In the present study microarray data from the original patient tumor was compared with that of serial passages of the ASPS xenograft tumor. As can be seen in Figure 4A, approximately 79 % of transcripts identified as altered in primary disease were similarly regulated in the first xenograft tumor harvested 7 months following transplantation into NOD.SCID\NCr mice. This percentage was observed to approach 90% for the remaining xenograft passages; this difference may be due to loss of normal human cells, present in the original patient tumor, during subsequent serial passage in mice. These results suggest that the ASPS tumor maintained as a xenograft retains the ASPS transcriptomic profile observed in the original patient. To emphasize this point a detailed examination of 55 genes was conducted using quantitative RT-PCR and results indicated remarkable stability with serial passage in immunocompromised mice (Figure 4B). Genes known to be involved in angiogenesis (ANGPTL2, HIF-1α, MDK, c-Met, TIMP-2 and VEGF), were consistently elevated in ASPS xenograft tumors during 5 serial passages, encompassing 32 months.
Finally, the highly vascular nature of ASPS observed clinically (1) is maintained in the ASPS xenograft model. Numerous large vessels were observed entering the ASPS xenograft tumors (Figure 6A). Additionally, the endothelial network characteristic of ASPS tumors (1) was evident in the xenograft tumors (Figure 6B), and it stains intensely with antibodies to CD34 (Figure 6B). In an attempt to ascertain whether the ASPS capillary network was accessible from the general circulation, we employed intravenous infusion of FITC-Dextran into mice bearing ASPS tumors. The fluorescent tracer readily located to the capillary network surrounding the nests of ASPS cells (Figure 6C) indicating that this network is accessible. Taken together these results demonstrate the presence of a vasculature in ASPS xenograft tumors commonly seen clinically in ASPS (1); furthermore, this vasculature in the ASPS xenograft tumors is accessible and, besides serving as a vehicle for delivery of therapeutics, may, in itself, serve as a therapeutic target.
It is apparent from previous work with both primary and metastatic ASPS tumors (10) and in the presently described ASPS xenograft model, that angiogenesis- related genes are a highly visible, up-regulated subset of genes and an attractive therapeutic target. Several of the up-regulated genes in the ASPS transcriptome are under control of HIF: VEGF/c-Met (21), Midkine (22) and Angiopoietin-2 (23), thus providing a specific rationale for designing a study employing inhibitors of HIF and VEGF. A recent publication (24) using RNA from 3 frozen ASPS tumors identified 18 angiogenesis-related genes which were upregulated > 1.5 fold and 8 genes upregulated >3 fold. Although there appear to be differences between these results and ours, using RNA prepared from fresh ASPS tumor tissue, a pattern of up-regulated genes involved in angiogenesis is evident. It would appear that the results of our therapeutic study targeting both HIF and VEGF (Figure 7; Table 1) support further efforts to focus on the vasculature of ASPS by targeting angiogenesis-related genes. It is apparent from our results that combination therapy employing topotecan and bevacizumab was superior to either drug alone. Several important observations evolved from this study. First, it is noteworthy that bevacizumab as a single agent was effective only for as long as treatment was administered. Cessation of treatment, at day 125, was accompanied by rapid growth of the ASPS xenograft tumor (Figure 7). Second, the schedule employed for topotecan appeared to be important in the therapeutic result. Using a daily, low dose schedule of administration designed to target HIF (19) was more effective than high-dose intermittant administration of the drug. Ongoing work examining the dependency of HIF inhibition upon topotecan scheduling demonstrates that the combination of bevacizumab and daily topotecan, but neither drug alone, results in induction of tumor cell apoptosis and inhibition of tumor growth in U251-HRE xenografts (G. Melillo, personal communication). Although the results of our therapeutic study are striking, their interpretation is complicated. Immunohistochemical staining for nuclear HIF-1 α failed to reveal a difference between control tumors and tumors of all treatment groups isolated immediately following the third cycle of treatment (day 125 post tumor implantation). The range of HIF positive cells found in the control group overlapped with that observed in the experimental groups primarily because HIF positive cells can only be observed in hypoxic areas of the tumors and similar representative hypoxic areas were not present in the sampled tissues. Additionally, the observation that the lower, frequently administered regimen of topotecan was more effective than the intermittent, high dose regimen may be a consequence of the known anti-angiogenic effects of scheduling. Employing a frequently administered, low dose of cyclophosphamide, given to mice bearing the Lewis lung carcinoma (25) resulted in suppression of endothelial cell recovery leading to reduced tumor vasculature for support of tumor growth. This approach, termed metronomic chemotherapy (26), by virtue of frequent drug administration without extended intervals between treatment, also resulted in eradication of drug-resistant tumors and has been the subject of several extensive reviews (27, 28). Employing low-dose metronomic chemotherapy, using anti-vascular agents, would seem to be one approach to treatment of highly vascular, drug-resistant tumors such as ASPS. In summary, these results describe the establishment of an in-vivo model for ASPS reflecting the known clinical characteristics of this rare soft tissue sarcoma. This tumor model will allow investigation into the tumor biology of the disease as well as provide a means of evaluating novel ASPS therapeutics.
