SUMMARY
The isolation of human induced pluripotent stem cells (iPSCs)1-3 offers a novel strategy for modeling human disease. Recent studies have reported the derivation and differentiation of disease-specific human iPSCs4-7. However, a key challenge in the field is the demonstration of disease-related phenotypes and the ability to model pathogenesis and treatment of disease in iPSCs. Familial dysautonomia (FD) is a rare but fatal peripheral neuropathy caused by a point mutation in IKBKAP8 involved in transcriptional elongation9. The disease is characterized by the depletion of autonomic and sensory neurons. The specificity to the peripheral nervous system and the mechanism of neuron loss in FD are poorly understood due to the lack of an appropriate model system.
Here we report the derivation of patient specific FD-iPSCs and the directed differentiation into cells of all three germ layers including peripheral neurons. Gene expression analysis in purified FD-iPSC derived lineages demonstrates tissue specific mis-splicing of IKBKAP in vitro. Patient-specific neural crest precursors express particularly low levels of normal IKBKAP transcript suggesting a mechanism for disease specificity. FD pathogenesis is further characterized by transcriptome analysis and cell based assays revealing marked defects in neurogenic differentiation and migration behavior. Finally, we use FD-iPSCs for validating the potency of candidate drugs in reversing aberrant splicing and ameliorating neuronal differentiation and migration. Our study illustrates the promise of iPSC technology for gaining novel insights into human disease pathogenesis and treatment.
Familial dysautonomia (FD), also known as hereditary sensory and autonomic neuropathy III (HSAN-III; Riley-Day Syndrome), is a fatal autosomal recessive disease characterized by the degeneration of sensory and autonomic neurons8,10. The pathogenesis of the disease and its specificity to the peripheral nervous system are poorly understood. Most FD patients carry a point mutation in the I-κ-B kinase complex-associated protein (IKBKAP) gene resulting in a tissue specific splicing defect with various levels of exon 20 skipping and reduced levels of normal IKAP protein8,11. Reduced IKAP protein levels are associated with a defect in cell motility9. However, it is unclear whether decreased cell motility is functionally related to the specific peripheral neuron pathology observed in FD. Based on the splicing defect, in vitro drug screens have been performed in patient lymphoblast cell lines to identify compounds that improve normal IKBKAP splicing and increase IKAP protein levels12. However, lack of access to tissues affected by FD, such as neural crest precursors and peripheral neurons, has hampered functional validation of candidate drugs and a systematic analysis of disease pathogenesis. Here we demonstrate the potential of human iPSC technology to model FD pathogenesis and treatment (Suppl. Fig. 1).
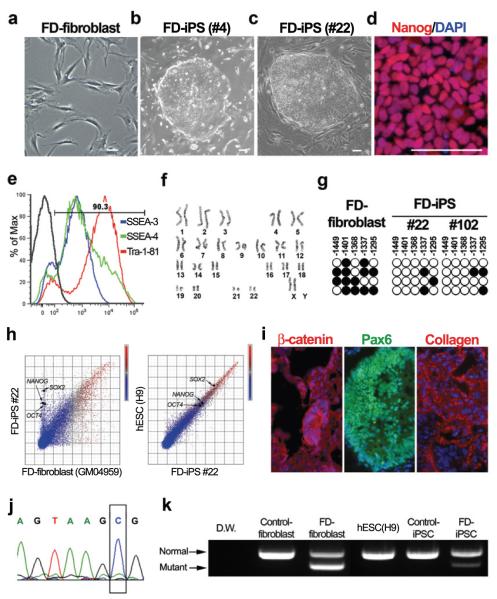
We obtained fibroblasts from a 10 year old female FD patient and established two independent clones of FD-iPSCs (FD#4, FD#22) following transduction with lentiviral vectors encoding for OCT4, SOX2, KLF4 and cMYC (Fig. 1a-c). Additional iPSCs clones were derived from two other FD patients (16 year old male and 12 year old female FD patient) and from non-affected control fibroblasts using the same procedure. The vector system used for iPSC generation allowed convenient monitoring of transgene expression by individual fluorescence tags via P2A technology13. All FD- and control iPSCs used in this study showed silencing of the four transgenes, exhibited characteristic human ES cell morphology, expressed pluripotency markers including Nanog, SSEA3, SSEA4, Tra-1-81 and had a normal karyotype (Fig. 1b-f; Suppl. Fig. 2-3). Pluripotent properties of FD-iPSCs were further assessed by monitoring methylation status of the Nanog promoter (Fig. 1g), by global transcriptome analysis in comparison to FD-fibroblasts, hESCs (line H9) and control-iPSC lines (Fig. 1h, Suppl. Fig. 3), and by teratoma formation upon intramuscular injection of undifferentiated FD-iPSCs into NOD-SCID mice (Fig. 1i). Genetic identity was confirmed by DNA fingerprinting analysis for microsatellite markers (Suppl. Table 1). The genetic defect was confirmed as a homozygous 2507+6T>C mutation in the IKBKAP gene (Fig. 1j) known to induce tissue specific skipping of exon 20. Gene expression analysis for IKBKAP revealed that FD-fibroblasts and FD-iPSCs express both the wild-type and mutant IKBKAP transcript, although the ratio of normal to mutant transcript was higher in FD-iPSCs compared with FD-fibroblasts (Fig. 1k).
