Abstract
Cells respond to oxidants and electrophiles by activating receptor/transcription factor nuclear factor erythroid 2-related factor 2 (Nrf2) to coordinate the induction of cytoprotective genes critical for defense against oxidative and other stresses. Activation involves blocking the ubiquitination-proteasomal degradation of Nrf2. Modification of cysteine thiol groups by inducers in the linker region of Kelch-like ECH-associated protein-1 (Keap1), which congregates Nrf2 into the Keap1/Cul3 E3 complex for ubiquitination, is important but not sufficient for activation of Nrf2. Here we show that evolutionarily conserved cysteine residues of Nrf2 are critical for Nrf2 regulation. FlAsH (an arsenic-based fluorophore) and phenylarsine oxide (PAO) potently induce Nrf2 target genes and bind to Nrf2 in vitro and in vivo. Binding is inhibited by prototypical inducers arsenic and tert-butylhydroquinone. PAO affinity pull-down and mutation of individual cysteine to alanine reveal that Cys235, Cys311, Cys316, Cys414, and Cys506 are critical for binding, and binding is modulated by intramolecular interactions. To corroborate the functions of cysteine residues, Nrf2 wild-type or mutants are expressed in Nrf2 knockout cells to reconstitute Nrf2 regulation. Nrf2 mutants have reduced t1/2 that inversely correlates with increased binding to Keap1 and polyubiquitination of mutant proteins. It is remarkable that the mutants fail to respond to arsenic for Nrf2 activation and gene induction. Furthermore, mutations at Cys119, Cys235, and Cys506 impede binding of Nrf2 to endogenous antioxidant response element and to coactivator cAMP response element-binding protein-binding protein/p300. The findings demonstrate that Nrf2 cysteine residues critically regulate oxidant/electrophile sensing, repress Keap1-dependent ubiquitination-proteasomal degradation, and promote recruitment of coactivators, such that chemical sensing, receptor activation, and transcription activation are integrated at the receptor molecule.
Living organisms are constantly exposed to oxidant and electrophilic chemicals from both environmental and endogenous sources that impose harm to the body. All life forms have also evolved ways of sensing and responding to oxidative stimuli. The cap “n” collar basic leucine zipper protein (CNC bZip) Nrf2 belongs to a group of specialized transcription factors known as xenobiotic-activated receptors (XARs). XARs sense a specific chemical environment in the cell and coordinate adaptive responses to the stimulus by controlling the transcription of cytoprotective enzymes/proteins to ultimately maintain cellular homeostasis (Ma, 2008). In eukaryotes, the Nrf2/Keap1 transcription system is conserved through evolution from fish to rodents and humans and plays a critical role in cellular defense against oxidative and electrophilic insults.
Nrf2 regulates both the basal and inducible expressions of a battery of cytoprotective genes through a common cis-acting DNA element called antioxidant response element (ARE) (Nguyen et al., 2003). Nrf2 target genes encode enzymes/proteins involved in a wide range of cellular functions (Kobayashi et al., 2004; Kensler et al., 2007; Ma, 2008). Consistent with the magnitude of detoxification and antioxidant functions, mice deficient in Nrf2 are susceptible to a variety of chemical and oxidative lesions, such as B[a]P-induced cancer (Ramos-Gomez et al., 2001), spontaneous autoimmune dysfunction and leukoencephalopathy (Ma et al., 2006; Hubbs et al., 2007), ovotoxicant-induced premature ovarian failure (Hu et al., 2006), pulmonary disorders (Cho et al., 2002; Marzec et al., 2007), diabetes (He et al., 2009), and chemical toxicity (Chan et al., 2001; Leung et al., 2003; He et al., 2006, 2007, 2008). On the other hand, activation of Nrf2 by chemoprotective agents confers substantial protection against cancer and chronic diseases (Dinkova-Kostova et al., 2005, 2006; Sussan et al., 2009). Constitutive activation of Nrf2 in mice due to the loss of Nrf2-suppressor Keap1 causes lethality in a young age from an esophageal lesion (Wakabayashi et al., 2003). Tumor cells exhibit high frequencies of mutation in Keap1, resulting in elevated Nrf2 activity, an advantage to tumor cells for resistance to tumor surveillance and anticancer therapeutics (Ohta et al., 2008). This double-edged sword nature of Nrf2 entails a tight control of Nrf2 in the body.
Under basal conditions, Nrf2 protein is rapidly turned over through a specific ubiquitin-26S proteasome pathway with a t1/2 of ∼20 min (He et al., 2006). Only barely detectable activities of Nrf2 are expressed to maintain basal expression of ARE genes in many cell types. Ubiquitination of Nrf2 is controlled by the Keap1/Cul3-dependent ubiquitin ligase in which Keap1 binds and congregates Nrf2 into the E3 complex for ubiquitination of Nrf2. In the presence of an inducer, Nrf2 protein is stabilized, and t1/2 is extended to a magnitude longer. Activated Nrf2 translocates into the nucleus with Keap1 and is deubiquitinated; Keap1 may assist Nrf2 for nuclear translocation and subsequent signaling events. Nuclear Nrf2 dimerizes with small Maf and binds to ARE cis-elements of target genes. Recruitment of coactivator CBP/p300 through the transactivation domain of Nrf2 marks transcription of the genes.
How Nrf2 senses and is activated by oxidant and electrophilic inducers remains elusive. Research in the past has been exclusively focused on Keap1 because Keap1 contains ∼25 cysteine residues and binds inducers more avidly than Nrf2 that has only 7 cysteines. Indeed, extensive analyses of inducer-Keap1 cysteine thiol interaction in several laboratories using mass spectrometry and mutagenesis have identified a number of cysteine residues that are highly reactive to inducers and are important for the repression of Nrf2 activity under basal conditions or activation of Nrf2 in the presence of an inducer (Dinkova-Kostova et al., 2002; Eggler et al., 2005; Hong et al., 2005). Highly reactive cysteines were consistently found in the linker region of Keap1 including Cys273, Cys288, and Cys297. Mutation of Keap1 cysteines revealed Cys151, Cys273, and Cys288 as critical residues for regulating Nrf2. Expression of the mutants in Keap1 null mice or cells further identified Cys273 and Cys288 as required for suppression of Nrf2 and Cys151 important in facilitating Nrf2 activation (Yamamoto et al., 2008; He and Ma, 2010). The findings reveal that individual Keap1 cysteines perform distinct functions in Nrf2 signaling.
Despite considerable progress in understanding inducer-Keap1 interaction, a number of observations on Nrf2 activation are not readily explained by the inducer-Keap1 model. In vitro analysis of the interaction between human Keap1 and Nrf2 Neh2, a domain required for Keap1-binding but with no cysteine residues, revealed that prototypical inducers modified Keap1 cysteines as expected but failed to dissociate Nrf2 Neh2 from Keap1 (Eggler et al., 2005). In cell-based assays, treatment with tert-butylhydroquinone (tBHQ) or mutation of Cys273 and Cys288 of Keap1 did not disrupt but rather increased the association between Nrf2 and Keap1 (He et al., 2006; Kobayashi et al., 2006), whereas treatment with arsenic dissociated Nrf2 from Keap1 in the nucleus (He et al., 2006). Finally, a study in a zebrafish model revealed that additional factors other than Keap1 are required for activation of Nrf2 by certain inducers (Kobayashi et al., 2009). These findings suggest that additional mechanisms of sensing and transducing oxidant/electrophilic signals are critical for activation of Nrf2.
Nrf2 contains seven highly conserved cysteine residues (Fig. 1). In this study, we found that FlAsH and phenylarsine oxide (PAO), two potent ARE inducers, bind to Nrf2, and binding is reversed by tBHQ and arsenic. Mutation of Nrf2 cysteines enhanced Nrf2-Keap1 association, ubiquitination of Nrf2, and proteasomal degradation of Nrf2 resulting in markedly shortened t1/2. Treatment with arsenic failed to activate Nrf2 mutants or induce Nrf2 target gene Nqo1 in Nrf2 knockout (KO) cells expressing the cysteine mutants. Mutations at Cys119, Cys235, and Cys506 reduced the binding of Nrf2 to endogenous ARE and to coactivator CBP/p300. To our knowledge, this report is the first study demonstrating multiple and critical roles of Nrf2 cysteine residues for inducer-sensing and Keap1-dependent ubiquitination/degradation of Nrf2 as well as transactivation by Nrf2.
Fig. 1.
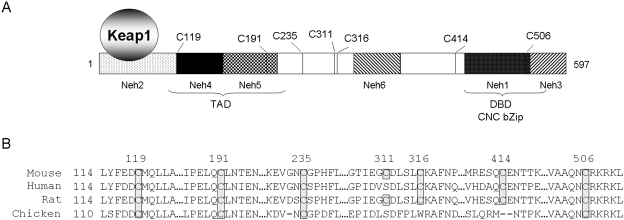
Cysteine residues in Nrf2 are conserved across species. A, cysteine residues and domain structure of Nrf2. B, conservation of Nrf2 cysteine residues across mouse, human, rat, and chicken. Numbers at the top represent cysteine positions in mouse Nrf2. Cysteine residues are boxed and shaded: rat Nrf2 has seven cysteine residues at the same positions as for mouse Nrf2; human Nrf2 has six cysteine residues at 119, 199, 242, 326, 422, and 515 positions; and chicken Nrf2 has four cysteine residues at 115, 181, 224, and 492.
