Abstract
Background
Idiopathic congenital talipes equinovarus (ICTEV) is a congenital limb deformity. Based on extended transmission disequilibrium testing, Gli-Kruppel family member 3 (Gli3) has been identified as a candidate gene for ICTEV. Here, we verify the role of Gli3 in ICTEV development.
Methods
Using the rat ICTEV model, we analyzed the differences in Gli3 expression levels between model rats and normal control rats. We used luciferase reporter gene assays and ChIP/EMSA assays to analyze the regulatory elements of Gli3.
Results
Gli3 showed higher expression levels in ICTEV model rats compared to controls (P < 0.05). We identified repressor and activator regions in the rat Gli3 promoter. The Gli3 promoter also contains two putative Hoxd13 binding sites. Using EMSA, the Hoxd13 binding site 2 was found to directly interact with Hoxd13 in vitro. ChIP assays of the Hoxd13-Gli3 promoter complex from a developing limb confirmed that endogenous Hoxd13 interacts with this region in vivo.
Conclusion
Our findings suggest that HoxD13 directly interacts with the promoter of Gli3. The increase of Gli3 expression in ICTEV model animal might result from the low expression of HoxD13.
Background
Idiopathic congenital talipes equinovarus (ICTEV) is a congenital limb deformity that affects 0.3-7% of live births worldwide [1]. It is defined by fixation of the foot in cavus, adductus, varus and equinus (inclined inwards, axially rotated outwards and pointing downwards) with related soft tissue abnormalities [2]. The mechanism underlying the development of ICTEV remains unclear, and neurological, muscular, bony, connective tissue and vascular mechanisms have all been proposed as contributing factors [2-5]. Although some studies have suggested that both genetic and environmental factors lead to ICTEV, twin studies, ethnic studies and pedigree analyses suggest a genetic basis for the condition [6]. Furthermore, studies have suggested that a significant ICTEV risk can be attributed to unknown disorder-causing genes [1]. Overall, little is known about the pathogenesis of human ICTEV.
Many candidate genes for this disorder have been proposed because the molecular and cellular components of vertebrate limb bud development are well known. Specialized regions of the developing limb bud, such as the zone of polarizing activity (ZPA), the apical ectodermal ridge (AER) and the non-ridge ectoderm, direct and coordinate the development of the limb bud along the anterior-posterior (AP), dorsal-ventral (DV) and proximal-distal (PD) axes in a pattern conserved for tetrapods [7]. Distal limb development along the AP (thumb to little finger) axis is governed primarily by the ZPA. The major signaling molecule with polarizing potential in the ZPA is Sonic hedgehog (Shh) [8], which plays a central role in pattern formation in the embryo [9] and is a key signal in establishing different digit fates along the AP axis of the vertebrate limb bud [10]. In Drosophila, hedgehog (Hh) signaling is mediated by the Cubitus interruptus (Ci) protein, a zinc finger transcription factor. In birds and mammals, Ci homologs constitute the three member Gli family (Gli1, Gli2, and Gli3). All three Gli genes are expressed in the developing limb, but only Gli3, a direct intracellular mediator of Shh [11-13], is necessary for limb patterning [14,15]. With a C-terminal repressor region and an N-terminal activator region, Gli3 is a bipotential transcription factor that can activate or repress some of the same target genes [16]. During vertebrate limb development, Shh signaling prevents the processing of the full-length Gli3 (Gli3-190) to a short form (Gli3-83) that functions as a strong repressor. In both mouse and chick limb buds, the repressor form of Gli3 is present in an anterior-posterior gradient with the highest levels in the anterior part of the limb bud where Shh signaling is at its lowest levels. The genetic data of the Shh, Gli3 and double-compound mutants indicate that the phenotype in the absence of Shh is caused by an excess in the Gli3R form that suppresses gene expression, cell survival and distal progression of limb bud development. Gli3 and Shh reciprocally restrict each other to control the normal limb morphogenesis [17].
A third family of transcription factors involved in limb development is the HOX family, which is evolutionarily conserved and plays a fundamental role in patterning the AP axis of developing embryos. Each HOX protein mediates cellular events during limb morphogenesis [18,19]. The physical position of the Hox gene within each cluster corresponds closely to their temporal and spatial expression patterns during development. Thus, genes at the 3' end of the clusters, such as HOXD1, are expressed early in the anterior and proximal regions, whereas genes at the 5' end, such as HOXD13, are expressed later in posterior and distal regions [20]. In early limb bud development, the Gli3 anterior expression overlaps the HoxD posterior expression, indicating that a genetic interaction between a 5' HoxD member and Gli3 regulates digit formation [21]. Biochemical and transfection analyses provide support for the physical interaction of the 5' HoxD protein and Gli3 protein via the homeodomain [21]. This interaction can convert the truncated Gli3 repressor form into an activator of its target promoters [21].
Most Gli3 research focuses on digit abnormalities and deformities. Gli3 mutations cause limb development disorders, such as Greig cephalopolysyndactyly syndrome (GCPS) [22], Pallister-Hall syndrome (PHS) [23], postaxial polydactyly type A (PAP-A) [24] and preaxial polydactyly type IV [25]. Association analyses suggest that Gli3 and HoxD13 are associated with ICTEV [26-28]. To confirm a role for Gli3 in ICTEV, we analyzed Gli3 expression in ICTEV model rat embryos. To understand how a change in the expression of Gli3 affects ICTEV, we investigated the interaction between Gli3 and HoxD13.
Methods
ICTEV patients and normal controls
Flexor hallucis longus was from 20 ICTEV patients (13 men and 7 women) aged 4-12 years (mean 6.7 years) and peripheral blood was from 84 ICTEV patients (50 men and 34 women) aged 3-12 years (mean 6.2 years). All patients were recruited from the Department of Pediatric Orthopedic Surgery, Second Affiliated Hospital of China Medical University. Informed consent was obtained from the patients and the study was approved by the China Medical University Ethics Committee. The probands showed the typical ICTEV phenotype (a fixation of the foot in adduction, supination and varus). Flexor hallucis longus and lung tissue were from nine normal cadavers (5 men and 4 women) aged 5-11 years (mean 7.5 years) as controls.