Figure 1. Establishment of FD-iPSCs from patient fibroblasts.
(a-b) FD patient fibroblasts (a) were converted into FD-iPSCs (b, c) following lentiviral transduction with OCT4, SOX2, KLF4, and cMYC. d, Nanog protein expression in FD-iPSC line. e, Flow-cytometry analysis of FD-iPSCs for pluripotent surface markers. f, Karyotype analysis of FD-iPSCs. g, Bisulfite sequencing analysis of NANOG promoter in FD-fibroblast and FD-iPSC clones. h, Global gene expression patterns were compared among FD-fibroblast, FD-iPSCs and human ESCs. i, Teratoma from FD-iPSCs showed three germ-layer differentiation as illustrated by the presence of endodermal epithelia expressing β-catenin, Pax6+ neuroectodermal precursors and mesodermal collagen+ cells. j, Sequencing result showed the 2507+6T>C mutation of IKBKAP in FD-iPSC. k, Analysis of IKBKAP RT-PCR products in mRNA derived from normal and FD-specific fibroblasts and pluripotent stem cells. All scale bars correspond to 50μm.
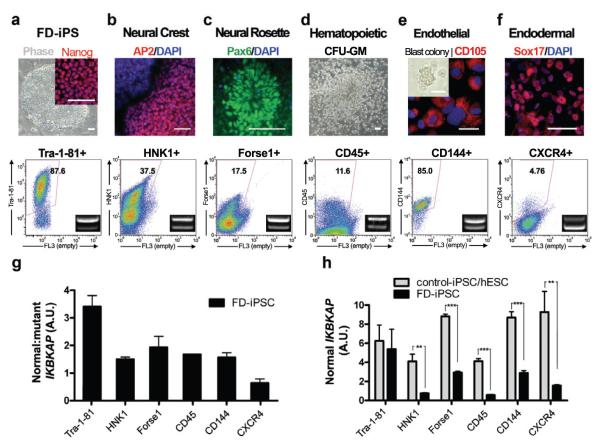
The pluripotent nature of iPSCs offers the opportunity for probing the tissue specificity of the IKBKAP splicing defect in vitro. FD- and control-iPSCs (2 clones each) were differentiated into five major tissues comprising all three embryonic germ layers. No reactivation of transgenes was observed in differentiated progeny (Suppl. Fig. 2). Following directed differentiation towards CNS14, PNS15, hematopoietic16, endothelial17, and endodermal18 precursors, iPSC derivatives were further enriched via prospective isolation using lineage specific surface markers (Fig. 2a-f). Early neural differentiation was characterized by the formation of neural rosette structures in both FD- and control-iPSCs (Suppl. Fig. 4). No major differences were observed in the efficiency of differentiation towards any of the five lineages comparing FD- versus control-iPSCs (data not shown). However, marked differences were observed for the ratio of normal:mutant transcript among the five FD-iPSCs derived tissues (insets, Fig. 2a-f). The mutant transcript predominated particularly in CXCR4+ purified endodermal precursors (Sox17+) and, to a lesser extent, in neural crest precursors (Fig. 2g). The disease symptoms in FD are thought to be caused by loss of sufficient levels of normal IKBKAP transcript. Therefore we next performed qRT-PCR analysis determining absolute levels of normal spliced IKBKAP transcript in each of the five FD- or control-iPSC derived cell populations (Fig. 2h). These data revealed particularly low levels of normal IKBKAP transcript in FD-iPSC derived neural crest precursors in vitro compatible with the tissue specific human pathology observed in FD patients. High levels of mutant IKBKAP expression in endodermal precursors are of interest in view of the debilitating gastrointestinal problems observed in FD; though such symptoms are thought to be a consequence of a primary defect in the peripheral nervous system.