ACKNOWLEDGEMENTS
We thank Mr. Scott Lawrence for FITC-Dextran image analysis.
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract N01-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does the mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This research was supported by the Developmental Therapeutics Program in the Division of Cancer Treatment and Diagnosis of the National Cancer Institute.
Footnotes
The authors declare no conflict of interest.
REFERENCES
- 1.Christopherson WM, Foote FW, Jr, Stewart FW. Alveolar soft-part sarcomas: structurally characteristic tumors of uncertain histogenesis. Cancer. 1952;5(1):100–111. doi: 10.1002/1097-0142(195201)5:1<100::aid-cncr2820050112>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 2.Shipkey FH, Lieberman PH, Foote FW, Jr., et al. Ultrastructure of alveolar soft part sarcoma. Cancer. 1964;17:821–830. doi: 10.1002/1097-0142(196407)17:7<821::aid-cncr2820170702>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 3.Vistica DT, Krosky PM, Kenney S, et al. Immunohistochemical discrimination between the ASPL-TFE3 fusion proteins of alveolar soft part sarcoma. J Pediatr Hematol Oncol. 2008;30:46–52. doi: 10.1097/MPH.0b013e31815d1d6f. [DOI] [PubMed] [Google Scholar]
- 4.Wu Z, Irizarry RA. Stochastic models inspired by hybridization theory for short oligonucleotide arrays. J Comput Biol. 2005;12(6):882–893. doi: 10.1089/cmb.2005.12.882. [DOI] [PubMed] [Google Scholar]
- 5.Seabright M. A rapid banding technique for human chromosomes. Lancet. 1971;11:971–972. doi: 10.1016/s0140-6736(71)90287-x. [DOI] [PubMed] [Google Scholar]
- 6.Telenius H, Pelmear AH, Tunnacliffe A, et al. Cytogenetic analysis by chromosome painting using DOP-PCR amplified flow-sorted chromosomes. Genes Chromosomes Cancer. 1992;4(3):257–263. doi: 10.1002/gcc.2870040311. [DOI] [PubMed] [Google Scholar]
- 7.Telenius H, Carter NP, Bebb CE, et al. Degenerate oligonucleotide-primed PCR: general amplification of target DNA by a single degenerate primer. Genomics. 1992;13:718–725. doi: 10.1016/0888-7543(92)90147-k. [DOI] [PubMed] [Google Scholar]
- 8.Schrock E, du Manoir S, Veldman T, et al. Multicolor spectral karyotyping of human chromosomes. Science. 1996;273:494–497. doi: 10.1126/science.273.5274.494. [DOI] [PubMed] [Google Scholar]
- 9.Plowman J, Dykes DJ, Hollingshead M, et al. Human tumor models in NCI drug development. In: Teicher B, editor. Anticancer Drug Development Guide: Preclinical Screening, Clinical Trials, and Approval. Humana Press Inc; New Jersey: 1997. pp. 101–129. [Google Scholar]
- 10.Stockwin LH, Vistica DT, Kenney S, et al. Gene Expression Profiling of Alveolar Soft-Part Sarcoma. submitted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heimann P, Devalck C, Debusscher C, et al. Alveolar soft-part sarcoma: further evidence by FISH for the involvement of chromosome band 17q25. Genes Chromosomes Cancer. 1998;23:194–197. [PubMed] [Google Scholar]
- 12.Ladanyi M, Lui MY, Antonescu CR, et al. The der(17)t(X;17)(p11;q25) of human alveolar soft part sarcoma fuses the TFE3 transcription factor gene to ASPL, a novel gene at 17q25. Oncogene. 2001;20:48–57. doi: 10.1038/sj.onc.1204074. [DOI] [PubMed] [Google Scholar]
- 13.NCI and NCBI's SKY/M-FISH and CGH database. 2001 http://www.ncbi.nlm.nih.gov/sky/skyweb.cgi.
