Abstract
The oxidized abasic lesion, 5′-(2-phosphoryl-1,4-dioxobutane) (DOB) is produced concomitantly with a single strand break by a variety of DNA damaging agents that abstract a hydrogen atom from the C5'-position. Independent generation of the DOB lesion in DNA reveals that it reversibly forms interstrand cross-links (ICLs) selectively with a dA opposite the 3′-adjacent nucleotide. Product studies and the use of monoaldehyde models suggest that ICL formation involves condensation of the dialdehyde with the exocyclic amine. Mechanistic studies and inspection of molecular models indicate that the local DNA environment and proximity of the exocyclic amine determines the selectivity for reaction with dA. Proximity control of the electrophile's reactivity is distinct from that of structurally similar freely diffusing molecules. ICL formation by a DOB lesion that is adjacent to a single strand break is potentially significant because the product constitutes a "clustered" or "complex" lesion. Clustered lesions can lead to highly deleterious double strand breaks upon nucleotide excision repair.
Keywords: DNA damage, reaction mechanism, interstrand cross-link
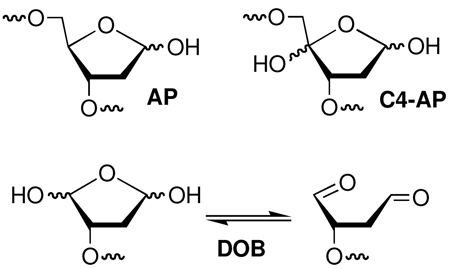
DNA is the cytotoxic target of a number therapeutic agents, and is also at the root of carcinogenesis. The chemical basis of these biological effects is not clear in many situations because a myriad of products are formed when DNA is exposed to various endogenous and exogenous damaging agents.1 In addition to single and double strand breaks some agents produce interstrand cross-links, which are an important family of lesions because they are absolute blocks of replication and transcription.2,3 By far the largest class of DNA lesions are those that result from modification of the sugar or nucleobase of an individual nucleotide. More than 50 modified nucleotides have been characterized.4,5 Many of these lesions decrease the fidelity of polymerases, resulting in mutations. However, a small number exhibit unusual chemical reactivity that may have significant biological consequences. For instance, two lesions have been discovered to irreversibly inhibit one or more of the DNA repair enzymes that have evolved to excise damage from the biopolymer.6–8 In two other examples, it was observed that the abasic site (AP) formed by formal hydrolysis of the nucleotide's glycosidic bond, and the C4'-oxidized abasic site (C4-AP) that is produced by ionizing radiation, bleomycin, and several other antitumor agents form interstrand cross-links.9–12 The latter produces high yields of interstrand cross-links (ICLs) in a sequence dependent manner. We report that the oxidized abasic site, dioxobutane (DOB) also forms interstrand cross-links and that its nucleotide selectivity and chemistry are distinct from those reported for AP and C4-AP.
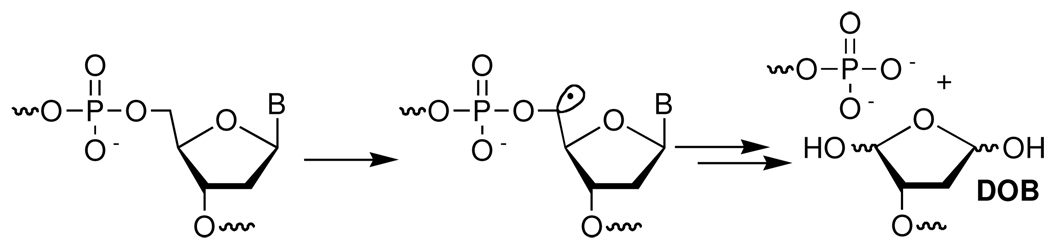
5'-(2-Phosphoryl-1,4-dioxobutane) (DOB) is a product of DNA oxidation.13,14 Although the detailed mechanism of its formation is not understood, it is believed to arise from C5'-oxidation under O2 deficient conditions (Scheme 1).15,16 DOB is unique amongst oxidized abasic lesions in that its formation is accompanied by a strand break at the adjacent nucleotide. Consequently, DNA damage containing DOB constitutes a tandem lesion, which is defined as two contiguously damaged nucleotides. Tandem lesions are a subset of clustered (or complex) lesions, which are defined as two damage sites within 1–2 helical turns. Clustered lesions, particularly those in which the lesions are in close proximity to one another, are known to adversely affect DNA repair.17–21 They were even postulated to lead to double strand breaks during misrepair.22 Recently, this was unambiguously demonstrated during nucleotide excision repair of an interstrand cross-link adjacent to a single strand break that resulted from the chemical promiscuity of the C4'-oxidized abasic site (C4-AP).23
Scheme 1.
The structure of DOB boded well for the likelihood that it too may be very reactive. For instance, the presence of the β-phosphate suggested that it is a potential source of 2-butene-1,4-dial, a product of furan metabolism, which alkylates DNA.24–27 Independent generation of DOB in oligonucleotides from a photochemical precursor revealed that the 1,4-dicarbonyl compound forms a stable cyclic adduct with Tris, and served as the basis of a sensitive method for its selective detection.28,29 The structurally related 1,4-dicarbonyl containing lesion, C4-AP, reacts readily with simple amines and dA and dC in duplex DNA.11,12,30,31 Hence, it seemed plausible that DOB would also react with DNA nucleobases.
