Abstract
Recently with the rapid improvements in high-throughout genotyping techniques, researchers are facing the very challenging task of analyzing large-scale genetic associations, especially at the whole-genome level, without an optimal solution. In this study, we propose a new approach for genetic association analysis that is based on a variable-sized sliding-window framework and employs principal component analysis to find the optimum window size. With the help of the bisection algorithm in window-size searching, our method is more computationally efficient than available approaches. We evaluate the performance of the proposed method by comparing it with two other methods—a single-marker method and a variable-length Markov chain method. We demonstrate that, in most cases, the proposed method outperforms the other two methods. Furthermore, since the proposed method is based on genotype data, it does not require any computationally intensive phasing program to account for uncertain haplotype phase.
Background
Currently, with the availability of large-scale genotyping technologies, the genotyping cost of genome-wide association (GWA) studies has been largely reduced and a boom of large-scale GWA studies is underway. Nevertheless, the success of most association studies is based on the linkage disequilibrium (LD) between the functional mutations and markers in a local region of the genome. Varieties of statistical approaches that rely on LD pattern have been developed to map functional variants (Spielman et al. 1993; Olson et al. 1994; Rannala and Reeve 2001; Ardlie et al. 2002). The most straightforward approach of LD-based association analysis is the single-marker analysis, which tests each single nucleotide polymorphism (SNP) for association with the disease. However, many studies have shown that this simple method may be inefficient in most cases because of the limited genetic information used in finding the functional mutations. We need methods that could better use information of multi-markers jointly. An alternative approach of the single-marker analysis is multiple-marker analysis based on either haplotypes or genotypes (Morris and Kaplan 2002; Clayton et al. 2004; Seaman and Müller-Myhsok 2005). This approach still has the disadvantage that large degrees of freedom are always involved in the test statistic due to the large number of haplotypes. For mapping complex disease genes, it is still hard to make the verdict on which of the two methods is more powerful (Sevice et al. 1999; Barton 2000; Maclean et al. 2000; Zöllner and von Haeseler 2000; Akey et al. 2001; Morris and Kaplan 2002; Wessel and Schork 2006). Under certain disease models and certain LD patterns one method outperforms the other, so it is likely that there is no single best approach to detect the common risk factors. In practice, researchers have employed both single-marker and multiple-marker analysis in genetic association studies. If conducting a multiple-marker analysis, a researcher has to determine how many neighboring SNPs should be included in the analysis.
Recent studies have suggested that the human genome can be partitioned into blocks with limited haplotype diversity within each block (Gabriel et al. 2002). Therefore, most of the genetic variation can be captured by a limited number of haplotypes and haplotype association tests are performed within each predefined block (Gabriel et al. 2002). For haplotype block approaches, there are several different criteria that have been proposed to predetermine the blocks, but it is still not clear which one is the best (Perola et al. 2002; Zhang and Li 2003; Zhang et al. 2004; Zhu et al. 2004 ). Furthermore, it is hard to determine the boundaries of the blocks and it usually will result in many single-marker blocks, which shows no advantage over the single-marker analysis. Considering the reasons mentioned above, haplotype block approaches may not be the most efficient method to conduct the association studies (Zhao et al. 2003).
The sliding-window approach is another strategy of multiple-marker analysis. In this approach, a genome region under study is divided into windows and a multiple-marker association test is performed in each window. There are two groups of sliding-window methods: uniform-sized sliding-window approaches and variable-sized sliding-window approaches (Clayton et al 1999; Bourgain et al. 2000; Toivonen et al. 2000; Mathias et al. 2006; Yang et al. 2006; Yi et al. 2007; Huang et al. 2007). For the uniform-sized sliding-window approaches, it is hard to decide the optimal window size under different scenarios. It will become more problematic when the uniform-sized sliding-window approaches are performed over a large genome region or over the whole genome, where the LD patterns certainly vary frequently. Therefore, the variable-sized sliding-window approaches with a variable window size decided by the underlying LD pattern perform more efficiently in large scale data analysis. The problem for the variable-sized sliding-window approach is in finding the optimal window size.