A rat ICTEV model
Forty pregnant Wistar rats were obtained from the experimental animal center of our university. The ICTEV phenotype in rat embryos was induced by administration of 135 mg/kg all-trans-retinoic acid (ATRA) on gestation day 10 (GD10), as previously described [29,30]. All studies were performed with the approval of the experimental animal committee at our university (SCXK 2008-0005).
RNA isolation and expression analysis
The Gli3 gene is expressed as an 8.5 kb mRNA in tissues such as the postnatal testis, myometrium, placenta and lung [31]. We investigated whether the gene was also expressed in the hindlimb. RNA was therefore extracted from the flexor hallucis longus of ICTEV patients and normal controls using a Tissue RNA kit (Tiangen, China) according to the manufacturer's protocol. RNA was also extracted from lung tissues of normal cadavers as a positive control. Using a reverse transcription kit (Promega, USA) according to the manufacturer's instructions, cDNA was generated. For Gli3, the forward and reverse primers were 5'-TTT TCC AAC ACA GAG GCC TAT TC-3' and 5'-ATC TTG GAC CTC TTG TTG TGC AT-3', respectively. Human β-actin forward and reverse primers were 5'-TCA CCC ACA CTG TGC CCA TCT ACG A-3' and 5'-CAG CGG AAC CGC TCA TTG CCA ATG G-3', respectively. PCR reactions contained 40 ng cDNA, 1.2 mM MgCl2, 20 mM dNTPs, 320 nM of each primer and 2U Taq polymerase in a 25 μl total volume. PCR reactions had an initial denaturing step at 94°C for 5 minutes, followed by 35 cycles of denaturing at 94°C for 45 sec, annealing at 58°C for 45 sec and extension at 72°C for 60 sec, and ended with a final extension at 72°C for 10 min.
Gli3 expression in ICTEV rat models and normal control rats
Real-time PCR (RT-PCR) was performed to evaluate differences in RNA expression levels. The ICTEV model rats were dissected at GD15, GD17, GD19 and GD21. Five embryos with clubfoot were selected and the hindlimbs of the embryos were stored at -70°C. We also dissected control rats at GD15, GD17, GD19 and GD21 and five embryos were selected and their hindlimbs were similarly stored. RNA was extracted from the hindlimbs using a Tissue RNA kit (Tiangen, China) and cDNA was synthesized using a reverse transcription kit (Promega, USA) according to the manufacturer's instructions. Real-time PCR amplification was performed in a 25 μl reaction mixture, which included 12.5 μl SYBR Premix Ex Taq, 9.5 μl deionized water, 0.5 μl (initial concentration 10 μM) of each primer and 2 μl cDNA, according to the manufacturer's instructions (Takara Biotechnology). Amplification was performed by one round of pre-denaturation at 95°C for 10 s, a step-cycle mode of 40 rounds of denaturation at 95°C for 5 s and an annealing and extension step at 58°C for 20 s. The primers used for Gli3 were: forward: 5'-TTT TCC AAC ACA GAG GCC TAT TC-3' and reverse: 5'-ATG CAC AAC AAG AGG TCC AAG AT-3'. The primers of β-actin were: forward: 5'-TCC TTC CTG GGT ATG GAA TC- 3' and reverse: 5'-GCA CTG TGT TGG CAT AGA GG-3'.
To evaluate Gli3 protein expression level, we performed western blot analyses. Cytoplasmic protein was extracted from the embryonic hindlimbs of five rats presenting with clubfoot and normal control rat embryonic hindlimbs at GD15 and GD17 using a cytoplasmic and nuclear protein extract kit (Activ Motif, USA) according to the manufacturer's instructions. Protein concentration was determined spectrophotometrically (Unico, USA) at 280 nm[32]. Sample buffer (Beyotime, China) was added to the cytoplasmic protein and the solution was loaded onto a 6% polyacrylamide gel. Following protein separation, the polyacrylamide gel was electro-blotted onto a PVDF membrane (Millipore, MA, USA). Non-specific binding sites were blocked by soaking the membrane in a 3% BSA (Sigma, Poole, Dorset, UK) in TBST buffer (20 mM Tris-buffered saline, 0.047% Tween at pH 7.4) blocking solution overnight at 4°C. The membrane was incubated for 3 hours with rabbit anti-human Gli3 polyclonal antibody (Santa Cruz, USA, 1:50 dilution) at room temperature and washed for 40 min in TBST buffer. The membrane was then incubated for 2 hours at room temperature with a goat anti-rabbit IgG horseradish peroxidase-conjugated antibody (Antibodies Incorporated, USA. 1:4000 dilution). Protein bands were visualized using modified enhanced chemiluminescence (Tiangen, China).
To determine differences in Gli3 levels and localization, we used immunohistochemistry assays. The ICTEV model rats and control rats were dissected at GD19. We selected five embryos presenting with clubfoot from the ICTEV rats and five normal embryos from normal control rats. The hindlimbs of the embryos were embedded in paraffin and fixed in 4% paraformaldehyde overnight at 4°C. The tissues were sectioned into 4-μm slices and labeled with Gli3 antibody (Santa Cruz, USA, 1:50 dilution). The normal control hindlimbs were labeled with Phosphate buffered saline as negative controls. A standard immunohistochemistry protocol was used (Maxim Biotech, Inc., USA).
Gli3 mutation analysis
Genomic DNA was extracted from 84 fresh blood samples obtained from the ICTEV patients using a Blood DNA kit (Tiangen, China) according to the manufacturer's protocol. A previous study investigated mutations in Gli3 exons 9, 10, 11 and 12 [26]. We therefore designed primers (shown in Table 1) to amplify the remaining Gli3 exons 1-8, 13 and 14 and the 5' flanking sequence. Mutations in Gli3 were detected by denaturing gradient gel electrophoresis (DGGE), a sensitive method to separate alleles based on differences in melting behavior [33].
Table 1.