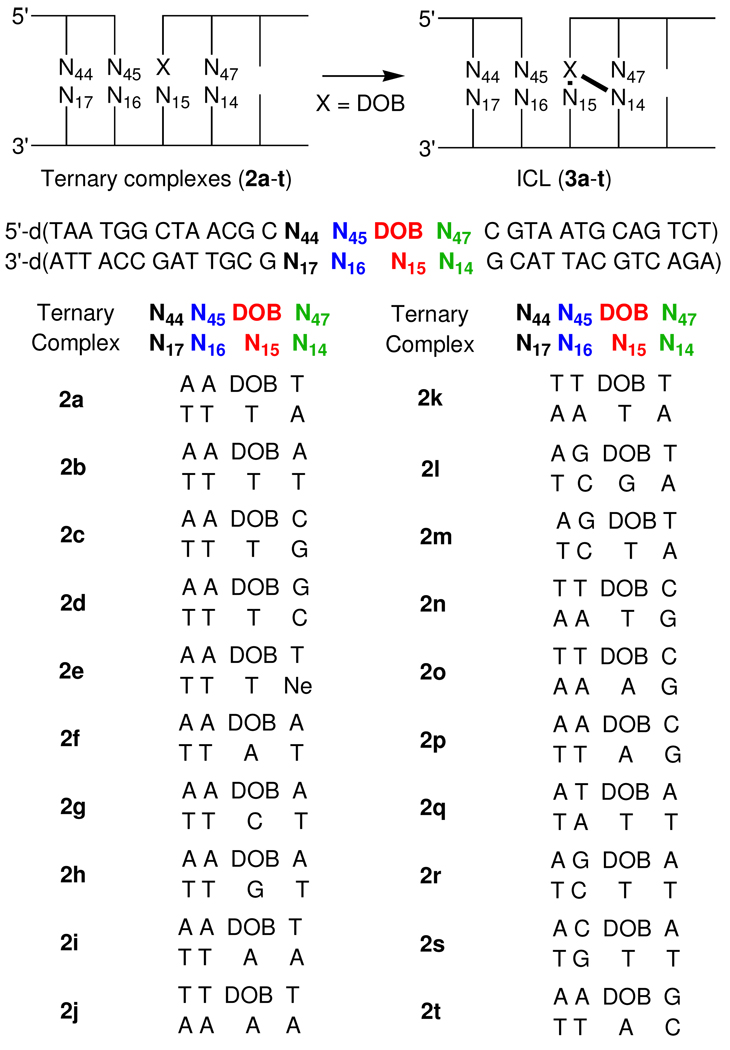
Results and Discussion
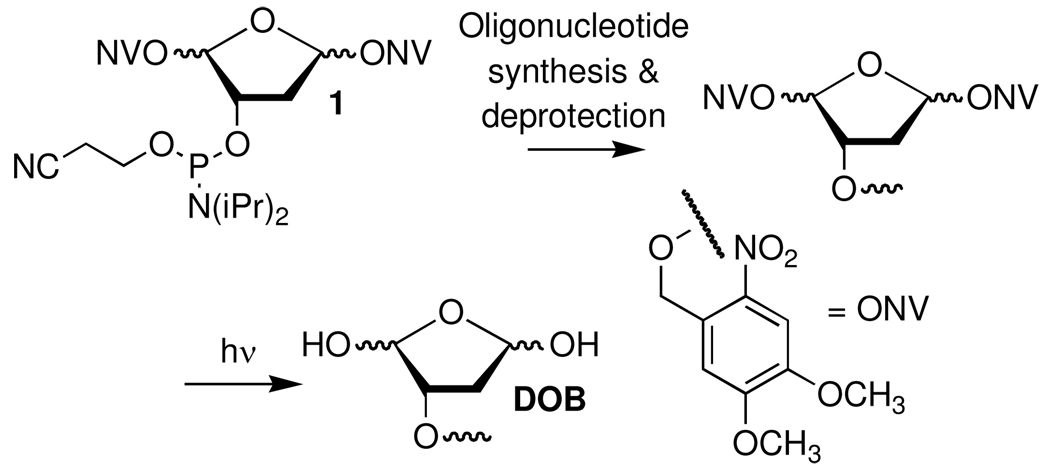
Oligonucleotides containing the photolabile DOB precursor were prepared as previously described from 1 (Scheme 2).28 Following hybridization with the template and flanking oligonucleotides to form the ternary complexes, the chemically labile DOB lesion was generated as needed via photolysis (60 min) in a Rayonet Photoreactor® at λmax = 350 nm from the bis-o-nitrobenzyl precursor. Hybridization was carried out prior to photochemical generation of the oxidized abasic site in order to minimize adventitious cleavage of the DOB lesion. The flanking sequence was varied (2a–t) to determine the effect of local DNA structure on DOB's reactivity with each of the native nucleotides that contain one or more nucleophilic nitrogen atoms. Native gel analysis showed that the ternary complexes containing DOB produced upon irradiation of the precursor (Scheme 2) remained hybridized under the subsequent incubation conditions (data not shown).
Scheme 2.
Preferential interstrand cross-linking by DOB with dA14
Examination of molecular models and prior studies on the reactivity of AP and C4-AP suggested that dN14 of the ternary complex would be the preferred reaction position.9,11,12 Consequently, we examined the reactivity of DOB in a series of ternary complexes in which unreactive thymidine was incorporated at positions dN15 and dN16 while the nucleotide was varied at dN14 (Table 1). Incubation in phosphate buffered (10 mM, pH 7.2) saline (100 mM) at 37 °C for 24 h yielded >30% ICL (3) when dA was present at position 14, but less then 1% when any other nucleotide was incorporated at this site. In the two instances (2a, 2c) in which ICLs were detected hydroxyl radical cleavage experiments revealed that cross-linking occurred exclusively at dN14.32,33 The lack of reaction with nebularine (Ne) was an initial indication that the exocyclic amine of dA was involved in cross-linking. This is similar to what was observed in the cross-linking reactions of C4-AP.11,12 As discussed below, the lack of reactivity with Ne provides insight into the structure of the ICL obtained from DOB.
Table 1.
Interstrand cross-link formation at dN14 (3)
| Substrate | Yield 3 (%)a | ICL Location | ||
|---|---|---|---|---|
| 2a | A DOB T | 32.6 ± 1.8 | A14 |  |
| T T A | ||||
| 2b | A DOB A | - | - | |
| T T T | ||||
| 2c | A DOB C | 0.8 ± 0.1 | G14 | |
| T T G | ||||
| 2d | A DOB G | - | - | |
| T T C | ||||
| 2e | A DOB T | - | - | |
| T T Ne | ||||
Yields are the average of at least 3 replicates.