Figure 2. FD-iPSC derived cell lineages model the tissue specificity of FD IKBKAP splicing defect.
a, Undifferentiated iPSCs were defined by NANOG expression and quantitative analysis of TRA1-81. b-f, FD-iPSCs were directed towards specific lineages and purified by flow-cytometric sorting for appropriate surface markers. The cell types analyzed included AP2+ neural crest precursors purified based on HNK1+ expression (b), FACS purified Forse1+ neural rosette cells (Pax6+) (c), hematopoietic cells that were harvested from colony forming unit culture and sorted with CD45 marker (d), FACS isolated CD144+ (VE-cadherin) endothelial cells (CD105+) from blast colony culture (e), FACS isolated CXCR4+ endodermal precursors (Sox17+) from ActivinA-induced differentiation culture (f). IKBKAP RT-PCR products of FACS purified lineages (bottom panel). The ratio of normal:mutant splicing (g) and expression of normal IKBKAP transcript (h) are shown. n = 3 – 6; **, P < 0.01; ***, P < 0.001. All values are mean ± s.d. All scale bars correspond to 50μm.
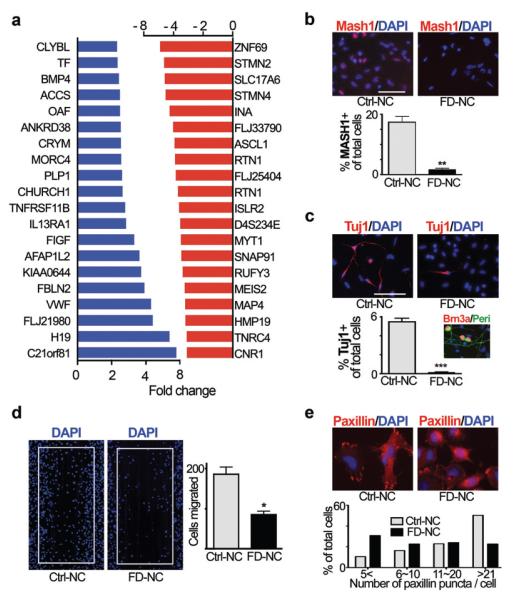
Directed differentiation of patient-specific iPSCs towards neural crest lineages, the primary tissue affected in FD, enables modeling functional aspects of pathogenesis in vitro. Neuropathological studies have demonstrated a dramatic depletion of autonomic and sensory neurons in FD patients. However, the cause for peripheral neuron loss in FD remains unclear, and there are currently no mouse models that faithfully replicate the disease. Mouse modeling in other neural crest disorders has identified a number of potential disease mechanisms such as impaired neural precursor cell migration in Hirschsprungs’ disease19 or degeneration of established neural crest tissues in Treacher Collins syndrome20. We explored the involvement of candidate disease mechanisms in FD by performing comparative transcriptome analysis of FACS purified FD-iPSC versus control-iPSC derived neural crest precursors (Fig. 3a, Suppl. Table 2; raw data of microarray data available at GEO http://www.ncbi.nlm.nih.gov/geo/ accession number: GSE17043. Among 48,000 probe sets, 35 transcripts were significantly increased and 54 transcripts decreased in FD-iPSC versus control iPSC or hESC derived neural crest precursors (fold-change ≥ 2 fold; corrected p-value of 0.05). Differential expression of key transcripts from the microarray study was confirmed by qPCR analysis for neural crest precursors of both FD-iPSC clones (Suppl. Fig. 5). Importantly, among the 20 most decreased transcripts in FD neural crest precursors were many genes involved in peripheral neurogenesis and neuronal differentiation. Examples include SLC17A6 (VGLUT2) expressed in sensory neurons, the neuronal intermediate filaments MAP4 and INA, and STMN2 (SCG-10; a known marker of autonomic neurons21). Of particular