Gli3 primers for DGGE
| Primer sequence | Annealing Temp (°C) | Product size (bp) | |
|---|---|---|---|
| Promoter 1 F | *AGCTTGCAGTTCCCTTGC | 60 | 684 |
| R | GTCCGACAATTTCTAACATCGA | ||
| Promoter 2 F | *GCCTTCGATGTTAGAAATTGTCG | 62 | 668 |
| R | TGGGCTGCTGGTAATCCCTGTGC | ||
| Exon 1 F | *TTTTGGAAAGTTGATGGCTCT | 60 | 229 |
| R | GGCTGCTGGTAATCCCTGT | ||
| Exon 2 F | *AATTGCTCCTTAAAGTAGTT | 60 | 335 |
| R | CATAGCTCCTGAACAAGTG | ||
| Exon 3 F | *CAATGTTGCTTTGTGAAT | 60 | 234 |
| R | TAAAAGCCAGCATCTCGT | ||
| Exon 4 F | *CCCCTTGTATCTGGTTTT | 57 | 316 |
| R | GTCTACTTTATACACGTCCC | ||
| Exon5 F | *ATTGCTGATGTGGGTTGT | 60 | 297 |
| R | GTTGCCTTTGCCATTTCC | ||
| Exon 6 F | *TAGGCAAGTAGCAATAAATAG | 60 | 303 |
| R | ACATAATGGATTCAGGAAAA | ||
| Exon 7 F | *GCTCAGCGTTTAAGTGAT | 61 | 268 |
| R | GCATCGACCTGTCCCTCT | ||
| Exon 8 F | *GGGATTGGAGAATTATCAG | 60 | 299 |
| R | AGTCTTGGGAGGAGTGGG | ||
| Exon13 F | *TTCCTTTCCACTTGACCCC | 60 | 391 |
| R | AAAACCCTGAGCAGATGCA | ||
| Exon 14-1 F | *TGTGAGGCAGGCAATGTG | 53 | 325 |
| R | GGAGAAGCAGGGCGAGAT | ||
| Exon 14-2 F | *ATCTCGCCCTGCTTCTCC | 55 | 489 |
| R | GCTGCTGAGGCTGCTGAA | ||
| Exon 14-3 F | *AGAGGATGAGCCTGAAGACG | 56 | 486 |
| R | GCTGCTCGTACCCTGCTT | ||
| Exon 14-4 F | *CTGATGCCAACCTGAACG | 56 | 479 |
| R | TCCACTGGTGCCACTTCC | ||
| Exon 14-5 F | *TGGTCGTCCACCCGCAGAA | 58 | 490 |
| R | GGCCCTTGGTAGATGTTGATGT | ||
| Exon 14-6 F | *AGATGCTTGGGCAGATTAG | 60 | 350 |
| R | TGAACCAGCTTTCGTGTC | ||
| Exon 14-7 F | *CGCTGTGCTCTAATCTGC | 60 | 491 |
| R | TATTGATTTCCGTTGGTTG |
*GC clamp was added at the 5' end of the forward primers
GC clamp: cgcccgccgcgccccgcgcccggcccgccgcccccgcccg
Identification of the rat Gli3 promoter
To obtain a candidate rat Gli3 promoter sequence, four forward primers were designed upstream of the rat Gli3: Gli3(-1107), 5'-GTG ACC TGC CTG TGC CTG TA-3'; Gli3(-532), 5'-TTA ACC TCT GCG TTA CAA CC-3'; Gli3(-388), 5'-ATC AGA GGG TCT CAG CGT TAG-3'; and Gli3(-128), 5'-CTC CTC AGG CAG AAG ATG CA -3'. Four products were obtained when rat genomic DNA was amplified with each of these primers and the Gli3 reverse primer 5'-ACA CCA CAG TGC CAT CAA A -3'. The amplified fragments were verified by sequencing and cloned into a PMD-18T vector. The clones were digested with KpnI and HindIII and cloned into the KpnI and HindIII sites of the pGL3-luciferase vector (Promega, USA).
L6GNR4 rat myoblastocytes (Cell Laboratory, Chinese Academy of Sciences, Shanghai) were cultured in Dulbecco's Modified Eagle's Medium supplemented with 10% fetal calf serum, 100 units/mL penicillin, 100 μg/mL streptomycin and then incubated in 5% CO2 at 37°C. L6 cells were transfected using a liposome transfection kit (Invitrogen, USA). In each transfection, 12 μg reporter plasmid and 0.1 μg pRL-TK (internal control for transfection efficiency) were used per 6-cm dish. Cells were incubated for 48 hours at 37°C. After transfection, the cells were washed with PBS, lysed with 1× passive lysis buffer and assayed for luciferase activity using the Dual-Luciferase Reporter Assay System (Promega, USA).
Analysis of the rat Gli3 5' region
To search for regulatory elements in the Gli3 transcriptional control regions, we analyzed the genomic sequence 1000 bp upstream of the transcription start site using P-Match http://www.gene-regulation.com.
Chromatin immunoprecipitation assays (ChIP)
ChIP assays were performed according to the manufacturer's instructions (Active Motif, USA). Hindlimbs of GD14 rats were homogenized in PBS on ice. Hoxd13 antibody (8 μg) (Santa Cruz, USA) was used for the immunoprecipitation, and 8 μg of the Sox9 antibody (Santa Cruz, USA) was used as a negative control. DNA was extracted as recommended by protocol. We used 2.5 μL of each sample as a template for PCR using the following primers:
Gli3 Site1: ACACTGAGGGCCCTGGGTAG
Gli3 Site1 rev: GTGCCGAAAGGGTTGTAACG
Gli3 Site2: AACCCTTTCGGCACACTTCTG
Gli3 Site2 rev: GTGGCTCTCAACCTTCCTAACG
PCR reactions had an initial denaturing step at 94°C for 5 minutes, followed by 35 cycles of denaturing at 94°C for 35 sec, annealing at 59°C for 35 sec and extension at 72°C for 35 sec, and ended with a final extension at 72°C for 10 min.