Reactivity at dN15 and dN16 of the ternary complexes containing DOB were probed in a similar manner. Although no cross-link products were observed when dA (2q), dC (2r), dG (2s), or dT (2b) was incorporated at dN16 (data not shown), modest yields of cross-links were observed (Table 2) with an opposing dA (2f). However, positioning dC or dG at dN15 yielded less than 1% of the corresponding cross-link product. The preference for DOB cross-linking with dA14 was further illustrated by examining the effects of local sequence on cross-linking site (Table 3). The ICL yield varied between ∼22% and 38%, but hydroxyl radical cleavage revealed that cross-linking occurred exclusively at dA14 in all instances, even when 4 contiguous dAs were present (2j, Table 3).32 The yields of DOB cross-links are comparable to those involving C4-AP and considerably greater than ICLs resulting from reaction of AP.9,11,12
Table 2.
Interstrand cross-linking at the nucleotide opposite DOB (N15).
| Substrate | Yield 3 (%)a | |
|---|---|---|
| 2f | A DOB A | 3.2 ± 0.1 |
| T A T | ||
| 2g | A DOB A | 1.0 ± 0.2 |
| T C T | ||
| 2h | A DOB A | 0.6 ± 0.1 |
| T G T | ||
| 2b | A DOB A | - |
| T T T | ||
Yields are the average of at least 3 replicates.
Table 3.
Selective cross-linking of DOB at dA14.
| Substrate |  |
Yield 3 (%)a | ICL Location |
|---|---|---|---|
| 2i | A A DOB T | 21.9 ± 0.7 | A14 |
| T T A A | |||
| 2a | A A DOB T | 32.6 ± 1.8 | A14 |
| T T T A | |||
| 2j | T T DOB T | 38.8 ± 0.9 | A14 |
| A A A A | |||
| 2k | T T DOB T | 38.3 ± 1.0 | A14 |
| A A T A | |||
Yields are the average of at least 3 replicates.
The preference for DOB cross-linking with dA14 was evident even when the oxidized abasic site was presented with alternate nucleotides in this position and others (Table 4). ICL yields greater than a few percent were only obtained when dA was present at dN14. Specifically, reaction was only observed with dC (and in low yield) when this nucleotide was present at dN15 (2g, Table 2). In addition, the two instances in which cross-linking was detected at dG14 (2c, Table 1) and dG15 (2h, Table 2) occurred in <1% yield.
Table 4.
Attempted cross-linking of DOB at nucleotides other than dA.
 | |||
|---|---|---|---|
| Substrate | Yield 3 (%)a | ICL Location | |
| 2l | A G DOB T | 30.5 ± 1.0 | A14 |
| T C G A | |||
| 2m | A G DOB T | 23.4 ± 1.7 | A14 |
| T C T A | |||
| 2n | T T DOB C | 2.2 ± 0.1 | n.d. |
| A A T G | |||
| 2o | T T DOB C | 1.7 ± 0.7 | n.d. |
| A A A G | |||
| 2p | A A DOB C | 3.5 ± 0.3 | A15 |
| T T A G | |||
| 2s | A C DOB A | 0 | - |
| T G T T | |||
| 2t | A A DOB G | 0 | - |
| T T A G | |||
Yields are the average of at least 3 replicates. n.d. = Not determined
Structural characterization of the interstrand cross-link product formed between DOB and dA
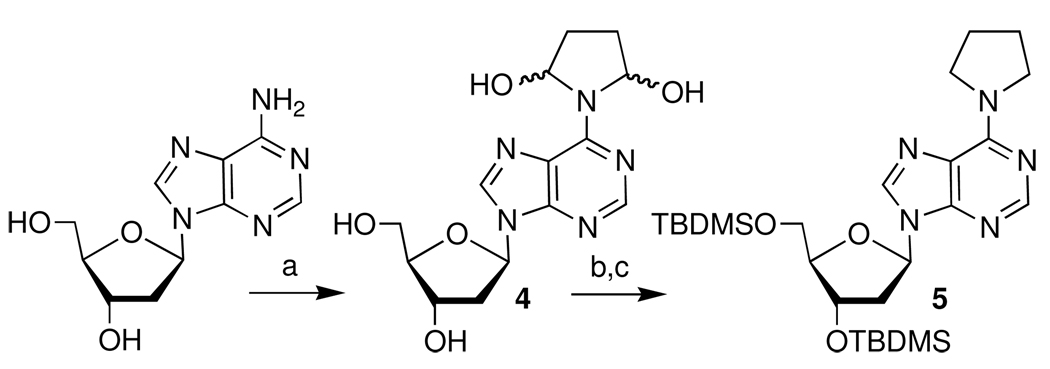
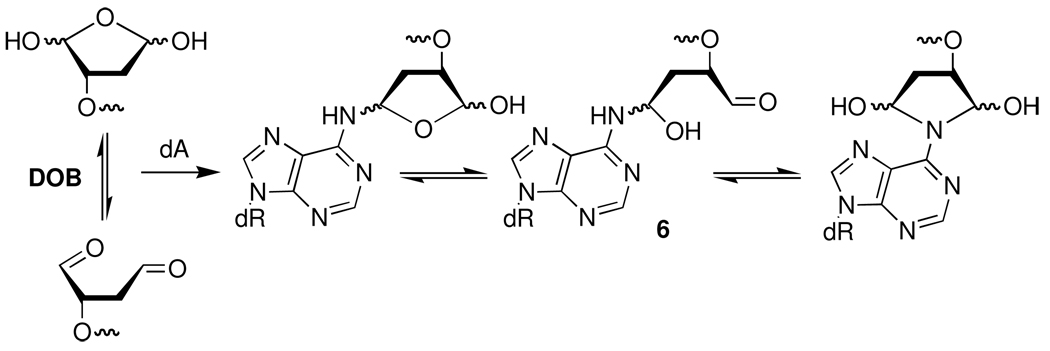
The ICL proved to be chemically labile and was not stabilized by reaction with NaCNBH3. In these respects, the cross-link was similar to one of the ICLs produced from C4-AP in which the lesion containing strand was uncleaved, but distinct from that isolated from the reaction of an AP site with dG.9,11,12 The ICL containing an uncleaved C4-AP strand also only forms when a dA is opposite a 3'-adjacent thymidine. These similarities suggested that the structure of the DOB-dA cross-link might involve condensation with the N6-amino group of dA, which is how C4-AP reacts with this nucleotide.12 The lack of reaction with Ne is consistent with this proposal. Although the ICL decomposition kinetics are discussed below, the cross-link product was sufficiently stable to enable its isolation and analysis by ESI-MS.32 ESI-MS of the product obtained from 2k showed that it resulted from condensation with the complement without loss of water. Additional information regarding the structure of the cross-link was obtained by reacting monomeric dA with 1,4-butanedial as a model for DOB in the polymer.34 Adduct 4 was isolated by reverse phase HPLC as a mixture of diastereomers that were contaminated with dA due to decomposition of the product during concentration. 1H NMR analysis was difficult due to the presence of multiple diastereomers and dA. However, MS data were consistent with the proposed structure.32 Further support for this assignment was obtained by converting 4 to previously reported 5 (Scheme 4).35 Based upon the lability of the ICL and these data obtained from the DNA and monomer, we propose that the cross-link product with reported in Table 1–Table 4 results from condensation of the DOB lesion with the N6-amino group of dA (6, Scheme 5) in the opposing strand and may exist as an equilibrium mixture.