Materials and Methods
Reagents and Cell Lines.
Arsenic chloride (As3+), β-mercaptoethanol (β-ME), cycloheximide (CHX), PAO, and tBHQ were purchased from Sigma-Aldrich (St. Louis, MO). Affigel 10 gel was from Bio-Rad Laboratories (Hercules, CA). 4-Amino-phenylarsine oxide (p-aminophenyl arsenoxide, dihydrate) was purchased from Toronto Research Chemicals, Inc., (Toronto, ON, Canada). MG132 was from BIOMOL Research Laboratories (Plymouth Meeting, PA). FlAsH and 2,3-dimercaptopropanol (Bal) were from Invitrogen (Carlsbad, CA).
Mouse hepa1c1c7 cells were provided by Dr. J. P. Whitlock, Jr. (Stanford University, Stanford, CA). The cells were cultured in α-minimal essential medium with 10% fetal bovine serum and 5% CO2. Mouse Nrf2 wild-type (WT) and KO embryonic fibroblast cells were derived from Nrf2 WT and KO mice (Bi et al., 2004). The cells were immortalized by expressing SV40 large T-antigen following an established procedure (Takenouchi et al., 2007). The cells were cultured in Dulbecco's modified Eagle's medium with 10% fetal bovine serum and 5% CO2. Penicillin (100 U/ml) and streptomycin (100 μg/ml) were added to the media to prevent contamination. Cos7 cells were purchased from the American Type Culture Collection (Manassas, VA) and were cultured in Dulbecco's modified Eagle's medium with 10% fetal bovine serum.
Plasmid Construction, Point Mutation, and Cell Transfection.
Full-length mouse Nrf2 cDNA coding sequence was subcloned into the pCMV-HA vector to generate pCMV-HANrf2. N-terminal deletion mutants of Nrf2 were generated by PCR and subcloned into pCMV-HA vector. Single mutations at Cys119, Cys191, Cys235, Cys414, and Cys506 and a double mutant at Cys311 and Cys316 were generated using the Quik-Change Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). Mouse full-length Keap1 was cloned into the pcDNA3.1V5His vector (Invitrogen). All plasmid constructs and mutations were confirmed by DNA sequencing. Primers for constructing the plasmids and mutations are available upon request. Transfection was performed with Lipofectamine Plus (Invitrogen).
RNA Preparation and Northern Blotting.
Total RNA was isolated from cells using the a total RNA isolation kit (QIAGEN, Valencia, CA). Total RNA of 3 μg each was fractionated in a 1.2% formaldehyde agarose gel, transferred to a supercharged nylon membrane, and blotted with the digoxigenin-labeled riboprobe prepared with the digoxigenin-labeling reagents (Roche Applied Science, Indianapolis, IN). Plasmid constructs for riboprobes of mouse Nqo1, Nrf2, and Actin were verified by sequencing. Northern signals were visualized by chemiluminescence using a digoxigenin RNA detection kit with CDP Star as a substrate (Roche Applied Science).
Cell Fraction and Immunoblotting.
Nuclear and cytoplasmic factions were prepared using the nuclei EZ-PREP reagents from Sigma-Aldrich. Cells at 90% confluence in 10-cm dishes were washed with ice-cold phosphate-buffered saline (PBS) and lysed with ice-cold Nuclei EZ-PREP lysis buffer containing protease and phosphatase inhibitors (1 mM phenylmethylsulfonyl fluoride, 1 mM Na3VO4, 1 mM NaF, and 1 μg/ml each of aprotinin, leupeptin, and pepstatin A). Cell lysate was centrifuged at 500g for 5 min at 4°C to give rise to nuclei and cytosol. Nuclei pellet was washed once with the lysis buffer and resuspended in a radioimmunoprecipitation assay buffer.
For immunoblotting, cells were lysed on ice with radioimmunoprecipitation assay buffer containing protease and phosphatase inhibitors for 30 min. Cell lysate was sonicated briefly and was centrifuged at 14,000g for 20 min to remove cell debris. Lysate of 10 to 20 μg was fractionated on 10% SDS-polyacrylamide gel electrophoresis (PAGE), transferred to polyvinylidene difluoride membrane (Bio-Rad), and blocked with 5% nonfat milk in PBS plus 0.05% Tween 20. The membrane was blotted with primary antibody at 4°C overnight with shaking, followed by incubation with horseradish peroxidase-conjugated secondary antibodies for 1 h at room temperature. Protein bands were visualized using enhanced chemiluminescence detection reagents from GE Healthcare (Chalfont St. Giles, Buckinghamshire, UK). Actin was blotted as loading control.
In Vitro Transcription and Translation.
The TnT quick-coupled transcription/translation system (Promega, Madison, WI) was used for in vitro transcription and translation of Nrf2 and mutant proteins. Proteins were produced with or without biotin tRNA and were detected by SDS-PAGE with streptavidin-horseradish peroxidase, or specific antibodies followed by chemiluminescence detection.
Cycloheximide Chase.
Twenty-four hours after transfection with plasmids, cells were split from one 10-cm dish to five 35-cm dishes and were cultured overnight. The cells were then treated with MG132 (15 μM) for 2 h followed by washing with PBS three times to remove MG132. CHX was added, and cells were collected 0, 30, 60, 90, and 120 min after treatment. Thirty micrograms of total protein was fractionated on SDS-PAGE and blotted with anti-Nrf2 and anti-actin antibodies.
Purification of Nrf2.
pNrf2/PET28c was transformed into the bacteria BL21(DE3) cells (Novagen, La Jolla, CA). The bacteria were first grown in 10 ml of Luria broth medium at 37°C overnight and then transferred to a 4-l Luria broth with absorbance at λ600 nm adjusted to between 0.2 and 0.3. Culture was continued at 37°C with vigorous shaking until optical density at 600 nm reached between 0.5 and 0.6. Isopropyl-β-d-thiogalactoside was added, and culture was continued for another 24 h with gentle agitation at 15°C. Nrf2 was purified using the nickel-nitrilotriacetic acid-agarose affinity chromatography (Qiagen). Purified Nrf2 was concentrated with Centricon 30 (Millipore, Bedford, MA) and was confirmed for purify by SDS-PAGE and mass spectrometry.
PAO Bead Conjugation and Pull-Down.
Ten milligrams of 4-amino-phenylarsene oxide was dissolved in 3.05 ml of methanol and mixed with 1.22 ml of Affigel (50% slurry) for 2 h at room temperature. Aminoethanol (100 μl) was then added to block the remaining active binding sites. The mixture was washed three times with methanol and three times with PBS and was then resuspended in 0.6 ml of PBS. Control Affigel was prepared by mixing 1.22 ml of Affigel slurry (50%) with 3.05 ml of methanol and 1 ml of aminoethanol, followed by shaking at room temperature for 2 h.
Measurement of Protein-Free Thiols.
Protein-free thiol groups were measured as described previously with modifications (Aitken and Learmonth, 1996; Nishikimi et al., 2001). Purified proteins (10 μg in 100 μl) were incubated with As3+ or PAO for 30 min. The proteins were mixed with 10 μg of bovine serum albumin and 10% ice-cold trichloroacetic acid containing 1 mM dithiothreitol. The suspension was centrifuged at 14,000g for 5 min at 4°C. Precipitate was resuspended in 400 μl of the Ellman's reagent or the buffer [0.5 M potassium phosphate buffer, pH 7.4, containing 0.2 mM 5,5′-dithiobis-(2-nitrobenzoic acid) and 5 mM EDTA]. The mixture was incubated at 4°C for 30 min and was centrifuged at 14,000g for 5 min to remove insoluble materials. Absorbance of supernatant was measured at λ412 nm.
Coimmunoprecipitation and Detection of Ubiquitination.
Plasmids pCMV-HANrf2 and pcDNA3.1-V5Keap1 or pCW-ubiquitin-myc were cotransfected into Nrf2 KO cells using Lipofectamine Plus. Coimmunoprecipitation was performed as described previously (He et al., 2006) and in the figure legends. For IP, cell lysates or cell fractions were precleared with protein G-agarose (Invitrogen) for 1 h at 4°C, followed by incubation with IP antibodies at 4°C overnight with shaking. Immune complexes were precipitated by incubation with protein-G agarose at 4°C for 1 h and a brief centrifugation. The precipitates were washed extensively with PBS plus 0.05% Tween 20 and were subjected to fractionation by SDS-PAGE. Protein bands were detected by immunoblotting with specific antibodies as described in figure legends. Antibodies against Nrf2, Keap1, CBP/p300, and actin were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Antibody against the HA tag was obtained from BAbCO (Berkeley Antibody Company, Richmond, CA). Anti-ubiquitin antibody was from Zymed Laboratories Inc. (South San Francisco, CA). Anti-V5 was from Invitrogen.
Chromatin Immunoprecipitation Assay.