Browning (2006) proposed a variable-sized sliding-window approach based on a variable-length Markov chain model, which automatically adapts to the LD pattern between markers. Browning argued that this approach can be thought of as haplotype testing with sophisticated windowing that accounts for extent of LD to reduce both the degrees of freedom and number of tests. Li et al. (2007) also proposed a variable-sized sliding-window approach in which the maximum size of a sliding window is determined by local haplotype diversity and a regularized regression analysis is used to tackle the problem of multiple degrees of freedom in the haplotype test. However, both Browning’s and Li et al.’s methods require phased data as input. Even though haplotype phasing programs are now available, it is still very time-consuming to phase a large number of markers.
In this study, we proposed a novel method for multiple-marker association analysis of genotype data. Based on the variable-sized sliding-window frame, we decide the optimal window size by the local LD pattern via Principal Component Analysis (PCA). Then we use a score test based on a logistic model to test association within a window. Simulation studies are used to compare the power of the proposed approach with that of the single-marker association test and the haplotype clustering method based on variable-length Markov chains by Brownings (2007). Our simulation studies demonstrate that the proposed method provides better performance than the single-marker association test and Browning’s method in most of the scenarios. Our method is much faster computationally than Browning’s and Li et al.’s methods because our method is based on genotypes and thus does not need to estimate haplotypes.
Methods
Optimum Window-Size Searching Procedure
Consider a case-control sample with total M individuals and assume each individual has been genotyped at N SNPs. Let Gi = (gi1, gi2 , … ,giN )T ( i = 1, 2,…, M ) denote the multi-marker genotype of the i th individual, where gij denote the genotype of the i th individual at the j th SNP and gij code as 0, 1, or 2 (the number of minor allele). Let yi denote the trait value of individual i (1 for cases and 0 for controls).
In the sliding-window frame, a window, denoted as , is a set of neighboring SNPs {b,b + 1,b + 2,…,b +l − 1 }. A variable-sized sliding window which begins with SNP b , denoted as Ωb , is a collection of windows with l ranging from s to Γb , where s and Γb are predefined smallest and largest window sizes, respectively (in our simulation studies, we use s = 4 and Γb = 35 ).
In this study, we apply the PCA to define the optimum window size. The optimal window size for windows beginning with SNP b is defined as the maximum window size among windows such that c0 proportion of the total information can be explained by the first k Principal Components (PCs), where c0 and k are predefined. In our searching procedure, we start with a window , l = s = k + 1 , so at least the window length is longer than k , the number of the important PCs.
To carry out the PCA, we let , a l × l matrix, denote the sample variance-covariance matrix of genotypic numerical codes, where and . Let be the eigenvector corresponding to the j th largest eigenvalue of the sample variance-covariance matrix Thus in window , the total variance in the original data set explained by the j th PC is . Let , the proportion of the total variability explained by the first k PCs. The following three steps show a natural way to search for the optimum window size.
Step 1: Among a set of windows Ω , conduct PCA on the genotypes within the window , a window begins at SNP b and with s = l = k + 1 as the shortest window size.
Step 2: Calculate C , the proportion of the total variability explained by the first k PCs for window . If C > c0, we let l = l + 1 , which enlarges the window size by including one more SNP and we continue to carry out step 3. Otherwise, we say that l is the best window size for the windows that begin at SNP b .
Step 3: Repeat step 2.
Adapt Bisection Method to Modify the Optimum Window-Size Searching Procedure
As we can imagine, our previous optimum window-size searching procedure is very computational demanding for the genome-wide analysis, especially when the window size gets larger. Therefore it is necessary for us to relieve the computational burden of our sliding-window method. In mathematics, the bisection method is a root-finding algorithm that works by repeatedly dividing an interval into half and then selecting the subinterval in which the root exists.
By adapting the bisection method, we modified our optimum window-size searching procedure as follows:
Step 1: let l = [ (s + Γ)/ 2 ], where s and Γ are the predefined smallest and largest window sizes among a set of windows Ωb , and [a] is the largest integer that is less than or equal to a.
Step 2: Among Ωb , firstly we conduct PCA within the window , a window begins at SNP b and the window size is l .
Step 3: Calculate C for this window . If C > c0 , we let s = l , which enlarges the window size by including more SNPs and we continue to carry out step 4. Otherwise, we let Γ = l , which shortens the window size by excluding more SNPs but does not change the start position of this window.
Step 4: Repeat step 1 to step 3 until Γ −s ≤ 1 .
By employing the bisection algorithm in the optimal window-size searching process, the computational burden is significantly relieved.
