Abstract
The 90-kDa heat shock protein (Hsp90) assists in the proper folding of numerous mutated or overexpressed signal transduction proteins that are involved in cancer. Consequently, there is considerable interest in developing chemotherapeutic drugs that specifically disrupt the function of Hsp90. Here, we investigated the extent to which a novel novobiocin-derived C-terminal Hsp90 inhibitor, designated KU135, induced antiproliferative effects in Jurkat T-lymphocytes. The results indicated that KU135 bound directly to Hsp90, caused the degradation of known Hsp90 client proteins, and induced more potent antiproliferative effects than the established N-terminal Hsp90 inhibitor 17-allylamino-demethoxygeldanamycin (17-AAG). Closer examination of the cellular response to KU135 and 17-AAG revealed that only 17-AAG induced a strong up-regulation of Hsp70 and Hsp90. In addition, KU135 caused wild-type cells to undergo G2/M arrest, whereas cells treated with 17-AAG accumulated in G1. Furthermore, KU135 but not 17-AAG was found to be a potent inducer of mitochondria-mediated apoptosis as evidenced, in part, by the fact that cell death was inhibited to a similar extent by Bcl-2/Bcl-xL overexpression or the depletion of apoptotic protease-activating factor-1 (Apaf-1). Together, these data suggest that KU135 inhibits cell proliferation by regulating signaling pathways that are mechanistically different from those targeted by 17-AAG and as such represents a novel opportunity for Hsp90 inhibition.
Members of the 90-kDa heat shock protein (Hsp90) family are commonly overexpressed in cancer cells and play critical roles in promoting survival by chaperoning client proteins associated with all six of the acquired cancer capabilities (Hanahan and Weinberg, 2000; Isaacs et al., 2003; Blagg and Kerr, 2006). A growing number of natural product, synthetic, and semisynthetic Hsp90 inhibitors are being developed that largely target the N-terminal ATP-binding pocket and have been shown to cause potent antiproliferative effects (Roe et al., 1999; Whitesell and Lindquist, 2005; Avila et al., 2006a,b). However, the potential clinical utility of several of the N-terminal inhibitors as anticancer drugs has been dampened significantly due to concerns about their adverse hepatotoxic effects (Egorin et al., 1998) and tendency to induce expression of cytoprotective Hsp90 and Hsp70 proteins (Chiosis et al., 2003; Whitesell et al., 2003; Powers and Workman, 2007; Schmitt et al., 2007). More recently, the observation was made that Hsp90 contains a previously unrecognized C-terminal ATP-binding domain (Marcu et al., 2000a,b), which has led several groups to pursue the development of specific C-terminal Hsp90 inhibitors as potential anticancer drug modalities (Burlison et al., 2006, 2008; Le Bras et al., 2007; Donnelly et al., 2008; Radanyi et al., 2009). Both N-terminal and C-terminal Hsp90 inhibitors can exert an antiproliferative response, in some instances, by stimulating apoptosis (Isaacs et al., 2003; Georgakis and Younes, 2005; Whitesell and Lindquist, 2005), although the underlying mechanisms are not well understood.
Apoptotic cell death is mediated by a family of cysteine proteases that cleave after aspartate residues (caspases). In general, the activation of caspases can occur by two distinct signaling pathways. Within the extrinsic (receptor-mediated) pathway, ligand (e.g., FasL and tumor necrosis factor-α) binding to a corresponding death receptor (e.g., Fas and tumor necrosis factor-R1) leads to recruitment of FADD and procaspase-8 molecules to the cytosolic side of the cell membrane to form the death-inducing signaling complex (Kischkel et al., 1995). Activation of procaspase-8 occurs at the death-inducing signaling complex, and active caspase-8, in turn, can activate caspase-3 directly or by first cleaving and activating the BH3-only protein Bid to truncated Bid, which, in turn, can engage the intrinsic or mitochondria-mediated apoptotic pathway (Li et al., 1998; Luo et al., 1998). The intrinsic (mitochondria-mediated) pathway, however, is most often initiated by cytotoxic stress, including growth factor withdrawal, DNA damage, γ-radiation, and heat. In response to these types of stimuli, mitochondrial outer membrane permeabilization (MOMP) generally occurs, resulting in the release of cytochrome c, Smac (also known as DIABLO), and Omi into the cytosol, in which they work together to activate procaspase-9 within the Apaf-1 apoptosome complex (Li et al., 1997). Once activated, caspase-9 can cleave and thereby activate caspase-3, which, in turn, is responsible for proteolytic dismantling of the cell.
The aim of the current study was to investigate the extent to which a novobiocin-derived C-terminal Hsp90 inhibitor, KU135, could induce an antiproliferative response and how such an effect compared with that exhibited by the established N-terminal inhibitor 17-allylamino-demethoxygeldanamycin (17-AAG). The results indicated that KU135 binds directly to Hsp90 and exhibits more potent antiproliferative effects than 17-AAG. Both 17-AAG and KU135 stimulated the degradation of known Hsp90 client proteins, whereas only 17-AAG triggered a pronounced increased expression of Hsp70 and Hsp90 proteins. KU135 was found to induce both G2/M cell cycle arrest and mitochondria-mediated apoptosis, whereas 17-AAG predominantly induced G1 cell cycle arrest.
Materials and Methods
Cell Culture.
Wild-type (clones E6.1 and A3) and caspase-8-deficient (clone I 9.2) Jurkat T-lymphocytes were cultured in RPMI 1640 medium (Invitrogen, Carlsbad, CA) supplemented with 10% (v/v) heat-inactivated fetal bovine serum (HyClone, Logan, UT), 2% (w/v) glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin at 37°C in a humidified 5% CO2 incubator. For Apaf-1-deficient, Bcl-2-overexpressing, and Bcl-xL-overexpressing Jurkat cells, 1 mg/ml G-418 (Geneticin; Invitrogen) was substituted for penicillin and streptomycin. Cells were maintained in an exponential growth phase and were replated in fresh complete nonselective medium for all experiments. Cells were treated with KU135 (2.5 nM to 50 μM), novobiocin (25 nM to 500 μM) (Sigma, St. Louis, MO), 17-AAG (0.5 nM to 10 μM) (InvivoGen, San Diego, CA), etoposide (2.4 nM to 47 μM) (Sigma), or DMSO (≤0.5% final concentration).
Cell Proliferation Assay.
Cellular metabolic activity/viability was assessed using the CellTiter 96 AQueous One Solution Cell Proliferation Assay (Promega, Madison, WI) according to the manufacturer's instructions. This approach used the tetrazolium compound 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt (MTS) that is bioreduced by metabolically active/viable cells into a colored formazan product that is soluble in tissue culture medium. In brief, 3 × 104 cells/well were cultured in 96-well plates in 100 μl of complete growth medium for 24 or 48 h. At the end of incubation, 20 μl of MTS solution was added to each well and incubated for 1 to 4 h at 37°C. The amount of soluble formazan from cellular reduction of MTS was assessed by measurement of absorbance at 490 nm VICTOR3V Multilabel Reader (PerkinElmer Life and Analytical Sciences, Waltham, MA). Data were analyzed using nonlinear regression and sigmoidal dose-response curves (Prism; GraphPad Software Inc., San Diego, CA) from which IC50 values were calculated.