Electrophoretic mobility shift assays (EMSA)
Nucleoprotein was extracted from E14.5 rat embryo limbs using a cytoplasmic and nuclear protein extract kit (Activ Motif, USA) according to the manufacturer's instructions Oligonucleotides and their complementary strands were used to evaluate binding of HoxD13 to the Gli3 HoxD13-binding site (5'-CTAGG-3'). The double-stranded wild-type (tttacaCTAGGattcataaccatagcataattacagcta) and mutated (tttacaGCTAAattcataaccatagcataattacagcta) sequences were labeled with biotin according to standard protocols (Pierce, USA). A 100-fold excess of unlabeled probe was used as a specific competitor. The DNA-binding ability of the different proteins was monitored by EMSA on a 10% non-denaturing polyacrylamide gel. DNA binding bands were detected using a chemiluminescence system (Pierce, USA).
Results
Gli3 is not expressed in the flexor hallucis longus of ICTEV patients
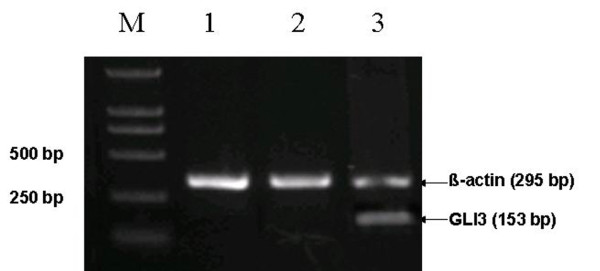
To investigate the expression profiles of Gli3 in flexor hallucis longus, Semi-quantitative RT-PCR was performed using total RNA from the flexor hallucis longus of patients and cadavers as well as the lung tissues of cadavers. No Gli3 expression was detected in the flexor hallucis longus; however, expression was detected in control lung tissue (Fig. 1).
Figure 1.
Semi-quantitative RT-PCR analysis of Gli3 mRNA expression in flexor hallucis longus and lung tissue. Gli3 expression was not found in the flexor hallucis longus. Lanes 1 is an RT-PCR sample from an ICTEV patient flexor hallucis longus, lane 2 is a normal control flexor hallucis longus RT-PCR and lane 3 is a normal control lung RT-PCR. β-actin expression is shown as a control for all samples. Lane M is a size marker.
Identification of rat ICTEV model
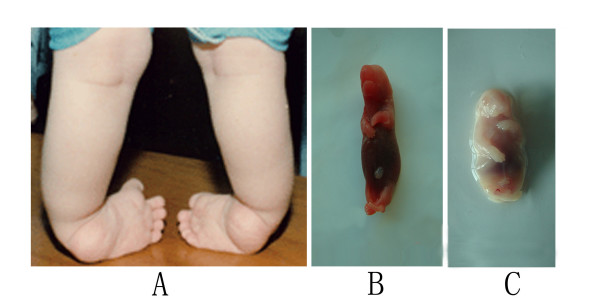
The embryos were harvested from ATRA-treated pregnant rats at GD15, 17, 19 and 21. We found embryos with clubfoot at GD15. A comparison between ICTEV and normal embryos at GD 21 is shown in Fig 2. Hind limb abnormality was observed in embryos of the ICTEV model rat, similar to that seen in ICTEV patients. A total of 74 embryos with clubfoot were found out of 245 embryos (30.2%), indicating that the ICTEV model was successfully established.
Figure 2.
Comparison of ICTEV and normal embryos at GD21. (A) Foot of an ICTEV patient, showing cavus, adductus, varus and equines. (B) The hindlimbs of normal embryos at GD21 presented with normal development. (C) The hindlimbs of ICTEV embryos at GD21 presented with dysplasia of the foot, showing cavus and adductus as observed in ICTEV patients.
ICTEV association with Gli3 is not due to genetic mutations
Unlike other disorders associated with Gli3, no mutations were observed in the 5' flanking sequence or in Gli3 exons 1-8, 13 and 14 in the ICTEV patients.
Gli3 has higher expression in ICTEV model rats than in control rats
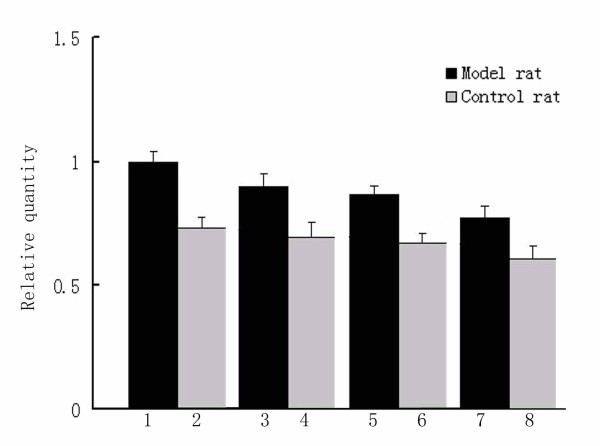
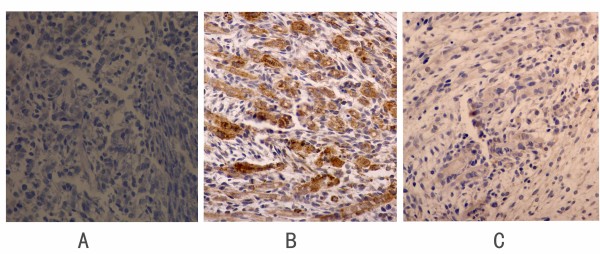
To study Gli3 earlier in development, we used a rat model for ICTEV. Expression of Gli3 was higher in ICTEV model rat embryonic hindlimbs compared to normal control rat embryonic hindlimbs (Fig. 3). Similarly, Gli3 protein showed increased abundance in ICTEV model rat embryonic hindlimbs compared to normal control rat embryos (Fig. 4). For both ICTEV model rats and normal control rats, Gli3 RNA levels and Gli3 protein abundance decreased during embryonic development (Figs. 3 and 4). Using immunohistochemistry, the Gli3 protein in ICTEV rats was localized to the foot muscle of the rats (Fig. 5).
Figure 3.
