Abstract
Four major glutamate receptor 2 (GluR2) transcripts differing in size (∼4 and ∼6 kilobases) due to alternative 3′ untranslated regions (UTRs), and also containing alternative 5′UTRs, exist in the brain. Both the long 5′UTR and long 3′UTR repress translation of GluR2 mRNA; repression by the 3′UTR is relieved after seizures. To understand the mechanism of translational repression, we used rabbit reticulocyte lysates as an in vitro translation system to examine the expression profiles of firefly reporter mRNAs bearing alternative combinations of GluR2 5′UTR and 3′UTR in the presence of inhibitors of either translational elongation or initiation. Translation of reporter mRNAs bearing the long GluR2 3′UTR was insensitive to low concentrations of the translation elongation inhibitors cycloheximide (0.7–70 nM) and anisomycin (7.5–750 nM), in contrast to a reporter bearing the short 3′UTR, which was inhibited. These data suggest that the rate-limiting step for translation of GluR2 mRNA bearing the long 3′UTR is not elongation. Regardless of the GluR2 UTR length, translation of all reporter mRNAs was equally sensitive to desmethyl-desamino-pateamine A (0.2–200 nM), an initiation inhibitor. Kasugamycin, which can facilitate recognition of certain mRNAs by ribosomes leading to alternative initiation, had no effect on translation of a capped reporter bearing both short 5′UTR and short 3′UTR, but increased the translation rate of reporters bearing either the long GluR2 5′UTR or long 3′UTR. Our findings suggest that both the long 5′UTR and long 3′UTR of GluR2 mRNA repress translation at the initiation step.
AMPA receptors, a subfamily of ionotropic glutamate receptors mediating the majority of fast excitatory neurotransmission in the central nervous system (Dingledine et al., 1999), are ligand-gated ion channels formed by heteromeric or homomeric combinations of GluR1, GluR2, GluR3, and GluR4 subunits (Köhler et al., 1994; Rosenmund et al., 1998). Although most well known examples of gene regulation involve transcription, the efficiency of mRNA translation can be regulated in a transcript-specific manner by structural motifs residing in the 5′- or 3′-UTR, alternate or additional 5′-UTR AUG codons (or their cognate short open reading frames), RNA binding proteins, or the nucleotide context of the initiator AUG (Gray and Wickens, 1998). Control of translation and trafficking of GluR2 subunits is involved in long-term synaptic potentiation (Isaac et al., 2007; Gainey et al., 2009). Multiple GluR2 transcripts exist in hippocampus that have alternative 5′- and 3′-untranslated regions (UTRs) (Köhler et al., 1994; Myers et al., 1998; Irier et al., 2009). GluR2 mRNAs with at least two different 3′UTRs, long (∼2750 bases) and short (∼750 bases), exist in the brain (Köhler et al., 1994) but encode the same protein. GluR2 transcription is regulated by positive and negative regulatory elements in the promoter region(Myers et al., 1998; Huang et al., 1999). Translation is repressed by GU repeats located in the long 5′-UTR, and also by unknown elements in the long 3′UTR (Myers et al., 2004; Irier et al., 2009). Seizures reduce overall GluR2 mRNAs level but derepress the translation of GluR2 mRNAs bearing the long 3′UTRs in rat hippocampus (Irier et al., 2009). These findings suggest that GluR2 mRNAs with alternative combinations of GluR2 5′- and 3′-UTRs are subject to transcript-specific regulation of translation, but the regulated step of translation is unknown.
In general, eukaryotic mRNA translation occurs in four consecutive phases: initiation, elongation, termination, and ribosome recycling. In the initiation phase, a 43S preinitiation complex is formed by interaction of 40S ribosomal subunits with initiator methionyl transfer RNA (tRNA). This preinitiation ribosomal complex then binds to mRNA at the 5′ cap structure (methylated GTP, m7GpppN) in a process facilitated by translation factor complex eIF4, and begins to scan through the 5′UTR, unwinding the secondary structure until it encounters the first eligible AUG codon, which is then oriented onto the P (peptidyl) site of the scanning ribosome. Once this start signal is recognized, the larger 60S subunit joins the 40S to form an 80S initiation complex that is ready to accept appropriate aminoacyl-tRNA into the A (aminoacyl) site on the ribosome, thus starting the elongation phase of translation (Kozak, 1989; Pestova et al., 2001; Sonenberg and Hinnebusch, 2009) .