Chromatin immunoprecipitation was performed using anti-Nrf2 antibodies as described previously (He et al., 2006). Nrf2 WT and cysteine mutants were expressed in Nrf2 KO cells. Cells were treated with MG132 at 15 μM for 2 h. DNA proteins were cross-linked by incubating cells with 1% formaldehyde at 37°C for 10 min. Excess formaldehyde was quenched with 0.125 M glycine at room temperature for 5 min. Cells were collected in 1 ml of a lysis buffer (5 mM PIPES, pH 8.0, 85 mM KCl, and 0.5% Igepal CA-630) with protease inhibitors and were centrifuged to pellet nuclei. The nuclei were resuspended in the lysis buffer, repelleted, and resuspended in a nuclei lysis buffer (50 mM Tris/HCl, pH 8.0, 10 mM EDTA, and 1% SDS) with protease inhibitors. Chromatin was sonicated to an average size of 200 to 1000 base pairs using a tapered microtip at 40% power output (10 times, 10 s each). Cell debris was removed by centrifugation at 14,000g for 10 min at 4°C. Sheared chromatin was diluted in an IP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris/HCl, pH 8.0, and 167 mM NaCl), precleared with protein G containing salmon sperm DNA (240 μg of sperm DNA in 1 ml of solution), and immunoprecipitated with anti-Nrf2 antibody. DNA-protein complexes were eluted from the protein G-agarose beads with 200 μl of an elution buffer (50 mM NaHCO2 and 1% SDS) and were reverse cross-linked by incubating with 8 μl of 5 M NaCl at 65°C overnight. The DNA samples were purified and analyzed by real-time PCR using SYBR Green PCR master mix (Applied Biosystems, Foster City, CA) performed on a Bio-Rad iCycler following standard procedures. Real-time PCR results were normalized using 1% input as an internal control. Relative DNA amounts were calculated from CT values for each sample by interpolating into a standard curve obtained using a series dilution of standard DNA samples run under the same conditions. The primer sets used for real-time PCR were as reported before (He et al., 2006).
Statistical Analysis.
Quantification of protein bands was performed using the ImageQuant program (GE Healthcare). Quantitative data represent means and standard deviations from three different samples. Statistical analysis was performed with one-way analysis of variance followed by t test using the Microsoft Excel program (Microsoft, Redmond, WA). p Values of <0.05 were considered statistically significant.
Results
Binding of FlAsH to Nrf2.
Mouse Nrf2 contains seven cysteine residues (Fig. 1A). Cys119 and Cys191 are located in the transcription activation domain (TAD) C-terminal to the Keap1-binding motif, Cys414 and Cys506 in the CNC bZip DNA-binding domain (DBD) at the carboxyl end, and Cys235, Cys311, and Cys316 in the middle region between TAD and DBD. Moreover, the cysteine and surrounding residues are highly conserved across species from chicken to mouse, rat, and human (Fig. 1B): Cys119, Cys191, Cys235, and Cys506 are conserved in all species; Cys316 and Cys414 in mouse, rat, and human; and Cys311 in mouse and rat (replaced by serine in chicken and human), suggesting a role of the residues in Nrf2 function that is preserved through evolution. Previous studies indicated that binding of ARE inducers to Keap1 cysteine thiols is important but not sufficient for activation of Nrf2, underpinning additional mechanisms other than Keap1 for Nrf2 activation (Eggler et al., 2005; He et al., 2006; Kobayashi et al., 2009). Therefore, we hypothesized that Nrf2 cysteine residues play critical roles in Nrf2-inducer interaction and other functions.
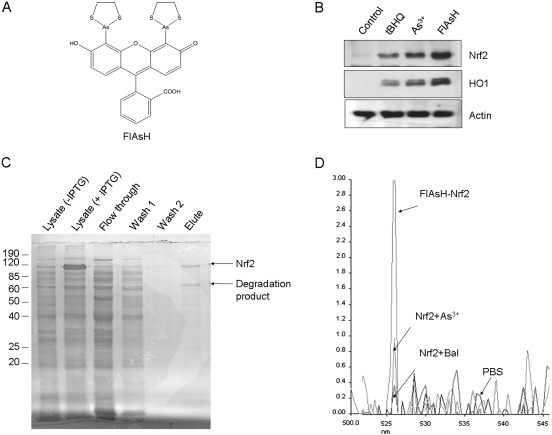
Arsenic, a thiol-reactive metalloid, is a prototypical activator of Nrf2. To examine whether inducers, such as arsenic, bind to Nrf2, we used an arsenic-based fluorophore FlAsH as a sensitive probe for the arsenic-Nrf2 interaction, because FlAsH fluoresces upon binding with protein cysteine thiols and potently activates Nrf2 (Fig. 2A). FlAsH (10 μM, 5 h) activated Nrf2 and induced HO1 to higher levels than either As3+ (10 μM, 5 h) or tBHQ (30 μM, 5 h) (Fig. 2B). Nrf2 was expressed in bacteria and purified to apparent homogeneity as judged by SDS-PAGE (Fig. 2C), immunoblotting, and mass spectrometry (data not shown). Upon incubation with purified Nrf2, FlAsH produced a single major fluorescent peak at ∼526 nm (Fig. 2D), indicating binding of FlAsH to Nrf2 thiol groups. Preincubation of Nrf2 with As3+ or Bal, a thiol-reactive agent, but not PBS, reduced the fluorescent signal to near or at the background level, indicating that As3+ or Bal directly competes with FlAsH for binding to Nrf2 thiols. Thus, FlAsH binds to the cysteine thiols of purified Nrf2, and As3+ probably binds to Nrf2 in a similar fashion.
Fig. 2.
Binding of FlAsH to purified Nrf2. A, structure of FlAsH. B, activation of Nrf2 by FlAsH. Hepa1c1c7 cells were treated with tBHQ (30 μM), As3+ (10 μM), and FlAsH (10 μM) for 5 h. Cell lysates were immunoblotted for Nrf2, HO1 and actin. C, purification of Nrf2. Mouse Nrf2 was expressed from pNrf2/ET28c in BL21(DE3) cells and purified using nickel beads. Proteins were fractionated in 10% SDS-PAGE and stained with Coomassie blue. D, competition for binding to Nrf2 among FlAsH, arsenic, and Bal. FlAsH (2 μM) was incubated with PBS, purified Nrf2 (10 μg), or purified Nrf2 preincubated for 30 min with As3+ (50 μM) or Bal (50 μM). Fluorescent intensity was measured using a fluorescence spectrophotometer with excitation at 508 nm.
Binding of PAO to Nrf2.
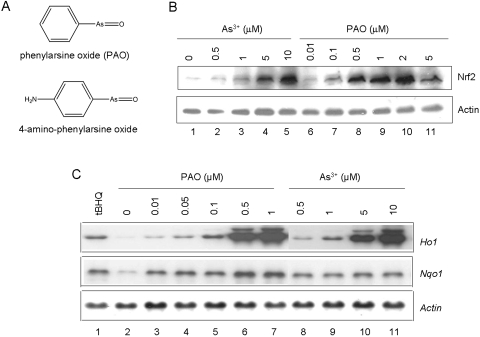
PAO binds to protein vicinal cysteine thiols to form stable structures. Moreover, PAO derivative 4-amino-phenylarsine oxide can be coupled to Affigel to generate affinity beads that have high affinity toward protein vicinal thiols (Fig. 3A). We found that PAO is a magnitude more potent than As3+ in activating Nrf2 (Fig. 3B; EC50 of 0.25 versus 2.5 μM). Activation by PAO was reduced at 5 μM because of increased toxicity of PAO at high concentrations. Likewise, PAO induced Ho1 with a higher potency than that of As3+ (Fig. 3C). Nqo1 was also induced by PAO.
Fig. 3.
Activation of Nrf2 by PAO. A, structure of PAO and 4-amino-phenylarsine oxide. B, activation of Nrf2. Hepa1c1c7 cells were treated with arsenic or PAO at concentrations indicated for 5 h. Cell lysates were immunoblotted for Nrf2 and actin. C, induction of Ho1 and Nqo1. Cells were treated with tBHQ (30 μM), PAO, or arsenic for 5 h. Total RNA was blotted for mRNA expression of Ho1, Nqo1, and Actin.
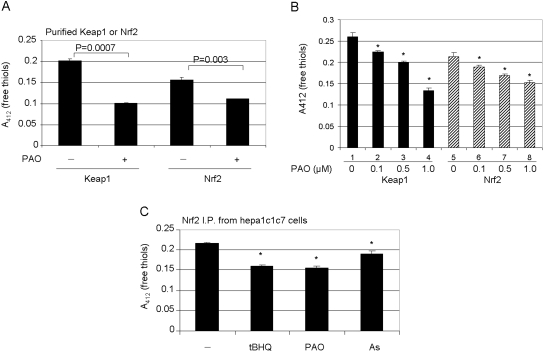
Binding of PAO to Nrf2 cysteine thiols was first examined by measuring protein-free thiol content. Keap1, which is known to bind oxidants and electrophilic inducers through its thiol groups, was used as a positive control. Purified Nrf2 or Keap1 was incubated with PAO. The overall free thiol measurement in Nrf2 is less than that in Keap1 in the absence of treatment because of its lower cysteine content than that of Keap1 (Fig. 4A). PAO significantly reduced free thiol contents of purified Nrf2 and Keap1. Similar results were observed with As3+ and tBHQ (data not shown). In a separate experiment, reactivity to low concentrations of PAO between Nrf2 and Keap1 was compared (Fig. 4B). PAO at concentrations as low as 0.1 and 0.5 μM significantly reduced the free thiol contents of both proteins. The data indicate that both Keap1 and Nrf2 bind to low concentrations of PAO in a similar manner. Binding of PAO to endogenous Nrf2 was examined. Cells were incubated with PAO or other inducers, followed by immunoprecipitation with anti-Nrf2 and measurement of the free thiol content of precipitated Nrf2. The data showed that PAO, as well as tBHQ and As3+, significantly reduced the free thiol contents of the precipitates from treated cells (Fig. 4C). This result is consistent with the findings from purified Nrf2. Taken together, the findings suggest that PAO, As3+, and tBHQ bind to Nrf2 thiol groups both in vitro and in vivo.