Flow Cytometry for Cell Death, Cell Cycle, and Mitochondrial Membrane Potential Measurements.
Phosphatidylserine exposure on the outer leaflet of the plasma membrane was detected using the annexin V-fluorescein isothiocyanate (FITC) Apoptosis Detection Kit II (BD Pharmingen, San Diego, CA) according to the manufacturer's instructions. In brief, 106 cells were pelleted after KU135 or 17-AAG treatment and washed with PBS. Next, the cells were resuspended in 100 μl of binding buffer containing annexin V-FITC and propidium iodide. Before flow cytometric analysis, 400 μl of binding buffer was added to the cells. Necrotic cells never accounted for more than 1% of total cells. For cell cycle analysis, 106 cells were pelleted after KU135 or 17-AAG treatment and washed with cold PBS. Cells were fixed in 1 ml of ice-cold 70% ethanol and incubated at room temperature (22°C) for 30 min. Next, the fixed cells were pelleted, resupended in 500 μl of PI/RNase staining buffer (BD Pharmingen), incubated at room temperature (22°C), and analyzed by flow cytometry. For mitochondrial membrane potential (ΔΨ) determination, the MitoProbe DiIC1(5) Kit (Invitrogen) was used. Cells (106) were pelleted after drug treatment, washed once with PBS, and resuspended in 1 ml of warm PBS. Next, 5 μl of 10 μM DiIC1(5) were added to the cells and incubated in a humidified 5% CO2 incubator at 37°C for 15 min. Cells were pelleted, resuspended in 500 μl of PBS, and analyzed by flow cytometry.
Western Blotting.
Pelleted cells (5 × 106) were resuspended and lysed in 200 μl of ice-cold lysis buffer (10 mM Tris/HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 1 mM EDTA, and 0.1% Nonidet P-40) supplemented with a mixture of protease inhibitors (Complete Mini EDTA-Free; Roche Applied Science, Indianapolis, IN). Protein concentrations were determined using the bicinchoninic acid assay (Pierce, Rockford, IL), and equal amounts were mixed with Laemmli loading buffer. Western blot analysis was carried out as described previously (Robertson et al., 2002). The antibodies used were rabbit anti-Akt (pan) (clone C67E7; Cell Signaling Technology, Danvers, MA), rabbit anti-Bak, NT (Millipore Corporation, Billerica, MA), mouse anti-β-actin (clone AC-15; Sigma), mouse anti-caspase-2 (clone 35; BD Pharmingen), rabbit anti-caspase-3 (clone 8G10; Cell Signaling Technology), rabbit anti-caspase-9 (Cell Signaling Technology), mouse anti-cdc2 p34 (Santa Cruz Biotechnology, Santa Cruz, CA), mouse anti-cytochrome c (clone 7H8.2C12; BD Pharmingen), rat anti-GRP94 (clone 9G10; Assay Designs, Ann Arbor, MI), rabbit anti-Hif-1α (Novus Biologicals, Littleton, CO), mouse anti-Hsp70 (Hsp72) (clone C92F3A-5; Assay Designs), rat anti-Hsp90α (clone 9D2; Assay Designs), mouse anti-Hsp90β (clone K3705; Assay Designs), rabbit anti-phospho-Akt (Ser473) (clone 193H12, Cell Signaling Technology), and mouse anti-TRAP (Affinity BioReagents, Rockford, IL).
Novobiocin Affinity Column Chromatography.
Novobiocin-Sepharose was prepared as described previously (Marcu et al., 2000b). In brief, 3 g of epoxy-activated Sepharose 6B (Sigma) was washed and swollen in 100 ml of distilled water for 1 h at room temperature (25°C). The resin was washed further with coupling buffer (300 mM sodium carbonate, pH 9.5). The gel was mixed with 400 mg of novobiocin (sodium salt; Sigma) in 10 ml of coupling buffer and incubated at 37°C with gentle rotation for 20 h. Excess ligand was washed away with coupling buffer, and the remaining epoxy-active groups were blocked with 1 M ethanolamine in coupling buffer for 12 h at 30°C with gentle shaking. The gel was washed sequentially with coupling buffer, 500 mM NaCl in coupling buffer, distilled water, 500 mM NaCl in 100 mM sodium acetate, pH 4.0, and again in distilled water. The resin was subsequently equilibrated in 25 mM HEPES, pH 8.0, containing 1 mM EDTA, 10% ethylene glycol, and 200 mM KCl and stored in the dark at 4°C.
Rabbit reticulocyte lysate was incubated in the presence of an ATP-regenerating system (10 mM creatine phosphate and 20 U/ml creatine phosphokinase) for 5 min at 37°C, after which the reticulocyte lysate was applied to the novobiocin column that had been pre-equilibrated with 10 mM Tris-HCl buffer, pH 7.4. The novobiocin column was then washed with 20 volumes of 10 mM Tris-HCl, pH 7.4, buffer containing 150 mM NaCl. Subsequently, bound proteins were eluted with increasing concentrations of KU135 or 10 mM novobiocin. All experiments were carried out under dark conditions at 4°C. The eluents were separated via SDS-PAGE and analyzed with Hsp90 antibodies.
Proteolytic Fingerprinting of Hsp90.
Proteolytic fingerprinting was performed using a modified method described previously (Yun et al., 2004). In brief, 50% (v/v) rabbit reticulocyte lysate, an ATP-regenerating system (10 mM creatine phosphate and 20 U/ml creatine phosphokinase), and 75 mM KCl was incubated in the presence of drug or vehicle for 5 min at 37°C. After the incubation period, the reaction mixtures were chilled on ice and digested with 125 μg/ml N-tosyl-l-phenylalanine chloromethyl ketone-treated trypsin (Worthington Biomedical, Lakewood, NJ) for 6 min (Hartson et al., 1999). Reactions were terminated by immediate boiling in Laemmli loading buffer. Subsequently, the samples were separated by SDS-PAGE and Western blotted using antibodies specific to the N terminus of Hsp90 (PA3-013; Affinity BioReagents) or the C terminus of Hsp90 (clone AC88; Assay Designs).
Surface Plasmon Resonance Analysis of KU135 Binding to Hsp90.
Insect Sf9 cells overexpressing Hsp90β were cultured and harvested by the Baculovirus/Monoclonal Antibody Core Facility at Baylor College of Medicine (Houston, TX). Hsp90β was extracted and purified (>98% pure) as described previously (Grenert et al., 1997; Owen et al., 2002) but without the initial DEAE-cellulose chromatography step. The surface of an SSOO COOH1 surface plasmon resonance (SPR) sensor chip mounted in a SensiQ SPR instrument (ICX Nomadics, Arlington, VA) was activated by treatment with N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride and N-hydroxysuccinimide for preferential cross-linking of the protein's N terminus to the surface. For Hsp90 immobilization, 250 μl of Hsp90 (7 mg/ml) in 20 mM sodium bicarbonate buffer, pH 8, containing 150 mM NaCl was injected at a flow rate of 10 μl/min, giving 2800 response units of protein stably immobilized to the surface of the flow cell, representing ∼0.08 picomoles Hsp90. Unreactive groups were then quenched with 1 M ethanolamine, pH 8, and the surface washed with buffer containing 10 mM Pipes, pH 7.4, 300 mM NaCl, 2% DMSO, and 0.01% Igepal CA-630 followed by equilibration in assay buffer. KU135 or novobiocin was diluted in assay buffer containing 10 mM Pipes, pH 7.4, 300 mM NaCl, 2% DMSO, and 0.00075% Igepal CA-630 and injected over the surface of the derivatized chip at a flow rate of 25 μl/min at 25°C at the indicated concentrations. All measurements were done in triplicate. SPR binding curves were analyzed using QDAT software (ICX Nomadics). To calculate the Kd, binding constants were fitted using the equations: Req = KA(A)Rmax/(KA(A) +1) and regraphed using Origin software (OriginLab Corp, Northampton, MA) for Scatchard analysis and to demonstrate saturation of binding. Novobiocin's binding isotherm was globally fit for total and nonspecific binding using the equation Y = (Bmax(X))/(KD + X)) + NS(X) to fit total binding and the equation Y = NX(X) to fit nonspecific binding.