interest were the significant differences in ASCL1 of (MASH1) expression between FD- and control-iPSC given the essential role of Mash1 for the development of autonomic neurons during mouse development21. Differential expression of MASH1 was further confirmed at the protein level by quantifying the percentages of MASH1-immunoreactive cells (Fig. 3b). The marked differences in neurogenesis were also reflected by a reduction in the percentage of spontaneously differentiating Tuj1+ neurons in FD-iPSC derived neural crest precursor cultures (Fig. 3c). Peripheral neurogenesis was not completely blocked, as FD-iPSC derived Brn3A+/Peripherin+ neurons were observed upon further in vitro differentiation (inset of Fig. 3c). These data indicate a defect in the propensity of FD-iPSC derived neural crest precursors to undergo neuronal differentiation. Previous studies in IKBKAP depleted or patient-specific fibroblasts indicated that reduced levels of normal IKBKAP are associated with defects in cell migration9,22. Using a wound healing (“scratch”) assay we observed a significant decrease in the migration of FD-iPSC versus control-iPSC derived neural crest precursors (Fig. 3d). FD-iPSC derived neural crest precursors also showed a reduction in the number of Paxillin+ focal adhesions critical for cell spreading and migration (Fig. 3e).
Figure 3. Molecular and functional characterization of FD-iPSC derived neural crest precursor cells.
a, List of the unselected top increased (blue) and top decreased (red) genes comparing FD-iPSC versus [C14 and H9] derived neural crest cells as assessed by microarray analysis. b-c, Representative images and quantifications of MASH1 (b) and Tuj1 (c) expression in spontaneously differentiated neural crest cells derived from FD-iPSC and C14-iPSC. n = 4; **, P < 0.01; ***, P < 0.001. c, inset, Representative image of putative sensory neuron progeny (Brn3a+/Periperin+) from FD-iPSC derived neural crest cells. d-e, Representative images of wound healing assay (fixed at 48 hrs after scratching) and paxillin staining (fixed at 1.5 hrs after plating) of FD-iPSC and C14-iPSC derived neural crest cells. n = 4; *, p < 0.05. Quantification of paxillin staining was performed by counting the number of Paxillin puncta marking focal adhesion complex. All values are mean ± s. d. All scale bars correspond to 50μm.
Our study reveals defects in IKBKAP splicing, neurogenesis, and migration of FD-iPSC derived neural crest precursors. Results were validated in multiple independent FD-iPSC clones (see Suppl. Fig. 6) and compared to both non-affected control-iPSC and control hESC cultures. These data demonstrate that FD-iPSC derived neural crest precursors represent a powerful model system to probe disease pathogenesis, though larger sets of FD-iPSC lines may be useful to further validate disease phenotype.
