Abstract
Background
Carboxylesterase is a multifunctional superfamily and ubiquitous in all living organisms, including animals, plants, insects, and microbes. It plays important roles in xenobiotic detoxification, and pheromone degradation, neurogenesis and regulating development. Previous studies mainly used Dipteran Drosophila and mosquitoes as model organisms to investigate the roles of the insect COEs in insecticide resistance. However, genome-wide characterization of COEs in phytophagous insects and comparative analysis remain to be performed.
Results
Based on the newly assembled genome sequence, 76 putative COEs were identified in Bombyx mori. Relative to other Dipteran and Hymenopteran insects, alpha-esterases were significantly expanded in the silkworm. Genomics analysis suggested that BmCOEs showed chromosome preferable distribution and 55% of which were tandem arranged. Sixty-one BmCOEs were transcribed based on cDNA/ESTs and microarray data. Generally, most of the COEs showed tissue specific expressions and expression level between male and female did not display obvious differences. Three main patterns could be classified, i.e. midgut-, head and integument-, and silk gland-specific expressions. Midgut is the first barrier of xenobiotics peroral toxicity, in which COEs may be involved in eliminating secondary metabolites of mulberry leaves and contaminants of insecticides in diet. For head and integument-class, most of the members were homologous to odorant-degrading enzyme (ODE) and antennal esterase. RT-PCR verified that the ODE-like esterases were also highly expressed in larvae antenna and maxilla, and thus they may play important roles in degradation of plant volatiles or other xenobiotics.
Conclusion
B. mori has the largest number of insect COE genes characterized to date. Comparative genomic analysis suggested that the gene expansion mainly occurred in silkworm alpha-esterases. Expression evidence indicated that the expanded genes were specifically expressed in midgut, integument and head, implying that these genes may have important roles in detoxifying secondary metabolites of mulberry leaves, contaminants in diet, and odorants. Our results provide some new insights into functions and evolutionary characteristics of COEs in phytophagous insects.
Background
Carboxylesterase (COE, EC 3.1.1.1) is a multigene family and occurs in animals, plants, insects, and microbes [1-4]. COEs are mainly attributed to B esterases, which were essentially irreversibly inhibited by organophosphate insecticides (OPs). Based on sequence similarity and substrate specificity, insect COE genes can be subdivided into eight subfamilies: α-esterase (ae), β-esterase (be), juvenile hormone esterase (jhe), gliotactins (gli), acetylcholinesterases (ace, AChE), neurotactins (nrt), neuroligins (nlg), and glutactin (glt) class [3]. α-esterases, β-esterases, acetylcholinesterases and juvenile hormone esterase account for the majority of the catalytically active COEs [3]. Gliotactins, neurotactins, neuroligins, and glutactin classes are generally considered to be noncatalytic but have a variety of functions essential to development and neurogenesis [5].
COEs have a broad range of functions; the key role is hydrolyzing esters of carboxylic acids. Carboxylesterases are also a class of the metabolic enzymes involved in insecticide resistance, which are implicated in the resistance of insects to OPs, carbamates, and pyrethroids through gene amplification, upregulation and coding sequence mutations [6]. Furthermore, COEs also play important roles in allelochemical metabolism and tolerance, although the roles were validated only at the biochemical level in a few cases [6]. In addition, carboxylesterases can serve as noncatalytic adhesive proteins involved in cell-to-cell interactions [5] and participate in other functions, such as pheromone degradation in moths [7] and hydrolysis of the neurotransmitter acetylcholine and juvenile hormone (JH) [8,9].
Studies on insect carboxylesterases have been mainly focused on mediating insecticide resistance [6,10]. Relatively, the mechanism of degrading plant allelochemicals is still unclear, and only some biochemical evidence confirmed that COEs were related to detoxification of the secondary metabolites of plants. Carboxylesterases can be induced by phenolic glycosides in Papilio Canadensis [11], and its activity was positively correlated with the survival rate of the gypsy moth, suggesting that esterase may be responsible for glycoside metabolism [12]. In the tobacco cutworm, Spodoptera litura, sublethal doses of the widely occurring plant glycoside rutin resulted in a significant increase in midgut carboxylesterase activity [13]. It was also found that COEs can be induced by indole alkaloid gramine in Sitobion avenae, and the increase of COE activity was positively correlated with dietary gramine concentrations, suggesting that COEs were involved in gramine detoxification [14]. In addition, quercetin, rutin and 2-tridaconone can also induce the activities of COEs in insects [15,16].
Herbivorous animals encounter a wide variety of secondary products in the plants on which they feed. They must therefore have developed mechanisms to metabolically inactivate some of the potentially toxic plant chemicals that they ingest. Silkworm is phytophagous insect, and specifically feeds on mulberry, which also encounters a mass of allelochemicals from its host plant. Because the silkworm grows well on mulberry leaves, the toxicities and defensive activities of these leaves against herbivorous insects have been overlooked. However, a recent study revealed that mulberry latex rich in sugar-mimic alkaloids was highly toxic to caterpillars [17]. Some alkaloids contained in mulberry leaves are potential inhibitors of mammalian digestive glycosidases but not inhibitors of silkworm midgut glycosidases, suggesting that the silkworm has enzymes specially adapted to enable it to feed on mulberry leaves [18]. In addition, β-fructofuranosidase was characterized in the silkworm genome, which has been no direct experimental evidence that this gene is encoded in the genome of animals [19]. Bmsuc1 played an important role in avoiding the toxic effects of 1,4-dideoxy-1,4-imino-D-arabinitol (D-AB1) and 1-deoxynojirimycin (DNJ) that are present in extremely high concentrations in the mulberry latex. In the "animal-plant warfare", silkworm has developed the mechanisms to metabolically inactivate those potentially toxic chemicals, such as detoxification enzyme carboxylesterase, cytochrome P450 monooxygenases (P450) and glutathione S-transferase (GST), etc. Thus, silkworm can be used as a model of the insect-plant interaction.
B. mori is an economically important insect and the Lepidoptera model for the study of pest control in agriculture. Recently, the fine genome map of the silkworm has been assembled. Totally, 87% of the scaffold sequences were anchored to all 28 chromosomes and 14,623 genes were predicted [20]. In addition, carboxylesterases are a functionally important superfamily, which play important roles in insecticide resistance, allelochemical tolerance, and developmental regulation. Previous studies on silkworm carboxylesterases mainly focused on isozyme polymorphism [21-23]. Herein, we present the identification and genomic analysis of silkworm COEs using the newly assembled 9× genome sequence. We have searched available EST data for each silkworm COE to confirm active transcription and examined the expression patterns using the genome-wide microarray of the silkworm [24]. Studying the expressions and evolutionary aspects of such large family of COEs will help us understand its functional versatilities.
Results and Discussion
Annotation and phylogeny of B. mori COEs
Drosophila melanogaster, Anopheles gambiae and Apis mellifera COEs were retrieved from GenBank and used for blast search against the new assembly of the silkworm genome to characterize the COE superfamily in B. mori. Through genomic analysis and gene prediction, 76 putative COE genes were identified in the silkworm genome (Additional file 1). This indicated that the B. mori genome contained more COE members compared with D. melanogaster (35), An. mellifera (24), and Ap. gambiae (51) (Table 1).
Table 1.