Real-time PCR analysis of Gli3 expression in the hindlimb of ICTEV model rat and normal control rat embryos. Compared to the normal control, Gli3 mRNA expression was significantly enhanced. With birth drawing near, Gli3 expression in both model and normal rats had the tendency to decrease. Column 1, 3, 5 and 7 show Gli3 relative expression in ICTEV model rat at GD15, GD17, GD19 and GD21, respectively. Column 2, 4, 6 and 8 show Gli3 relative expression in normal control rat embryos at GD15, GD17, GD19 and GD21, respectively.
Figure 4.
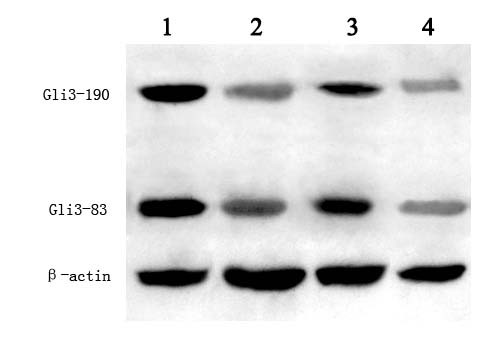
Western blot results. Western blots were used to determine Gli3 protein abundance in nuclear extracts from the hindlimb tissue of ICTEV model rat and normal control rat embryos. Compared to the normal control, Gli3 expression was significantly enhanced in model rats. With birth drawing near, Gli3 protein expression in both model and normal rats had a tendency to decrease. Lane 1 is the protein of ICTEV model rat embryos at GD15, lane 2 is the protein of normal rat embryos at GD15, lane 3 is the protein of ICTEV model rat embryos at GD17 and lane 4 is the protein of normal rat embryos at GD17.
Figure 5.
Immunohistochemistry results. Immunohistochemistry revealed that Gli3 expression in the hindlimb tissue of ICTEV model rat embryos was higher than that in normal control rat embryos at GD19 (400×). (A) Hindlimb tissue of a normal control rat embryo labeled with PBS. (B) Hindlimb tissue of an ICTEV model rat embryo labeled with Gli3 antibody. (C) Hindlimb tissue of a normal control rat embryo labeled with Gli3 antibody.
Gli3 promoter identification
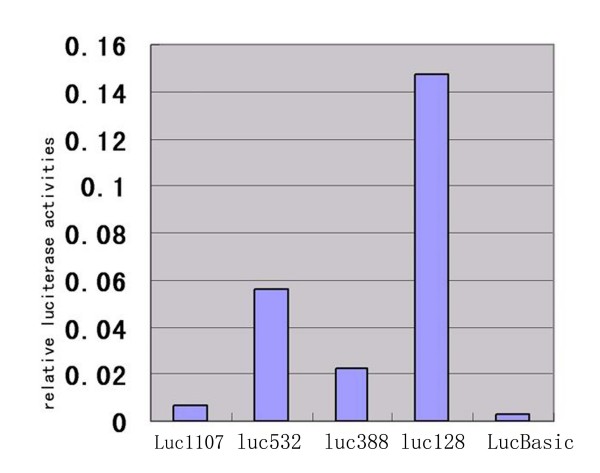
To determine the promoter region of Gli3, we generated four luciferase reporter constructs (Luc1107, Luc532, Luc388 and Luc128) with fragments upstream of Gli3 as the promoters. All the luciferase reporter constructs showed higher promoter activity compared to the pGL3-basic reporter vector. Relative to the pGL3-basic reporter vector, Luc128, Luc388, Luc532 and Luc1107 increased luciferase activity 2-fold, 20-fold, 8-fold and 53-fold, respectively (Fig. 6). These data suggest that there are positive regulatory elements in the regions from -1107 to -532, -388 to -128 and from -128 to -48 upstream of Gli3 and negative regulatory elements in the region from -532 to -388 upstream of Gli3.
Figure 6.
Relative activities of rat Gli3 promoter regions in a luciferase reporter construct. Activities were measured in L6 cells. Findings suggested positive regulatory elements existed in the regions from -1107 to -532, -388 to -128 and from -128 to -48 upstream of Gli3 and negative regulatory elements in the region from -532 to -388 upstream of Gli3.
HoxD13 binding sites in the 5' region of the rat Gli3
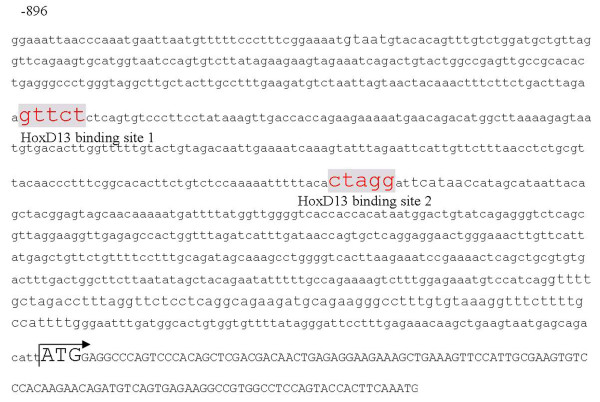
To identify the transcription factors that directly regulate Gli3 expression, we analyzed the genomic region upstream of the transcription start site of the Gli3 gene with the transcription binding-site prediction program P-Match. Two putative HoxD13 binding sites, the HoxD13 binding site 1 (-667): gttct and the HoxD13 binding site 2 (-477):ctagg were identified, suggesting that Hoxd13 could directly bind to these sites to regulate Gli3 expression (Fig. 7).
Figure 7.
Potential binding sites. The rat Gli3 proximal promoter region contains two potential HoxD13 binding sites (gray boxes). The transcription start site is indicated by an arrow. The Gli3 protein translation start site (ATG) is indicated.