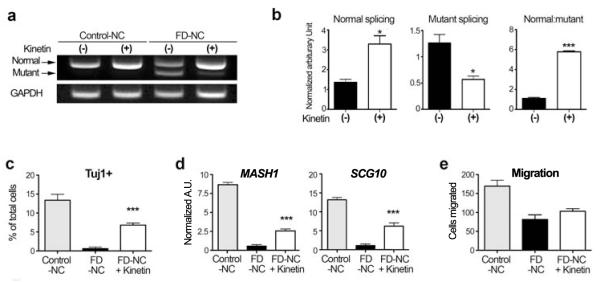
Another key promise of human iPSC research is the use of patient-specific cells in drug discovery. Our study has demonstrated three easily quantifiable parameters of the disease state: a defect in IKBKAP splicing, decreased rate of neurogenesis, and reduced migration in FD-iPSC derived neural crest precursors. We next tested whether exposure to candidate drugs can impact any of these three parameters. The plant hormone kinetin was previously shown to reduce levels of the mutant IKBKAP splice form in FD-derived lymphoblast cell lines12. Epigallocatechin Gallate (ECGC) and tocotrienol have also been reported to affect splicing and absolute levels of IKBKAP 23,24. Exposure of FD-iPSC derived neural crest precursors to kinetin resulted in a dramatic reduction of the mutant IKBKAP splice form (Fig. 4a). In contrast, no significant improvements in IKBKAP splicing were observed upon ECGC or tocotrienol exposure (Suppl. Fig. 7). The kinetin-mediated decrease in mutant IKBKAP was associated with an increase in normal IKBKAP levels and the ratio of normal:mutant transcript (Fig. 4b). No significant increase in normal IKBKAP transcript levels were observed upon kinetin treatment of neural crest precursors derived from control iPSCs (Suppl. Fig. 8). While short-term (1 day or 5 day) kinetin treatment of FD-iPSC derived neural crest precursors had dramatic effects on IKBKAP splicing, it did not result in a significant increase in the expression of neurogenic markers or improve migration behavior. (Suppl. Fig. 9 & Suppl. Fig. 10). We next tested the impact of continuous (28 day) kinetin treatment of FD-iPSCs starting at the pluripotent stage 1 day prior to differentiation (Suppl. Fig. 11). Importantly, continuous kinetin treatment induced a significant increase in the percentage of differentiating neurons and in the expression of key peripheral neuron markers such as MASH1 and SCG-10 at the neural crest precursor stage (Fig. 4c,d). No significant increase was observed in FD-iPSC neural crest precursor cell migration (Fig. 4e) suggesting incomplete restoration of disease phenotype. These data demonstrate the feasibility of exploring disease mechanism and action of candidate drugs in FD-iPSCs. While future studies will be required to define the developmental windows of kinetin action in greater detail, our data indicate that early stage long-term treatment may be particularly beneficial in FD patients.
Figure 4. Validating kinetin as a candidate compound for treating FD-iPSC derived neural crest cells.
a, Gel image of RT-PCR products for IKBKAP splicing rescued by kinetin treatment in control and FD-iPSC derived neural crest. b, Quantification of band intensity of FD-iPSC derived neural crest cells normalized by GAPDH and the ratio of normal and mutant spliced IKBKAP transcripts. n = 5; *, p < 0.05; ***, P < 0.001. c-d, MASH1 (c) and Tuj1 (d) expression in neuronal differentiation with kinetin treated neural crest cells derived from FD-iPSC . n = 4; ***, P < 0.001. (E-F) Results of wound healing assay (e) and paxillin staining (f) of kinetin treated FD-iPSC derived neural crest cells. n = 4 - 6; ***, P < 0.001. All values are mean ± s. d.
Since the initial reports on human iPSC isolation1-3, a number of patient-specific iPSC lines have been developed4-7. Modeling human disease in iPSCs has remained challenging due to difficulties in directing iPSC fate and identifying disease related behaviors. Previous reports have demonstrated the derivation of patient specific motoneurons and dopamine neurons as an initial step in modeling ALS, SMA and Parkinson’s disease respectively4,5,7. While the study using SMA-iPSCs reported a potentially disease-related loss of cholinergic neurons5, the other studies did not observe7 or not report4 any disease related phenotypes. Interestingly, both SMA and FD are early onset disorders suggesting that iPSC technology may be particularly suited to model developmental disorders. Here we extend the approach to modeling disease in the neural crest lineage and provide clear evidence of disease related behaviors in FD-iPSCs. While FD is a rare genetic disorder with an incidence of 1/3600 live birth among the Ashkenazi Jewish population (~ 600 registered patients world-wide25), it provides a platform to demonstrate the potential of iPSC technology for exploring disease mechanism in a human genetic disorder. Studying aberrant tissue specific IKBKAP splicing in iPSCs will be particularly attractive given the pluripotent nature and unlimited number of cells available for mechanistic studies and for high-throughput drug discovery. Our data on defects in neurogenic propensity and migration of FD-iPSC derived neural crest precursors provide novel insights into disease pathogenesis and suggest alternative functional assays for the identification and validation of candidate drugs. Finally, kinetin treatment illustrates the potential of iPSC technology for modeling therapeutic action in human neural disease in vitro using patient specific cells.