One approach to determining whether initiation or elongation is the rate-limiting step in protein expression is to treat cells with low concentrations of modulators of these processes (Lodish and Jacobson, 1972; Walden et al., 1981; Chen and Sarnow, 1995). If elongation is the rate-limiting step, then translation rate should be sensitive to a low concentration of elongation inhibitors. On the contrary, if translation of an mRNA is insensitive to low concentrations of translation elongation inhibitors or is sensitive to translation initiation modulators, then the rate-limiting step for such mRNAs is probably initiation. Taking this approach with rabbit reticulocyte lysates as an in vitro translation system, we report that translational repression caused by the long 5′UTR and, surprisingly, also the long 3′UTR is mediated at the initiation step.
Materials and Methods
GluR2 5′ and 3′UTR Constructs and in Vitro Reporter mRNA Synthesis.
The firefly luciferase reporter mRNAs bearing alternative GluR2 5′- and 3′-UTRs were constructed using methods described previously (Irier et al., 2009). In brief, these constructs contain a firefly luciferase coding region flanked by alternative combinations of GluR2 5′- and 3′-UTRs (see Fig. 1A; for Luciferase protein coding region, designated SS if flanked by short 5′UTR and short 3′UTR of GluR2, SL if flanked by short 5′UTR and long 3′UTR of GluR2, or LS if flanked by long 5′UTR and short 3′UTR of GluR2). These constructs were linearized at the 3′-end using the BstEII restriction site. In vitro synthesis of 5′-capped mRNAs from the linearized reporter constructs was performed using T3 RNA polymerase following the instructions provided in the T3 mMESSAGE mMACHINE (Ambion, Austin, TX). The resulting mRNAs were quantified and quality-checked in the RNA Nano Chip apparatus using a model 2100 Bioanalyzer (Agilent, Waldbronn, Germany). Reporter mRNAs were stored in RNase-free water as small aliquots of 100 μM stocks at −80°C. As an additional control for obtaining luciferase reporter mRNAs with full-length GluR2 long 3′UTRs after the in vitro synthesis of reporter mRNAs, the reporter mRNAs bearing long GluR2 3′UTRs (SL) were reverse-transcribed to cDNAs using Thermoscript reverse transcriptase (Invitrogen, Carlsbad, CA) and amplified using primer pairs specific to short (3022 forward and 3119 reverse), long proximal (3891 forward and 4015 reverse), and long distal (4403 forward and 4597 reverse) regions of GluR2 3′UTR. The number of PCR cycles required to reach threshold were compared for the three primer pairs.
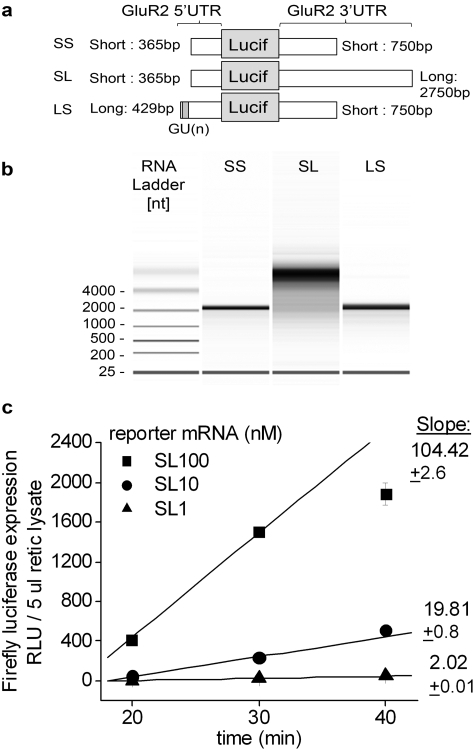
Fig. 1.