Scheme 4a.
aKey: a) 1,4-Butanedial, Phosphate buffer (50 mM, pH 7.2) b) NaCNBH3, H2O, AcOH (pH 5.0) c) t-BuMe2SiCl, imidazole, DMF
Scheme 5.
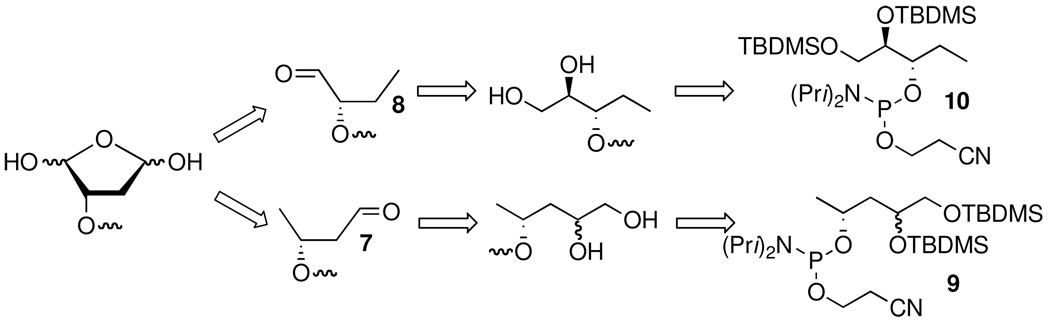
Additional inferential support for the biscondensation product (6) was obtained by examining the ability of oligonucleotides containing model compounds 7 or 8 to form cross-links. These model compounds position an individual carbonyl group in approximately the same location as each aldehyde from DOB. Oligonucleotides containing these monoaldehydes were prepared via solid phase oligonucleotide synthesis from the protected vicinal diol phosphoramidites (9, 10) (Scheme 6).
Scheme 6.
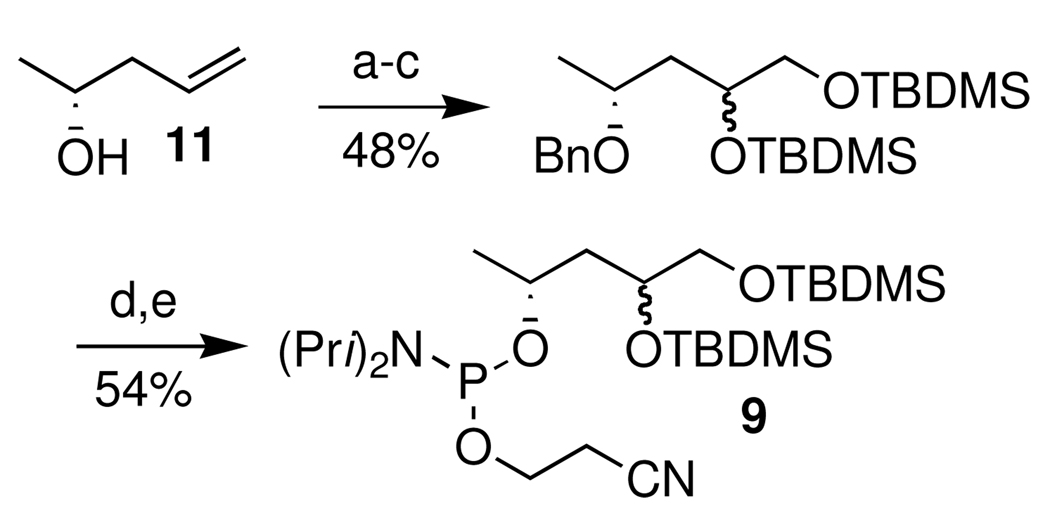
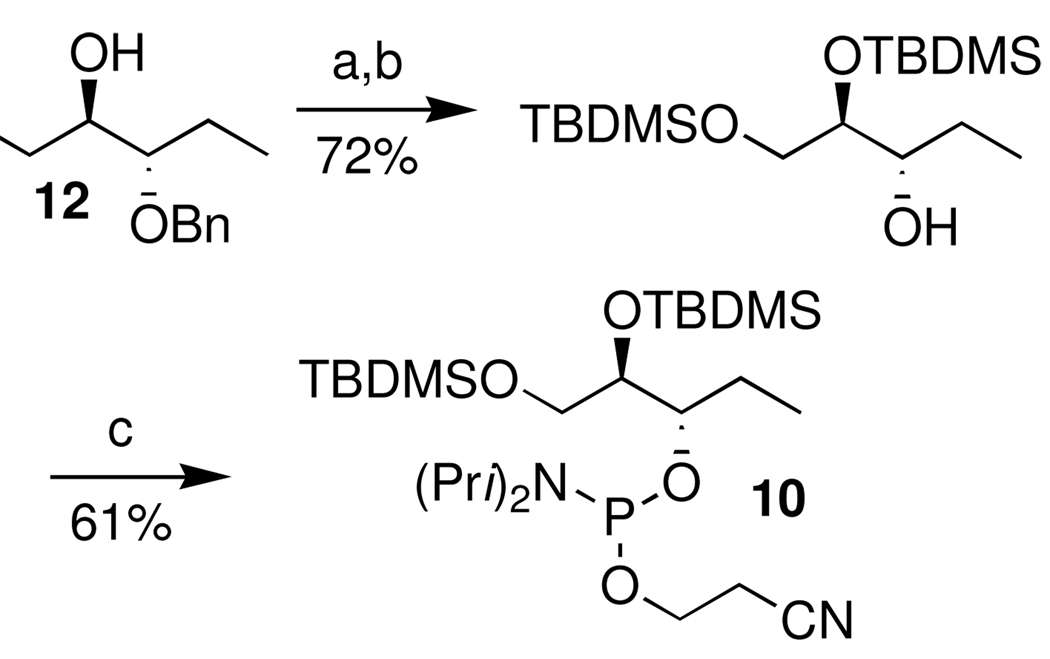
Sodium periodate treatment of the oligonucleotides containing vicinal diols was used to release the labile aldehydes (7, 8) immediately prior to their use.28,36,37 The respective phosphoramidites were synthesized using routes (Scheme 7 and Scheme 8) that started from a known substrate. The proper configuration at the ultimate stereogenic center in 7 and 8 is incorporated in these starting materials. For instance, the synthesis of the C1-aldehyde analogue (7) started from homoallylic alcohol 11 (Scheme 7). DNA containing 8 was prepared from previously reported 12 (Scheme 8), which was obtained from ascorbic acid.38–40 The modified phosphoramidites were coupled using standard reagents, but longer coupling times were employed.32 The vicinal diol containing oligonucleotides were deprotected via standard methods and purified by denaturing polyacrylamide gel electrophoresis.32
Scheme 7a.