Methods Summary
iPSC generation and differentiation
FD- and control fibroblasts were obtained from Coriell and reprogrammed towards iPSCs using a lentiviral four factor protocol (Oct4, Sox2, Klf4 and Myc) described recently13. Undifferentiated hESCs and iPSC lines were maintained on mitotically inactivated MEFs in serum-replacement medium in the presence of 4 ng/ml FGF215. For neural induction, hESCs were plated on a confluent layer of irradiated stromal cells (MS-5) as described previously15,26. After 3-4 weeks of differentiation hESC progeny was triturated into single cells following Accutase dissociation (Innovative cell technologies) and labeled with antibodies for flow cytometry. Protocols for directed differentiation into all the other individual lineages are described in the Full Methods section.
Karyotype analysis and Teratoma formation
Standard G-banding analysis was performed by MSKCC molecular cytogenetics core laboratory as described previously27. For teratoma formation FD-iPSCs were injected into the hindlimb muscle of NOD-SCID IL2Rgc null mice (Jackson Laboratory) and analyzed histologically 4-6 weeks after injection. Animal studies were approved by MSKCC Institutional Animal Care and Use Committee.
Molecular analyses
Total RNA was extracted using the RNeasy kit and DNAse I treatment (Qiagen) and reverse transcribed (Superscript, Invitrogen). Primer sequences, cycle numbers, and annealing temperatures (Suppl. Table 3) for IKBKAP correspond to those used by others12 and quantification of PCR bands was performed using ImageJ software (NIH) and ratio of normal:mutant transcript was determined. Quantitaive RT-PCR analysis was performed as described previously28. Gene expression microarray analysis was carried out by the SKI genomics core laboratory using the human Illumina-12 and Illumina-6 platforms. Bisulfite sequencing analysis of Nanog promoter was performed according to3.
Immunocytochemistry
Cells were fixed in 4% paraformaldehyde, permeabilized with Triton-X100 containing buffer and stained with primary antibodies (list of antibodies provided in the Full Methods section). Appropriate fluorescent labeled secondary antibodies (Molecular Probes) and DAPI counter-staining was used for visualization.
Supplementary Material
ACKNOWLEDGEMENTS
We thank J. Hendrikx, M. Leversha, and C. Zhao for technical help. The work was supported by grants from the Starr Foundation and NYSTEM, by the New York Stem Cell Foundation (NYCSF, Druckenmiller fellowships to G.L. and C.A.F.) and by the Starr Scholar fellowship to S.M.C.
REFERENCES
- 1.Yu J, et al. Induced Pluripotent Stem Cell Lines Derived from Human Somatic Cells. Science. 2007 doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, et al. Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell. 2007 doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 3.Park IH, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 4.Dimos JT, et al. Induced Pluripotent Stem Cells Generated from Patients with ALS Can Be Differentiated into Motor Neurons. Science. 2008 doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- 5.Ebert AD, et al. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature. 2009;457:277–280. doi: 10.1038/nature07677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park IH, et al. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soldner F, et al. Parkinson’s disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136:964–977. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Slaugenhaupt SA, et al. Tissue-specific expression of a splicing mutation in the IKBKAP gene causes familial dysautonomia. Am. J. Hum. Genet. 2001;68:598–605. doi: 10.1086/318810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Close P, et al. Transcription impairment and cell migration defects in elongator-depleted cells: implication for familial dysautonomia. Mol. Cell. 2006;22:521–531. doi: 10.1016/j.molcel.2006.04.017. [DOI] [PubMed] [Google Scholar]
- 10.Axelrod FB, Goldberg JD, Ye XY, Maayan C. Survival in familial dysautonomia: Impact of early intervention. J. Pediatr. 2002;141:518–523. doi: 10.1067/mpd.2002.127088. [DOI] [PubMed] [Google Scholar]