Comparison of the gene number for COEs in B. mori, D. melanogaster, Ap. Mellifera and An. gambiae
| Class/clades | B. mori | D. melanogaster | Ap. mellifera | An. gambiae |
|---|---|---|---|---|
| intracellular catalytic class | ||||
| A clade, α-esterase | 42 | 1 | 0 | 0 |
| B clade, α-esterase | 13 | 0 | 8 | 0 |
| C clade, α-esterase | 0 | 12 | 0 | 16 |
| secreted catalytic class | ||||
| D clade, JHE | 4 | 0 | 1 | 4 |
| E clade, integument esterase | 2 | 3 | 1 | 0 |
| F clade, JHE | 0 | 2 | 0 | 5 |
| G clade, β-esterase | 2 | 3 | 3 | 5 |
| H clade, uncharacterized | 1 | 1 | 1 | 1 |
| I clade, glutactin | 0 | 4 | 0 | 9 |
| neurodevelopmental class | ||||
| J clade, AChE | 2 | 1 | 2 | 2 |
| K clade, uncharacterized | 1 | 1 | 1 | 1 |
| L clade, gliotactin | 1 | 1 | 1 | 1 |
| M clade, neuroligin | 6 | 4 | 5 | 5 |
| N clade, neurotactin | 2 | 2 | 1 | 2 |
| total | 76 | 35 | 24 | 51 |
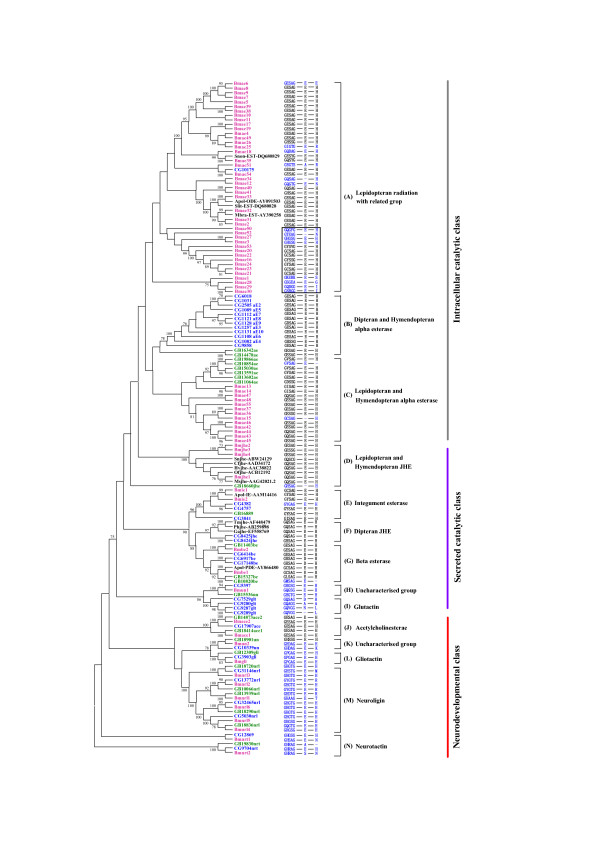
The neighbor-joining tree of COEs in B. mori, D. melanogaster, Ap. mellifera, and some related species was reconstructed (Figure 1). It can be seen from Figure 1 that the topology of phylogenetic tree was very similar to those obtained in previous studies [5]. Insect carboxylesterases can be divided into fourteen clades and three major classes (intracellular catalytic, secreted catalytic, and neurodevelopmental classes) based on the phylogenetic tree. The gene numbers of neurodevelopmental class and secreted catalytic class were alike in the four organisms, especially, the orthologous genes in neurodevelopmental class can be unambiguously defined (Figure 1, Table 1). Thus, this class of COEs might be involved in essential steps in conserved physiological pathways and subject to function constraints. While α-esterases were independently expanded in D. melanogaster, Ap. mellifera, and An. gambiae, the silkworm α-esterases experienced an obvious species-specific expansion: 55 α-esterase members were identified. This suggested that this class of COEs may play important roles in the adaptation of these insects to their specific biological niches rather than fulfilling general housekeeping functions.
Figure 1.
Neighbor-joining tree of amino acid sequences from B. mori, Ap. mellifera, and D. melanogaster, and some known COEs in other Lepidopteran insects. Bootstrap values > 70% are shown. The catalytic triad was predicted by blastp searching NCBI conserved domain database (CDD) http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml using COE amino acid sequences as queries. Thus, the catalytic residues (S200, E327 and H440) and GXSXG consensus sequence around the active site serine were presented. Short dash: absence of the catalytic residue. Red: B. mori; Green: Ap. mellifera; Blue: D. melanogaster; Black: other Lepidoptera. Snon (Sesamia nonagrioides), Apol (Antheraea polyphemus), Slit (Spodoptera littoralis), Mbra (Mamestra brassicae), Sn (Sesamia nonagrioides), Cf (Choristoneura fumiferana), Hv (Heliothis virescens), Of (Omphisa fuscidentalis), Ms (Manduca sexta), Tm (Tenebrio molitor), Ph (Psacothea hilaris), Ga (Gryllus assimilis).
Expansion of intracellular catalytic class in the silkworm
The intracellular catalytic class of COEs belongs to α-esterases, which function to detoxify xenobiotics and some members are related to organophosphorus insecticides (OPs) resistance in insects [5,25,26]. This class (clades A-C) includes 55, 16, 13, and 8 esterases in B. mori, An. gambiae, D. melanogaster and Ap. mellifera, respectively (Figure 1, Table 1). Intracellular catalytic esterases in the silkworm were in clades A and C, while the corresponding esterases in D. melanogaster and Ap. mellifera were mainly located in the clades B and C, respectively. All the other sequences in clade A came from Lepidopteran insects except for the D. melanogaster CG10175 that shared 37.7% amino acid identity with Bmae54. Furthermore, most of the silkworm α-esterases were located in this clade.
Intracellular catalytic carboxylesterases have some common characteristics, such as conserved catalytic triad S200, E327 and H440, the numbering of which is that of torpedo californica AChE [27]. The catalytic triad of COEs was predicted by blastp searching NCBI conserved domain database (CDD) using COE amino acid sequences as queries (Figure 1). The results indicated that most of the intracellular catalytic carboxylesterases in the silkworm had GESAG consensus sequences similar to Drosophila. However, 15 of 55 silkworm α-esterases changed one or more residues of the catalytic triad. Most of the variants of the catalytic triad in the silkworm α-esterases were phylogenetically related to the variants of GESAG, which formed a cluster boxed on the phylogenetic tree. Similarity analysis of amino acid sequences indicated that α-esterases with substitutions in catalytic triad shared < 30% identity with other α-esterases, and most of them were only about 20%. These results indicated that those rapidly evolved α-esterases in the silkworm might lose their hydrolyzing functions.
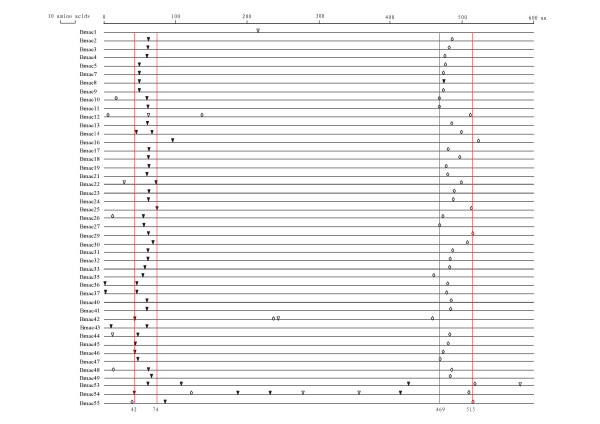
The intron number and location of silkworm α-esterase were analyzed. In total, 111 introns were found in 45 α-esterases with putative complete coding sequence (Figure 2). For intron number, most of the α-esterases (30 out of 45) contained only two introns. While Bmae1 had only one, others contained three or more introns, especially, Bmae54 contained eight introns. The intron locations were relatively conserved for the α-esterases. The first conserved one was located between the 42th and 74th amino acids whereas the second one lay between the 469th and 515th amino acids. In addition, almost all of the first and second introns were phase 0 and phase 2, respectively. Thus, the gene structure of the silkworm α-esterases was relatively conserved.
Figure 2.
Location of introns of the silkworm α-esterases. Inverted black triangle: phase 0; Inverted white triangle: phase 1; White diamond: phase 2. Only those COE genes with putative complete coding sequences are shown. The vertical lines were the boundary of conservative distribution of introns.