In vitro transcription of luciferase reporters bearing alternative combinations of GluR2 UTRs. A, schematic illustration of firefly luciferase reporter mRNAs bearing alternative combinations of GluR2, 5′- and 3′UTRs. The luciferase coding region is common to all reporters. The dark gray box indicates the position of GU repeats on the long 5′UTR (Myers et al., 2004). B, the quality and amount of each in vivo-transcribed firefly reporter were evaluated with an Agilent 2100 Bioanalyzer, which produces a gel-like image. The expected sizes of the transcripts is SS, 2760; SL, 4710; and LS, 3140 base pairs. C, the activity of luciferase expressed from the firefly reporter mRNAs bearing the short 5′- and long 3′-UTRs of GluR2 (SL) was proportional to both mRNA amount and incubation time in rabbit reticulocyte lysate translation mix.
In Vitro Translation of Reporters in Rabbit Reticulocyte Lysates.
Commercially available rabbit reticulocyte lysates were used according to the instructions provided with Retic Lysate IVT (Applied Biosystems, Foster City, CA). In brief, a 50-μl final translation assay mix was assembled in a 0.5-μl thin-walled PCR reaction tube on ice, and the components were added in the following order: drug or (vehicle), 34 μl of rabbit reticulocyte lysate, 20× Translation Mix(-met), 1 mM methionine, 0.8 U of RNase inhibitor, reporter mRNA, and RNase-free water. The translation mix was incubated at 30°C for up to 40 min using a digitally monitored heat block. A 5-μl sample from the final translation mix was removed at 10-min intervals and flash frozen in a 96-well plate (Microlite white flat-bottomed plates; Thomas Scientific, Swedesboro, NJ) that was placed on dry ice. The luciferase activity, defined as recorded luminescence units (RLU) measured at 570 nm, was measured from the individual wells using the luciferase assay substrate (Dual-Luciferase Reporter Assay System; Promega, Madison, WI). The RLU from the 5-μl reticulocyte samples without reporter mRNAs (0.05–0.5 RLU) constituted background, which was consistent for each 96-well plate used. The lowest experimental sample RLUs at 20 min were at least 100-fold higher than background. To determine reporter mRNA stability, each firefly reporter mRNA was extracted from the rabbit reticulocyte lysates at time 0 and 40 min using a standard phenol-chlorophorm method. The extracted mRNA was dissolved in a 20-μl in vitro reverse transcription reaction that contained Thermoscript reverse transcriptase (Invitrogen), and the reaction was run for 50 min at 50°C. The resulting cDNAs were quantified by quantitative real-time PCR analysis (SyberGreen Supermix; Applied Biosystems) using primer sets that amplified UTR-specific regions along with standard dilutions of the luciferase reporter cDNA.
Drugs Used.
DMDA-PatA was kindly made available to us by Dr. Daniel Romo (Department of Chemistry, Texas A&M University, College Station, TX). Cycloheximide and anisomycin were purchased from Sigma-Aldrich (St. Louis, MO). Kasugamycin was purchased from BIOMOL Research Laboratories (Plymouth Meeting, PA). DMDA-PatA and anisomycin were dissolved in dimethyl sulfoxide.
Statistics.
For the statistical analyses of the data sets for each drug treatment shown in Fig. 2, the one-way analysis of variance was used with the post test Bonferroni multiple comparison test.
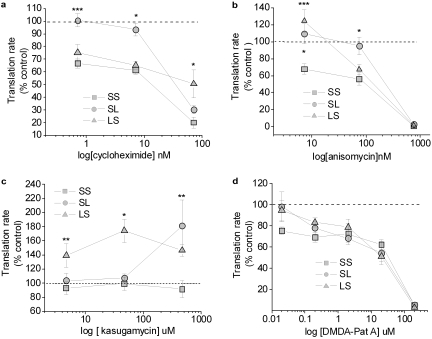
Fig. 2.