Score Test
After we find the optimum window size for a window, we can apply any appropriate test statistic to test for association in this window. In our study, we use the score test statistic based on a logistic model to test for association. Consider , a window beginning at SNP b with the optimum window size l . Let Gi, , and yi denote the genotype, the first k PCs of the genotype, and the trait value (1 for cases and 0 for controls) of the i th individual, where i = 1, 2 ,…,M . Let pi denote the probability of disease given genotype Gi . Suppose that the k PCs follow a logistic model , where β = (β1,…,βk)t ; then the score test statistic is given by (Clayton et al. 2004)
where , , , and M is the sample size. The statistic T2 asymptotically follows the χ 2 distribution with k degrees of freedom.
In this score test, we use PCA to reduce the degrees of freedom from l to k . According to our experience, we can reduce the degrees of freedom greatly while the first k PCs can still explain more than 90% of the total variability. Since the proposed method can reduce the number of degrees of freedom greatly and also keep the majority of the information, the power of the test can be increased.
Comparison of Methods and Adjustment for Multiple Testing
We compare the power of the proposed method (TPCSW) with the single-marker association test (TSingle) and variable-length Markov chain test (Tbeagle) proposed by Brownings (2007). We propose to use permutation tests to adjust for multiple testing. For the Tbeagle, we use its inbuilt multiple-testing correction via permutation. For the TSingle or TPCSW, the permutation procedure is as follow. Suppose that there are L SNPs (windows). Let pi denote the p-value of the test in the i th SNP (window) ( i = 1,…, L ). For each permutation, we randomly shuffle the case and control status and recalculate the test statistics and p-values based on the permuted data. Let pij denote the p-value of the test in the i th SNP (window) and the j th permutation and . Suppose that we perform J permutations. Then, the adjusted p-value of the test in the i th SNP (window) is given by . In this study, we use 1000 permutations to evaluate the adjusted p-values.
Simulation Setup
To evaluate the performance of the proposed method, we conduct simulation studies under variety of scenarios. We generate haplotypes using the ms program by Hudson (2002). In the ms program, we use a mutation rate of 2.5×10−8 per nucleotide per generation, a recombination rate of 10−8 per pair of nucleotides per generation, and an effective population size of 10,000. These choices were also adopted in Nordborg and Tavare (2002), Kimmel and Shamir (2006), and Feng et al. (2007). Using the ms program, we first generate a haplotype pool with 10,000 haplotypes (1,000 SNPs) and a genotype can be generated by randomly choosing two haplotypes from the pool.
When generating data to evaluate the type I error, the genotype of each individual is composed of two haplotypes randomly chosen from the haplotype pool. We randomly assign one individual as a case or a control independent of the genotypes. There are four sample sizes: 600, 800, 1000, and 1200 (half cases and half controls), and the proportion of the total variability explained by the first k ( k = 3 ) PCs is c0 = 95%. For each scenario, we generate 1,000 replicated samples to evaluate the type I error rate.
For power comparison, we consider two sets of disease models. In the first set, we consider four three-locus disease models, denoted as model L1 to model L4 , which are similar to those used by Millstein et al. (2006) in their simulation studies. We randomly choose three SNPs with minor allele frequencies between 0.1 and 0.33 as the three disease loci (the genotypes of the disease loci are kept in the data set for analysis). A logistic model is used to relate genotypes at the disease loci to the trait. Let p = pr(affected/genotype) and x1, x2 , and x3 be the numerical codes of the genotypes at the three disease loci. The relationship between p and x1, x2, x3 is given by the logistic model .
Assume that the overall population prevalence is 10%. Then the value of β0 can be determined by the values of the other parameters. The four different models are determined by different values of the parameters. The values of the parameters are given in Table 1. In models L1 to L4 , xk = 0, 1, or 2 corresponds to genotypes akak , Ak ak , or Ak Ak at the k th disease loci ( k = 1, 2,3), an additive coding of the genotypes.
Table 1.
Values of the parameters in the four three-locus disease models
| Models | Parameters |
|---|---|
| L 1 | β123 = log(3) |
| L 2 | β1 = log(1.5) ; β123 = log(3) |
| L 3 | β1 = log(1.5) ; β2 = log(0.65) ; β123 = log(3) |
| L4 | β1 = β2 = β3 = log(1.5) |
Disease prevalence is set to be 0.1. Except for β0 , all other parameters not in the table are zero and the value of β0 can be determined by the values of the other parameters.