Lepidopteran odorant-degrading enzyme (ODE) and antennal esterase play an important role in inactivating pheromone because degradation of odorant molecules is a necessity to avoid the continuous stimulation of the receptors [28,29]. Those known ODEs and antennal esterases of Lepidopteran insects were also included in the phylogenetic analysis. The results indicated that some of the odorant-degrading esterase were clustered with silkworm α-esterases in clade A. Silkworm Bmae33, Antheraea polyphemus ODE (Apol-ODE) and Spodoptera littoralis ODE (Slit-EST) might be orthologous genes, and Bmae33 shared 73.1% of amino acid identity with Apol-ODE and 64.6% with Slit-EST. Bmae35 was phylogenetically closely related to Sesamia nonagrioides ODE (Snon-EST); they shared 48.6% identity. In addition, Bmae32 was homologous with Mamestra brassicae antennal-specific esterase (Mbra-EST); they had 58.8% identity. Thus, existence of putative ODEs in Lepidoptera was one of the reasons that α-esterases were obviously expanded in the silkworm compared with Dipteran and Hymendopteran insects.
Secreted catalytic class
Juvenile hormone esterase (JHE), integument esterase, β-esterase, and glutactin belong to the secreted catalytic class. In this class, uncharacterized esterases were also identified. JHEs play important roles in development, metamorphosis, diapause and reproduction in insects, which can hydrolyze and regulate the titre of juvenile hormone (JH) [8]. Four putative JHEs were identified in the silkworm genome (Figure 1). Silkworm JHEs showed a moderate expansion compared with D. melanogaster (2), Ap. mellifera (1), An. gambiae (5) [5,30], and Aedes aegypti (10) [31]. The phylogenetic tree indicated that Lepidopteran and Ap. mellifera JHEs were clustered together, while Dipteran and Coleopteran JHEs formed another cluster. Thus, this provides support for the hypothesis that there are two separate origins of JHE in the insect esterases [30].
BmJHE1 was phylogenetically related to CfJHE, HvJHE, OfJHE and MsJHE, and shared higher identities with them (49.6% - 60.8%). Thus, they might be orthologous genes (Figure 1). However, BmJHE1 and the other three BmJHEs showed lower identities (42.4% - 44%) than those among BmJHE2, BmJHE3 and BmJHE4 (72.0% - 74.6%). So, we predicted that Bmjhe2, jhe3 and jhe4 were recently duplicated by Bmjhe1. JHEs contain some specific characteristics, such as a GQSAG core catalytic motif required for JH-specific esterase activity [5], a particular amphipathic helix as for a characteristic of Lepidopteran JHEs [32]. The analysis of catalytic triad indicated that BmJHE1 contained the specific GQSAG motif while BmJHE2 and BmJHE4 had the GESAG, and BmJHE3 had GESSG. Only BmJHE1 and BmJHE4 were identified to have the three Args (R 174, 181, 185) along one face of an amphipathic helix. In addition, BmJHE1 has been verified to have JH-specific esterase activity in vivo [33,34]. Based on sequence characteristics and known functional data, we speculated that BmJHE1 was the only one physiologically functional JHE in the silkworm, and that those new duplicated JHEs might have evolved other functions.
In insects, most of pheromone molecules are strongly hydrophobic and therefore tend to adhere to waxy surfaces. Both male and female may need to remove the pheromones from their integument so that they can better identify and control the signal, respectively [35]. During this signaling, integument esterase plays important role in inactivation of pheromones [29,35]. In total, two putative integument esterases were identified in the silkworm (Figure 1). One and three integument esterases were identified in Ap. mellifera and D. melanogaster, respectively, whereas none was found in An. gambiae (Table 1). Previous study found that Apol-IE in Ant. polyphemus was distributed in adult antennae and legs, suggesting that it may have the function of degrading pheromone [29]. Bmie1 shared 62.6% identity with Apol-IE at amino acid level; they were phylogenetically related and may be orthologous genes. In addition, identities among silkworm and Drosophila integument esterases were about 44%. Thus, integument esterases among insect species showed higher similarities and played similar roles in degrading pheromone or detoxifying xenobiotics entered into integument.
Although the number of β-esterase genes is not big in insects, these enzymes have multiple functions, including metabolic resistance to OPs and carbamates in Hemipteran insects [5], reproductive function in Diptera [5], and pheromone signaling in Lepidoptera [36]. In total, two β-esterases were found in the silkworm, three in Ap. mellifera and D. melanogaster and five in An. gambiae. Bmbe2 shared 43.6% and 39.1% amino acid identities with GB11403 and CG6414, respectively, and they were clustered together on the phylogenetic tree and may be 1 : 1 : 1 orthologs (Figure 1). Bmbe1 is the ortholog of Apol-PDE (pheromone-degrading enzyme) in Ant. polyphemus, which plays important role in validated rapid inactivation of sex pheromone [36]. Thus, Bmbe1 may have similar function to Apol-PDE, involved in pheromone signaling.
Glutactin is a novel Drosophila basement membrane related glycoprotein located in the envelope of the developing nervous system; it may play a role in intercellular ordering and adhesion [37-39]. Four and nine glutactins were found in D. melanogaster and An. gambiae, respectively, while no glutactin gene was found in the silkworm and Ap. mellifera. However, uncharacterized clade (H) was phylogenetically related to Drosophila glutactin clade. Bmun1 shared 36.2% and 32.3% amino acid identities with CG5397 and GB15536, respectively, and the three genes may be 1 : 1 : 1 orthologs. Whether Bmun1 had the function of glutactin or how this function was substituted in the silkworm remains to be determined.
Conserved neurodevelopmental class
In neurodevelopmental class, orthologs among insects can be easily identified (Figure 1). Thus, these genes were generated by duplication events occurred before insect radiation and might have experienced purifying selection process after speciation. Acetylcholinesterases (AChEs) were the only enzymes that perform catalytic function in the neurodevelopmental class. Furthermore, they may be the important target of OPs [30]. In Drosophilidae and Muscidae, only one ace gene was identified, while two ace genes were found in other Dipteran, Hymenoptera, and Lepidopteran insects. On the phylogenetic tree, ace clade can be obviously divided into ace1 and ace2 subclades (Figure 1). In the silkworm, BmAChE1 and BmAChE2 shared only 31.5% amino acid identity and 54.5% and 32.6% amino acid identities with AmAChE1, 31.9% and 60.0% with AmAChE2, 29.1% and 50.4% with DmAChE2, respectively. However, either BmAChE1 or BmAChE2 showed higher conservation with other Lepidopteran insect AChE1 or AChE2 (Additional file 2). BmAChE1 shared 72.9% - 98.6% amino acid identities with orthologous gene from Bombyx mandarina, Helicoverpa assulta, Helicoverpa armigera, Cydia pomonella, and Plutella xylostella, and BmAChE2 showed the 91.2% - 99.2% identities with the corresponding orthologs from species above.
The major function of AChE is hydrolysis of the neurotransmitter acetylcholine bounded at cholinergic synapses in the central nervous system of insects [40], conferring target site resistance to OP and carbamat insecticides [5]. In silkworm, both BmAChE1 and BmAChE2 contained the catalytic triad and disulfide bridges (C88-C115, C275-C286 and C423-C542 in Torpedo AChE [40], Additional file 2). In addition, an inhibition assay indicated that both of them can be inhibited by eserine and paraoxon [41]. However, the alignment of known Lepidopteran AChEs indicated that AChE1 had protrudent C-terminal compared with AChE2, and AChE2 (position at 150-230 aa) had an insertion of 18 incontinuous amino acids like the hydrophilic insertion in DipteranAChE, which had 31 residues [42]. Thus, BmAChE1 and BmAChE2 showed obvious differentiation in sequence. Previous studies indicated that the AChE2 in those insects with two ace gene system had cell-to-cell communication/adhesive properties [5]. Thus, we speculated that BmAChE2 could substitute the function of glutactin due to the loss of glutactin gene in the silkworm and that BmAChE1 had the function of hydrolyzing acetylcholine.