Fig. 4.
Binding of PAO to free thiol groups of Nrf2. A, binding of PAO to purified Keap1 or Nrf2. Purified Keap1 and Nrf2 were incubated with PAO for 2 h. The proteins were precipitated with trichloroacetic acid, and protein-free thiol groups were measured with the Ellman's reagent at λ412 nm. B, concentration-dependence. Keap1 or Nrf2 was incubated with increasing concentrations of PAO, and free cysteine thiol groups were measured. C, binding to endogenous Nrf2. Hepa1c1c7 cells were treated with tBHQ (30 μM), PAO (1 μM), or arsenic (10 μM) for 5 h. Nrf2 was immunoprecipitated with anti-Nrf2 antibody. The precipitate was treated with Ellman's reagent, and free thiol groups of Nrf2 were measured at λ412 nm. Data represent means and standard deviations from three samples. ∗, p < 0.05.
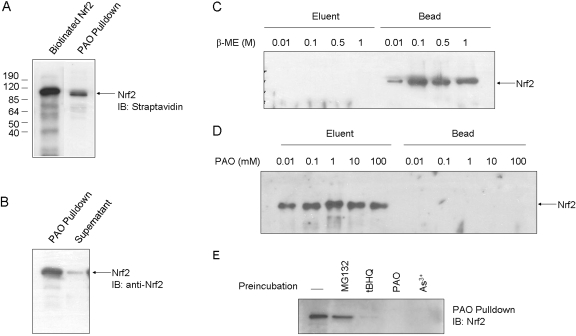
Binding of PAO to Nrf2 was further analyzed by using PAO affinity pull-down. Nrf2 was produced by in vitro transcription and translation. PAO affinity beads pulled down Nrf2 (Fig. 5A), whereas Affigel beads without PAO did not (data not shown). PAO pull-down was efficient with >90% of input Nrf2 precipitated (Fig. 5B). Binding between PAO and Nrf2 was tight because thiol-reactive agent β-ME failed to elute Nrf2 from PAO beads at concentrations as high as 1 M (Fig. 5C, compare Elute and Bead). On the other hand, free PAO at concentrations from 0.01 to 100 mM effectively eluted most of the bead-bound Nrf2 (Fig. 5C, compare Elute and Bead). Specificity of Nrf2-PAO binding was examined by incubating Nrf2 with inducers before PAO pull-down (Fig. 5D). PAO, As3+, or tBHQ effectively blocked Nrf2 pull-down by PAO beads, whereas MG132, which does not bind to Nrf2 directly, did not affect pull-down. Next, in vivo binding of PAO to Nrf2 was examined (Fig. 6). Cells were treated with MG132 to increase the amount of Nrf2 protein. Nrf2 was pulled down by PAO beads from the cells effectively (Fig. 6A). As a control, Keap1 was shown to be pulled down equally well with or without MG132, because Keap1 protein is constitutively expressed and is not affected by MG132. Affigel without PAO did not pull down Nrf2 or Keap1 from the cells as expected (data not shown). Treatment of cells with As3+ (10 μM) or tBHQ (30 μM) for 5 h activated Nrf2 and prevented pull-down of Nrf2 by PAO beads (Fig. 6B). As a negative control, MG132 induced Nrf2 but did not affect pull-down. Together these data revealed that PAO potently activates Nrf2 by directly binding to Nrf2 cysteine thiol groups, and As3+ and tBHQ bind to Nrf2 cysteine residues similarly to PAO both in vitro and in intact cells.
Fig. 5.
PAO pull-down of Nrf2 in vitro. A, pull-down of Nrf2 in vitro. Biotinylated Nrf2 was produced in vitro with TnT reticulocyte lysate and was pulled down by PAO beads. B, efficiency of PAO pull-down. Nrf2 in 1 or 10 μl of TnT lysate was pulled down by PAO; >90% of input Nrf2 was pulled down by PAO beads. C, elution by β-ME. Nrf2 in 10 μl of TnT lysate was precipitated by PAO beads. The pellet was incubated with β-ME from 0.01 to 1 M to elute Nrf2. D, elution by free PAO. Nrf2 was precipitated by PAO beads and was eluted from the beads by free PAO at concentrations from 0.01 to 100 mM. E, inhibition of PAO pull-down by tBHQ, PAO, and As3+ in vitro. Nrf2 in 10 μl of TnT lysate was incubated with MG132 (15 μM), tBHQ (30 μM), PAO (1 μM), or As3+ (10 μM) for 2 h, followed by incubation with PAO beads overnight. Precipitated Nrf2 was immunoblotted with anti-Nrf2 antibody.
Fig. 6.
PAO pull-down of Nrf2 in vivo. A, pull-down of endogenous Nrf2. Hepa1c1c7 cells were treated with MG132 (15 μM) for 2 h to increase the protein level of Nrf2. Cell lysate (1 mg) was precleaned with Affigel without PAO and then incubated with PAO beads at 4°C overnight. PAO pull-down precipitates and supernatants were immunoblotted for Nrf2 and Keap1. B, inhibition of PAO pull-down by tBHQ, PAO, and As3+ in vivo. Cells were treated with MG132 (15 μM), As3+ (1 or 10 μM), or tBHQ (30 μM) for 5 h. Cell lysate was blotted for Nrf2 or actin (top and middle), and PAO pull-down blotted for Nrf2 (bottom).
Effect of Intramolecular Interaction.
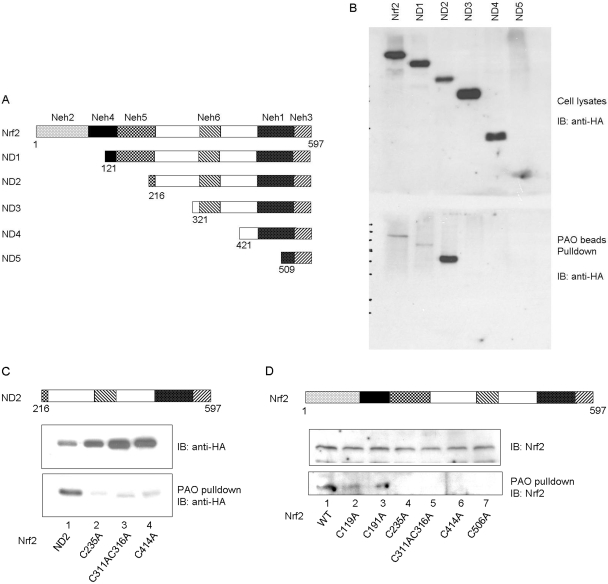
We determined the relative importance of cysteine residues in PAO binding. Consecutive N-terminal deletions of Nrf2 were made (Fig. 7A) and expressed in Cos7 cells. Expression of mutant proteins was comparable, except for ND5, the expression of which was difficult to assess because of its much smaller molecular mass than others (Fig. 7B, top). Full-length Nrf2 and ND1 (lack of Cys119) were pulled down by PAO beads at similarly low levels; on the other hand, ND2 (lack of Cys119 and Cys191) showed strong pull-down (Fig. 7B, bottom). This result suggests that the N-terminal portion of Nrf2 (aa1–200) suppresses PAO binding, and suppression potentially involves Cys191 more than Cys119 because the loss of Cys119 in ND1 did not affect suppression. PAO pull-down was largely diminished in ND3 (lack of Cys235, Cys311, and Cys316 in addition to Cys119 and Cys191), ND4 (lack of six cysteines except Cys506), and ND5 (lack of all cysteines) (Fig. 7B and data not shown). Together, these results imply that Cys235, Cys311, and Cys316 are necessary for strong binding, whereas C-terminal cysteines Cys414 and Cys506 alone do not support binding. However, this conclusion does not exclude the possibility that Cys414 and Cys506 play a role in binding in the context of whole Nrf2 molecule as described later in mutational studies. On the other hand, the amino terminal portion of Nrf2 inhibits PAO binding. Thus, binding of PAO to individual cysteine residues varies and binding is probably modulated by the overall Nrf2 structure and intramolecular interactions.
Fig. 7.
PAO pulldown of deletion mutants. A, N-terminal deletion of Nrf2. B, binding of PAO to deletion mutants. Nrf2 and deletion mutants were expressed in Cos7 cells and were pulled down by PAO beads. Top, immunoblotting of cell lysate with anti-HA antibody; bottom, immunoblotting of PAO pull-down with anti-HA. C, binding of PAO to cysteine mutants of Nrf2 ND2 fragment. D, binding of PAO to cysteine mutants of full-length Nrf2.