For the second set of disease models, we consider a single-locus disease model. Let p be defined same as the above and x be the additive code of the genotype at the disease locus. The relationship between p and x is given by the logistic model . Assume that the overall population prevalence is 10%. Then the value of β0 can be determined by the values of the other parameters. We consider four different disease models (denoted by L5 to L8 ) based on the above logistic model with different disease models corresponding to different values of β1 and intervals of the minor allele frequency (MAF) of the disease locus. The values of β1 and the intervals of the MAF of the disease locus for the four disease models are given in Table 2. When the interval of the MAF at the disease locus is given, we randomly choose a SNP with MAF in the interval as the disease locus (the genotypes of the disease locus are kept in the data set for analysis).
Table 2.
Values of the parameters in the four single-locus disease models
| single locus disease model |
odds ratio | MAF interval |
|---|---|---|
| L 5 | β1 = log(2.3) | [0.049, 0.05] |
| L 6 | β1 = log(2) | [0.09, 0.10] |
| L 7 | β1 = log(1.7) | [0.19, 0.20] |
| L 8 | β1 = log(1.5) | [0.29, 0.30] |
Disease prevalence is set to be 0.1. Except for β0 , all other parameters not in the table are zero and the value of β0 can be determined by the values of the other parameters.
Results
Throughout the simulation studies, we set c0 , the proportion of the total variability explained by the first k PCs ( k = 3 ), as 95% for TPCSW. The results of type I error rates are given in Table 3. With 1,000 replicated samples, the standard deviations for the type I error rate are and for the nominal levels of 0.05 and 0.01. The 95% confidence intervals are (0.036, 0.064) and (0.004, 0.016) for the nominal levels of 0.05 and 0.01. The results shown in Table 3 illustrate that the estimated type I errors of all the three methods are within the 95% confidence intervals, which indicate that the estimated type I errors are not significantly different from the nominal levels.
Table 3.
Type I error rates of the three methods: TSingle, Tbeagle, and TPCSW.
| Sample size |
Methods | Significance level 5% |
Significance level 1% |
|---|---|---|---|
| 600 | TSingle | 0.048 | 0.007 |
| Tbeagle | 0.052 | 0.006 | |
| TPCSW | 0.043 | 0.013 | |
| 800 | TSingle | 0.039 | 0.008 |
| Tbeagle | 0.062 | 0.012 | |
| TPCSW | 0.039 | 0.014 | |
| 1000 | TSingle | 0.045 | 0.01 |
| Tbeagle | 0.061 | 0.006 | |
| TPCSW | 0.042 | 0.008 | |
| 1200 | TSingle | 0.042 | 0.011 |
| Tbeagle | 0.058 | 0.007 | |
| TPCSW | 0.047 | 0.009 |
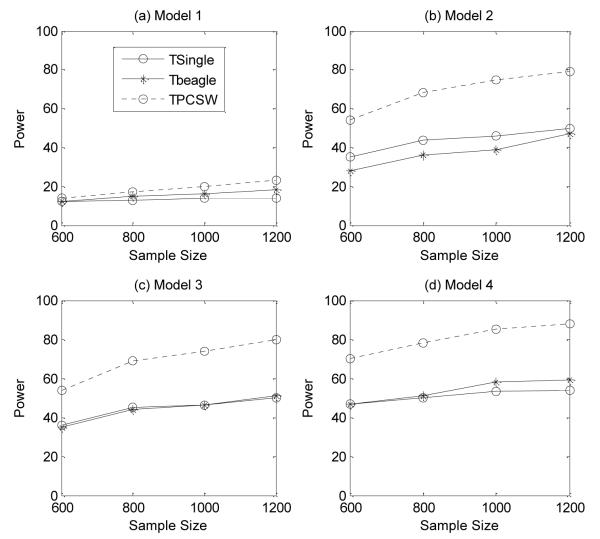
The power comparison results of the three methods under the four three-locus disease models are shown in Figure 1. From Figure 1, we can see that our method consistently outperforms the other two methods in terms of the detection power at various sample sizes under the four disease models. The power of the other two methods, Tbeagle and TSingle, are very similar.
Figure 1.
Power comparison among the three methods in four three-locus disease models. The power is calculated under the assumption of a 5% significance level and the permutation test is used to correct for multiple testing.