Expression profiles of the 5′ capped firefly reporter mRNAs bearing alternative combinations of GluR2 5′ and 3′UTRs in rabbit reticulocyte lysate. At indicated concentrations, effects of translation elongation inhibitors cycloheximide (A) and anisomycin D (B), and translation initiation modulators kasugamycin (C) and DMDA-pateamine A (D) on the in vitro translation rates of the firefly reporters were determined. The translation rates of individual firefly reporters were determined by measuring firefly luciferase activities as recorded luminescence units (RLU) from a 5-μl reticulocyte lysate sample collected at the indicated time points. Slopes of RLU from the 5-μl samples collected between 20 and 40 min were calculated for the drug-treated samples and normalized to that of vehicle-treated (water for cycloheximide and kasugamycin; dimethyl sulfoxide for anisomycin D and DMDA-pateamine A) samples and presented as percentage of control. Translation rates of the controls were set to 100% (dotted line). ∗, P < 0.01 for SS versus SL at 7 nM cycloheximide, 7.5 and 75 nM anisomycin D; for SS versus LS at 75 nM anisomycin D; for SS versus LS at 7.5 nM anisomycin D; and for SS versus LS at 46 μM kasugamycin. ∗∗, P < 0.05 for SS versus SL at 460 μM kasugamycin and for SS versus LS at 4.6 to 460 μM kasugamycin. ∗∗∗, P < 0.001 for SS versus SL at 0.7 nM cycloheximide and for SS versus LS at 7.5 nM anisomycin D (analysis of variance, post hoc Bonferroni). n = 5–6 per reporters per drug treatment; error bars, standard error.
Results
Effects of Elongation Inhibitors on Translation of GluR2 Reporter mRNAs.
To study the mechanisms of transcript-specific regulation of GluR2 translation, we employed a set of reporter constructs previously designed in our laboratory (Irier et al., 2009) (Fig. 1A). In brief, these constructs contain a firefly luciferase coding region flanked by alternative combination of GluR2 5′- and 3′-UTRs. In vitro synthesis of mRNAs from linearized reporter constructs yielded good quality mRNAs as determined by the Agilent 2100 bioanalyzer (Fig. 1B). As an additional quality measure, the in vitro synthesized reporter mRNAs were reverse-transcribed to cDNAs and amplified by Q-RT-PCR analysis using primers specific to short and long 3′UTR of GluR2. These experiments demonstrated that the reporter mRNAs bearing long 3′UTRs of GluR2 are full-length (Supplemental Fig. 1A). The rate of firefly luciferase expression by rabbit reticulocyte lysate was proportional to the amount of reporter mRNA added (Fig. 1C and Supplemental Fig. 1, B and C). More than half of the reporter mRNA added was recovered after 40-min incubation at 30°C (Supplemental Fig. 1D).
Using rabbit reticulocyte lysates as in vitro translation system, we examined the expression of firefly reporter mRNAs bearing alternative combinations of GluR2 5′- and 3′-UTRs (SS, SL, and LS) in the presence or absence of translation elongation inhibitors cycloheximide (0.7–70 nM) or anisomysin D (7.5–75 nM). At low concentrations, cycloheximide has been shown to inhibit the elongation of nascent peptide by blocking the translocation of peptidyl-tRNA on the translating (80S) ribosomes (Baliga et al., 1968; Munro et al., 1968; Lodish, 1971). In rabbit reticulocyte lysates, the translation of reporter mRNAs bearing a short 5′- and a short 3′-UTR (SS) was inhibited by low concentrations of cycloheximide (0.7–7 nM), whereas translation of the reporter bearing GluR2 short 5′ and long 3′UTR (SL) was insensitive (Fig. 2A). Moreover, translation of reporters bearing a long 5′- and a short 3′-UTR (LS) was also inhibited by low concentrations of cycloheximide. At higher concentrations of cycloheximide (≥70 nM), expression of all three reporter mRNAs was equally inhibited (data not shown). Likewise, anisomycin also inhibits the elongation phase of translation by blocking a peptidyl transferase, a protein that catalyzes translocation of peptidyl-tRNA on translating ribosomes (Grollman, 1967). In the translation mix treated with a relatively high concentration of anisomycin (750 nM), the translation rate of luciferase reporters bearing all three combinations of GluR2 UTRs were equally inhibited; however, the lower concentrations of anisomycin (7.5–75 nM) significantly repressed the expression of SS, but not SL, reporter (Fig. 2B), providing further evidence that translation elongation is not the rate-limiting step for the expression of the SL reporter mRNA bearing a long 3′UTR of GluR2. The LS reporter exhibited intermediate sensitivity to anisomycin (Fig. 2B).