Generally, gliotactin, neuroligin, and neurotactin are noncatalytic adhesive proteins involved in cell-to-cell interactions [5]. Single gliotactin gene was identified in the B. mori genome as D. melanogaster and Ap. mellifera (Figure 1), and these orthologous genes shared about 52% amino acid identities. Like D. melanogaster, two putative neurotactin genes were found in the silkworm. CG9704, GB19830 and Bmnrt2 were orthologous genes, which showed about 40% amino acid identities. Compared with gliotactin and neurotactin, neuroligin genes were obviously duplicated in the B. mori, D. melanogaster and Ap. mellifera, corresponding to 6, 4 and 5 duplicates, respectively. The neuroligin clade (M) contained three pairs of 1 : 1 : 1 orthologs. This indicated that most of the neuroligins were duplicated before radiation of the three species. In addition, except for the known families, uncharacterized group was also found in clade (M). Bmun2, CG10339 and GB18901 were 1 : 1 : 1 orthologous genes, and Bmun2 shared 40.1% and 40.9% identities with CG10339 and GB18901, respectively.
Genomic distribution of BmCOEs
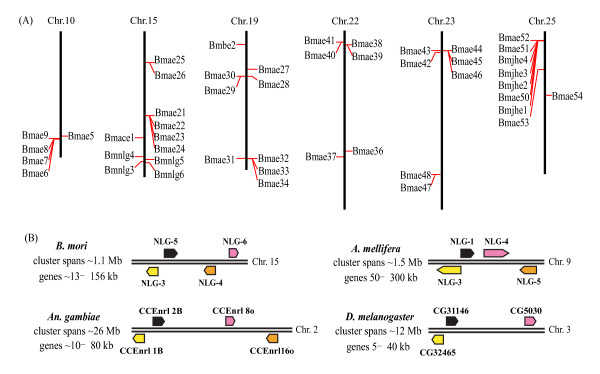
In the fine genome map of the silkworm, most of the silkworm COEs could be mapped to chromosomes. Totally, 73 out of 76 silkworm COEs were distributed on 19 chromosomes and there are 13 BmCOE clusters on different chromosomes (Additional file 1 and 3). About 55% of BmCOE genes were tandem arranged in the silkworm genome. Moreover, BmCOE genes were not evenly distributed on chromosomes and about 62% COEs were massively located on six chromosomes (Figure 3a).
Figure 3.
The cluster organization of BmCOEs and microsynteny of neuroligins. (A) The cluster organization of BmCOEs in the silkworm genome. Only those genes involved in clusters of five or more COE genes on the same chromosomes are shown. (B) microsynteny of neuroligins among B. mori, D. melanogaster, An. gambiae, and Ap. Mellifera. The infromation for D. melanogaster, An. gambiae, and Ap. Mellifera came from [25]. The arrows represent gene localization and transcriptional orientation.
Generally, tandem arranged genes showed higher similarities one another and could be classified into a common family. For instance, Bmae6 - 9 were located on chromosome 10 and shared 89.1% - 95.7% amino acid identities. Although the Bmae5 is away from the gene cluster on the same chromosome, it showed high similarities (about 82% identity) to the genes in the cluster (Figure 3a). Most of the other tandem arranged COEs also showed 40% - 50% amino acid identities among members in a cluster. The tandem arranged COEs might be created by local duplications. Tandem arranged COEs tend to form a family, however, some are not the case. For example, Bmnlg1 and Bmae2 were tandem arranged but shared only 17% identity identities, and located in a mixed gene cluster with three alpha-esterases (Bmae50 - 52) and four JHEs (Bmjhe1 - 4). Thus, these tandem arranged COEs might be created by other mechanisms, not by local duplication or they are very old duplicates.
Neuroligins showed remarkable conservation of microsynteny among the Ap. mellifera, D. melanogaster, and An. gambiae genomes [25]. In the silkworm genome, six neuroligins were found; four of six were located on the chromosome 15, which spanned about 1.1 Mb (Figure 3b). The four silkworm neuroligins showed similar gene arrangement to those in Ap. mellifera, D. melanogaster, and An. gambiae. Thus, such microsynteny of neuroligins was present in these four organisms. The only difference is that the location of the NLG4 and NLG6 in Ap. mellifera and An. gambia was changed in B. mori.
Expression profiles of BmCOEs
ESTs analysis
In order to detect the expression of the B. mori COEs, we searched the silkworm dbEST database downloaded from GenBank using the putative coding sequences as queries. The results indicated that 47 COEs matched at least one EST and most of the transcriptionally active genes were specifically expressed in tissues.
Microarray-based gene expression profiles in multiple tissues
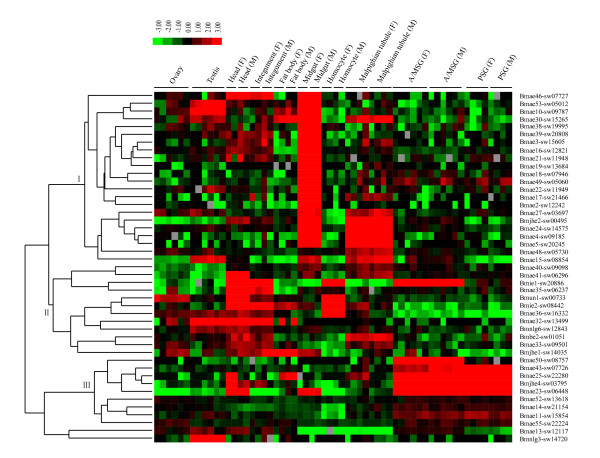
Based on the silkworm genome-wide microarray dataset http://silkworm.swu.edu.cn/microarray[24], expressions of BmCOEs in multiple tissues on the day three of the fifth instar were surveyed. It was found that 63 COE genes contained the oligonucleotide probes, whereas only 45 COEs showed expression signals, with signal value >400 at least in one tissue. The signal values of those expressed COEs were clustered to analyze the expression profiles (Figure 4). The results indicated that almost all of the COEs showed tissue specificity, including midgut-, head and integument-, and silk gland-specific expressions.
Figure 4.
Tissue expression patterns of the silkworm COEs in different larvae tissues based on microarray data. Hierarchical clustering with the average linkage method was performed using Cluster software http://genome-www.stanford.edu/clustering/. F: female; M: male. A/MSG: anterior/median silk gland; PSG: posterior silk gland.
The catalytic triad is the foundation of carboxylesterases that perform catalytic function. For the D. melanogaster and Ap. mellifera α-esterases, only GB10854 gene lost the His of the catalytic triad. Fifteen α-esterases were found in the silkworm but they mutated in the catalytic triad, which may result in loss of the hydrolase activity. Based on the ESTs/microarray datasets, most of these α-esterases (11/15) had expression evidence (Additional file 4). Thus, these α-esterases may acquire new functions. Previous studies indicated that esterase gene amplification is one of the important mechanisms resistant to organophosphorus and carbamate insecticides [43]. The amplified esterases can produce broad spectrum insecticide resistance through rapid-binding (sequestration) mechanism. So we supposed that these amplified α-esterases may play roles in sequestering secondary metabolites of mulberry leaves or insecticide contaminants in diet. In addition, due to mutation of the catalytic triad and no expression evidence, Bmae6 and Bmae28 may be pseudogenes.
Midgut-specific expression genes
Twenty-two BmCOE genes were found to be predominantly expressed in the midgut and their expression levels did not significantly differ between male and female (Figure 4, group I). Except for Bmjhe2, all the midgut-specific genes belonged to α-esterases. It should be noted that Bmae3, ae18, ae27, and ae30 mutated in the catalytic triad were also included in this class. Thus, most of the midgut-specific expression genes should have hydrolyzing function. The potential hazard faced in sericulture is the occasional contamination of the mulberry leaves by air-borne insecticides that have been used in neighboring fields. Midgut is the first barrier of xenobiotics peroral toxicity, in which COEs can eliminate insecticides, such as OPs, carbamate insecticides, on mulberry leaves.