To further dissect the role of individual cysteine residues in PAO binding, each cysteine residue was replaced with alanine by site-directed mutagenesis. We first examined ND2 that showed strong binding. Mutating Cys235 or Cys311/Cys316 together to alanine abolished PAO pull-down of ND2, confirming critical roles of the residues in PAO binding (Fig. 7C). We then mutated each cysteine residue of full-length Nrf2. Mutation of Cys119 or Cys191 did not significantly affect PAO pull-down (Fig. 7D). Mutation of Cys235, or Cys311/Cys316 together, abolished PAO binding (Fig. 7D), which is in agreement with the experiment on ND2 (Fig. 7, B and C). We were surprised to find that mutation of Cys414 in either ND2 or full-length Nrf2 or Cys506 in full-length Nrf2 also blocked PAO pull-down of Nrf2 (Fig. 7B). Thus, Cys414 and Cys506 contribute to PAO binding in the context of whole Nrf2 molecule, even though they do not support binding in ND3 and ND4; the results support the notion that intramolecular interaction affects inducer-Nrf2 binding.
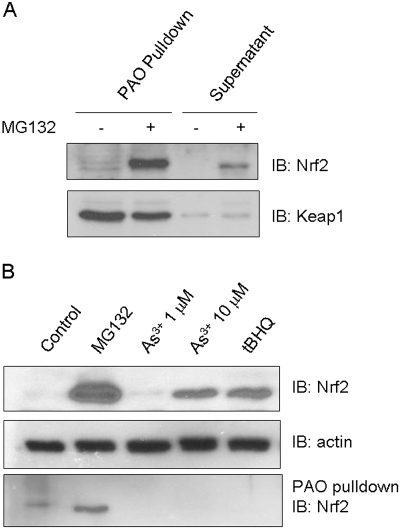
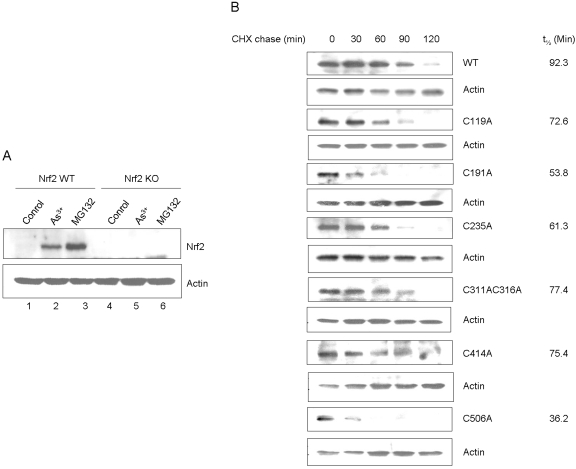
Role in Keap1-Dependent Proteasomal Degradation.
To corroborate the functional impact of cysteine residues on Nrf2, WT and Nrf2 KO cells were derived from mice and were immortalized by expressing SV40 T antigen. The cells were characterized for expression and activation of Nrf2 (Fig. 8A). Treatment with either As3+ or MG132 activated Nrf2 in WT cells as expected. No detectable expression or activation of Nrf2 by the inducers was observed in KO cells, confirming the loss of Nrf2. Reconstitution of KO cells with wild-type Nrf2 restored expression and function of Nrf2 (data not shown; see results below). We found that expression of cysteine mutants in KO cells was low and variable, which can be due to decreased protein expression, increased turnover, or both. Because Nrf2 is known to be labile, we examined the t1/2 values of the mutants (Fig. 8B). CHX chase experiments revealed that Nrf2 WT expressed in Nrf2 KO cells had a t1/2 of 92 min, which is longer than that of the endogenous Nrf2 protein (He et al., 2006); extension of t1/2 of plasmid-expressed Nrf2 is probably a result of overexpression of plasmid Nrf2 compared with the endogenous protein. It is noteworthy that all cysteine mutants exhibited shortened t1/2 compared with WT. In particular, the t1/2 values of C506A (36 min), C191A (53 min), C235A (61 min), and C119 (72 min) were 39, 58, 66, and 78% of the WT, respectively. Thus, Nrf2 cysteine mutants had faster turnover than wild type.
Fig. 8.
Effect of cysteine mutations on the half-life of Nrf2. A, characterization of immortalized Nrf2 KO cell line. Embryonic fibroblasts derived from Nrf2 WT and KO mice, respectively, were immortalized by expressing the SV40 T-antigen. The cells were treated with As3+ (10 μM) or MG132 (15 μM) for 5 h. Stabilization of Nrf2 protein was examined by immunoblotting with anti-Nrf2 antibody. B, half-life of cysteine point mutants. Nrf2 WT and cysteine point mutants were expressed in Nrf2 KO cells. The cells were treated with MG132 (15 μM) for 2 h, followed by CHX chase for 0, 30, 60, 90, and 120 min. Cell lysate was blotted for expression of Nrf2. Actin was blotted as loading control.
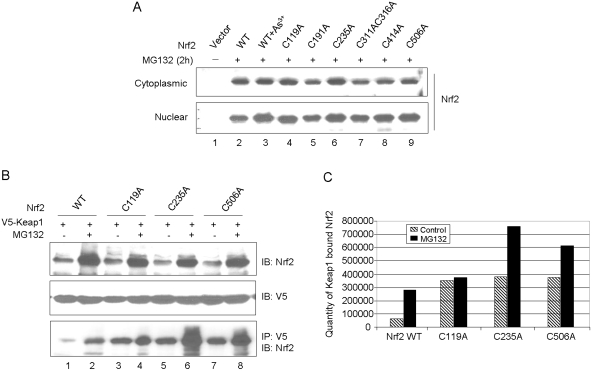
Treatment of KO cells expressing WT or cysteine mutants with MG132 significantly increased the protein levels in both the cytoplasm and nucleus, and the levels were similar among the proteins (Fig. 9A). The results imply that both WT and mutant proteins were degraded through the 26S proteasome-mediated proteolysis, and mutation of the cysteines did not affect their nuclear translocation. We further analyzed the degradation of C119A, C235A, and C506A because they have shorter t1/2 than other mutants (Fig. 8B) and are highly conserved (Fig. 1).
Fig. 9.
Cysteine mutants are degraded through the Keap1-dependent ubiquitin-proteasome pathway. A, stabilization of cysteine mutants by MG132. Nrf2 WT and cysteine point mutants were expressed in Nrf2 KO cells. Cells were treated with MG132 (15 μM, 2 h). Cytoplasmic and nuclear fractions were blotted for Nrf2 expression. B, binding of cysteine mutants with Keap1. V5-Keap1 and Nrf2 WT or Nrf2 cysteine mutant were coexpressed in Nrf2 KO cells. The cells were treated with vehicle or MG132 (15 μM) for 4 h. Cell lysate was blotted with anti-Nrf2 or anti-V5 (top and middle) or immunoprecipitated with anti-V5 followed by immunoblotting with anti-Nrf2 (bottom). C, quantification of coimmunoprecipitated Nrf2 in B.
We examined the interaction between Keap1 and Nrf2 mutants as a potential mechanism for increased proteasomal degradation of Nrf2 mutants. Nrf2 WT or cysteine mutants were coexpressed with V5-Keap1 in Nrf2 KO cells. As expected, expression of Nrf2 and mutants was increased by MG132 (Fig. 9B, top), but that of V5-Keap1 was not (middle). Immunoprecipitation with anti-V5 followed by immunoblotting with anti-Nrf2 revealed that, in either the absence or presence of MG132, the amounts of cysteine mutants coprecipitated with Keap1 were significantly higher than that of wild-type (Fig. 9, B, bottom, and C). Furthermore, more C506A and C235A were bound to Keap1 than C119A, which is in agreement with shorter t1/2 values of Cys235 and Cys506 than that of Cys119.
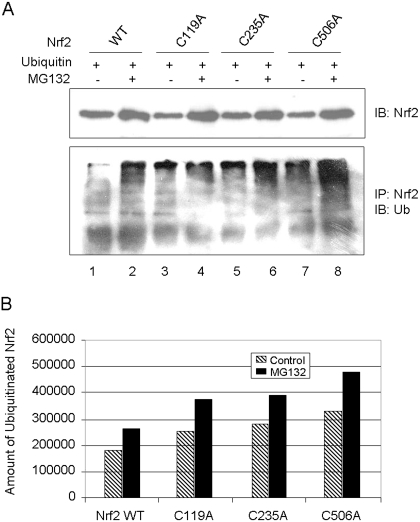
The observation that cysteine mutants show faster turnover and bind more to Keap1 prompted us to examine ubiquitination of the cysteine mutants. Nrf2 WT or cysteine mutants were coexpressed with ubiquitin in KO cells. The proteins were immunoprecipitated with anti-Nrf2 and blotted with anti-ubiquitin. Cells were treated with MG132 to boost the protein level of Nrf2 (Fig. 10A, top). Nrf2 WT was ubiquitinated, and ubiquitinated Nrf2 was significantly increased by MG132 as expected (Fig. 10, A, bottom lanes 1 and 2, and B). Ubiquitination of cysteine mutants was markedly increased either with or without MG132 compared with the wild type. It is noteworthy that the extent of ubiquitination of the proteins was in the order of C506A > C235A > C119A > WT either in the absence or presence of MG132 (Fig. 10, A and B), which is inversely correlated with the t1/2 values of the proteins (Fig. 8B). Taken together, the results indicate that mutation of Nrf2 cysteine residues significantly increases proteasomal degradation of the proteins by enhancing Keap1-binding and Keap1-dependent ubiquitination of mutant proteins, resulting in shortening of the t1/2 values of the mutants.