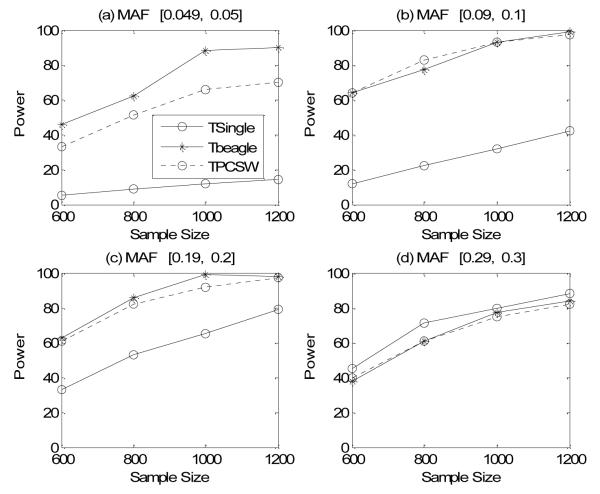
The power comparison results of the three methods under the four single-locus disease models are shown in Figure 2. When MAF is low, i.e. [0.045, 0.05], Tbeagle is the most powerful one and the proposed method and TSingle have similar power. When MAF is high, i.e. [0.29, 0.30], all three methods have similar power. When MAF is in the middle, i.e. between 0.05 and 0.3, the proposed method and Tbeagle have similar power and both methods are more powerful than TSingle. Overall, we can conclude that, except for the case of low MAF, our method is always one of the most powerful methods.
Figure 2.
Power comparison among the three methods in the four single-locus disease models. The power is calculated under the assumption of a 5% significance level and the permutation test is used to correct for multiple testing.
Discussion
In this article, we have proposed a genotype-based method through PCA to find optimal window sizes of variable-sized sliding windows to detect disease associations. We use intensive simulation studies to evaluate the performance of the proposed method. The simulation results show that in most cases our method outperforms the commonly used single-marker association test and Browning’s variable-length Markov chain method. Our method is capable in phase unknown situation while Browning’s variable-length Markov chain method is based on phase known situation, so intensive computation to phase the data is first required. Our method significantly outperforms the other two methods in multi-locus disease models.
There is a common problem for variable-size sliding-window approaches because they are usually based on haplotype data which demands a computationally intensive method to phase the genotype data first. Our method tackled the common disadvantage for variable-size sliding-window approaches by finding the optimal window size using genotype-based method. Therefore our method has the potential to be applied to genome-wide association studies.
To improve our methodology, there is still one thing that needs further consideration, that is, how to choose the values of the parameters k and c0 , the number of PCs we used and the proportion of the total variability explained by the first k PCs. In this study we set c0 = 95% and k = 3 . Through intensive simulation studies, we conclude that c0 = 95% and k = 3 are good choices in most cases. However, it is hard to find optimal values for the two parameters; therefore it should be one of our future steps of this method.
In our simulation studies, the disease-related SNPs are not removed from the genotype data before analysis. In this case, it seems that the single-marker analysis should give the best results. However, even if the disease-related SNPs are kept in the genotype data, several studies have shown that multiple-marker methods may be more powerful than the single-marker analysis (Zhao et al. 2000; Zhang et al. 2003). Following example may explain partially why multiple-marker methods can be more powerful. Consider a case-control study with 1000 cases and 1000 controls. Suppose that the frequencies of the disease allele in cases and controls are 0.2 and 0.15, respectively. Then, the p-value of the allelic chi-square test is 3.2×10−3 . Consider five markers around the disease locus (include the disease locus) and assume that a mutation occurred at haplotype 11111 many years ago. The frequency of haplotype 11111 in cases should be higher than that in controls. Suppose frequencies of haplotype 11111 in cases and controls are 0.05 and 0.00, respectively. Then, the p-value of the allelic chi-square test to test association of haplotype 11111 (haplotype 11111 as one allele and all other haplotypes as another allele) is 8.0×10−13 and the p-value after adjustment for multiple testing (at most 32 haplotypes) is 2.56×10−11 . This example shows that disease-marker association may not be detectable as first-order association between a single marker and the disease locus but may be detected by extended marker haplotypes.
In summary, it is shown that the proposed method is simpler, faster and more powerful than the recently developed method—Browning’s variable-length Markov chain method. The computational efficiency and power compared to its peers make our method an attractive choice in detecting disease associated SNP(s) in genome-wide association studies.