Effects of Initiation Modulators on Translation of GluR2 Reporters.
We first tested the effects of kasugamycin, an antifungal aminoglycoside antibiotic that inhibits translation initiation in prokaryotes by binding to a specific region between the 30S ribosome and 16S rRNA within the 70S ribosome, thereby impeding the binding of initiator tRNA (fMet-tRNA) to the ribosome (Schluenzen et al., 2006; Schuwirth et al., 2006). In rabbit reticulocyte lysates, none of the luciferase reporter mRNAs bearing alternative combinations of GluR2 5′- and 3-′UTR was inhibited by kasugamycin at the concentrations tested; on the contrary, and to our surprise, the expression of the SL and LS reporters were selectively potentiated by kasugamycin (Fig. 2C), with no effect on the SS reporter over the concentration range examined. Kasugamycin has been shown to accelerate the translation of some prokaryotic mRNAs by a mechanism involving creation of a novel ribosomal initiation complex (Kaberdina et al., 2009); a similar mechanism could operate in eukaryotes.
We then studied the effects of DMDA-PatA, a simplified structural analog of marine natural product pateamine A (Romo et al., 2004). DMDA-PatA is thought to inhibit translation initiation of 5′-capped mRNAs by disrupting the interaction of the eIF4F protein complex with eIF4A (Low et al., 2005; Bordeleau et al., 2006), both of which are essential protein factors during ribosome scanning at the 5′UTR of capped mRNAs leading to a final 80S ribosome assembly at the start codon (Pestova et al., 2001; Kapp and Lorsch, 2004). In rabbit reticulocyte lysates, the translation of reporters bearing all three combinations of GluR2 UTRs was approximately equally sensitive to inhibition by DMDA-pateamine A (Fig. 2D), suggesting that initiation influences translation rate of these luciferase reporters bearing alternative combinations of GluR2 5′- and 3′-UTRs.
Discussion
The major finding of this study is that alternative GluR2 3′UTRs determine the sensitivity to inhibition of translation by two well characterized elongation inhibitors, cycloheximide and anisomycin D. The long 3′UTR of GluR2 imparts relative insensitivity to these elongation inhibitors compared with the short 3′UTR, which strongly suggests that elongation is not the rate-limiting step for translation of GluR2 bearing the long 3′UTR. Furthermore, kasugamycin, which can promote formation of an alternative 61S ribosomal complex that improves initiation of translationally restricted mRNAs (Kaberdina et al., 2009), increased the translation of reporters bearing either a long 5′ or long 3′UTR. Translation rate of the reporter mRNAs bearing both short 5′UTR and short 3′UTR were not altered by the kasugamycin treatment. These results demonstrated for the first time that kasugamycin could potentiate the expression of translationally restricted GluR2 mRNAs. Results from translation inhibitors studies taken together suggest that initiation is a rate-limiting step for translation of GluR2 mRNAs bearing the long 3′UTR. Thus, the site of action for translational repression of GluR2 transcripts bearing the long 3-UTR (Irier et al., 2009) in vivo, and its derepression after seizures, is likely to involve the initiation phase of translation. The finding that the initiation inhibitor DMDA-pateamine A reduces the translation rate of reporters bearing the short 3′UTR of GluR2 suggests that the rates of elongation and initiation of this transcript are likely to be approximately the same in the reticulocyte lysate system. DMDA-Pateamine A disrupts the eIF4F complex by impairing the interaction between eIF4A and eIF4G, which is required in the process of 5′UTR scanning by the preinitiation 48S complex (Kaberdina et al., 2009). Moreover, regardless of the 5′UTR length or structure, some mRNAs are susceptible to inhibition by eIF4A mutants (Svitkin et al., 2001). The observation that DMDA-Pateamine A inhibits indiscriminately all the reporters bearing GluR2 UTRs may be due to a direct effect on the 5′UTR scanning process, which is common among all GluR2 transcripts during translation initiation. Alternatively, as mentioned above, the rates of elongation and initiation of the transcript bearing short 5′UTR and short 3′UTR may be approximately the same, such that this transcript would be sensitive to inhibitors of both elongation and initiation.