Silkworm is the phytophagous insect, and specifically feeds on mulberry, which also encounters a mass of allelochemicals from its host plant. A recent study revealed that mulberry latex rich in sugar-mimic alkaloids was highly toxic to caterpillars [17]. That silkworm is less affected by sugar-mimic alkaloid is due to insensitivity of B. mori glycosidases and β-fructofuranosidase [18,19]. So, during the evolution of the silkworm adapted to host plant mulberry, many mechanisms have been selected. COEs can be induced by multiple allelochemicals, such as phenolic glycosides, quercetin, rutin. Indeed, mulberry leaves contain these secondary metabolites [44]. The fact that a large numbers of BmCOEs were predominantly expressed in the midgut may suggest that these genes might also play important roles in tolerating the relevant allelochemicals. Like the COEs, UDP-glucosyltransferases were also expanded in the silkworm relative to Dipteran and Hymenopteran insects [45]. Thus, these expanded superfamilies may represent the characteristic of phytophagous insect.
Some of midgut carboxylesterases were expressed not only in the midgut, but also in the other tissues such as malpighian tubule, integument, head, fat body and testis. Especially, one cluster in group (I) including seven COE genes was found to be predominantly expressed in the malpighian tubule. The function of insect malpighian tubule is similar to mammalian kidney, which plays important roles in defending against insecticides such as DDT and metabolism of plant secondary and other molecules [46]. Thus, these COEs predominantly expressed in the malpighian tubule may be important detoxification enzymes that eliminate insecticides and allelochemicals. Simultaneously, these midgut carboxylesterases expressed in other tissues had important roles in protecting silkworms from xenobiotic damages.
Head and integument-specific expression genes
The head and integument specific group (II) included α-esterase, jhe, β-esterase, integument esterase, and neuroligin genes. Except for the common expression characteristic, these genes were also expressed in other tissues. For example, Bmae32 and Bmnlg6 showed male-predominant expression in the reproductive system, whereas Bmun1 was a female-predominant expression gene. Bmie1 and Bmie2 were also expressed in homocyte, and the former showed higher expression level in the anterior/median silk gland (A/MSG) and the latter was also expressed in fat body, midgut, and malpighian tubule.
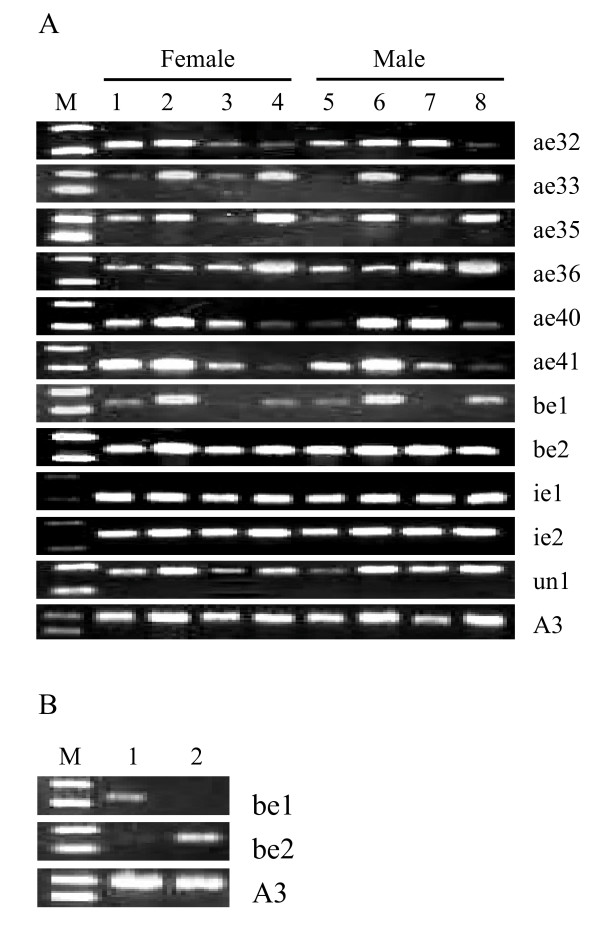
ODEs/PDEs, antennal and integument esterases play important roles in degrading odorants/pheromones, such as volatile acetate compounds; these enzymes belong to α-esterase, β-esterase and integument esterase [7,29]. In addition, for the herbivorous insects, odorant/pheromone-degrading esterase may have a role in the degradation of plant volatiles with an ester functional group [47]. Previous studies showed that Apol-ODE and Apol-PDE could be detected in adult male antennae, but not in female antennae and other control tissues [36]. Apol-IE, Mbra-EST, Slit-EST and Snon-EST were expressed in male and female antennae whereas Slit-EST and Snon-EST showed higher expression in legs [28,29,47]. For the silkworm, only larvae fetch mulberry leaves and also encounter a mass of volatile allelochemicals, such as hexyenyl acetate, 3-hexenyl acetate and 2-hexenyl acetate [48]. Based on the microarray data, it was found that BmCOEs phylogenetically related to odorant/pheromone-degrading esterases were specifically expressed in the head and integument of the silkworm larvae (Figure 4, group II). Thus, we employed RT-PCR technique to analyze the expression patterns in larva antenna and maxilla, on which important olfactory sensilla are distributed [48], and adult antennae and leg to presume the function of head and integument-specific expression esterases, excluding Bmnlg6 and Bmjhe1. The results indicated that most of the head and integument-specific expression esterases were expressed in larva antennae and maxilla (Figure 5a). Except for the Bmae35, ae40 and un1, others did not show obvious expression differences between female and male. The antenna and maxilla of the silkworm larvae are important olfactory organs. Thus, we predicted that these esterases would play important roles in degrading the volatile acetate allelochemicals and adaptive evolution of silkworm with mulberry leaves. In addition, The pheromone of B. mori is a blend of alcohol (bombykol) and aldehyde (bombykal) [49]. Antennal specific aldehyde oxidase (AOX) can degrade bombykal [7]. Bombykol was inactivated firstly by conversion, including the oxidation of the pheromones by oxidases and/or dehydrogenases to the corresponding fatty acids, and secondly by degradation [50]. Thus, odorant/pheromone-degrading like esterases in the silkworm may indirectly play role in degrading the pheromones. Furthermore, Bmae33, ae35 and Bmbe1 showed lower or no expression in adult antennae, suggesting that they may have different functions from their corresponding orthologous genes. In addition, it was observed that some head and integument-specific esterases were also expressed in adult antennae and legs. So, the functions of these esterases expressed both in the silkworm larvae and adult moths need to be further determined.
Figure 5.
Expression patterns of the silkworm COEs validated by RT-PCR. (a) Expression of the silkworm COEs in special tissues. M: DL2000 marker, 1 and 5: larvae antennae, 2 and 6: larvae maxilla, 3 and 7: adult antennae, 4 and 8: adult leg. (b) Expression of β-esterases in adult accessory gland. M: DL2000 marker, 1: male accessory gland, 2: female accessory gland.
Silk gland-specific expression genes
In the silk gland specific group (III), nine genes were found, including α-esterase and JHE (Figure 4). Bmae11, ae43, and ae50 were only expressed in the silk gland, especially Bmae50 only in the anterior/median silk gland. However, Bmjhe4 was also expressed in the head of females, Bmae25 in the head of females and fat body in males, and Bmae23 in the head and midgut in females/males, and fat body in males. Silkworm is an economically important insect, which can efficiently synthesize silk proteins. Silk gland in the silkworm is an organ specialized for the synthesis and secretion of silk proteins. Due to the fact that COEs have the activities of hydrolase and ethyl ester synthase [51], we speculated that silk-gland specifically expressed COEs may participate in synthesis of silk proteins and detoxify the xenobiotics entered into silk gland.
Other specific expression genes
In adult, the male accessory glands (MAGs) of many insect species can produce and secrete a number of reproductive proteins that are expressed exclusively or abundantly in the MAGs [52]. The previous studies indicated that EST-6 (CG6917) in Drosophila is expressed in the male genitalia and transferred to the female during mating, influencing egg-laying behavior and possibly receptivity to remating [53]. In An. gambiae, Agbe1d and be4d homologous to Drosophila EST-6 were also specifically expressed in the MAGs [52]. The expression patterns of two silkworm β-esterases in adult accessory glands were investigated. The results indicated that Bmbe1 and Bmbe2 were specifically expressed in the female accessory glands and MAGs, respectively (Figure 5b). Thus, Bmbe2 may have similar function to Drosophila EST-6. In addition, silkworm Bmae10, ae15, ae30, ae32, and ae53 and Bmnlg3 and nlg6 were expressed in male testis, but not in ovary. We supposed that these differential expression genes in reproductive system may play important roles in spermatogenesis or detoxification to avoid damage of xenobiotics.