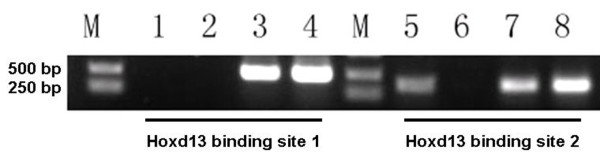
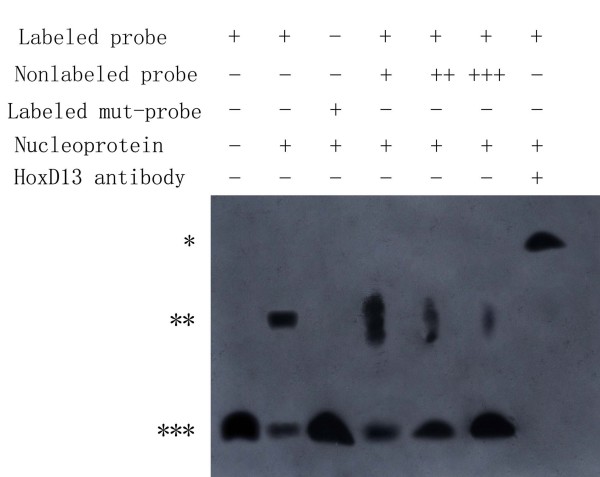
To verify the in vivo binding of Hoxd13 to the Gli3 promoter, we used ChIP. Cross-linked and sheared chromatin from rat GD14 embryonic hindlimbs were immunoprecipitated with an anti-Hoxd13 antibody and analyzed by PCR. These ChIP analyses revealed that, in vivo, Hoxd13 efficiently bound only to binding site 2 (Fig. 8). Additionally, nucleoprotein containing Hoxd13 was incubated with a fragment of -483 ~ -446 bp upstream of Gli3 containing site 2. As shown in Fig. 9, strong DNA binding was observed in the presence of the Hoxd13 protein. A competition experiment and supershift existence in the presence of the Hoxd13 antibody demonstrated the specificity of such binding. The result indicated Hoxd13 is bound to site 2 in vitro.
Figure 8.
ChIP results. The ChIP assay of putative HoxD13 binding sites 1 and 2 in the Gli3 promoter. Only HoxD13 binding site 2 binds HoxD13 in vivo. In lanes 1 and 5, chromatin from rat embryonic hindlimbs was immunoprecipitated with the HoxD13 antibody. In lanes 2 and 6, the Sox9 antibody was used as a negative control. Lanes 3 and 7 show the enzymatic shearing before immunoprecipitation. Genomic DNA was used as a positive control (lanes 4 and 8). Lane M is a size marker.
Figure 9.
EMSA results. Electrophoretic mobility shift assay (EMSA) using oligonucleotide probes containing the HoxD13 binding site 2. The *** denotes unbound biotin-labeled DNA, ** denotes Hoxd13-DNA complexes and * denotes Hoxd13-antibody-Hoxd13-DNA complexes.
Discussion
The mammalian Gli gene family encodes zinc finger transcription factors and plays a role in developmental regulation and human diseases [34]. One member of this family, Gli3, was identified as a candidate gene for ICTEV [26]. To investigate the mechanism of this association, we investigated Gli3 mRNA and protein expression patterns in ICTEV patients. Gli3 was not expressed in the flexor hallucis longus of ICTEV patients, suggesting that although Gli3 is important for correct limb development, it is no longer expressed in postnatal limbs.
To investigate the role of Gli3 role in ICTEV earlier in development, we established an ICTEV rat model and detected Gli3 expression in rat embryonic hindlimbs. There are some concerns regarding the rat model of ICTEV induced with ATRA in pregnant rats. The rat model has some limitations. After induction with ATRA, not all fetal rats were exhibited with symptoms of ICTEV. Those that developed the clubfoot also developed malformations such as rima oculi, spina bifida, cranial deformation and anal atresia. Although the rat model of ICTEV induced with ATRA is not ideal and is controversial, it is currently the best rat model of ICTEV. We tested different dosages of ATRA in preliminary experiments and found that 135 mg/kg was the dose at which the highest percentage (about 30%) of clubfeet in the lower limbs was achieved. We only selected the fetal rats with clubfeet as the experimental subjects to ensure the most legitimate comparison to patients with ICTEV. We found that both Gli3 mRNA and protein expression levels were higher in the ICTEV model rats compared to normal control rats. Furthermore, with birth drawing near, the Gli3 gene expression in both of the normal control group and the model group was gradually downregulated (Figs. 3 and 4), indicating that the Gli3 gene is a very important gene in regulating the limb development and that its role of regulating limb development may disappear after birth. This is also consistent the inability to detect the Gli3 gene in the flexor hallucis longus of adults (Fig. 1), which may be because it is not expressed in the lower limb post-birth. The occurrence of ICTEV may be caused by changes in the expression level of this critical gene that is related to limb development. Immunohistochemical results suggest that Gli3 protein expression in the muscles around ankles of model fetal rats was remarkably higher than in the control group, and the affected sites of patients with ICTEV were also in the ankle. The location of this muscle was identical to that of flexor hallucis longus; therefore, it is reasonable to conclude that the occurrence of ICTEV is associated with changes in the expression level of Gli3. To identify the underlying reason for the change in Gli3 expression, we first looked for mutations in the GLI3 promoter and coding regions of ICTEV patients. A previous study investigating mutations in GLI3 exons 9, 10, 11 and 12 identified only one polymorphism [26]. To complete this study, we sequenced the remaining exons and the promotor, but did not detect any polymorphisms. While a larger sample size might reveal polymorphisms, these data suggest that the Gli3 mutation is not the root of its association with ICTEV.
The changes in Gli3 gene expression may be caused by changes in some transcription factors that regulate the GLI3 gene, ultimately leading to the development of ICTEV. Accordingly, future studies should focus on the regulation of the GLI3 gene and its function.
Alternatively, Gli3 could be differentially regulated in ICTEV model rat embryos compared to normal control embryos. To determine the transcription factors that bind to the promoter region of Gli3, we constructed a series of rat Gli3 truncated promoters and cloned them into reporter gene constructs. These analyses suggested that the promoter has both positive and negative regulatory elements. Putative HoxD13 binding sites 1 and 2 in Gli3's upstream 5' region were located in positive and negative regulatory elements, respectively. HoxD13, however, only bound to site 2 in vivo. In vitro competition experiments also revealed a specific affinity of Hoxd13 to this site. Thus, Hoxd13 can directly regulate the expression of Gli3 during limb formation. We cannot, however, exclude the possibility that Hoxd13 interacts with the Gli3 promoter regions as a part of a protein complex. In ICTEV patients, HOXD13 has lower expression compared to healthy subjects [27]. Thus, the decrease in HOXD13 expression maybe led to a change in the expression of GLI3, which manifests as ICTEV.