aKey: a) NaH, BnBr, THF b) OsO4, NMO, acetone/H2O c) TBDMSCl, imidazole, DMF d) Pd(OH)2/C, H2, EtOAc e) Cyanoethyl N,N-diisopropylchlorophosphoramidite, DIPEA, CH2Cl2
Scheme 8a.
aKey: a) TBDMSCl, imidazole, DMF b) Pd(OH)2/C, H2, EtOAc c)Cyanoethyl N,N-diisopropylchlorophosphoramidite, DIPEA, CH2Cl2
The reactivity of each model compound was examined in 2 sequence contexts, each of which contained a dA at dN14 (Table 5). No cross-linking was detected in 13a,b and less than 1% was observed in 14a,b. These are far less than the 32.6% and 38.8% ICL yields measured for the analogous sequences (2a, 2j) containing DOB. These results indicate that ICL formation by DOB with dA does not involve Schiff base formation from a single carbonyl group, as is observed for cross-linking by AP with dG.9
Table 5.
Interstrand cross-link formation by DOB model compounds.
| Substrate | Yield ICL (%)a | |
|---|---|---|
| 13a | A A 7 T | 0 |
| T T T A | ||
| 13b | A A 8 T | 0 |
| T T T A | ||
| 14a | T T 7 T | 0.7 ± 0.1 |
| A A A A | ||
| 14b | T T 8 T | 0.8 ± 0.1 |
| A A A A | ||
Yields are the average of at least 3 replicates.
Interstrand cross-link formation between DOB and dA is reversible
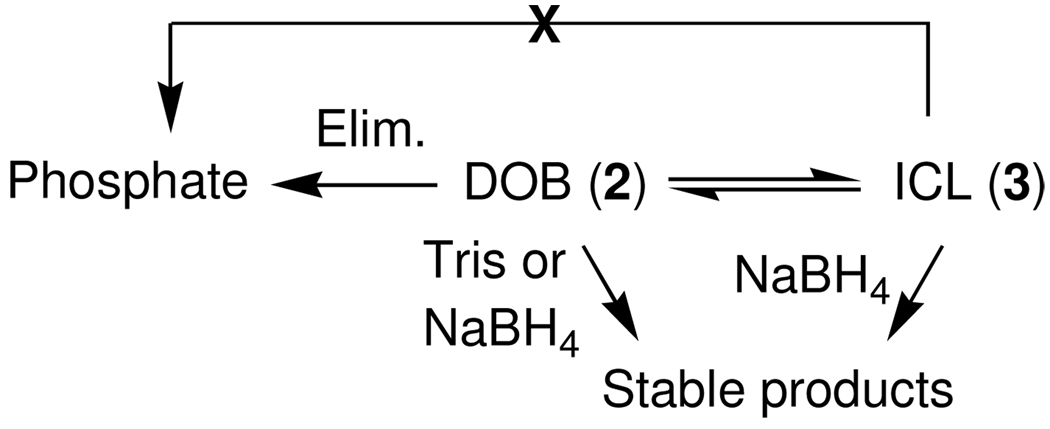
It was mentioned above that the ICL from DOB is unstable. Studies on the reaction of dA with 1,4-butanedial suggested that the cross-linking should be reversible. However, other decomposition pathways for the ICL were possible. One or more of these could result in release of a 5'-phosphate containing oligonucleotide and a template that may be alkylated by the remnants of the DOB lesion. The outcome of such an alkylation would be identical to reaction of DNA with butene-1,4-dial, a known metabolite of furan.15,24–27,41 This was investigated by following the decomposition of isolated 3i. Because DOB undergoes partial elimination during denaturing polyacrylamide gel electrophoresis to produce DNA containing 5'-phosphate termini, it was necessary to treat the samples in such a way so as to enable one to distinguish between this artifact and direct formation of oligonucleotides containing the phosphate end group from the ICLs. Consequently, the ICL was incubated in Tris buffer in order to capture released DOB as the previously characterized adduct (Scheme 9).28,29 In addition, aliquots removed from a sample of 3'-32P-3i as a function of time were quenched with NaBH4, which converts any remaining ICL to DOB and stabilizes the latter by reducing it to the diol. By employing these chemical treatments it was assumed that any oligonucleotide containing 5'-phosphate formed during incubation of 3'-32P-3i would result from ICL decomposition that does not involve formation of DOB. None of the phosphate product was detected by denaturing polyacrylamide gel electrophoresis, indicating that reversion to DOB was the sole pathway for ICL decomposition (data not shown).
Scheme 9.