Bmjhe1 was expressed in the anterior/middle/posterior silk glands in the 4th larval instar and on day 10 in the 5th larval instar just before pupation [33]. In addition, a previous study detected the expression of Bmjhe1 only at day 0 in the 5th instar in the fat body of seven tissues [54]. In this study, we observed that Bmjhe1 was expressed in midgut, ovary, fat body, and malpighian tubule, and the highest expression level in fat body at day 3 in the 5th instar. Thus, expression of carboxylesterase genes showed not only tissue but also developmental stage specificities.
Conclusion
A comprehensive search was conducted for potential COE genes in the silkworm genome. B. mori contains 76 COE genes, the largest number of COE genes among insects investigated. Relative to Dipteran and Hymenopteran insects, silkworm has experienced a significant expansion for α-esterases. The expanded α-esterases were predominantly expressed in midgut, head and integument, and silkgland, respectively, suggesting that they may participate in allelochemical tolerance and synthesis of silk proteins. Generally, α-esterases contain the conserved catalytic triad and showed catalytic function. However, 15 of 55 silkworm α-esterases mutated at the essential catalytic residue sites, implying that they may acquire some new functions. On the basis of tissue microarray, the putative odorant/pheromone-degrading esterases and related genes predominantly expressed in head and integument of the silkworm larvae were detected. RT-PCR verified that these genes were also expressed in the larvae antenna and maxilla, suggesting that they play important roles in detoxifying plant volatiles. In addition, Bmbe2 were specifically expressed in the adult MAGs, which may have a similar function of Drosophila EST-6, influencing egg-laying behavior and possibly receptivity to remating. In sum, our results provide some new insight into annotation and evolutionary characteristics of the silkworm COEs.
Methods
Identification of the B. mori COE genes
COEs amino acid sequences of D. melanogaster, An. gambiae, and Ap. mellifera were downloaded from the GenBank http://www.ncbi.nlm.nih.gov/. We searched the silkworm genome for candidate COEs genes using the tblastn program with the silkworm 9×genome database [19]. Genomic sequences that show even weak sequence similarity to any query sequence and its flanking regions (1 kb or more long) were extracted. Putative COE genes within the extracted sequences were predicted using BGF software [55] and Fgenesh+ http://www.softberry.com/.
Phylogenetic analysis
Multiple sequence alignments of COEs amino acids were aligned using Clustal X [56]. Positions that have a high percentage of gaps (>70%) were manually trimmed. Phylogenetic tree was reconstructed using the neighbor-joining method in which distance was estimated by JTT amino acid matrix implemented in MEGA 4.0 program [57]. The pairwise deletion option was used in the NJ tree reconstruction and the accuracy of the tree topology was assessed by bootstrap analysis with 100 resampling replicates.
Expression analysis with ESTs and microarray data
The putative coding sequences of BmCOEs were used as queries to perform Blastn searches against the silkworm EST database downloaded from GenBank. A 95% or greater identity and minimum cut-off E-value (≤e-20) were employed to discriminate between duplicate genes.
A genome-wide 69-mer oligonucleotides microarray with 22,987 probes has previously been customized for the silkworm [24]. Sixty-three of the 76 BmCOEs identified in this study were found to have probes on the microarray. The expression patterns of these genes have been surveyed for the 9 representative sample types of Chinese silkworm strains (Dazao) on day 3 of the fifth instar, including silk gland, testis, ovary, fat body, midgut, integument, hemocyte and malpighian tubule, and head. The detailed experimental process, quality control, consistency in replication and data analysis for these experiments have been described in previous report [24].
Gene expression by reverse transcriptase-polymerase chain reaction (RT-PCR)
Each tissue was dissected and stored in liquid nitrogen before pulverizing. Total RNA was isolated using Trizol Reagent (Invitrogen) according to the manufacturer's instructions. The contaminating genomic DNA was digested with Rnase-free Dnase I (Promega) for 15 min at 37°C. The RNA samples were eluted with Rnase-free water and stored at -80°C. The first strand of cDNA was synthesized using M-MLV reverse transcriptase following the manufacturer's instructions (Promega, USA).
RT-PCR primers were designed on the basis of the coding sequences of the silkworm COEs (Additional file 5). The silkworm cytoplasmic actin (A3) gene (accession No. U49854) was used as an internal control. PCR amplification reactions were performed in 25 μl volumes containing normalized cDNA, 0.2 mM of each primer, 2 mM MgCl2, 0.25 mM dNTP, 1× buffer and 2.5 units of Taq DNA polymerase (Promega). The PCR cycling program had an initial denaturation step of 95°C for 4 min, followed by 25 cycles of 94°C for 30 s, 30 s annealing (temperatures listed in Additional file 5), 40 s extension (72°C), and a final extension at 72°C for 10 min. The amplification products were analyzed on 1.2% agarose gels.
Authors' contributions
QYY and CL made the study design. QYY did the data collection and analysis, and drafted the manuscript. WLL did partial data analysis. ZZ revised the manuscript. ZZ and ZHX supervised the study. All authors read and approved the final manuscript.
Supplementary Material
Summary of the silkworm COE genes. NP indicates genes with no automatic prediction in silkworm. (N): missing N-terminal region; (C): missing C-terminal region. Chr.: chromosome. UN represents unknown chromosome locations.
Sequence alignment of Lepidopteran AChEs. Three intrachain disulfide bridges are drawn between conserved Cys. The asterisks represent the catalytic triad (S200, E327 and H440).
Chromosome distribution of all silkworm COEs. Arrows showed the transcriptional orientation.
The phylogeny and expression patterns of BmCOEs. Bootstrap values > 70% are shown. The solid black circles corresponding to cDNA, EST, tissue microarray and RT-PCR column indicate that these genes have expression evidence. The genes with grey font are that they had no probes in microarray dataset. The underlined α-esterases mean that they had substitutions in the catalytic triad.
Primers used in RT-PCR study. Primers used in RT-PCR study.
Contributor Information
Quan-You Yu, Email: quanyouyu@126.com.
Cheng Lu, Email: lucheng@swu.edu.cn.
Wen-Le Li, Email: lilideairen@live.cn.
Zhong-Huai Xiang, Email: xbxzh@swu.edu.cn.
Ze Zhang, Email: zezhang@swu.edu.cn.
Acknowledgements
This study was supported by the Hi-Tech Research and Development (863) Program of China (2006AA10A117), a grant from National Science Foundation of China (No. 30671587), and the Programme of Introducing Talents of Discipline to Universities (B07045).