Transcript-Specific Alternative Initiation of GluR2 mRNA Translation by Kasugamycin.
A recent report suggests that kasugamycin can facilitate translation of leaderless mRNAs (mRNAs starting with a 5′-AUG codon only) by forming 61S ribosomal particles that function as an alternative initiation complex in prokaryotes (Kaberdina et al., 2009). Our observation that kasugamycin selectively increases the expression of reporter mRNAs bearing either of the long UTRs of GluR2 raises the possibility that it favors mRNAs bearing conformationally structured UTRs. To our knowledge, this is the first demonstration that kasugamycin selectively potentiates the translation of eukaryotic mRNAs. The mechanism of induction in eukaryotes is currently unknown; however, the subunits comprising 61S ribosomal particles are evolutionarily conserved among prokaryotes and eukaryotes (Wilson and Nierhaus, 2005; Kaberdina et al., 2009). Whether 61S-like ribosomal particles are formed in eukaryotes in the presence of this aminoglycoside and constitute alternative translational machinery would be interesting to determine. Nevertheless, the effects of kasugamycin present further evidence that GluR2 mRNAs can have distinct translation patterns as a result of alternative 5′- and 3′UTRs. However, we note that the effects of these inhibitors and the conclusions drawn from these experiments are limited to in vitro rabbit reticulocyte lysate translation systems, and the translation regulation of endogenous GluR2 mRNAs by these inhibitors should be explored in vivo.
In eukaryotes, ribosomes bind most transcripts at the 5′-cap and scan along the 5′UTR until an appropriate AUG codon is encountered (Kozak, 1989). However, alternative modes of translational initiation have been described previously (Chen and Sarnow, 1995; Jackson, 2005; Sonenberg and Hinnebusch, 2009). For example, initiation of fibroblast growth factor-2 mRNA occurs at an internal ribosomal entry site (IRES) of 5′UTR in neurons of specific brain regions (Vagner et al., 1995; Audigier et al., 2008). Secondary structures within the IRES and IRES-transactivating proteins are thought to be involved in initiation (Sonenberg and Hinnebusch, 2009). GluR2 long 5′UTRs (≥429 bases from the start codon) exhibit a high GC content and an imperfect GU repeat region that mediates translational repression. The GU repeat in the GluR2 long 5′UTRs is predicted to form stable secondary structures that stall the ribosome scanning process, which suppresses translation of GluR2 in vitro in rabbit reticulocyte lysates and in vivo in Xenopus oocytes and cultured primary neurons (Myers et al., 2004). Understanding the mechanisms underlying the selective effects of kasugamycin on translationally restricted GluR2 mRNAs may present a potential target for therapeutic interventions.
Approximately half of the GluR2 mRNAs in rat hippocampus bear a long 3′UTR (Irier et al., 2009), and approximately 75% bear the short 5′UTR (H. A. Irier, unpublished observations), suggesting that a considerable fraction of native GluR2 transcripts are translationally regulated by the long 3′UTR alone. Owing to its essential role in AMPA receptor function, the GluR2 subunit has been of considerable interest. Our results present additional evidence that in addition to transcriptional regulation, expression of the GluR2 subunit is highly regulated at the level of mRNA translation by the presence of alternative UTRs. Understanding how the physiological properties of glutamate receptor channels are regulated at the translational level should be relevant to the late phase of long-term potentiation, learning, and the response to seizures, as well as other situations in which AMPA receptor phenotype is remodeled.
Supplementary Material
Acknowledgments
We thank Dr. Daniel Romo for making DMDA-Pateamine A available to us.
 The online version of this article (available at http://molpharm.aspetjournals.org) contains supplemental material.
The online version of this article (available at http://molpharm.aspetjournals.org) contains supplemental material.
This work was supported by the National Institutes of Health National Institute of Neurological Disorders and Stroke [Grant R01-NS036604].