Acknowledgments
This work was supported by NIH grant R01 GM069940 and the Overseas-Returned Scholars Foundation of Department of Education of Heilongjiang Province (1152HZ01).
Reference
- 1.Akey J, Li J, Xiong M. Haplotypes vs single marker linkage disequilibrium tests: what do we gain? Eur J Hum Genet. 2001;9:291–300. doi: 10.1038/sj.ejhg.5200619. [DOI] [PubMed] [Google Scholar]
- 2.Ardlie KG, Kruglyak L, Seielstad M. Patterns of linkage disequilibrium in the human genome. Nat Rev Genet. 2002;3:299–309. doi: 10.1038/nrg777. [DOI] [PubMed] [Google Scholar]
- 3.Barton NH. Estimating multilocus linkage disequilibria. Heredity. 2000;84:373–389. doi: 10.1046/j.1365-2540.2000.00683.x. [DOI] [PubMed] [Google Scholar]
- 4.Bourgain C, Genin E, Quesneville H, Clerget-Darpoux F. Search for multifactorial disease susceptibility genes in founder populations. Ann Hum Genet. 2000;64:255–265. doi: 10.1046/j.1469-1809.2000.6430255.x. [DOI] [PubMed] [Google Scholar]
- 5.Browning SR. Multilocus association mapping using variable-length markov chains. Am J Hum Genet. 2006;78:903–913. doi: 10.1086/503876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Browning BL, Browning SR. Efficient multilocus association mapping for whole genome association studies using localized haplotype clustering. Genet Epidemiol. 2007;31(5):365–375. doi: 10.1002/gepi.20216. [DOI] [PubMed] [Google Scholar]
- 7.Clayton D, Chapman J, Cooper J. Use of unphased multilocus genotype data in indirect association studies. Genet Epidemiol. 2004;27:415–428. doi: 10.1002/gepi.20032. [DOI] [PubMed] [Google Scholar]
- 8.Clayton D, Jones H. Transmission disequilibrium tests for extended marker haplotypes. Am J Hum Genet. 1999;65:1161–1169. doi: 10.1086/302566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng T, Zhang S, Sha Q. Two-stage association tests for genome-wide association studies based on family data with arbitrary family structure. Eu J Hum Genet. 2007;15:169–1175. doi: 10.1038/sj.ejhg.5201902. [DOI] [PubMed] [Google Scholar]
- 10.Gabriel SB, Schaffner SF, Nguyen H, et al. The structure of haplotype blocks in the human genome. Science. 2002;296(5576):2225–9. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 11.Huang BE, Amos CI, Lin DY. Detecting haplotype effects in genome-wide association studies. Genet Epidemiol. 2007;31:803–812. doi: 10.1002/gepi.20242. [DOI] [PubMed] [Google Scholar]
- 12.Hudson RR. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformations. 2002;18:337–338. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]
- 13.Kimmel G, Shamir R. A fast method for computing high significance disease association in large population-based studies. Am J Hum Genet. 2006;79:481–492. doi: 10.1086/507317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y, Sung W, Liu JJ. Association mapping via regularized regression analysis of single-nucleotide—polymorphism haplotypes in variable-sized sliding windows. Am J Hum Genet. 2007;80:705–715. doi: 10.1086/513205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Millstein J, Conti D, Gilliland F, Gauderman W. A testing framework for identifying susceptibility genes in the presence of epistasis. Am J Hum Genet. 2006;78:15–27. doi: 10.1086/498850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maclean CJ, Martin RB, Sham PC, Wang H, Straub RE, Kendler KS. The trimmed-haplotype test for linkage disequilibrium. Am J Hum Genet. 2000;66:1062–1075. doi: 10.1086/302796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mathias RA, Gao P, Goldstein JL, Wilson AF, Pugh EW, Furbert-Harris P, Dunson GM, Malveaux FJ, Togias A, Barnes KC, Beaty TH, Huang S-K. A graphical assessment of p-values from sliding window haplotype tests of association to identify asthma susceptibility loci on chromosome 11q. BMC Genet. 2006;7:38. doi: 10.1186/1471-2156-7-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morris RW, Kaplan NL. On the advantage of haplotype analysis in the presence of multiple disease susceptibility alleles. Genet Epidemiol. 2002;23:221–233. doi: 10.1002/gepi.10200. [DOI] [PubMed] [Google Scholar]