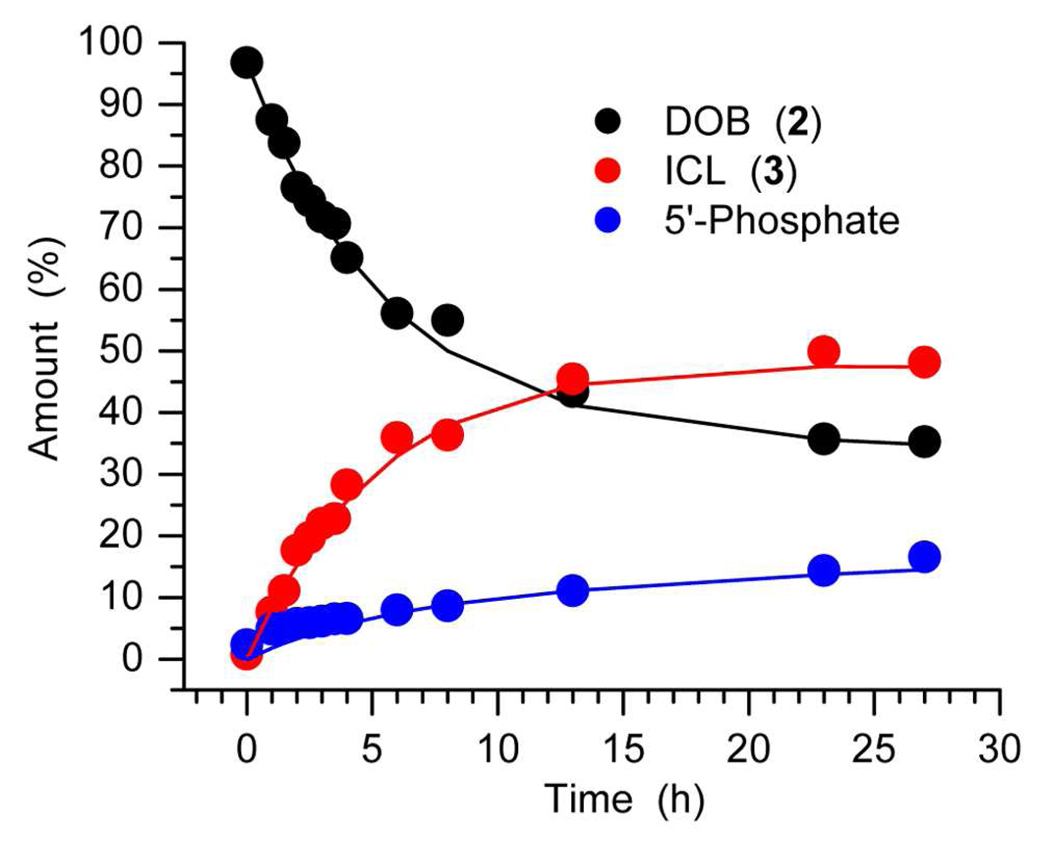
The rate constant for reversion of the ICL (3i) to DOB was initially determined by following its disappearance to less than 50% conversion, and treating the decomposition as an irreversible process. Fitting the decomposition to a first-order decay yielded a rate constant for its decomposition, k−1 = 1.7 × 10−5 s−1 (t1/2 = 11.2 h) under these buffer conditions (10 mM phosphate, pH 7.2).32 A more elaborate analysis was carried out by monitoring the growth of ICL (3i), DOB elimination product ("Phosphate", Eqn. 1), and DOB disappearance (2i, Figure 1). The concentrations of the 3 species as a function of time were fit to a kinetic scheme (Eqn. 1) in which ICL formation (k1) and elimination (k2) are in competition with one another, and cross-link formation is reversible. The rate constants for each step were determined using an iterative fitting procedure.42 The rate constant determined for ICL decomposition using this method is only slightly higher (k−1 = 1.9 × 10−5 s−1) than that calculated above. The fitting procedure also determines that the rate constant for ICL formation, k1 = 2.6 × 10−5 s−1, is only slightly faster than its decomposition, but that DOB undergoes elimination ∼5-times more slowly (k2 = 5.6 × 10−6 s−1). The DOB decomposition rate constant is ∼3-fold slower than that originally reported, but was measured at 5-fold lower buffer concentration.28
Figure 1.
Kinetic fitting of DOB reactivity as a function of time.
 |
(1) |
Why is DOB cross-linking to dA preferred?