Subcellular Fractionation.
Cells (106) were washed with PBS, resuspended in 50 μl of buffer (140 mM mannitol, 46 mM sucrose, 50 mM KCl, 1 mM KH2PO4, 5 mM MgCl2, 1 mM EGTA, and 5 mM Tris, pH 7.4) supplemented with a mixture of protease inhibitors (Complete Mini-EDTA Free) and permeabilized with 3 μg of digitonin (Sigma) on ice for 10 min. Plasma membrane permeabilization was monitored by trypan blue staining, and cell suspensions were centrifuged at 12,000g for 10 min at 4°C. Supernatant and pellet fractions were subjected to Western blot analysis.
Determination of Bak Oligomerization.
Cells (106) were harvested, washed, and resuspended in 80 μl of 100 mM EDTA/PBS, incubated with the cross-linking agent bismaleimidohexane (1 mM) for 30 min at room temperature (22°C), quenched with 100 mM dithiothreitol for 15 min, pelleted, and processed for Western blotting.
Results
A Novobiocin-Derived C-Terminal Hsp90 Inhibitor, KU135, Induces a Potent Antiproliferative Response in Jurkat T Lymphocytes.
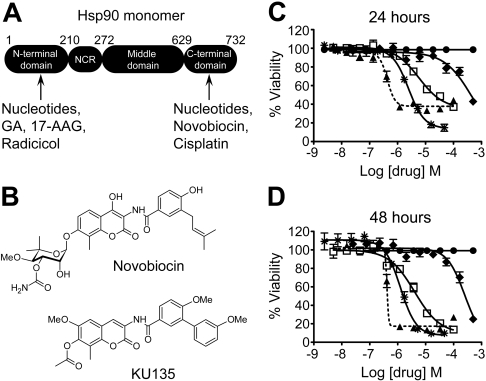
As mentioned previously, a new possibility for Hsp90 inhibition was raised when the antibiotic novobiocin was found to bind a previously unidentified C-terminal ATP-binding site (Fig. 1A), albeit with poor affinity (IC50, ∼700 μM). KU135 is part of a library of structurally related novobiocin derivatives (Fig. 1B) that will be discussed in detail in a forthcoming manuscript (H. Zhao, A. C. Donnelly, B. Reddy, G. A. Vielhauer, J. M. Holzbeierlein, B. S. J. Blagg, in preparation). However, during a previous study (Burlison et al., 2008), it was observed that modification of novobiocin by the inclusion of biaryl substituents led to compounds that exhibited improved activity relative to novobiocin. Subsequent modification of the sugar side chain (noviose) led to a very active series of compounds, including KU135 as one of the most potent of the group (Fig. 1B). This compound was prepared at greater than 98% purity (high-performance liquid chromatography peak area) in 14 steps with final purification by normal phase chromatography followed by recrystallization.
Fig. 1.
KU135, a novel analog of novobiocin, causes antiproliferation. A, Hsp90 contains four functional domains, and the numbering (1–732) reflects the approximate number of amino acids that comprise each region. NCR refers to the negatively charged region that links the N-terminal and middle domains. The regions in which nucleotides, small-molecule inhibitors, and/or cisplatin bind Hsp90 are shown. B, the structures of novobiocin and KU135. C, cells (3 × 104/well) were cultured in a 96-well plate in the absence or presence of DMSO (●), KU135 (2.5 nM to 50 μM) (▲), 17-AAG (0.5 nM to 10 μM) (□), etoposide (2.4 nM to 47 μM) (∗), or novobiocin (25 nM to 500 μM) (♦) for 24 or 48 h and processed for cell proliferation determination by the MTS assay.
We began our studies by evaluating the extent to which wild-type Jurkat T-lymphoblastoid leukemia cells (clone E6.1) were sensitive to KU135. The potency of KU135 was compared with that of novobiocin, 17-AAG, and the anticancer drug etoposide, which damages DNA by inhibiting topoisomerase II and is a well established inducer of mitochondria-mediated apoptosis. Wild-type Jurkat cells were incubated with increasing concentrations of KU135 (2.5 nM to 50 μM), novobiocin (25 nM to 500 μM), 17-AAG (0.5 nM to 10 μM), and etoposide (2.4 nM to 47 μM), or DMSO (0.5% final concentration) for 24 or 48 h, and cell viability was determined by the MTS proliferation assay. As illustrated in Fig. 1, C and D, all four compounds (i.e., KU135, novobiocin, 17-AAG, and etoposide) inhibited cell proliferation in a concentration- and time-dependent manner. In particular, the IC50 values for KU135, etoposide, 17-AAG, and novobiocin at 48 h after treatment were found to be 416 nM and 1.3, 4, and 252 μM, respectively. Thus, although all four drug treatments inhibited Jurkat cell proliferation, KU135 was ∼3, ∼10, and ∼600 times more potent than etoposide, 17-AAG, and novobiocin, respectively.
KU135 Inhibits Cell Division and Induces Apoptotic Cell Death in Wild-Type Jurkat Cells.
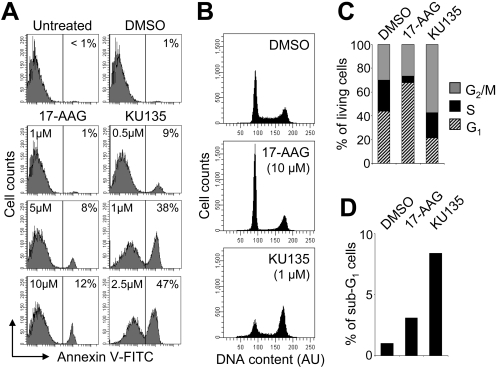
Next, we were interested in determining whether the decrease in viability observed in response to KU135 administration using the MTS assay was due to an induction of apoptosis. Indeed, as shown in Fig. 2A, wild-type Jurkat cells underwent apoptosis in a concentration-dependent manner when incubated with KU135 for 24 h. In particular, we found that treatment with 1 and 2.5 μM KU135 induced 38 and 47% apoptosis, respectively (Fig. 2A). The level of KU135-induced apoptosis did not increase further when cells were incubated with concentrations of KU135 greater than 2.5 μM (data not shown). By comparison, and in agreement with previous findings (Rahmani et al., 2003), 17-AAG exhibited a much milder apoptotic effect in Jurkat cells, even at the highest concentration of 10 μM, where only 12% of cells had externalized phosphatidylserine. Combined, these data suggest that the more potent antiproliferative effect of KU135 compared with 17-AAG (Fig. 1) is due, at least in part, to the ability of KU135 to induce apoptotic cell death.