This study provides a theoretical basis for the pathogenesis of ICTEV. Future studies will investigate how HoxD13 regulates Gli3 during limb development and will provide a clearer mechanism for the pathogenesis of ICTEV.
Conclusion
Our study indicates that HoxD13 directly interacts with the promoter of Gli3 and Gli3 mRNA and protein expression levels were increased in the ICTEV model rats. These findings suggest that HoxD13 is a transcription factor of Gli3. Low expression of HOXD13 might lead to increase GLI3 expression level during limb formation, which likely plays a key role in ICTEV pathogenesis.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
DHC is the guarantor of the study. He designed the study and was the primary writer of the manuscript. MHR and CKL designed and wrote the study and critically revised the study for its content. XZ and NZ initiated and monitored the study. CLJ supervised and critically revised the study for its content. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Contributor Information
DongHua Cao, Email: dhcao427@sina.com.
ChunLian Jin, Email: chunlianjin@126.com.
MeiHong Ren, Email: renmeihong@163.com.
ChangKun Lin, Email: cklin@mail.cmu.edu.cn.
Xuan Zhang, Email: zhangxuan1997@163.com.
Ning Zhao, Email: ningzhao78@hotmail.com.
Acknowledgements
This study was supported by a grant from the National Key Research Project of China (No. 30973140).
References
- Dietz FMD. The genetics of idiopathic clubfoot. Clin Orthop Relat Res. 2002;401:39–48. doi: 10.1097/00003086-200208000-00007. [DOI] [PubMed] [Google Scholar]
- Cardy AH, Barker S, Chesney D, Sharp L, Maffulli N, Miedzybrodzka Z. Pedigree analysis and epidemiological features of idiopathic congenital talipes equinovarus in the United Kingdom: a case-control study. BMC Musculoskelet Disord. 2007;8:62. doi: 10.1186/1471-2474-8-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebbeck TR, Dietz FR, Murray JC, Buetow KH. A single gene explanation for the probability of having idiopathic talipes equinovarus. Am J Hum Genet. 1993;53:1051–1063. [PMC free article] [PubMed] [Google Scholar]
- Wynne-Davies R. Genetic and environmental factors in the etiology of talipes equinovarus. Clin Orthop. 1972;84:9–13. doi: 10.1097/00003086-197205000-00003. [DOI] [PubMed] [Google Scholar]
- Chapman C, Stott NS, Port RV, Nicol RO. Genetics of clubfoot in Maori and Pacific people. J Med Genet. 2000;37:680–683. doi: 10.1136/jmg.37.9.680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miedzybrodzka Z. Congenital talipes equinovarus (clubfoot): a disorder of the foot but not the hand. J Anat. 2003;202:37–42. doi: 10.1046/j.1469-7580.2003.00147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capdevila J, Izpisúa Belmonte JC. Patterning mechanisms controlling vertebrate limb development. Annu Rev Cell Dev Biol. 2001;17:87–132. doi: 10.1146/annurev.cellbio.17.1.87. [DOI] [PubMed] [Google Scholar]
- Riddle RD, Johnson RL, Laufer E, Tabin C. Sonic hedgehog mediates the polarizing activity of the ZPA. Cell. 1993;75:1401–1416. doi: 10.1016/0092-8674(93)90626-2. [DOI] [PubMed] [Google Scholar]
- Weed M, Mundlos S, Olsen BR. The Role of Sonic Hedgehog in Vertebrate Development. Matrix Biology. 1997;16:53–58. doi: 10.1016/S0945-053X(97)90072-X. [DOI] [PubMed] [Google Scholar]
- Scherz PJ, McGlinn E, Nissim S, Tabin CJ. Extended exposure to Sonic hedgehog is required for patterning the posterior digits of the vertebrate limb. Dev Biol. 2007;15, 308:343–354. doi: 10.1016/j.ydbio.2007.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altaba ARI. Gli proteins encode context-dependent positive and negative functions: implications for development and disease. Development. 1999;126:3205–3216. doi: 10.1242/dev.126.14.3205. [DOI] [PubMed] [Google Scholar]
- Shin SH, Kogerman P, Lindström E, Toftgárd R, Biesecker LG. GLI3 mutations in human disorders mimic Drosophila Cubitus interruptus protein functions and localization. Proc Natl Acad Sci USA. 1999;96:2880–2884. doi: 10.1073/pnas.96.6.2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai P, Akimaru H, Tanaka Y, Maekawa T, Nakafuku M, Ishii S. Sonic hedgehog-induced activation of the Gli1 promoter is mediated by GLI3. Biol Chem. 1999;274:8143–8152. doi: 10.1074/jbc.274.12.8143. [DOI] [PubMed] [Google Scholar]
- Park HL, Bai C, Platt KA, Matise MP, Beeghly A, Hui CC, Nakashima M, Joyner AL. Mouse Gli1 mutants are viable but have defects in SHH signaling in combination with a Gli2 mutation. Development. 2000;127:1593–1605. doi: 10.1242/dev.127.8.1593. [DOI] [PubMed] [Google Scholar]
- Bai C Brian, Auerbach Wojtek, Lee Joon S, Stephen Daniel, Alexandra L. Gli2, but not Gli1, is required for initial Shh signaling and ectopic activation of the Shh pathway. Development. 2002;129:4753–4761. doi: 10.1242/dev.00115. [DOI] [PubMed] [Google Scholar]
- Ahn S, Joyner AL. Dynamic changes in the response of cells to positive hedgehog signaling during mouse limb patterning. Cell. 2004;118:505–516. doi: 10.1016/j.cell.2004.07.023. [DOI] [PubMed] [Google Scholar]