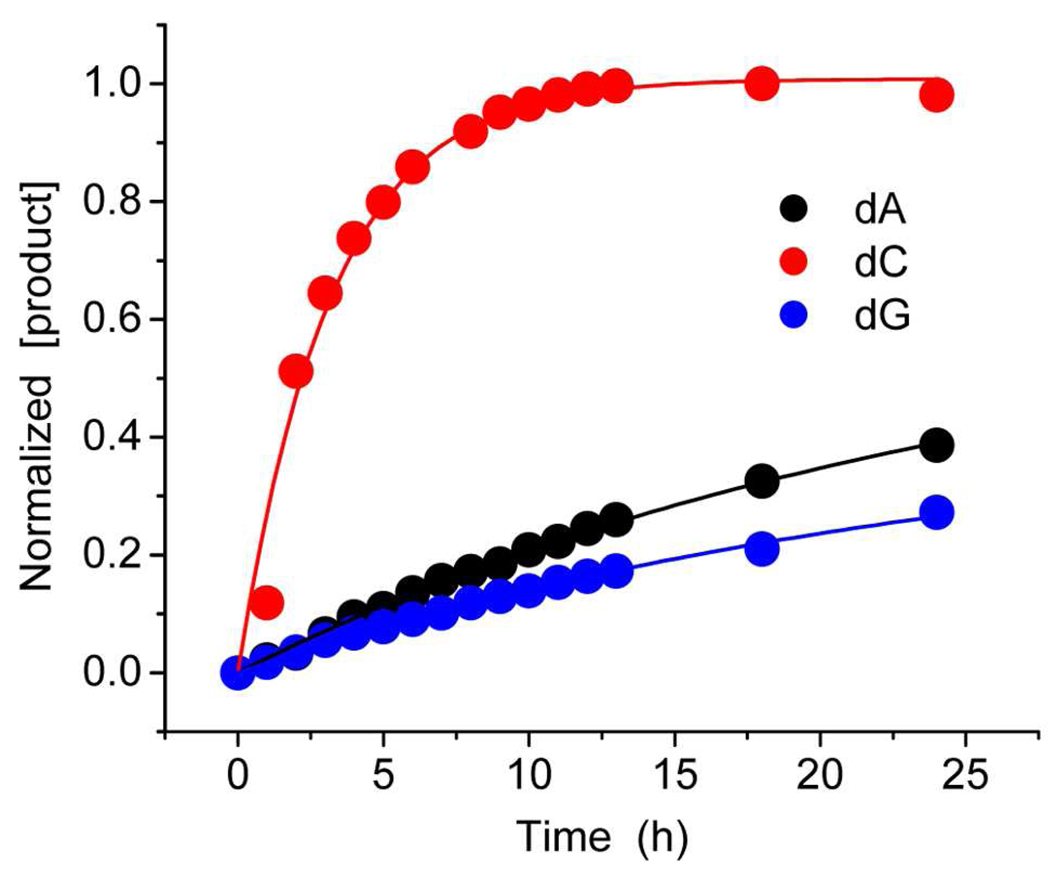
DOB is an example of a bis-electrophile. Other bis-electrophiles, including the C4'-oxidized abasic site (C4-AP) do not show such selectivity for dA.11,12 It is common for bis-electrophiles to form adducts with dA, dC, and dG.24,25,43–47 We sought to gain some insight into why DOB exhibits a preference for forming cross-links with dA by examining the reactions of the respective nucleosides with the DOB model compound, 1,4-butanedial (Scheme 4). Measuring the adduct growth rate (Figure 2) did not resolve the issue. Adduct formation between dG (kdG = 9.7 × 10−6 s−1) and 1,4-butanedial was almost as rapid as with dA (kdA = 1.2 × 10−5 s−1). Furthermore, the dialdehyde reacted with dC (kdC = 9.3 × 10−5 s−1) almost 10-times faster than it did with dA. This apparent discrepancy between reaction of the monomers with the model compound and that of DOB within the DNA was mitigated by measuring the decomposition rate constants for dC (k-dC = 7.6 × 10−5 s−1) and dA (k-dA = 9.2 × 10−6 s−1) adducts.32 Perhaps coincidentally, the equilibrium for the model reaction with dA (Keq = 1.3) is very similar to that for cross-linking between dA and DOB in 2i (Keq = 1.4). Overall, these data suggest that the inherent reactivity of dC (Keq = 1.2) and dA (Keq = 1.3) with 1,4-butanedial are quite similar, and that the equilibria for adduct formation with the model compound are almost identical. These data suggest that unlike o-quinone methides, reaction of 1,4-butanedial, and by inference DOB, with dA is not thermodynamically favored.48,49
Figure 2.
1,4-Butanedial adduct formation with nucleosides as a function of time.
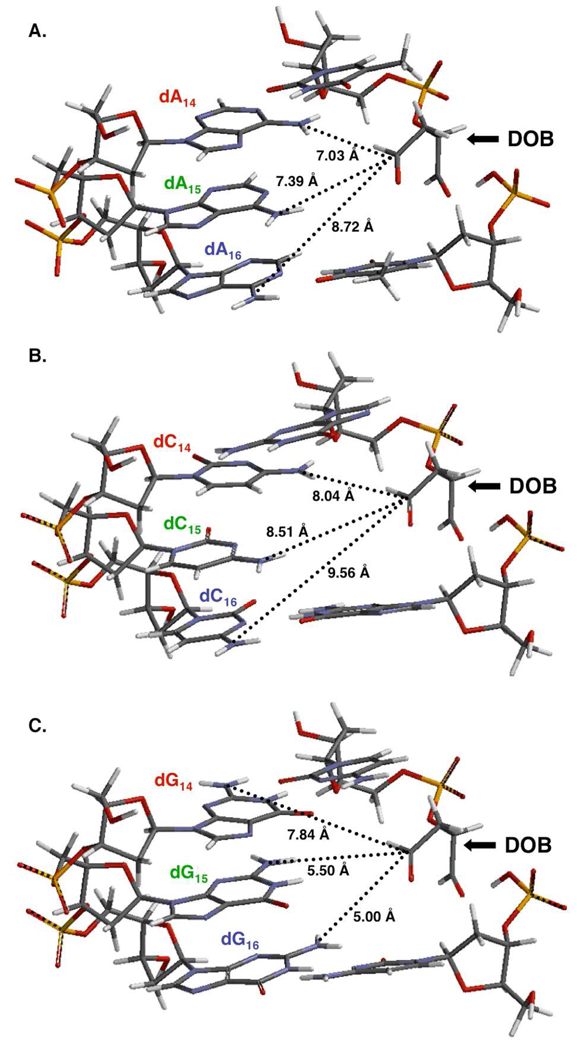
Since the inherent reactivity of the nucleosides did not explain the preferential cross-linking to dA in DNA, we considered whether proximity within the ternary complex could provide an explanation. Molecular models of three sequences were examined (Figure 3). The models were constructed from B-form DNA using Spartan. The DOB lesion was introduced by deleting the corresponding nucleotide without altering the positions of any of the remaining atoms. The models suggest that the N2-amino group of dG16 is the closest of the exocyclic amines to the DOB aldehyde, consistent with modeling of AP containing DNA.9 However, cross-linking is not detected at this nucleotide position (e.g. see ternary complex 2s) and suggests that the lack of reaction with dG in DNA correlates with the respective nucleoside's reduced reactivity with butanedial (Figure 2). The molecular models do provide a possible explanation for why reaction is not observed with dC in any of the ternary complexes. The shortest distance between DOB and the respective exocyclic amines of dC and dA occurs with the nucleotides at the dN14 position. However, the N6-amine of dA14 is ∼1 Å closer to the DOB aldehyde than is the N4-amine of dC14. The relatively closer proximity of the exocyclic amine of dA to the DOB carbonyl compared to that of dC is evident at dN15 and dN16 as well. Hence, we posit that the selectivity of DOB cross-linking is governed by its proximity to the respective nucleophiles of dC and dA in the ternary complexes. Closer proximity may increase the rate of ICL formation, but it also may correlate with cross-link stability. Although structural information is not available, the closer reactants are to one another, the less distorted and destabilizing the product DNA may be. One should note that these models suggest that the exocyclic amine in the opposing nucleotide (e.g. dN15) is less than 0.5 Å farther removed from the aldehyde than the respective nucleophile in dN14. However, reaction at dN15 (Table 2 and Table 4) is much less efficient than at dN14. We do not have an explanation for this, other than to suggest that the greater distance is sufficient to reduce the equilibrium for ICL formation. Anecdotal evidence in support of the hypothesis that cross-links involving dN15 are less stable is gleaned from the characterization of 3p by hydroxyl radical cleavage.32 The data are difficult to interpret because the ICL is much less stable compared to those involving cross-linking at dN14.