Fig. 2.
KU135 and 17-AAG exert different effects on apoptosis and cell cycle distribution. A, cells (5 × 105/ml) were cultured with DMSO or the indicated concentrations of 17-AAG or KU135 for 24 h and processed for apoptotic cell death determination by flow cytometric analysis of annexin V-FITC staining in a buffer containing propidium iodide. Increased annexin V-FITC fluorescence reflects phosphatidylserine externalization on the plasma membrane, and numbers refer to the percentage of cells undergoing apoptosis. B, cells (5 × 105/ml) were cultured with DMSO, 1 μM KU135, or 10 μM 17-AAG for 24 h, fixed in 70% ethanol, and analyzed for cell cycle status using PI/RNase staining buffer and flow cytometry. C, quantification of cell cycle distribution of viable/euploid cells as obtained in B. D, quantification of the apoptotic hypodiploid (sub-G1) cells as obtained in B. AU, arbitrary units.
Because 17-AAG induced significantly less apoptosis than KU135 and yet was found to have an IC50 value relatively similar to that of etoposide (Fig. 1), we speculated that the antiproliferative activity of 17-AAG might involve an effect on cell cycle status. As illustrated in Fig. 2, B and C, the distribution of nonapoptotic cells differed between KU135- and 17-AAG-treated cells at 24 h. Specifically, 66 and 26% of cells incubated with 17-AAG had accumulated in G1 and G2/M phases of the cell cycle, respectively. This was compared with KU135-treated cells in which 20 and 53% of cells had accumulated in G1 and G2/M phases of the cell cycle, respectively (Fig. 2, B and C). In agreement with the low level of 17-AAG-induced apoptosis that was observed compared with KU135 as assessed by annexin V positivity (Fig. 2A), cell cycle distribution analysis revealed a considerably smaller fraction of sub-G1 cells in response to 17-AAG than KU135 (Fig. 2, B and D). The percentage of KU135-treated cells containing hypodiploid DNA content was low compared with the amount of apoptosis that was observed using annexin V-FITC and propidium iodide staining as an endpoint; this can probably be explained by the fact that DNA fragmentation is a late apoptotic event relative to phosphatidylserine externalization. Overall, these data indicate that the antiproliferative effect of KU135 is mechanistically different from that exhibited by 17-AAG.
Binding of KU135 to Hsp90 and the Effect of KU135 and 17-AAG on the Levels of Hsp90 Isoforms and Associated Client Proteins.
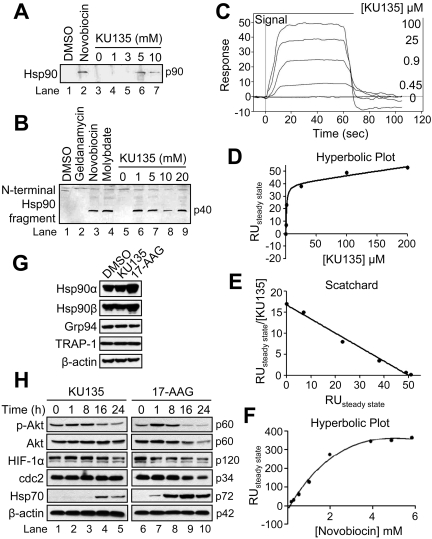
Next, using three complementary approaches, we tested the extent to which KU135 bound Hsp90. Initially, we performed novobiocin-binding experiments in which rabbit reticulocyte lysate was loaded onto a novobiocin-Sepharose column to bind endogenous Hsp90. Thereafter, the column was washed extensively with 10 mM Tris-HCl buffer, pH 7.4, containing 150 mM NaCl, and bound proteins were eluted with increasing concentrations of KU135 or 10 mM novobiocin. As shown in Fig. 3A, Hsp90 was eluted from the column with KU135 in a concentration-dependent manner (lanes 3–7). In contrast to novobiocin, which binds the C terminus of Hsp90 (lane 2), the N-terminal Hsp90 inhibitor geldanamycin was unable to elute Hsp90 from the novobiocin-Sepharose column (data not shown), which is consistent with previous studies by Neckers and coworkers (Marcu et al., 2000b). Next, proteolytic fingerprinting of Hsp90 in rabbit reticulocyte lysate was performed to determine whether binding of a C-terminal inhibitor, such as novobiocin, molybdate, and perhaps KU135, caused Hsp90 to adopt a conformation that differs from the tertiary structure it assumes upon binding of the N-terminal inhibitor geldanamycin (Fig. 3B) (Yun et al., 2004). In brief, rabbit reticulocyte lysate was incubated at 37°C for 5 min with DMSO, 20 mM geldanamycin, novobiocin, or molybdate (lanes 1–4, respectively), or 0, 1, 5, 10, and 20 mM KU135 (lanes 5–9), partially digested with trypsin, subjected to SDS-PAGE, and Western-blotted with an Hsp90 antibody against the N terminus of the protein. As shown in Fig. 3B, reticulocyte lysate incubated with KU135 produced a similar proteolytic fingerprint pattern as reported previously for novobiocin and molybdate (Yun et al., 2004). In contrast, incubation of reticulocyte lysate with geldanamycin (lane 2) resulted in no proteolytic fingerprinting. Combined, these data strongly suggest that KU135 binding of the C terminus of Hsp90 in reticulocyte lysate exposes a cleavage site necessary for tryptic digestion and production of a 40-kDa N-terminal fragment of Hsp90.
Fig. 3.
KU135 binds Hsp90 and exhibits effects different from 17-AAG on the expression levels of Hsp90 isoforms and known client proteins. A, rabbit reticulocyte lysate (50 μl) was applied to a novobiocin-Sepharose column after ATP regeneration and eluted with the indicated concentrations of KU135 and Western-blotted. DMSO and 10 mM novobiocin were used as controls. B, rabbit reticulocyte lysate (50 μl) was incubated at 30°C for 10 min with DMSO, 20 mM geldanamycin, novobiocin and molybdate (lanes 1–4, respectively) or 0, 1, 5, 10, and 20 mM KU135 (lanes 5–9). Subsequently, the reactions were chilled on ice and incubated for 6 min with 125 μg/ml trypsin and Western blotted. C to F, representative curves of SPR analysis of KU135 binding to Hsp90β injected at the indicated concentrations (C), hyperbolic replot of the amount of KU135 bound versus concentration of KU135 (D), a Scatchard plot of the binding of KU135 to Hsp90 (E), and hyperbolic replot of the amount of novobiocin bound versus concentration of novobiocin, corrected for nonspecific binding as described under Materials and Methods (F). G and H, cells (5 × 105/ml) were cultured with DMSO, 1 μM KU135, or 10 μM 17-AAG for 24 h, harvested, and lysed for Western blotting. β-Actin was used as a loading control. RU, relative units.