- 19.Nordborg M, Tavare S. Linkage disequilibrium: what history has to tell us? Trends Genet. 2002;18:83–90. doi: 10.1016/s0168-9525(02)02557-x. [DOI] [PubMed] [Google Scholar]
- 20.Olson JM, Wijsman EM. Design and sample size considerations in the detection of linkage disequilibrium with a marker locus. Am J Hum Genet. 1994;55:574–580. [PMC free article] [PubMed] [Google Scholar]
- 21.Perola M, Koivisto M, Varilo T, Hennah W, Ekelund J, Lugg M, Peltonen L, Ukkonen E, Mannila H. A method to find and compare the strength of haplotype block boundaries and its application in populations with different settlement histories. Am J Hum Genet. 2002;71(Suppl):219. [Google Scholar]
- 22.Rannala B, Reeve JP. High-resolution multipoint linkage-disequilibrium mapping in the context of a human genome sequence. Am J Hum Genet. 2001;69:159–178. doi: 10.1086/321279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seaman SR, Müller-Myhsok B. Rapid simulation of p-values for product methods and multiple-testing adjustment in association studies. Am J Hum Genet. 2005;76:399–408. doi: 10.1086/428140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sevice SK, Lang DW, Freimer NB, Sandkuijl LA. Linkage disequilibrium mapping disease genes by reconstruction of ancestral haplotypes in founder populations. Am J Hum Genet. 1999;64:1728–1738. doi: 10.1086/302398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spielman RS, McGinnis RE, Ewens WJ. Transmission test for linkage disequilibrium: the insulin gene region and insulin dependent diabetes mellitus (IDDM) Am J Hum Genet. 1993;52:506–516. [PMC free article] [PubMed] [Google Scholar]
- 26.Toivonen HT, Onkamo P, Vasko K, Ollikainen V, Sevon P, Mannila H, Herr M, Kere J. Data mining applied to linkage disequilibrium mapping. Am J Hum Genet. 2000;67:133–145. doi: 10.1086/302954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wessel J, Schork N. Generalized genomic distancebased regression methodology for multilocus association analysis. Am J Hum Genet. 2006;79:792–806. doi: 10.1086/508346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang HC, Lin CY, Cathy SJ Fann. A sliding-window weighted linkage disequilibrium test. Genet Epidemiol. 2006;30:531–545. doi: 10.1002/gepi.20165. [DOI] [PubMed] [Google Scholar]
- 29.Yi L, Sung W, Liu JJ. Association mapping via regularized regression analysis of single-nucleotide—polymorphism haplotypes in variable-sized sliding windows. Am J Hum Genet. 2007;80:705–715. doi: 10.1086/513205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang K, Qin Z, Ting C, Waterman MS, Liu JS, Sun F. Haplotype block partitioning and tag SNP selection using genotype data and their applications to association studies. Genome Res. 2004;14:908–916. doi: 10.1101/gr.1837404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang K, Li J. HaploBlockFinder: an efficient program for haplotype block identification. Bioinformatics. 2003;19:1–3. [Google Scholar]
- 32.Zhang S, Sha Q, Chen HS, Dong J, Jiang R. Transmission/disequilibrium test based on haplotype sharing for tightly linked markers. Am J Hum Genet. 2003;73:566–579. doi: 10.1086/378205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao H, Zhang S, Merikangas KR, et al. Transmission/disequilibrium tests using multiple tightly linked markers. Am J Hum Genet. 2000;67(4):936–946. doi: 10.1086/303073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao HG, Pfeiffer R, Gail MH. Haplotype analysis in population genetics and association studies. Pharmacogenomics. 2003;4:171–178. doi: 10.1517/phgs.4.2.171.22636. [DOI] [PubMed] [Google Scholar]
- 35.Zhu X, Zhang S, Kan D, Cooper R. Haplotype block definition and its application. Pacific Symposium on Biocomputing. 2004;9:152–163. doi: 10.1142/9789812704856_0015. [DOI] [PubMed] [Google Scholar]
- 36.Zöllner S, von Haeseler A. Coalescent approach to study linkage disequilibrium between single-nucleotide polymorphisms. Am J Hum Genet. 2000;66:615–628. doi: 10.1086/302766. [DOI] [PMC free article] [PubMed] [Google Scholar]