- 14.Ferrara N, Hillan KJ, Novatny W. Bevacizumab(Avastin), a humanized anti-VEGF monoclonal antibody for cancer therapy. Biochem Biophys Res Commun. 2005;333:328–335. doi: 10.1016/j.bbrc.2005.05.132. [DOI] [PubMed] [Google Scholar]
- 15.O'Leary JJ, Shapiro RL, Ren CJ, et al. Antiangiogenic effects of camptothecin analogues 9-amino-20(S)-camptothecin, topotecan, and CPT-11 studied in the mouse cornea model. Clinical Cancer Res. 1999;5:181–187. [PubMed] [Google Scholar]
- 16.Clements MK, Jones CB, Cumming M, et al. Antiangiogenic potential of camptothecin and topotecan. Cancer Chemother Pharmacol. 1999;44:411–416. doi: 10.1007/s002800050997. [DOI] [PubMed] [Google Scholar]
- 17.Azizi AA, Haberler C, Czech T, et al. Vascular-endothelial-growth factor(VEGF) expression and possible response to angiogenesis inhibitor bevacizumab in metastatic alveolar soft part sarcoma. Lancet Oncol. 2006:521–523. doi: 10.1016/S1470-2045(06)70729-X. [DOI] [PubMed] [Google Scholar]
- 18.Rapisarda A, Uranchimeg B, Scudiero DA, et al. Identification of small molecule inhibitors of hypoxia-inducible factor 1 transcriptional activation pathway. Cancer Res. 2002;62:4316–4324. [PubMed] [Google Scholar]
- 19.Rapisarda A, Uranchimeg B, Sordet O, et al. Topoisomerase 1 mediated inhibition of hypoxia-inducible factor 1: mechanism and therapeutic implications. Cancer Res. 2004;64:1475–1482. doi: 10.1158/0008-5472.can-03-3139. [DOI] [PubMed] [Google Scholar]
- 20.Rapisarda A, Zalek J, Hollingshead M, et al. Schedule-dependent inhibition of hypoxia-inducible factor-1 alpha protein accumulation, angiogenesis, and tumor growth by topotecan in U251-HRE glioblastoma xenografts. Cancer Res. 2004;64:6845–6848. doi: 10.1158/0008-5472.CAN-04-2116. [DOI] [PubMed] [Google Scholar]
- 21.Rocha S. Gene regulation under low oxygen: holding your breath for transcription. Trends Biochem Sci. 2007;32:389–397. doi: 10.1016/j.tibs.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 22.Reynolds PR, Mucenski ML, LeCras TD, et al. Midkine is regulated by hypoxia and causes pulmonary vascular remodeling. J Biol Chem. 2004;279:37124–37132. doi: 10.1074/jbc.M405254200. [DOI] [PubMed] [Google Scholar]
- 23.Giaccai AJ, Rankin EB. The role of hypoxia-inducible factors in tumorigenesis. Cell Death Differ. 2008;15:678–685. doi: 10.1038/cdd.2008.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lazar AJF, Das P, Tuvin D, et al. Angiogenesis-promoting gene patterns in alveolar soft part sarcoma. Clin Cancer Res. 2007;13:7314–7321. doi: 10.1158/1078-0432.CCR-07-0174. [DOI] [PubMed] [Google Scholar]
- 25.Browder T, Butterfield CE, Kraling BM, et al. Antiangiogenic scheduling of chemotherapy improves efficacy against experimental drug-resistant cancer. Cancer Res. 2000;60:1878–1886. [PubMed] [Google Scholar]
- 26.Hanahan D, Bergers G, Bergsland E. Less is more, regularly: metronomic dosing of cytotoxic drugs can target tumor angiogenesis in mice. J Clin Inves. 2000;105:1045–1047. doi: 10.1172/JCI9872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kerbel RS, Kamen BA. The anti-angiogenic basis of metronomic chemotherapy. Nat Rev Cancer. 2004;4:423–436. doi: 10.1038/nrc1369. [DOI] [PubMed] [Google Scholar]
- 28.Kerbel RS. Tumor Angiogenesis. N Engl J Med. 2008;358:2039–2049. doi: 10.1056/NEJMra0706596. [DOI] [PMC free article] [PubMed] [Google Scholar]