To further support that Hsp90 specifically binds KU135, the interaction of KU135 with immobilized Hsp90β was measured by surface plasmon resonance spectroscopy. Figure 3C shows representative curves for the binding of KU135 to Hsp90β. As illustrated in Fig. 3, D and E, the binding of KU135 to Hsp90 was saturable with a calculated Kd of 1 to 2 μM and a stoichiometry of 1 mole per mole of Hsp90β monomer. Thus, the high-affinity binding of KU135 correlates directly with its potency in whole-cell assays (Fig. 1 and 2). By comparison, the Kd for novobiocin binding of Hsp90 was mathematically determined to be 3 ± 1 mM, and the half-maximal saturation concentration of novobiocin determined by extrapolation from the hyperbolic fitted curve was just lower than 1 mM (Fig. 3F). Taken together, these results demonstrate that Hsp90 exhibits a binding affinity for KU135 that is more than 500 times greater than its affinity for novobiocin. Significantly, Hsp90 binding affinity for KU135 is comparable with that reported for geldanamycin and 17-AAG (Kd ∼1 μM) (Marcu et al., 2000b).
Currently, approximately 100 to 200 Hsp90-dependent client proteins have been identified. Many of these clients are oncogenic proteins (e.g., Akt, p-Akt, HIF-1α, cdc2, and Hsp70) that are involved in regulating signal transduction, cell growth, and apoptosis (Isaacs et al., 2003). In many instances, the molecular profile of Hsp90 inhibition includes depletion of numerous client proteins and induction of Hsp70 and/or Hsp90 in a concentration- and/or time-dependent manner (Banerji et al., 2005a,b). Because KU135 was found to be more potent than 17-AAG at inhibiting cell proliferation, we sought to investigate the extent to which this might correspond to differences in the expression profiles of the four different Hsp90 proteins (i.e., Hsp90α, Hsp90β, glucose-regulated protein of 94 kDa, and TRAP-1), the depletion of associated client proteins, and/or the induction of Hsp70.
As illustrated in Fig. 3G, Jurkat cells constitutively express Hsp90α, Hsp90β, glucose-regulated protein of 94 kDa, and TRAP-1, and incubation with 17-AAG but not KU135 for 24 h led to a considerable increase in the expression levels of Hsp90α and Hsp90β. In addition, both KU135 and 17-AAG caused significant alterations in the level of known Hsp90 client proteins (Fig. 3H). Specifically, treatment with 17-AAG or KU135 caused a time-dependent decrease in the level of p-Akt. We also found that incubation with KU135 or 17-AAG caused a decrease in the level of Hif-1α over 24 h, whereas only 17-AAG was found to affect the expression level of the cell cycle regulator cdc2. Significantly, Fig. 3H also illustrates that Hsp70 expression was induced to a far greater extent in cells treated with 17-AAG than in cells treated with KU135. Collectively, these findings demonstrate that both 17-AAG and KU135 effectively inhibit the chaperoning function of Hsp90. Furthermore, the fact that the expression of Hsp90α, Hsp90β, and Hsp70 were markedly increased only in response to 17-AAG might partially explain why KU135 was found to exert more potent cytotoxic effects than 17-AAG (Fig. 2).
KU135-Induced Apoptosis Occurs Independently of the Extrinsic Pathway.
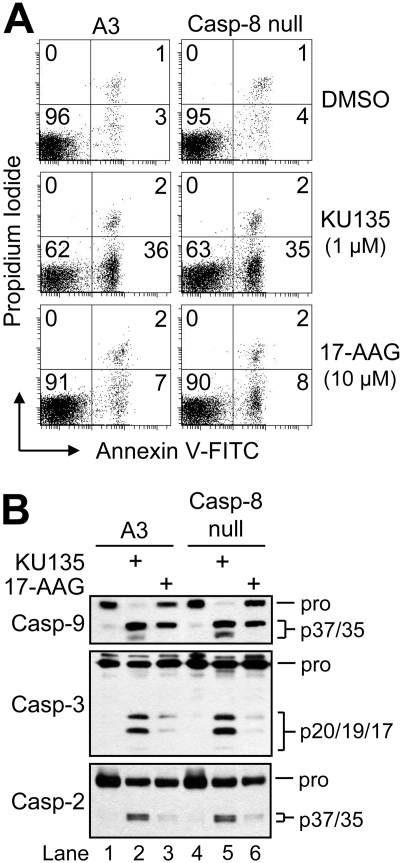
In light of the finding that KU135 was a potent inducer of apoptosis, we next investigated the extent to which this effect involved the extrinsic (receptor-mediated) pathway. To test this possibility, we used a clone of Jurkat cells lacking caspase-8 (I 9.2). The absence of this protein was confirmed previously by Western blot analysis, and this clone was found to be completely resistant to Fas-induced apoptosis (Shelton et al., 2009). As illustrated in Fig. 4A, cells lacking caspase-8 were as sensitive as control A3 cells to KU135-induced apoptosis at 24 h, as determined by annexin V-FITC and propidium iodide fluorescence. In addition, Western blot analysis of cell lysates obtained at 24 h after treatment revealed that KU135 (1 μM) caused robust processing of procaspase-9, -3, and -2 (Fig. 4B). Furthermore, caspase-3-like (DEVDase) activity was stimulated to the same extent in caspase-8-null cells as control A3 cells when incubated in the presence of KU135 for 24 h (data not shown). Although caspase-9 underwent proteolytic cleavage in response to 17-AAG (Fig. 4B, lanes 3 and 6), only the p37 inactive form of this caspase was observed (Srinivasula et al., 1998; Denault et al., 2007). Together, these findings confirm that 17-AAG is a relatively poor inducer of apoptosis (Fig. 2) and demonstrate that KU135-induced apoptosis occurs independently of the extrinsic (receptor-mediated) pathway.
Fig. 4.
KU135-induced apoptosis occurs independently of the receptor-mediated apoptotic pathway. A, cells (5 × 105/ml) were cultured in the presence or absence of DMSO, 1 μM KU135, or 10 μM 17-AAG, harvested, and processed for cell death determination by flow cytometric analysis of annexin V-FITC and propidium iodide staining. Quadrants are defined as live (bottom left), early apoptotic (bottom right), late apoptotic (top right), and necrotic (top left). Numbers refer to the percentage of cells in each quadrant. B, duplicate aliquots of cells in A were harvested and lysed for Western blotting. Casp, caspase.
KU135-Induced Apoptosis Relies Heavily on the Intrinsic Pathway.
We next investigated whether the intrinsic (mitochondria-mediated) pathway was responsible for KU135-induced cell killing. For these experiments, we took advantage of Apaf-1-deficient Jurkat cells (Franklin and Robertson, 2007) and cells in which we had overexpressed Bcl-2 or Bcl-xL (Shawgo et al., 2008). Apaf-1 is strictly required for apoptosome-mediated activation of initiator caspase-9 within the intrinsic pathway, whereas Bcl-2 and Bcl-xL are well characterized antiapoptotic proteins whose overexpression is known to inhibit intrinsic apoptosis by preventing MOMP and the release of intermembrane space proapoptotic proteins (e.g., cytochrome c and Smac) into the cytosol.