Fig. 10.
Increased ubiquitination of cysteine mutants. A, ubiquitination of Nrf2 mutants. Nrf2 WT or cysteine mutants were coexpressed with c-myc-ubiquitin in Nrf2 KO cells. The cells were then treated with vehicle or MG132 (15 μM) for 4 h. Cell lysate was blotted with anti-Nrf2 to show Nrf2 protein expression (top) or immunoprecipitated with anti-Nrf2 and then immunoblotted with anti-ubiquitin. B, quantification of ubiquitinated Nrf2 in A.
Role in Arsenic Responsiveness.
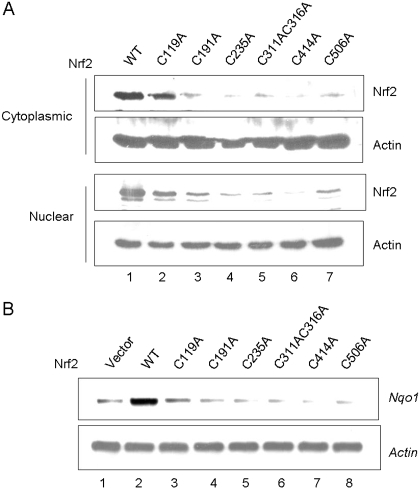
Binding of arsenic to Nrf2 cysteine residues may serve as a mechanism of inducer-sensing by Nrf2. To directly test the notion, Nrf2 WT and cysteine mutants were expressed in KO cells and were treated with As3+. Nrf2 KO cells transfected with the WT Nrf2 plasmid expressed Nrf2 protein at a very low level similar to that of Nrf2 WT cells in the absence of inducers (data not shown). As3+ increased the protein levels of WT (data not shown and Fig. 11A, lane 1), and to a lesser extent C119A (Fig. 11A, lane 2), in the cytoplasm, indicating the activation of the proteins. However, As3+ failed to increase the protein levels of C191A, C235A, C311A/C316A double mutation, C414A, and C506A, respectively (Fig. 11A, lanes 3–7). A similar trend was observed for the nuclear expression of the proteins (Fig. 11A, bottom). These results imply that As3+ failed to activate Nrf2 when the cysteine residues were mutated to alanine. We further analyzed arsenic responsiveness by examining induction of Nrf2 target genes. Expression of WT in KO cells was shown to restore the induction of Nqo1 in the presence of As3+ (Fig. 11B). On the contrary, induction was lost for all cysteine mutants. Together, the findings demonstrate that the cysteine residues of Nrf2 are critical for arsenic sensing and responsiveness by Nrf2.
Fig. 11.
Loss of arsenic-responsiveness in cysteine mutants. Nrf2 WT and cysteine mutants were expressed in Nrf2 KO cells. Cells were treated with As3+ (10 μM, 5 h). A, cytoplasmic and nuclear fractions were immunoblotted with anti-Nrf2 and anti-actin. B, induction of Nqo1 was examined by Northern blotting.
Impact on Transcription Activation.
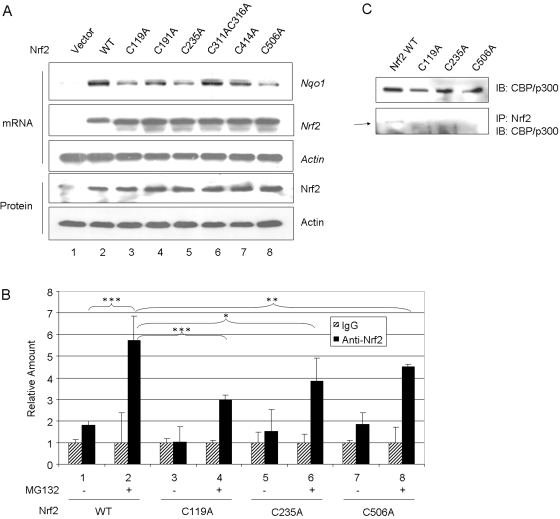
Loss of induction of Nqo1 can be due to loss of arsenic sensing, reduced t1/2, or both. Alternatively, mutation of the cysteines may also reduce the transcription activity of Nrf2. To distinguish the possibilities, Nrf2 WT and cysteine mutants were expressed in KO cells and were treated with both MG132 and As3+; this combined treatment activates Nrf2 by both blocking the 26S-proteasomal degradation of Nrf2 and by inhibiting Keap1-mediated ubiquitination of Nrf2. As controls, mRNA and protein levels of Nrf2 and mutants were shown to be expressed at comparable levels (Fig. 12A). Acting mRNA and protein were measured as the loading control. Indeed, the treatment increased the protein levels of Nrf2 and mutants to high and comparable levels. However, except for the C311A/C316A double mutant, in which induction of Nqo1 was comparable with that of WT, induction was largely reduced in C119A, C235A, and C506A mutants; induction in C191A and C414A was also reduced, but to a lesser extent. Thus, mutation of the cysteine residues, in particular Cys119, Cys235, and Cys506, impaired the transcription function of Nrf2 independently of Nrf2 turnover.
Fig. 12.
Effect of cysteine mutations on transcription activity of Nrf2. A, induction of Nqo1. Nrf2 WT and cysteine point mutants were expressed in Nrf2 KO cells. Induction of Nqo1 was examined by treating the cells with As3+ plus MG132 (15 μM) for 5 h. Northern blotting was performed for Nqo1, Nrf2, and Actin mRNA expressions, and immunoblotting was done for Nrf2 and actin protein expression. B, binding of cysteine mutants to endogenous ARE. Nrf2 and cysteine mutants were expressed in Nrf2 KO cells. After treatment with MG132 for 4 h, chromatin immunoprecipitation assay was performed. Nrf2-bound Nqo1 ARE was quantitated by real-time PCR. Data represent mean ± S.D. (n = 3). ∗, p > 0.05; ∗∗, p < 0.05; ∗∗∗, p < 0.001. C, coimmunoprecipitation of Nrf2 and mutants with endogenous CBP/p300. WT or cysteine mutants were expressed in KO cells treated with MG132. Cell lysate was immunoblotted with anti-CBP/p300 to show the protein expression (top) or immunoprecipitated with anti-Nrf2 and then blotted with anti-CBP/p300. Arrow, CBP/p300 protein band.
Chromatin immunoprecipitation was performed to examine the binding of C119A, C235A, and C506A to endogenous Nqo1 ARE enhancer. MG132 increased ARE binding of WT by ∼6-fold, C119A by 3-fold, Cys235 by 4-fold, and Cys506 by 4.5-fold over the IgG control (Fig. 12B). Therefore, mutation at Cys119, Cys235, and Cys506 reduced but did not totally abolish binding of the mutants to endogenous ARE.
Nrf2 recruits coactivators to mediate transcription activation. Of the coactivators, CBP/p300 seems to be essential. It is thus plausible to posit that Nrf2 cysteine residues affect the interaction between Nrf2 and CBP/p300 and thereby regulate target gene transcription by Nrf2. To test the hypothesis, WT, C119A, C235A, and C506A were expressed in KO cells, and interaction between Nrf2 and endogenous CBP/p300 was examined by coimmunoprecipitation. CBP/p300 and Nrf2 WT and mutants were shown to be expressed in the cells at comparable levels (Fig. 12C, top, and data not shown). Coimmunoprecipitation with anti-Nrf2 antibodies pulled down endogenous CBP/p300 from cells expressing wild-type but not C119A, C235A, or C506A mutants (Fig. 12C, bottom). The results indicate that Nrf2 Cys119, Cys235, and Cys506 are required for recruiting CBP/p300 that is necessary for the transactivation function of Nrf2.
Discussion
The Nrf2-Keap1-ARE pathway is a major mechanism by which cells defend against oxidative and electrophilic stresses. In this context, Nrf2 functions as an XAR to sense the chemical signals and mediate the induction of cytoprotective enzymes/proteins that protect cells from damage by the insults. Two features of Nrf2-mediated responses are apparent. First, because oxidants and electrophiles consist of a wide variety of structurally diverse chemicals, the mechanism of chemical-sensing by Nrf2 must allow high plasticity in chemical recognition. Second, coupling of chemical-sensing and gene transcription by Nrf2 permits host-environment response to occur rapidly and only as needed at a genomic level; it is reasonable to believe that this dual functionality requires considerable integration of the intrinsic receptivity and transcription activity of Nrf2 and its regulatory machinery at molecular levels (Ma, 2008; Ma and Lu, 2008).