References
- Satoh T, Hosokawa M. The mammalian carboxylesterases: from molecules to functions. Annu Rev Pharmacol Toxicol. 1998;38:257–288. doi: 10.1146/annurev.pharmtox.38.1.257. [DOI] [PubMed] [Google Scholar]
- Marshall SD, Putterill JJ, Plummer KM, Newcomb RD. The carboxylesterase gene family from Arabidopsis thaliana. J Mol Evol. 2003;57:487–500. doi: 10.1007/s00239-003-2492-8. [DOI] [PubMed] [Google Scholar]
- Ranson H, Claudianos C, Ortelli F, Abgrall C, Hemingway J, Sharakhova MV, Unger MF, Collins FH, Feyereisen R. Evolution of multigene families associated with insecticide resistance. Science. 2002;298:179–181. doi: 10.1126/science.1076781. [DOI] [PubMed] [Google Scholar]
- Bornscheuer UT. Microbial carboxyl esterases: classification, properties and application in biocatalysis. FEMS Microbiol Rev. 2002;26:73–81. doi: 10.1111/j.1574-6976.2002.tb00599.x. [DOI] [PubMed] [Google Scholar]
- Oakeshott JG, Claudianos C, Campbell PM, Newcomb RD, Russell RJ. In: Comprehensive molecular insect science. Gilbert LI, Iatrou K, Gill SS, editor. Vol. 5. London: Elsevier; 2005. Biochemical genetics and genomics of insect esterases; pp. 309–361. full_text. [Google Scholar]
- Li X, Schuler MA, Berenbaum MR. Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annu Rev Entomol. 2007;52:231–253. doi: 10.1146/annurev.ento.51.110104.151104. [DOI] [PubMed] [Google Scholar]
- Vogt RG. In: Comprehensive insect physiology, biochemistry, pharmacology and molecular biology. Gilbert LI, Iatrou K, Gill SS, editor. Vol. 3. London: Elsevier; 2005. Molecular basis of pheromone detection in insects; pp. 753–804. [Google Scholar]
- Riddiford LM, Hiruma K, Zhou X, Nelson CA. Insights into the molecular basis of the hormonal control of molting and metamorphosis from Manduca sexta and Drosophila melanogaster. Insect Biochem Mol Biol. 2003;33:1327–1338. doi: 10.1016/j.ibmb.2003.06.001. [DOI] [PubMed] [Google Scholar]
- Taylor P, Radic Z. The cholinesterases: From genes to proteins. Annu Rev Pharmacol Toxicol. 1994;34:281–320. doi: 10.1146/annurev.pa.34.040194.001433. [DOI] [PubMed] [Google Scholar]
- Hemingway J, Karunaratne SH. Mosquito carboxylesterases: a review of the molecular biology and biochemistry of a major insecticide resistance mechanism. Med Vet Entomol. 1998;12:1–12. doi: 10.1046/j.1365-2915.1998.00082.x. [DOI] [PubMed] [Google Scholar]
- Lindroth RL. Host plant alteration of detoxification activity in Papilio glaucus glaucus. Entomol Exp Appl. 1989;50:29–36. doi: 10.1007/BF00190125. [DOI] [Google Scholar]
- Lindroth RL, Weisbrod AV. Genetic variation in response of the gypsy moth to aspen phenolic glycosides. Biochem Syst Ecol. 1991;19:97–103. doi: 10.1016/0305-1978(91)90031-T. [DOI] [Google Scholar]
- Ghumare SS, Mukherjee SN, Sharma RN. Effects of rutin on the neonate sensitivity, dietary utilization and midgut carboxylesterase activity of Spodoptera litura (Fabricius) (Lepidoptera: Noctuidae) Proc Indian Acad Sci Anim Sci. 1989;98:399–404. [Google Scholar]
- Cai QN, Han Y, Cao YZ, Hu Y, Zhao X, Bi JL. Detoxification of gramine by the cereal aphid Sitobion avenae. J Chem Ecol. 2009;35:320–5. doi: 10.1007/s10886-009-9603-y. [DOI] [PubMed] [Google Scholar]
- Gao XW, Zhao Y, Wang X, Dong XL, Zheng BZ. Induction of carboxylesterase in Helicoverpa armigera by insecticides and plast allelochemicals. Acta Entamol Sinica. 1998;41(Suppl):5–11. In Chinese with English abstract. [Google Scholar]
- Mu SF, Pei L, Gao XW. Effects of quercetin on specific activity of carboxylesteras and glutathione S-transferase in Bemisia tabaci. Chinese Bulletin Entomol. 2006;43:491–495. In Chinese with English abstract. [Google Scholar]
- Konno K, Ono H, Nakamura M, Tateishi K, Hirayama C, Tamura Y, Koyama A, Kohno K. Mulberry latex rich in antidiabetic sugar-mimic alkaloids forces dieting on caterpillars. Proc Natl Acad Sci USA. 2006;103:1337–1341. doi: 10.1073/pnas.0506944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano N, Yamashita T, Yasuda K, Ikeda K, Kizu H, Kameda Y, Kato A, Nash RJ, Lee HS, Ryu KS. Polyhydroxylated alkaloids isolated from mulberry trees (Morusalba L.) and silkworms (Bombyx mori L.) J Agric Food Chem. 2001;49:4208–4213. doi: 10.1021/jf010567e. [DOI] [PubMed] [Google Scholar]
- Daimon T, Taguchi T, Meng Y, Katsuma S, Mita K, Shimada T. Beta-fructofuranosidase genes of the silkworm, Bombyx mori: insights into enzymatic adaptation of B. mori to toxic alkaloids in mulberry latex. J Biol Chem. 2008;283:15271–15279. doi: 10.1074/jbc.M709350200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International Silkworm Genome Consortium. The genome of a lepidopteran model insect, the silkworm Bombyx mori. insect Biochem Mol Biol. 2008;38:1036–1045. doi: 10.1016/j.ibmb.2008.11.004. [DOI] [PubMed] [Google Scholar]
- Yoshitake N. On the esterase types in the mid gut of the silkworm Bombyx mori L. J Seric Sci Jpn. 1963;32:289–291. [Google Scholar]
- Eguchi M, Sugimoto T. Changes in esterase zymograms of the silkworm Bombyx mori L. during development. J Sericult Sci Jpn. 1964;33:321–326. doi: 10.1016/0022-1910(65)90184-8. [DOI] [PubMed] [Google Scholar]
- Eguchi M, Yosuitaki N, Kai H. Type and inheritance of blood esterase in silkworm Bombyx mori L. Jpn J Genet. 1965;40:15–19. doi: 10.1266/jjg.40.15. [DOI] [Google Scholar]
- Xia QY, Cheng DJ, Duan J, Wang GH, Cheng TC, Zha XF, Liu C, Zhao P, Dai FY, Zhang Z, He NJ, Zhang L, Xiang ZH. Microarray-based gene expression profiles in multiple tissues of the domesticated silkworm, Bombyx mori. Genome Biol. 2007;8:R162. doi: 10.1186/gb-2007-8-8-r162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claudianos C, Ranson H, Johnson RM, Biswas S, Schuler MA, Berenbaum MR, Feyereisen R, Oakeshott JG. A deficit of detoxification enzymes: pesticide sensitivity and environmental response in the honeybee. Insect Mol Biol. 2006;15:615–36. doi: 10.1111/j.1365-2583.2006.00672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero FD. Cloning of a horn fly cDNA, HiαE7, encoding an esterase whose transcript concentration is elevated in diazinon-resistant flies. Insect Biochem Mol Biol. 2000;30:1107–15. doi: 10.1016/S0965-1748(00)00088-6. [DOI] [PubMed] [Google Scholar]
- de Carvalho RA, Torres TT, de Azeredo-Espin AM. A survey of mutations in the Cochliomyia hominivorax (Diptera: Calliphoridae) esterase E3 gene associated with organophosphate resistance and the molecular identification of mutant alleles. Vet Parasitol. 2006;140:344–351. doi: 10.1016/j.vetpar.2006.04.010. [DOI] [PubMed] [Google Scholar]
- Oakeshott JG, Devonshire AL, Claudianos C, Sutherland TD, Horne I, Campbell PM, Ollis DL, Russell RJ. Comparing the organophosphorus and carbamate insecticide resistance mutations in cholin- and carboxyl-esterases. Chem Biol Interact. 2005;157-158:269–275. doi: 10.1016/j.cbi.2005.10.041. [DOI] [PubMed] [Google Scholar]
- Maibeche-Coisne M, Merlin C, Francois MC, Queguiner I, Porcheron P, Jacquin-Joly E. Putative odorant-degrading esterase cDNA from the moth Mamestra brassicae: cloning and expression patterns in male and female antennae. Chem Senses. 2004;29:381–390. doi: 10.1093/chemse/bjh039. [DOI] [PubMed] [Google Scholar]
- Ishida Y, Leal WS. Cloning of putative odorant-degrading enzyme and integumental esterase cDNAs from the wild silkmoth, Antheraea polyphemus. Insect Biochem Mol Biol. 2002;32:1775–1780. doi: 10.1016/S0965-1748(02)00136-4. [DOI] [PubMed] [Google Scholar]