As illustrated in Fig. 5A, cells lacking Apaf-1 or overexpressing Bcl-2/Bcl-xL were resistant to KU135-induced apoptosis, having undergone only 3 to 6% apoptosis after incubation with KU135 (1 μM) for 24 h, whereas control-transfected cells had undergone 40% apoptosis at the same time point. In agreement with these findings, Western blot analysis of cell lysates obtained at 24 h after KU135 treatment revealed that extensive proteolytic processing of procaspase-9, -3, and -2 occurred in wild-type and control-transfected cells (Fig. 5B). Whereas proteolytic processing of these three caspases did not occur in Bcl-2- or Bcl-xL-overexpressing cells, some processing was observed in Apaf-1-deficient cells (Fig. 5B), despite their being resistant to KU135-induced apoptosis. However, the cleavage of caspase-9 in response to KU135 in the wild-type and control-transfected cells produced two fragments (p37/p35), whereas only one caspase-9 cleavage fragment (p37) was detected in the Apaf-1-deficient cells (Fig. 5B). This finding and the observation made with 17-AAG (Fig. 4B) are significant because apoptosome-dependent activation of caspase-9 yields the p35 fragment, whereas the p37 form of caspase-9 is generally believed to be produced by caspase-3-mediated cleavage and to be catalytically inactive (Srinivasula et al., 1998; Denault et al., 2007). However, only a trace amount of active caspase-3 was detected in both instances (Figs. 5B and 4B), suggesting that the generation of the p37 fragment of caspase-9 may be caspase-3-independent. Finally, caspase-2 also underwent some proteolytic cleavage in KU135-treated Apaf-1-deficient cells (Fig. 5B). However, whether proteolysis of caspase-2 in this context reflects its activation or whether caspase-2 is being cleaved as an “innocent bystander” remains to be determined. Notwithstanding, these data demonstrate that the antiproliferative effect of KU135 involves an ability to induce intrinsic (mitochondria-mediated) apoptosis.
Fig. 5.
KU135-induced apoptosis relies on the mitochondria-mediated apoptotic pathway. A, cells (5 × 105/ml) were cultured in the presence or absence of DMSO or 1 μM KU135 for 24 h, harvested, and processed for cell death determination by flow cytometric analysis of annexin V-FITC and propidium iodide staining. Quadrants are defined as live (bottom left), early apoptotic (bottom right), late apoptotic (top right), and necrotic (top left). Numbers refer to the percentage of cells in each quadrant. B, duplicate aliquots of cells in A were harvested and lysed for Western blotting. C, cells (5 × 105/ml) were cultured in the presence or absence of DMSO or 1 μM KU135 for 24 h, harvested, and processed for determination of Bak oligomerization by Western blotting. D and E, duplicate aliquots of cells in C were processed for subcellular fractionation into supernatant (s) and pellet (p) fractions (D) or for ΔΨ determination by flow cytometry (E). Reduced DiIC1(5) fluorescence is indicative of a loss of ΔΨ, and numbers refer to the percentage of cells that underwent a dissipation of ΔΨ. BMH, bismaleimidohexane; Cyt c, cytochrome c; Casp, caspase.
We next evaluated the extent to which mitochondrial endpoints typically associated with MOMP, including Bak/Bax activation, the release of mitochondrial intermembrane space proteins (e.g., cytochrome c), and the loss of ΔΨ, were inhibited in KU135-resistant cells. As illustrated in Fig. 5C, wild-type, control-transfected, and Apaf-1-deficient cells treated with 1 μM KU135 for 24 h activated Bak (Jurkat E6.1 cells do not express Bax), as evidenced by the presence of Bak dimers in these cells using the cross-linking agent bismaleidmidohexane. In agreement with these data, KU135 induced cytochrome c release in wild-type, control-transfected, and Apaf-1-deficient cells (Fig. 5D). Both Bak oligomerization and cytochrome c release were completely inhibited in Bcl-2- or Bcl-xL-overexpressing cells incubated under the same conditions (Fig. 5, C and D). Finally, we evaluated the different clones for KU135-induced changes in ΔΨ at 24 h (Fig. 5E). Wild-type and control-transfected cells incubated in the presence of 1 μM KU135 showed that 35% of cells had lost ΔΨ, whereas cells overexpressing Bcl-2 or Bcl-xL had retained ΔΨ. By comparison, Apaf-1-deficient cells, which were found to be resistant to KU135, despite activating Bak, releasing cytochrome c, and cleaving certain caspases, largely retained ΔΨ (Fig. 5E). Overall, these findings offer support of a model of KU135-induced apoptosis that requires the involvement of the mitochondrial pathway.
Discussion
Hsp90 represents a family of ATPase-containing molecular chaperones that comprise as much as 1 to 2% of total cellular protein under unstressed conditions. In response to various stressors, this amount can increase to 4 to 6% (Whitesell and Lindquist, 2005). Hsp90 is often present at elevated levels in cancer cells and functions to stabilize oncogenic proteins involved in signal transduction, growth, and apoptosis regulation (Isaacs et al., 2003). A possible explanation for Hsp90 overexpression in cancer cells is that malignant cells are in a persistent state of proteotoxic stress due, in part, to acidosis and/or the accumulation of mutated signaling molecules that would otherwise result in cell lethality. In this way, the presence of Hsp90 (and other heat shock proteins) is believed to compensate for the hostility of many cancer cell microenvironments (Whitesell and Lindquist, 2005; Solit and Rosen, 2006). As mentioned previously, there are currently between 100 and 200 reported cytosolic and nuclear client proteins for Hsp90, including protein kinases (e.g., Akt and Her2), transcription factors (e.g., mutant p53 and HIF-1α), chimeric signaling proteins (e.g., Bcr-Abl), and several proteins involved in apoptosis (e.g., Bid, Apaf-1, and RIP) (for an updated list see http://www.picard.ch/downloads/downloads.htm). Whereas many of the aforementioned Hsp90 client proteins are being pursued individually as targets for anticancer drug development, inhibition of Hsp90 would prevent the maturation and stabilization of numerous Hsp90 client proteins simultaneously, leading to their ultimate degradation within the ubiquitin-proteasome pathway (Whitesell et al., 1994; Schulte et al., 1995). In this regard, Hsp90 has emerged as an exciting novel target for cancer chemotherapeutic drug design because inhibition of its chaperoning function simultaneously leads to the destabilization and degradation of multiple oncogenic proteins.
Although one of the earliest Hsp90 inhibitors, 17-AAG, which targets the N-terminal ATP-binding pocket of Hsp90, has entered clinical trials for refractory malignancies, it has shown little clinical effect as monotherapy (Goetz et al., 2005; Nowakowski et al., 2006). Furthermore, even though 17-AAG is less toxic than its parent compound geldanamycin, its widespread use is still hindered by concerns regarding its hepatotoxicity (Egorin et al., 1998; Solit et al., 2007). A new approach to targeting Hsp90 began with the observation that the antibiotic novobiocin binds with low affinity to a C-terminal ATP-binding pocket (Marcu et al., 2000a,b). More potent analogs of novobiocin have been developed, and we report here that one such lead candidate inhibitor, designated KU135, binds directly to Hsp90 and suppresses cell proliferation by engaging signaling pathways that are mechanistically different from those affected by 17-AAG.
Indeed, KU135 was found to exert more potent antiproliferative effects than 17-AAG. Whereas 17-AAG predominantly induces cell cycle arrest, KU135 causes both cell cycle arrest and an induction of apoptosis. This is significant because an Hsp90 inhibitor exhibiting combined cytostatic and cytotoxic effects would be expected to eradicate tumor cells and thereby lower tumor cell burden, whereas a purely cytostatic inhibitor would only inhibit tumor cell growth/division. A possible explanation for why 17-AAG is not able to induce an apoptotic response similar to that of KU135 is that differences may exist in the number and types of client proteins that are affected by an N-terminal (17-AAG) versus a C-terminal (KU135) Hsp90 inhibitor. Another possibility is that the relative absence of cytotoxic activity with 17-AAG is due to the robust up-regulation of Hsp90α, Hsp90β, and Hsp70 that is observed compared with KU135.