- Bastida MF, Delgado MD, Wang B, Fallon JF, Fernandez-Teran M, Ros MA. Levels of Gli3 repressor correlate with Bmp4 expression and apoptosis during limb development. Dev Dyn. 2004;231(1):148–160. doi: 10.1002/dvdy.20121. [DOI] [PubMed] [Google Scholar]
- Albrecht AN, Kornak U, Böddrich A, Süring K, Robinson PN, Stiege AC, Lurz R, Stricker S, Wanker EE, Mundlos S. A molecular pathogenesis for transcription factor associated poly-alanine tract expansions. Hum Mol Genet. 2004;13:2351–2359. doi: 10.1093/hmg/ddh277. [DOI] [PubMed] [Google Scholar]
- D'Esposito M, Morelli F, Acampora D, Migliaccio E, Simeone A, Boncinelli E. EVX2, a human homeobox gene homologous to the even-skipped segmentation gene, is localized at the 5-prime end of HOX4 locus on chromosome 2. Genomics. 1991;10:43–50. doi: 10.1016/0888-7543(91)90482-T. [DOI] [PubMed] [Google Scholar]
- Davis AP, Witte DP, Hsieh-Li HM, Potter SS, Capecchi MR. Absence of radius and ulna in lacking Hoxa-11 and Hoxd-11. Nature. 1995;375:791–795. doi: 10.1038/375791a0. [DOI] [PubMed] [Google Scholar]
- Chen Y, Knezevic V, Ervin V, Hutson R, Ward Y, Mackem S. Direct interaction with Hoxd proteins reverses Gli3-repressor function to promote digit formation downstream of Shh. Development. 2004;131:2339–2347. doi: 10.1242/dev.01115. [DOI] [PubMed] [Google Scholar]
- Vortkamp A, Gessler M, Grzeschik K-H. GLI3 zinc-finger gene interrupted by translocations in Greig syndrome families. Nature. 1991;352:539–540. doi: 10.1038/352539a0. [DOI] [PubMed] [Google Scholar]
- Kang S, Graham JM Jr, Olney AH, Biesecker LG. GLI3 frameshift mutations cause autosomal dominant Pallister-Hall syndrome. Nat Genet. 1997;15:266–268. doi: 10.1038/ng0397-266. [DOI] [PubMed] [Google Scholar]
- Radhakrishna U, Wild A, Grzeschik KH, Antonarakis SE. Mutation in GLI3 in postaxial polydactyly type A. Nat Genet. 1997;17:269–271. doi: 10.1038/ng1197-269. [DOI] [PubMed] [Google Scholar]
- Radhakrishna U, Bornholdt D, Scott HS, Patel UC, Rossier C, Engel H, Bottani A, Chandal D, Blouin JL, Solanki JV, Grzeschik KH, Antonarakis SE. The phenotypic spectrum of GLI3 morphopathies includes autosomal dominant preaxial polydactyly type-IV and postaxial polydactyly type-A/B; no phenotype prediction from the position of GLI3 mutations. Am J Hum Genet. 1999;65:645–655. doi: 10.1086/302557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Jin CL, Liu LY, Zhao N, Zhang LJ, Ji SJ, Sun KL. Association and mutation analysis of GLI3 gene in idiopathic congenital talipes equinovarus. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2006;23:551–554. [PubMed] [Google Scholar]
- Wang LL, Jin CL, Liu LY, Zhang X, Ji SJ, Sun KL. Analysis of association between 5' HOXD gene and idiopathic congenital talipes equinovarus. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2005;22:653–656. [PubMed] [Google Scholar]
- Wang LL, Fu WN, Li-Ling J, Li ZG, Li LY, Sun KL. HOXD13 may play a role in idiopathic congenital clubfoot by regulating the expression of FHL1. Cytogenet Genome Res. 2008;121(3-4):189–95. doi: 10.1159/000138884. [DOI] [PubMed] [Google Scholar]
- Delgado Baeza E, Santos Alvarez I, Martos Rodriguez A. Retinoic acid induced clubfoot like deformity: pathoanatomy in rat embryos. J Pediatr Orthop B. 1999;8:12–18. doi: 10.1097/00009957-199901000-00003. [DOI] [PubMed] [Google Scholar]
- Li ZG, Ji H, Fu WN, Zhao YY, Jin CL, Ji SJ, Sun KL. Proteomic analysis of the ankle joint bone, ankle joint tissue and spinal cord of clubfoot-like deformity in rat fetuses. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2007;24(1):52–8. [PubMed] [Google Scholar]
- Ruppert JM, Kinzler KW, Wong AJ, Bigner SH, Kao F-T, Law ML, Seuanez HN, O'brien SJ, Vogelstein B. The GLI-Kruppel family of human genes. Molec Cell Biol. 1988;8:3104–3113. doi: 10.1128/mcb.8.8.3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalb VF Jr, Bernlohr RW. A new spectrophotometric assay for protein in cell extracts. Anal Biochem. 1977;82:362–371. doi: 10.1016/0003-2697(77)90173-7. [DOI] [PubMed] [Google Scholar]
- Hout AH van der, Ouweland AM van den, Luijt RB van der, Gille HJ, Bodmer D, Brüggenwirth H, Mulder IM, Vlies P van der, Elfferich P, Huisman MT, ten Berge AM, Kromosoeto J, Jansen RP, van Zon PH, Vriesman T, Arts N, Lange MB, Oosterwijk JC, Meijers-Heijboer H, Ausems MG, Hoogerbrugge N, Verhoef S, Halley DJ, Vos YJ, Hogervorst F, Ligtenberg M, Hofstra RM. A DGGE system for comprehensive mutation screening of BRCA1 and BRCA2: application in a Dutch cancer clinic setting. Hum Mutat. 2006;27:654–666. doi: 10.1002/humu.20340. [DOI] [PubMed] [Google Scholar]
- Li Y, Zhang H, Choi SC, Litingtung Y, Chiang C. Sonic hedgehog signaling regulates Gli3 processing, mesenchymal proliferation, and differentiation during mouse lung organogenesis. Developmental Biology. 2004;270:214–231. doi: 10.1016/j.ydbio.2004.03.009. [DOI] [PubMed] [Google Scholar]