- Bai H, Ramaseshadri P, Palli SR. Identification and characterization of juvenile hormone esterase gene from the yellow fever mosquito, Aedes aegypti. Insect Biochem Mol Biol. 2007;37:829–837. doi: 10.1016/j.ibmb.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas BA, Church WB, Lane TR, Hammock BD. Homology model of juvenile hormone esterase from the crop pest, Heliothis virescens. Proteins. 1999;34:184–196. doi: 10.1002/(SICI)1097-0134(19990201)34:2<184::AID-PROT4>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Tan A, Tanaka H, Tamura T, Shiotsuki T. Precocious metamorphosis in transgenic silkworms overexpressing juvenile hormone esterase. Proc Natl Acad Sci USA. 2005;102:11751–11756. doi: 10.1073/pnas.0500954102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamimura M, Takahashi M, Kikuchi K, Reza AM, Kiuchi M. Tissue-specific regulation of juvenile hormone esterase gene expression by 20-hydroxyecdysone and juvenile hormone in Bombyx mori. Arch Insect Biochem Physiol. 2007;65:143–151. doi: 10.1002/arch.20186. [DOI] [PubMed] [Google Scholar]
- Vogt RG, Riddiford LM. Pheromone binding and inactivation by moth antennae. Nature. 1981;293:161–163. doi: 10.1038/293161a0. [DOI] [PubMed] [Google Scholar]
- Ishida Y, Leal WS. Rapid inactivation of a moth pheromone. Proc Natl Acad Sci USA. 2005;102:14075–14079. doi: 10.1073/pnas.0505340102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darboux I, Barthalay Y, Piovant M, Hipeau-Jacquotte R. The structure-function relationships in Drosophila neurotactin show that cholinesterasic domains may have adhesive properties. EMBO J. 1996;15:4835–4843. [PMC free article] [PubMed] [Google Scholar]
- Grisaru D, Sternfeld M, Eldor A, Glick D, Soreq H. Structural roles of acetylcholinesterase variants in biology and pathology. Eur J Biochem. 1999;264:672–686. doi: 10.1046/j.1432-1327.1999.00693.x. [DOI] [PubMed] [Google Scholar]
- Olson PF, Fessler LI, Nelson RE, Sterne RE, Campbell AG, Fessler JH. Glutactin, a novel Drosophila basement membrane-related glycoprotein with sequence similarity to serine esterases. EMBO J. 1990;9:1219–1227. doi: 10.1002/j.1460-2075.1990.tb08229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toutant JP. Insect acetylcholinesterase: catalytic properties, tissue distribution and molecular forms. Prog Neurobiol. 1989;32:423–446. doi: 10.1016/0301-0082(89)90031-2. [DOI] [PubMed] [Google Scholar]
- Shang JY, Shao YM, Lan GJ, Yuan G, Tang ZH, Zhang CX. Expression of two types of acetylcholinesterase gene from the silkworm, Bombyx mori, in insect cells. Insect Sci. 2007;14:443–449. [Google Scholar]
- Weill M, Fort P, Berthomieu A, Dubois MP, Pasteur N, Raymond M. A novel acetylcholinesterase gene in mosquitoes codes for the insecticide target and is non-homologous to the ace gene in Drosophila. Proc Biol Sci. 2002;269:2007–2016. doi: 10.1098/rspb.2002.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemingway J. The molecular basis of two contrasting metabolic mechanisms of insecticide resistance. Insect Biochem Mol Biol. 2000;30:1009–1015. doi: 10.1016/S0965-1748(00)00079-5. [DOI] [PubMed] [Google Scholar]
- Hirakura K, Fujmoto Y, Fukai T, Nomura T. Two phenolic glycosides from the root bark of the cultivated mulberry tree. J Nat Prod. 1986;48:218–224. doi: 10.1021/np50044a004. [DOI] [Google Scholar]
- Huang FF, Chai CL, Zhang Z, Liu ZH, Dai FY, Lu C, Xiang ZH. The UDP-glucosyltransferase multigene family in Bombyx mori. BMC Genomics. 2008;9:563. doi: 10.1186/1471-2164-9-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow JA. Insights into the malpighian tubule from functional genomics. J Exp Biol. 2009;212:435–445. doi: 10.1242/jeb.024224. [DOI] [PubMed] [Google Scholar]
- Merlin C, Rosell G, Carot-Sans G, Francois MC, Bozzolan F, Pelletier J, Jacquin-Joly E, Guerrero A, Maibeche-Coisne M. Antennal esterase cDNAs from two pest moths, Spodoptera littoralis and Sesamia nonagrioides, potentially involved in odourant degradation. Insect Mol Biol. 2007;16:73–81. doi: 10.1111/j.1365-2583.2006.00702.x. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Uda Y, Ono Y, Nakagawa T, Suwa M, Yamaoka R, Touhara K. Highly selective tuning of a silkworm olfactory receptor to a key mulberry leaf volatile. Curr Biol. 2009;19:881–890. doi: 10.1016/j.cub.2009.04.035. [DOI] [PubMed] [Google Scholar]
- Kasang G, Kaissling KE, Vostrowsky O, Bestmann HJ. Bombykal, a second pheromone component of the silkworm moth Bombyx mori. Angew Chem Int Ed Engl. 1978;17:60. doi: 10.1002/anie.197800601. [DOI] [Google Scholar]
- Kasang G, Nicholls M, von Proff L. Sex pheromone conversion and degradation in antennae of the silkworm moth Bombyx mori L. Experientia. 1989;45:81–87. doi: 10.1007/BF01990456. [DOI] [Google Scholar]
- Holmes RS, Chan J, Cox LA, Murphy WJ, VandeBerg JL. Opossum carboxylesterases: sequences, phylogeny and evidence for CES gene duplication events predating the marsupial-eutherian common ancestor. BMC Evol Biol. 2008;8:54. doi: 10.1186/1471-2148-8-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dottorini T, Nicolaides L, Ranson H, Rogers DW, Crisanti A, Catteruccia F. A genome-wide analysis in Anopheles gambiae mosquitoes reveals 46 male accessory gland genes, possible modulators of female behavior. Proc Natl Acad Sci USA. 2007;104:16215–20. doi: 10.1073/pnas.0703904104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meikle DB, Sheehan KB, Phyllis DM, Richmond RC. Localization and longevity of seminal-fluid esterase-6 in mated female Drosophila melanogaster. J Insect Physiol. 1990;36:93–101. doi: 10.1016/0022-1910(90)90179-J. [DOI] [Google Scholar]
- Hirai M, Kamimura M, Kikuchi K, Yasukochi Y, Kiuchi M, Shinoda T, Shiotsuki T. cDNA cloning and characterization of Bombyx mori juvenile hormone esterase: an inducible gene by the imidazole insect growth regulator KK-42. Insect Biochem Mol Biol. 2002;32:627–635. doi: 10.1016/S0965-1748(01)00141-2. [DOI] [PubMed] [Google Scholar]
- Wang J, Xia QY, He XM, Dai MT, Ruan J, Chen J, Yu G, Yuan HF, Hu YF, Li RQ, Feng T, Ye C, Lu C, Wang J, Li SG, Wong GK, Yang HM, Wang J, Xiang ZH, Zhou ZY, Yu J. SilkDB: a knowledgebase for silkworm biology and genomics. Nucleic Acids Res. 2005;33:D399–D402. doi: 10.1093/nar/gki116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Summary of the silkworm COE genes. NP indicates genes with no automatic prediction in silkworm. (N): missing N-terminal region; (C): missing C-terminal region. Chr.: chromosome. UN represents unknown chromosome locations.
Sequence alignment of Lepidopteran AChEs. Three intrachain disulfide bridges are drawn between conserved Cys. The asterisks represent the catalytic triad (S200, E327 and H440).
Chromosome distribution of all silkworm COEs. Arrows showed the transcriptional orientation.
The phylogeny and expression patterns of BmCOEs. Bootstrap values > 70% are shown. The solid black circles corresponding to cDNA, EST, tissue microarray and RT-PCR column indicate that these genes have expression evidence. The genes with grey font are that they had no probes in microarray dataset. The underlined α-esterases mean that they had substitutions in the catalytic triad.
Primers used in RT-PCR study. Primers used in RT-PCR study.