Overall, the data presented here provide compelling evidence for the continued development of novobiocin-based C-terminal Hsp90 inhibitors as promising alternatives to N-terminal inhibitors, such as 17-AAG, for the treatment of human malignancies. KU135 is one such lead candidate novobiocin-derived compound. Experiments are currently ongoing in the laboratory to characterize the specific molecular ordering of the caspase activation cascade induced by KU135 and the molecular requirements necessary for MOMP to occur in response to this compound.
Acknowledgments
We thank Michael Hughes, Melinda Broward, and Dr. Scott Weir for their assistance with project coordination, Dr. Joyce Slusser for expert help with flow cytometry, and Dr. Kathy Roby for helpful discussion. We are grateful to the late Dr. Stanley Korsmeyer for the pSFFV-neo, pSFFV-Bcl-2, and pSFFV-Bcl-xL mammalian expression vectors. We also extend special thanks to Dr. Len Neckers for invaluable scientific input and ICX Nomadics for the use of their SensiQ SPR Instrument.
This work was supported by the National Institutes of Health National Cancer Institute [Grants R01-CA125392 and R01-CA120458]; the National Institutes of Health National Institute of Environmental Health Sciences [Grant T32-ES007079]; the National Institutes of Health National Center for Research Resources [Grant P20-RR016443]; the American Cancer Society [Grant IRG-09-062-01]; Oklahoma Agricultural Experiment Station [Project 1975]; and the Kansas Technology Enterprise Corporation through the Centers of Excellence Program.
Article, publication date, and citation information can be found at http://molpharm.aspetjournals.org.
doi:10.1124/mol.109.058545
- Hsp
- heat shock protein
- MOMP
- mitochondrial outer membrane permeabilization
- Apaf-1
- apoptotic protease activating factor-1
- 17-AAG
- 17-allylamino-demethoxygeldanamycin
- MTS
- 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt
- FITC
- fluorescein isothiocyanate
- DMSO
- dimethyl sulfoxide
- SPR
- surface plasmon resonance
- Δψ
- mitochondrial membrane potential
- PBS
- phosphate-buffered saline
- PAGE
- polyacrylamide gel electrophoresis
- Pipes
- piperazine-N,N′-bis(2-ethanesulfonic acid)
- TRAP
- tumor necrosis factor receptor-associated protein
- KU135
- 3-(3′,6-dimethoxybiphenyl-3-ylcarboxamido)-6-methoxy-8-methyl-2-oxo-2H-chromen-7-yl acetate.
References
- Avila C, Hadden MK, Ma Z, Kornilayev BA, Ye QZ, Blagg BS. (2006a) High-throughput screening for Hsp90 ATPase inhibitors. Bioorg Med Chem Lett 16:3005–3008 [DOI] [PubMed] [Google Scholar]
- Avila C, Kornilayev BA, Blagg BS. (2006b) Development and optimization of a useful assay for determining Hsp90's inherent ATPase activity. Bioorg Med Chem 14:1134–1142 [DOI] [PubMed] [Google Scholar]
- Banerji U, O'Donnell A, Scurr M, Pacey S, Stapleton S, Asad Y, Simmons L, Maloney A, Raynaud F, Campbell M, et al. (2005a) Phase I pharmacokinetic and pharmacodynamic study of 17-allylamino, 17-demethoxygeldanamycin in patients with advanced malignancies. J Clin Oncol 23:4152–4161 [DOI] [PubMed] [Google Scholar]
- Banerji U, Walton M, Raynaud F, Grimshaw R, Kelland L, Valenti M, Judson I, Workman P. (2005b) Pharmacokinetic-pharmacodynamic relationships for the heat shock protein 90 molecular chaperone inhibitor 17-allylamino, 17-demethoxygeldanamycin in human ovarian cancer xenograft models. Clin Cancer Res 11:7023–7032 [DOI] [PubMed] [Google Scholar]
- Blagg BS, Kerr TD. (2006) Hsp90 inhibitors: small molecules that transform the Hsp90 protein folding machinery into a catalyst for protein degradation. Med Res Rev 26:310–338 [DOI] [PubMed] [Google Scholar]
- Burlison JA, Avila C, Vielhauer G, Lubbers DJ, Holzbeierlein J, Blagg BS. (2008) Development of novobiocin analogues that manifest anti-proliferative activity against several cancer cell lines. J Org Chem 73:2130–2137 [DOI] [PubMed] [Google Scholar]
- Burlison JA, Neckers L, Smith AB, Maxwell A, Blagg BS. (2006) Novobiocin: redesigning a DNA gyrase inhibitor for selective inhibition of hsp90. J Am Chem Soc 128:15529–15536 [DOI] [PubMed] [Google Scholar]
- Chiosis G, Huezo H, Rosen N, Mimnaugh E, Whitesell L, Neckers L. (2003) 17AAG: low target binding affinity and potent cell activity—finding an explanation. Mol Cancer Ther 2:123–129 [PubMed] [Google Scholar]
- Denault JB, Eckelman BP, Shin H, Pop C, Salvesen GS. (2007) Caspase 3 attenuates XIAP (X-linked inhibitor of apoptosis protein)-mediated inhibition of caspase 9. Biochem J 405:11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly AC, Mays JR, Burlison JA, Nelson JT, Vielhauer G, Holzbeierlein J, Blagg BS. (2008) The design, synthesis, and evaluation of coumarin ring derivatives of the novobiocin scaffold that exhibit antiproliferative activity. J Org Chem 73:8901–8920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egorin MJ, Rosen DM, Wolff JH, Callery PS, Musser SM, Eiseman JL. (1998) Metabolism of 17-(allylamino)-17-demethoxygeldanamycin (NSC 330507) by murine and human hepatic preparations. Cancer Res 58:2385–2396 [PubMed] [Google Scholar]
- Franklin EE, Robertson JD. (2007) Requirement of Apaf-1 for mitochondrial events and the cleavage or activation of all procaspases during genotoxic stress-induced apoptosis. Biochem J 405:115–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgakis GV, Younes A. (2005) Heat-shock protein 90 inhibitors in cancer therapy: 17AAG and beyond. Future Oncol 1:273–281 [DOI] [PubMed] [Google Scholar]
- Goetz MP, Toft D, Reid J, Ames M, Stensgard B, Safgren S, Adjei AA, Sloan J, Atherton P, Vasile V, et al. (2005) Phase I trial of 17-allylamino-17-demethoxygeldanamycin in patients with advanced cancer. J Clin Oncol 23:1078–1087 [DOI] [PubMed] [Google Scholar]
- Grenert JP, Sullivan WP, Fadden P, Haystead TA, Clark J, Mimnaugh E, Krutzsch H, Ochel HJ, Schulte TW, Sausville E, et al. (1997) The amino-terminal domain of heat shock protein 90 (hsp90) that binds geldanamycin is an ATP/ADP switch domain that regulates hsp90 conformation. J Biol Chem 272:23843–23850 [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. (2000) The hallmarks of cancer. Cell 100:57–70 [DOI] [PubMed] [Google Scholar]