Previous studies on Nrf2 regulation exclusively focused on Keap1, because Keap1 binds oxidants and electrophilic inducers through its cysteine thiol groups and suppresses Nrf2 by promoting the ubiquitination-proteasomal degradation of Nrf2 in the absence of an inducing signal (Dinkova-Kostova et al., 2002; Eggler et al., 2005; Hong et al., 2005; Yamamoto et al., 2008; Kobayashi et al., 2009; He and Ma, 2010). Two conclusions can be drawn from these studies. First, different inducers may preferentially bind to disparate sets of cysteine residues to activate Nrf2, suggesting the existence of “cysteine codes” that contribute to the high plasticity of chemical recognition by Nrf2. Second, different cysteine residues may play discrete functions. Thus, Cys151 may play a role in Nrf2 activation by certain inducers, such as tBHQ, whereas Cys273 and Cys288 are critical for suppression of Nrf2 (Yamamoto et al., 2008; He and Ma, 2010); in light of these findings, binding of Cys273 and Cys288 to inducers is probably not required for their function. However, binding of inducers to human Keap1 does not seem to be sufficient to dissociate Keap1 and Nrf2 (Eggler et al., 2005). Furthermore, the antioxidant tBHQ does not dissociate but increases the binding between Keap1 and Nrf2, whereas metal inducers, such as As3+, Cr6+, and Cd2+, disrupt Keap1-Nrf2 association (He et al., 2006, 2007, 2008). In addition, genetic evidence supports the requirement of protein factors other than Keap1 in the activation of Nrf2 by certain inducers (Kobayashi et al., 2009). It is clear additional molecular steps and/or factors are required for Nrf2 activation in a cell-type and inducer-dependent fashion.
In this study, we provided evidence showing that the cysteine residues of Nrf2 play multiple and critical roles in oxidant/electrophile sensing, Keap1-dependent ubiquitination/proteasomal degradation of Nrf2, and transcription activation of ARE target genes by Nrf2. This novel interaction between inducers and Nrf2 through its cysteine thiols expands the means of inducer-sensing by the Nrf2 regulator system. Because the cysteines regulate multiple Nrf2 functions, and inducers directly modulate the residues, it is also rational to conclude that this inducer-Nrf2 cysteine interaction effectively integrates chemical-sensing and transcription function of Nrf2. It is noteworthy that metal-activated transcription factor 1, an XAR that mediates the induction of metallothioneins 1 and 2 by metals and oxidants, contains a stretch of conserved cysteine residues at its C-terminal end that are critical for metal-sensing and transcription activation of metal-activated transcription factor 1 (He and Ma, 2009). Thus, inducer-cysteine thiol interaction may broadly regulate transcriptional, adaptive responses to oxidative and electrophilic stresses.
The evidence supporting a critical role of Nrf2 cysteine residues in Nrf2 regulation is several-fold. First, the cysteine residues are highly conserved across species from chicken to mouse, rat, and human, suggesting a revolutionarily conserved function. Indeed, the Nrf2/Keap1 pathway is found to be conserved as a critical defensive mechanism across mammalian species. Second, Nrf2 binds FlAsH and PAO, which potently induce ARE genes, both in vitro and in intact cells. Moreover, binding is blocked by tBHQ and As3+, indicating that tBHQ and As3+ bind to Nrf2 cysteine thiols in a similar fashion to FlAsH and PAO. We are currently using mass spectrometry to identify cysteine adducts of arsenic, FlAsH, and PAO in Nrf2, which provides additional means of analyzing inducer-Nrf2 cysteine interactions at a protein level. Third, mutation of individual cysteine residues markedly shortens the t1/2 of mutant proteins, reduces or totally abolishes Nrf2 responsiveness to arsenic for activation, and blocks the binding of coactivator CBP/p300 to Nrf2 for gene transcription. Likewise, it can be deduced from these findings that Nrf2 cysteine residues perform multiple functions in Nrf2 signaling including suppression of Nrf2-Keap1 association and Keap1-dependent ubiquitination of Nrf2, inducer-recognition and binding, and recruitment of coactivators to Nrf2 TAD for transcription of target genes. Therefore, our study clearly defines inducer-Nrf2 cysteine residue interactions as critical mechanisms of chemical-sensing and Nrf2 activation in additional to those of Keap1 cysteines. A cysteine residue in the Neh5 domain of human Nrf2 (Cys199) seems involved in nuclear export of Nrf2 and is redox-sensitive, suggesting the regulation of Nrf2 nuclear localization by redox signals via Cys199 (Li et al., 2006).
Deletion and mutational analyses revealed three groups of cysteine residues in Nrf2 for inducer binding. First, the three cysteine residues in the middle region (Cys235, Cys311, and Cys316) are critical for strong binding, because ND2 showed strong binding to PAO, but binding was largely reduced in ND3, in which the three cysteines were removed, and mutation of the residues to alanine abolished PAO binding. Second, Cys414 and Cys506 located in the DBD region are not capable of strong binding to PAO alone but are required for PAO binding in the full-length Nrf2 because mutation of either cysteine also abolished PAO binding. Third, Cys119 and Cys191 in the TAD region seem to mediate weak binding to PAO because mutation of either residue diminished but did not abolish the binding. In addition, Cys119 and Cys191 may negatively regulate PAO binding by other cysteines because deletion of the N-terminal region that contains Cys119 and Cys191 markedly increased PAO binding. Although the mechanism of inhibition is currently unclear, we posit that intramolecular interactions between Cys119/Cys191 and other cysteines impede binding of PAO to other cysteines. Of the two cysteines, Cys191 is more likely involved in suppression than Cys119. Therefore, binding of inducers to individual cysteine residues of Nrf2 probably depends on the context and three-dimensional structure of Nrf2 and is regulated by intramolecular interactions.
Nrf2 cysteine residues exhibit variable effects on Nrf2 functions. Mutation of individual cysteine residue to alanine shortens the t1/2 of each mutant. In particular, the t1/2 of C506A was reduced to 39% of wild-type, followed by C191A (58%), C235A (66%), and C119 (78%). It is remarkable that shortening of the t1/2 inversely correlates with increased binding to Keap1 and ubiquitination of Nrf2, consistent with the notion that the cysteine residues negatively control Nrf2-Keap1 binding and ubiquitination of Nrf2 by Keap1. Among the cysteines, Cys506 is probably critical for Nrf2-Keap1 interaction.
All cysteine residues except for Cys119 seem important for both activation of Nrf2 and induction of Nqo1 by arsenic. C119A retained much of its capability to stabilize Nrf2 but failed to mediate the induction. We envision that most cysteine residues except for Cys119 are required for oxidant/electrophile sensing and Nrf2 activation, whereas Cys119 is important for subsequent functions. The findings also imply that activation of Nrf2 in response to inducers and gene transcription by Nrf2 are separable functions. Indeed, cotreatment of cells expressing wild-type or cysteine mutants of Nrf2 with MG132 and arsenic, which activates Nrf2 by both blocking proteasomal degradation of Nrf2 and by inhibiting ubiquitination of the protein through Keap1, restore Nqo1 induction in the C311A/C316A double mutant, and in C191A and C414A to a less extent, indicating that these cysteines are mainly involved in Nrf2 activation. However, induction in C119A-, C235A-, and C506A-expressing cells is markedly reduced compared with wild-type, even though the protein levels are high and comparable with that of wild-type, suggesting a role of the cysteine residues in downstream functions, such as transactivation, of Nrf2 in addition to Nrf2 activation. Consistent with this prediction, the three mutants had reduced binding to endogenous ARE and lost binding to coactivator CBP/p300.
To our knowledge, our findings reveal for the first time that Nrf2 cysteine residues mediate multiple functions of Nrf2 in response to inducers that include oxidant/electrophile-sensing by direct binding to inducers, repressing Nrf2-Keap1 interaction and Keap1-dependent ubiquitination of Nrf2, and recruiting coactivators. Given the importance of Nrf2-mediated defense in cell survival and the chemical complexity of oxidant/electrophilic insults, it is not surprising that Nrf2 used a dual sensor mechanism in which both Keap1 and Nrf2 recognize oxidants/electrophilic signals via their cysteine thiols to activate Nrf2 through an integrated signaling cascade. In this respect, our findings pave a new avenue to decipher the higher-order integration of signal transduction by the Nrf2-Keap1-ARE system that include Nrf2-Keap1 interaction under both basal and activated states, ubiquitination of Nrf2, and interaction of Nrf2 and coactivators.
This work was supported by the Intramural Research program of the National Institutes of Health National Institute for Occupational Safety and Health.
Article, publication date, and citation information can be found at http://molpharm.aspetjournals.org.
doi:10.1124/mol.109.058453
- CNC bZip
- cap “n” collar basic leucine zipper protein
- ARE
- antioxidant response element
- Bal
- 2,3-dimercaptopropanol
- β-ME
- β-mercaptoethanol
- CHX
- cycloheximide
- DBD
- DNA-binding domain
- HO1
- heme oxygenase 1
- Keap1
- Kelch-like ECH-associated protein-1
- MTF1
- metal-activated transcription factor 1
- Nrf2
- nuclear factor erythroid 2-related factor 2
- PAO
- phenylarsine oxide
- TAD
- transcription activation domain
- tBHQ
- tert-butylhydroquinone
- CBP
- cAMP response element-binding protein-binding protein
- XAR
- xenobiotic-activated receptor
- KO
- knockout
- WT
- wild type
- PBS
- phosphate-buffered saline
- PAGE
- polyacrylamide gel electrophoresis
- IP
- immunoprecipitation
- HA
- hemagglutinin
- Neh
- nuclear factor erythroid 2 Ech homology domain
- PCR
- polymerase chain reaction
- PIPES
- piperazine-N,N′-bis(2-ethanesulfonic acid)
- MG132
- N-benzoyloxycarbonyl (Z)-Leu-Leu-leucinal
- SV40
- simian virus 40.