Article, publication date, and citation information can be found at http://molpharm.aspetjournals.org.
doi:10.1124/mol.109.060343
- AMPA
- α-amino-3-hydroxy-5-methylisoxazole-4-propionate
- GluR
- glutamate receptor
- UTR
- untranslated region
- eIF
- eukaryotic initiation factor
- tRNA
- transfer RNA
- SS
- flanked by short 5′UTR and short 3′UTR of GluR2
- SL
- flanked by short 5′UTR and long 3′UTR of GluR2
- LS
- flanked by long 5′UTR and short 3′UTR of GluR2
- PCR
- polymerase chain reaction
- RLU
- relative light units
- DMDA-PatA
- desmethyl-desamino-pateamine A
- IRES
- internal ribosomal entry site.
References
- Audigier S, Guiramand J, Prado-Lourenco L, Conte C, Gonzalez-Herrera IG, Cohen-Solal C, Récasens M, Prats AC. ( 2008) Potent activation of FGF-2 IRES-dependent mechanism of translation during brain development. RNA 14: 1852– 1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baliga BS, Pronczuk AW, Munro HN. ( 1968) Regulation of polysome aggregation in a cell-free system through amino acid supply. J Mol Biol 34: 199– 218 [DOI] [PubMed] [Google Scholar]
- Bordeleau ME, Cencic R, Lindqvist L, Oberer M, Northcote P, Wagner G, Pelletier J. ( 2006) RNA-mediated sequestration of the RNA helicase eIF4A by Pateamine A inhibits translation initiation. Chem Biol 13: 1287– 1295 [DOI] [PubMed] [Google Scholar]
- Chen CY, Sarnow P. ( 1995) Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science 268: 415– 417 [DOI] [PubMed] [Google Scholar]
- Dingledine R, Borges K, Bowie D, Traynelis SF. ( 1999) The glutamate receptor ion channels. Pharmacol Rev 51: 7– 61 [PubMed] [Google Scholar]
- Gainey MA, Hurvitz-Wolff JR, Lambo ME, Turrigiano GG. ( 2009) Synaptic scaling requires the GluR2 subunit of the AMPA receptor. J Neurosci 29: 6479– 6489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray NK, Wickens M. ( 1998) Control of translation initiation in animals. Annu Rev Cell Dev Biol 14: 399– 458 [DOI] [PubMed] [Google Scholar]
- Grollman AP. ( 1967) Inhibitors of protein biosynthesis. II. Mode of action of anisomycin. J Biol Chem 242: 3226– 3233 [PubMed] [Google Scholar]
- Huang Y, Myers SJ, Dingledine R. ( 1999) Transcriptional repression by REST: recruitment of Sin3A and histone deacetylase to neuronal genes. Nat Neurosci 2: 867– 872 [DOI] [PubMed] [Google Scholar]
- Irier HA, Shaw R, Lau A, Feng Y, Dingledine R. ( 2009) Translational regulation of GluR2 mRNAs in rat hippocampus by alternative 3′ untranslated regions. J Neurochem 109: 584– 594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaac JT, Ashby M, McBain CJ. ( 2007) The role of the GluR2 subunit in AMPA receptor function and synaptic plasticity. Neuron 54: 859– 871 [DOI] [PubMed] [Google Scholar]
- Jackson RJ. ( 2005) Alternative mechanisms of initiating translation of mammalian mRNAs. Biochem Soc Trans 33: 1231– 1241 [DOI] [PubMed] [Google Scholar]
- Kaberdina AC, Szaflarski W, Nierhaus KH, Moll I. ( 2009) An unexpected type of ribosomes induced by kasugamycin: a look into ancestral times of protein synthesis? Mol Cell 33: 227– 236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapp LD, Lorsch JR. ( 2004) The molecular mechanics of eukaryotic translation. Annu Rev Biochem 73: 657– 704 [DOI] [PubMed] [Google Scholar]
- Köhler M, Kornau HC, Seeburg PH. ( 1994) The organization of the gene for the functionally dominant alpha-amino-3-hydroxy-5-methylisoxazole-4-propionic acid receptor subunit GluR-B. J Biol Chem 269: 17367– 17370 [PubMed] [Google Scholar]
- Kozak M. ( 1989) The scanning model for translation: an update. J Cell Biol 108: 229– 241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodish HF. ( 1971) Alpha and beta globin messenger ribonucleic acid. Different amounts and rates of initiation of translation. J Biol Chem 246: 7131– 7138 [PubMed] [Google Scholar]