- Hartson SD, Thulasiraman V, Huang W, Whitesell L, Matts RL. (1999) Molybdate inhibits hsp90, induces structural changes in its C-terminal domain, and alters its interactions with substrates. Biochemistry 38:3837–3849 [DOI] [PubMed] [Google Scholar]
- Isaacs JS, Xu W, Neckers L. (2003) Heat shock protein 90 as a molecular target for cancer therapeutics. Cancer Cell 3:213–217 [DOI] [PubMed] [Google Scholar]
- Kischkel FC, Hellbardt S, Behrmann I, Germer M, Pawlita M, Krammer PH, Peter ME. (1995) Cytotoxicity-dependent APO-1 (Fas/CD95)-associated proteins form a death-inducing signaling complex (DISC) with the receptor. EMBO J 14:5579–5588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bras G, Radanyi C, Peyrat JF, Brion JD, Alami M, Marsaud V, Stella B, Renoir JM. (2007) New novobiocin analogues as antiproliferative agents in breast cancer cells and potential inhibitors of heat shock protein 90. J Med Chem 50:6189–6200 [DOI] [PubMed] [Google Scholar]
- Li H, Zhu H, Xu CJ, Yuan J. (1998) Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell 94:491–501 [DOI] [PubMed] [Google Scholar]
- Li P, Nijhawan D, Budihardjo I, Srinivasula SM, Ahmad M, Alnemri ES, Wang X. (1997) Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell 91:479–489 [DOI] [PubMed] [Google Scholar]
- Luo X, Budihardjo I, Zou H, Slaughter C, Wang X. (1998) Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell 94:481–490 [DOI] [PubMed] [Google Scholar]
- Marcu MG, Chadli A, Bouhouche I, Catelli M, Neckers LM. (2000a) The heat shock protein 90 antagonist novobiocin interacts with a previously unrecognized ATP-binding domain in the carboxyl terminus of the chaperone. J Biol Chem 275:37181–37186 [DOI] [PubMed] [Google Scholar]
- Marcu MG, Schulte TW, Neckers L. (2000b) Novobiocin and related coumarins and depletion of heat shock protein 90-dependent signaling proteins. J Natl Cancer Inst 92:242–248 [DOI] [PubMed] [Google Scholar]
- Nowakowski GS, McCollum AK, Ames MM, Mandrekar SJ, Reid JM, Adjei AA, Toft DO, Safgren SL, Erlichman C. (2006) A phase I trial of twice-weekly 17-allylamino-demethoxy-geldanamycin in patients with advanced cancer. Clin Cancer Res 12:6087–6093 [DOI] [PubMed] [Google Scholar]
- Owen BA, Sullivan WP, Felts SJ, Toft DO. (2002) Regulation of heat shock protein 90 ATPase activity by sequences in the carboxyl terminus. J Biol Chem 277:7086–7091 [DOI] [PubMed] [Google Scholar]
- Powers MV, Workman P. (2007) Inhibitors of the heat shock response: biology and pharmacology. FEBS Lett 581:3758–3769 [DOI] [PubMed] [Google Scholar]
- Radanyi C, Le Bras G, Marsaud V, Peyrat JF, Messaoudi S, Catelli MG, Brion JD, Alami M, Renoir JM. (2009) Antiproliferative and apoptotic activities of tosylcyclonovobiocic acids as potent heat shock protein 90 inhibitors in human cancer cells. Cancer Lett 274:88–94 [DOI] [PubMed] [Google Scholar]
- Rahmani M, Yu C, Dai Y, Reese E, Ahmed W, Dent P, Grant S. (2003) Coadministration of the heat shock protein 90 antagonist 17-allylamino- 17-demethoxygeldanamycin with suberoylanilide hydroxamic acid or sodium butyrate synergistically induces apoptosis in human leukemia cells. Cancer Res 63:8420–8427 [PubMed] [Google Scholar]
- Robertson JD, Enoksson M, Suomela M, Zhivotovsky B, Orrenius S. (2002) Caspase-2 acts upstream of mitochondria to promote cytochrome c release during etoposide-induced apoptosis. J Biol Chem 277:29803–29809 [DOI] [PubMed] [Google Scholar]
- Roe SM, Prodromou C, O'Brien R, Ladbury JE, Piper PW, Pearl LH. (1999) Structural basis for inhibition of the Hsp90 molecular chaperone by the antitumor antibiotics radicicol and geldanamycin. J Med Chem 42:260–266 [DOI] [PubMed] [Google Scholar]
- Schmitt E, Gehrmann M, Brunet M, Multhoff G, Garrido C. (2007) Intracellular and extracellular functions of heat shock proteins: repercussions in cancer therapy. J Leukoc Biol 81:15–27 [DOI] [PubMed] [Google Scholar]
- Schulte TW, Blagosklonny MV, Ingui C, Neckers L. (1995) Disruption of the Raf-1-Hsp90 molecular complex results in destabilization of Raf-1 and loss of Raf-1-Ras association. J Biol Chem 270:24585–24588 [DOI] [PubMed] [Google Scholar]
- Shawgo ME, Shelton SN, Robertson JD. (2008) Caspase-mediated Bak activation and cytochrome c release during intrinsic apoptotic cell death in Jurkat cells. J Biol Chem 283:35532–35538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelton SN, Shawgo ME, Robertson JD. (2009) Cleavage of Bid by executioner caspases mediates feed forward amplification of mitochondrial outer membrane permeabilization during genotoxic stress-induced apoptosis in Jurkat cells. J Biol Chem 284:11247–11255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solit DB, Ivy SP, Kopil C, Sikorski R, Morris MJ, Slovin SF, Kelly WK, DeLaCruz A, Curley T, Heller G, et al. (2007) Phase I trial of 17-allylamino-17-demethoxygeldanamycin in patients with advanced cancer. Clin Cancer Res 13:1775–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solit DB, Rosen N. (2006) Hsp90: a novel target for cancer therapy. Curr Top Med Chem 6:1205–1214 [DOI] [PubMed] [Google Scholar]
- Srinivasula SM, Ahmad M, Fernandes-Alnemri T, Alnemri ES. (1998) Autoactivation of procaspase-9 by Apaf-1-mediated oligomerization. Mol Cell 1:949–957 [DOI] [PubMed] [Google Scholar]
- Whitesell L, Bagatell R, Falsey R. (2003) The stress response: implications for the clinical development of hsp90 inhibitors. Curr Cancer Drug Targets 3:349–358 [DOI] [PubMed] [Google Scholar]
- Whitesell L, Lindquist SL. (2005) HSP90 and the chaperoning of cancer. Nat Rev Cancer 5:761–772 [DOI] [PubMed] [Google Scholar]
- Whitesell L, Mimnaugh EG, De Costa B, Myers CE, Neckers LM. (1994) Inhibition of heat shock protein HSP90-pp60v-src heteroprotein complex formation by benzoquinone ansamycins: essential role for stress proteins in oncogenic transformation. Proc Natl Acad Sci U S A 91:8324–8328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun BG, Huang W, Leach N, Hartson SD, Matts RL. (2004) Novobiocin induces a distinct conformation of Hsp90 and alters Hsp90-cochaperone-client interactions. Biochemistry 43:8217–8229 [DOI] [PubMed] [Google Scholar]