- 11.Anderson SL, et al. Familial dysautonomia is caused by mutations of the IKAP gene. Am. J. Hum. Genet. 2001;68:753–758. doi: 10.1086/318808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Slaugenhaupt SA, et al. Rescue of a human mRNA splicing defect by the plant cytokinin kinetin. Hum. Mol. Genet. 2004;13:429–436. doi: 10.1093/hmg/ddh046. [DOI] [PubMed] [Google Scholar]
- 13.Papapetrou EP, et al. Stoichiometric and temporal requirements of Oct4, Sox2, Klf4 and cMyc expression for efficient human iPSC induction and differentiation. Proc Natl Acad Sci U S A. 2009 Jun 23; doi: 10.1073/pnas.0904825106. [ahead of publication] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elkabetz Y, et al. Human ES cell-derived neural rosettes reveal a functionally dinstinct early neural stem cell stage. Genes Dev. 2008;22:152–165. doi: 10.1101/gad.1616208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee G, et al. Isolation and directed differentiation of neural crest stem cells derived from human embryonic stem cells. Nat. Biotechnol. 2007;25:1468–1475. doi: 10.1038/nbt1365. [DOI] [PubMed] [Google Scholar]
- 16.Lee GS, Kim BS, Sheih JH, Moore M. Forced expression of HoxB4 enhances hematopoietic differentiation by human embryonic stem cells. Mol Cells. 2008;25:487–493. [PubMed] [Google Scholar]
- 17.Lu SJ, et al. Generation of functional hemangioblasts from human embryonic stem cells. Nat. Methods. 2007;4:501–509. doi: 10.1038/nmeth1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.D’Amour KA, et al. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat. Biotechnol. 2005;23:1534–1541. doi: 10.1038/nbt1163. [DOI] [PubMed] [Google Scholar]
- 19.Iwashita T, Kruger GM, Pardal R, Kiel MJ, Morrison SJ. Hirschsprung disease is linked to defects in neural crest stem cell function. Science. 2003;301:972–976. doi: 10.1126/science.1085649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones NC, et al. Prevention of the neurocristopathy Treacher Collins syndrome through inhibition of p53 function. Nature Med. 2008;14:125–133. doi: 10.1038/nm1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sommer L, Shah N, Rao M, Anderson DJ. The cellular function of MASH1 in autonomic neurogenesis. Neuron. 1995;15:1245–1258. doi: 10.1016/0896-6273(95)90005-5. [DOI] [PubMed] [Google Scholar]
- 22.Johansen LD, et al. IKAP localizes to membrane ruffles with filamin A and regulates actin cytoskeleton organization and cell migration. J. Cell Sci. 2008;121:854–864. doi: 10.1242/jcs.013722. [DOI] [PubMed] [Google Scholar]
- 23.Anderson SL, Qiu J, Rubin BY. EGCG corrects aberrant splicing of IKAP mRNA in cells from patients with familial dysautonomia. Biochem. Biophys. Res. Commun. 2003;310:627–633. doi: 10.1016/j.bbrc.2003.09.019. [DOI] [PubMed] [Google Scholar]
- 24.Anderson SL, Qiu J, Rubin BY. Tocotrienols induce IKBKAP expression: a possible therapy for familial dysautonomia. Biochem. Biophys. Res. Commun. 2003;306:303–309. doi: 10.1016/s0006-291x(03)00971-9. [DOI] [PubMed] [Google Scholar]
- 25.Axelrod FB. Familial dysautonomia: a review of the current pharmacological treatments. Expert. Opin. Pharmacother. 2005;6:561–567. doi: 10.1517/14656566.6.4.561. [DOI] [PubMed] [Google Scholar]
- 26.Perrier AL, et al. From the Cover: Derivation of midbrain dopamine neurons from human embryonic stem cells. Proc Natl Acad Sci U S A. 2004;101:12543–8. doi: 10.1073/pnas.0404700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Placantonakis D, et al. BAC transgenesis in human ES cells as a novel tool to define the human neural lineage. Stem Cells. 2009;27:521–532. doi: 10.1634/stemcells.2008-0884. [DOI] [PubMed] [Google Scholar]
- 28.Chambers SM, et al. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kennedy M, D’Souza SL, Lynch-Kattman M, Schwantz S, Keller G. Development of the hemangioblast defines the onset of hematopoiesis in human ES cell differentiation cultures. Blood. 2007;109:2679–2687. doi: 10.1182/blood-2006-09-047704. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.