- Lodish HF, Jacobson A. ( 1972) Translational control of protein synthesis in eucaryotic cells: is there tissue or species specificity of the translational apparatus? Dev Biol 27: 283– 285 [DOI] [PubMed] [Google Scholar]
- Low WK, Dang Y, Schneider-Poetsch T, Shi Z, Choi NS, Merrick WC, Romo D, Liu JO. ( 2005) Inhibition of eukaryotic translation initiation by the marine natural product pateamine A. Mol Cell 20: 709– 722 [DOI] [PubMed] [Google Scholar]
- Munro HN, Baliga BS, Pronczuk AW. ( 1968) In vitro inhibition of peptide synthesis and GTP hydrolysis by cycloheximide and reversal of inhibition by glutathione. Nature 219: 944– 946 [DOI] [PubMed] [Google Scholar]
- Myers SJ, Huang Y, Genetta T, Dingledine R. ( 2004) Inhibition of glutamate receptor 2 translation by a polymorphic repeat sequence in the 5′-untranslated leaders. J Neurosci 24: 3489– 3499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers SJ, Peters J, Huang Y, Comer MB, Barthel F, Dingledine R. ( 1998) Transcriptional regulation of the GluR2 gene: neural-specific expression, multiple promoters, and regulatory elements. J Neurosci 18: 6723– 6739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Kolupaeva VG, Lomakin IB, Pilipenko EV, Shatsky IN, Agol VI, Hellen CU. ( 2001) Molecular mechanisms of translation initiation in eukaryotes. Proc Natl Acad Sci U S A 98: 7029– 7036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romo D, Choi NS, Li S, Buchler I, Shi Z, Liu JO. ( 2004) Evidence for separate binding and scaffolding domains in the immunosuppressive and antitumor marine natural product, pateamine a: design, synthesis, and activity studies leading to a potent simplified derivative. J Am Chem Soc 126: 10582– 10588 [DOI] [PubMed] [Google Scholar]
- Rosenmund C, Stern-Bach Y, Stevens CF. ( 1998) The tetrameric structure of a glutamate receptor channel. Science 280: 1596– 1599 [DOI] [PubMed] [Google Scholar]
- Schluenzen F, Takemoto C, Wilson DN, Kaminishi T, Harms JM, Hanawa-Suetsugu K, Szaflarski W, Kawazoe M, Shirouzu M, Shirouzo M, et al. ( 2006) The antibiotic kasugamycin mimics mRNA nucleotides to destabilize tRNA binding and inhibit canonical translation initiation. Nat Struct Mol Biol 13: 871– 878 [DOI] [PubMed] [Google Scholar]
- Schuwirth BS, Day JM, Hau CW, Janssen GR, Dahlberg AE, Cate JH, Vila-Sanjurjo A. ( 2006) Structural analysis of kasugamycin inhibition of translation. Nat Struct Mol Biol 13: 879– 886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG. ( 2009) Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136: 731– 745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkin YV, Pause A, Haghighat A, Pyronnet S, Witherell G, Belsham GJ, Sonenberg N. ( 2001) The requirement for eukaryotic initiation factor 4A (elF4A) in translation is in direct proportion to the degree of mRNA 5′ secondary structure. RNA 7: 382– 394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagner S, Gensac MC, Maret A, Bayard F, Amalric F, Prats H, Prats AC. ( 1995) Alternative translation of human fibroblast growth factor 2 mRNA occurs by internal entry of ribosomes. Mol Cell Biol 15: 35– 44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walden WE, Godefroy-Colburn T, Thach RE. ( 1981) The role of mRNA competition in regulating translation. I. Demonstration of competition in vivo. J Biol Chem 256: 11739– 11746 [PubMed] [Google Scholar]
- Wilson DN, Nierhaus KH. ( 2005) Ribosomal proteins in the spotlight. Crit Rev Biochem Mol Biol 40: 243– 267 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.