References
- Aitken A, Learmonth M. (1996) Estimation of disulfide bonds using Ellman's reagent, in Protein Protocol Handbook (Walker JM. ed) pp 487–494, Humana Press, Inc, Totowa, NJ: [Google Scholar]
- Bi Y, Palmiter RD, Wood KM, Ma Q. (2004) Induction of metallothionein I by phenolic antioxidants requires metal-activated transcription factor 1 (MTF-1) and zinc. Biochem J 380:695–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K, Han XD, Kan YW. (2001) An important function of Nrf2 in combating oxidative stress: detoxification of acetaminophen. Proc Natl Acad Sci U S A 98:4611–4616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HY, Jedlicka AE, Reddy SP, Zhang LY, Kensler TW, Kleeberger SR. (2002) Linkage analysis of susceptibility to hyperoxia. Nrf2 is a candidate gene. Am J Respir Cell Mol Biol 26:42–51 [DOI] [PubMed] [Google Scholar]
- Dinkova-Kostova AT, Holtzclaw WD, Cole RN, Itoh K, Wakabayashi N, Katoh Y, Yamamoto M, Talalay P. (2002) Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc Natl Acad Sci U S A 99:11908–11913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkova-Kostova AT, Jenkins SN, Fahey JW, Ye L, Wehage SL, Liby KT, Stephenson KK, Wade KL, Talalay P. (2006) Protection against UV-light-induced skin carcinogenesis in SKH-1 high-risk mice by sulforaphane-containing broccoli sprout extracts. Cancer Lett 240:243–252 [DOI] [PubMed] [Google Scholar]
- Dinkova-Kostova AT, Liby KT, Stephenson KK, Holtzclaw WD, Gao X, Suh N, Williams C, Risingsong R, Honda T, Gribble GW, et al. (2005) Extremely potent triterpenoid inducers of the phase 2 response: correlations of protection against oxidant and inflammatory stress. Proc Natl Acad Sci U S A 102:4584–4589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggler AL, Liu G, Pezzuto JM, van Breemen RB, Mesecar AD. (2005) Modifying specific cysteines of the electrophile-sensing human Keap1 protein is insufficient to disrupt binding to the Nrf2 domain Neh2. Proc Natl Acad Sci U S A 102:10070–10075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Chen MG, Lin GX, Ma Q. (2006) Arsenic induces NAD(P)H-quinone oxidoreductase I by disrupting the Nrf2 x Keap1 x Cul3 complex and recruiting Nrf2 x Maf to the antioxidant response element enhancer. J Biol Chem 281:23620–23631 [DOI] [PubMed] [Google Scholar]
- He X, Chen MG, Ma Q. (2008) Activation of Nrf2 in defense against cadmium-induced oxidative stress. Chem Res Toxicol 21:1375–1383 [DOI] [PubMed] [Google Scholar]
- He X, Kan H, Cai L, Ma Q. (2009) Nrf2 is critical in defense against high glucose-induced oxidative damage in cardiomyocytes. J Mol Cell Cardiol 46:47–58 [DOI] [PubMed] [Google Scholar]
- He X, Lin GX, Chen MG, Zhang JX, Ma Q. (2007) Protection against chromium (VI)-induced oxidative stress and apoptosis by Nrf2. Recruiting Nrf2 into the nucleus and disrupting the nuclear Nrf2/Keap1 association. Toxicol Sci 98:298–309 [DOI] [PubMed] [Google Scholar]
- He X, Ma Q. (2009) Induction of metallothionein I by arsenic via metal-activated transcription factor 1: critical role of C-terminal cysteine residues in arsenic sensing. J Biol Chem 284:12609–12621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Ma Q. (2010) Critical cysteine residues of Keap1 in arsenic sensing and suppression of Nrf2. J Pharmacol Exp Ther doi: 10.1124/jpet.109.160465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong F, Sekhar KR, Freeman ML, Liebler DC. (2005) Specific patterns of electrophile adduction trigger Keap1 ubiquitination and Nrf2 activation. J Biol Chem 280:31768–31775 [DOI] [PubMed] [Google Scholar]
- Hu X, Roberts JR, Apopa PL, Kan YW, Ma Q. (2006) Accelerated ovarian failure induced by 4-vinyl cyclohexene diepoxide in Nrf2 null mice. Mol Cell Biol 26:940–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbs AF, Benkovic SA, Miller DB, O'Callaghan JP, Battelli L, Schwegler-Berry D, Ma Q. (2007) Vacuolar leukoencephalopathy with widespread astrogliosis in mice lacking transcription factor Nrf2. Am J Pathol 170:2068–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kensler TW, Wakabayashi N, Biswal S. (2007) Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol 47:89–116 [DOI] [PubMed] [Google Scholar]
- Kobayashi A, Kang MI, Watai Y, Tong KI, Shibata T, Uchida K, Yamamoto M. (2006) Oxidative and electrophilic stresses activate Nrf2 through inhibition of ubiquitination activity of Keap1. Mol Cell Biol 26:221–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi A, Ohta T, Yamamoto M. (2004) Unique function of the Nrf2-Keap1 pathway in the inducible expression of antioxidant and detoxifying enzymes. Methods Enzymol 378:273–286 [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Li L, Iwamoto N, Nakajima-Takagi Y, Kaneko H, Nakayama Y, Eguchi M, Wada Y, Kumagai Y, Yamamoto M. (2009) The antioxidant defense system Keap1-Nrf2 comprises a multiple sensing mechanism for responding to a wide range of chemical compounds. Mol Cell Biol 29:493–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung L, Kwong M, Hou S, Lee C, Chan JY. (2003) Deficiency of the Nrf1 and Nrf2 transcription factors results in early embryonic lethality and severe oxidative stress. J Biol Chem 278:48021–48029 [DOI] [PubMed] [Google Scholar]
- Li W, Yu SW, Kong AN.(2006) Nrf2 possesses a redox-sensitive nuclear exporting signal in the Neh5 transactivation domain. J Biol Chem 281:27251–27263 [DOI] [PubMed] [Google Scholar]
- Ma Q. (2008) Xenobiotic-activated receptors: from transcription to drug metabolism to disease. Chem Res Toxicol 21:1651–1671 [DOI] [PubMed] [Google Scholar]
- Ma Q, Battelli L, Hubbs AF. (2006) Multiorgan Autoimmune Inflammation, Enhanced Lymphoproliferation, and Impaired Homeostasis of Reactive Oxygen Species in Mice Lacking the Antioxidant-Activated Transcription Factor Nrf2. Am J Pathol 168:1960–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Lu AY. (2008) The challenges of dealing with promiscuous drug-metabolizing enzymes, receptors and transporters. Curr Drug Metab 9:374–383 [DOI] [PubMed] [Google Scholar]
- Marzec JM, Christie JD, Reddy SP, Jedlicka AE, Vuong H, Lanken PN, Aplenc R, Yamamoto T, Yamamoto M, Cho HY, et al. (2007) Functional polymorphisms in the transcription factor NRF2 in humans increase the risk of acute lung injury. FASEB J 21:2237–2246 [DOI] [PubMed] [Google Scholar]
- Nguyen T, Sherratt PJ, Pickett CB. (2003) Regulatory mechanisms controlling gene expression mediated by the antioxidant response element. Annu Rev Pharmacol Toxicol 43:233–260 [DOI] [PubMed] [Google Scholar]
- Nishikimi A, Kira Y, Kasahara E, Sato EF, Kanno T, Utsumi K, Inoue M. (2001) Tributyltin interacts with mitochondria and induces cytochrome c release. Biochem J 356:621–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T, Iijima K, Miyamoto M, Nakahara I, Tanaka H, Ohtsuji M, Suzuki T, Kobayashi A, Yokota J, Sakiyama T, et al. (2008) Loss of Keap1 function activates Nrf2 and provides advantages for lung cancer cell growth. Cancer Res 68:1303–1309 [DOI] [PubMed] [Google Scholar]
- Ramos-Gomez M, Kwak MK, Dolan PM, Itoh K, Yamamoto M, Talalay P, Kensler TW. (2001) Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in nrf2 transcription factor-deficient mice. Proc Natl Acad Sci U S A 98:3410–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussan TE, Rangasamy T, Blake DJ, Malhotra D, El-Haddad H, Bedja D, Yates MS, Kombairaju P, Yamamoto M, Liby KT, et al. (2009) Targeting Nrf2 with the triterpenoid CDDO-imidazolide attenuates cigarette smoke-induced emphysema and cardiac dysfunction in mice. Proc Natl Acad Sci U S A 106:250–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenouchi T, Iwamaru Y, Sato M, Yokoyama T, Shinagawa M, Kitani H. (2007) Establishment and characterization of SV40 large T antigen-immortalized cell lines derived from fetal bovine brain tissues after prolonged cryopreservation. Cell Biol Int 31:57–64 [DOI] [PubMed] [Google Scholar]
- Wakabayashi N, Itoh K, Wakabayashi J, Motohashi H, Noda S, Takahashi S, Imakado S, Kotsuji T, Otsuka F, Roop DR, et al. (2003) Keap1-null mutation leads to postnatal lethality due to constitutive Nrf2 activation. Nat Genet 35:238–245 [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Suzuki T, Kobayashi A, Wakabayashi J, Maher J, Motohashi H, Yamamoto M. (2008) Physiological significance of reactive cysteine residues of Keap1 in determining Nrf2 activity. Mol Cell Biol 28:2758–2770 [DOI] [PMC free article] [PubMed] [Google Scholar]