Figure 3.
Proximity of potential nucleophilic partners for DOB. (A) 5'-dT-DOB- dT/5'-(dA)3. (B) 5'-dGDOB-dG/5'-(dC)3. (C) 5'-dC-DOB-dC/5'-(dG)3. The models are obtained using Spartan.
Conclusions
5'-(2-Phosphoryl-1,4-dioxobutane) is an unusual DNA lesion that is produced by a variety of damaging agents.13,14 It is atypical in that it is formed concomitantly with a single strand break, and is itself a bis-electrophile. Single strand breaks are deleterious to cells. Furthermore, bis-electrophiles exhibit a rich chemistry that is derived from their ability to produce alkylated lesions that are often mutagenic. Previously, it was shown that AP and C4-AP abasic lesions react with nucleotides on the opposing strand to form interstrand cross-links.9,11,12 The latter forms two types of ICLs, one of which also contains a single strand break on the strand that originally contained the lesion. This "complex" lesion poses a significant challenge for DNA nucleotide excision repair, resulting is the formation of extremely deleterious double strand breaks.23
The DOB lesion exhibits reactivity that is in many respects similar to that of C4-AP, including high yields of interstrand cross-links.11,12 Interestingly, cross-links are not observed with the 3'-terminal nucleotide (dN45) in the strand that flanks the DOB lesion. The cross-linked product forms reversibly and predominantly with dA, as is the ICL obtained from C4-AP in which the lesion containing strand is uncleaved. High yields of the cross-links are obtained despite the reversibility because other reactions do not rapidly consume the DOB lesion. For instance, DOB does not readily form ICLs at other positions in the ternary complex, and elimination is slow compared to cross-link formation. The lability of the cross-links and the inability to stabilize them with reagents such as NaCNBH3 presents a challenge for their structural characterization and detection in cells. It may be possible to overcome the former by synthesizing a stable model cross-link. Structural characterization of the DOB cross-links will be particularly interesting if the precedent set by the respective C4-AP cross-link is repeated.23 Based upon the nucleotide excision repair of C4-AP cross-links, those formed by DOB (3) may also be especially deleterious because their misrepair could give rise to double strand breaks. Furthermore, the facile reaction of the 1,4-dicarbonyl containing DOB lesion with even the primary aromatic amine of adenine suggests that this molecule may readily form cross-links with DNA binding proteins. We hope to investigate these issues in due course.
Supplementary Material
Scheme 3.
Acknowledgment
We are grateful for support of this research by the National Institute of General Medical Sciences (GM-063028). We are grateful to Professor Jack Norton for assistance with analyzing the kinetics for cross-link formation and Dr. Shanta Dhar for carrying out preliminary experiments. We thank Aaron Jacobs and Jonathan Sczepanski for their comments on the manuscript.
Footnotes
Supporting Information Available. Complete description of experimental procedures, spectral data of new compounds, ESI-MS of modified oligonucleotides, sample autoradiograms of hydroxyl radical cleavage experiments, kinetic plots, and complete reference 34. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.von Sonntag C. The Chemical Basis of Radiation Biology. London: Taylor & Francis; 1987. [Google Scholar]
- 2.Noll DM, Mason TM, Miller PS. Chem. Rev. 2006;106:277–301. doi: 10.1021/cr040478b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schärer OD. Chem. Bio. Chem. 2005;6:27–32. doi: 10.1002/cbic.200400287. [DOI] [PubMed] [Google Scholar]
- 4.Cadet J, Douki T, Gasparutto D, Ravanat JL. Mutat. Res. 2003;531:5–23. doi: 10.1016/j.mrfmmm.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 5.Dizdaroglu M, Jaruga P, Birincioglu M, Rodriguez H. Free Rad. Biol. Med. 2002;32:1102–1115. doi: 10.1016/s0891-5849(02)00826-2. [DOI] [PubMed] [Google Scholar]
- 6.Kroeger KM, Hashimoto M, Kow YW, Greenberg MM. Biochemistry. 2003;42:2449–2455. doi: 10.1021/bi027168c. [DOI] [PubMed] [Google Scholar]
- 7.Hashimoto M, Greenberg MM, Kow YW, Hwang J-T, Cunningham RP. J. Am. Chem. Soc. 2001;123:3161–3162. doi: 10.1021/ja003354z. [DOI] [PubMed] [Google Scholar]
- 8.Makino K, Ide H. J. Biol. Chem. 2003;278:25264–25272. doi: 10.1074/jbc.M212847200. [DOI] [PubMed] [Google Scholar]
- 9.Dutta S, Chowdhury G, Gates KS. J. Am. Chem. Soc. 2007;129:1852–1853. doi: 10.1021/ja067294u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Regulus P, Duroux B, Bayle P-A, Favier A, Cadet J, Ravanat J-L. Proc. Nat. Acad. Sci. USA. 2007;104:14032–14037. doi: 10.1073/pnas.0706044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sczepanski JT, Jacobs AC, Greenberg MM. J. Am. Chem. Soc. 2008;130:9646–9647. doi: 10.1021/ja8030642. [DOI] [PubMed] [Google Scholar]
- 12.Sczepanski JT, Jacobs AC, Majumdar A, Greenberg MM. J. Am. Chem. Soc. 2009;131:11132–11139. doi: 10.1021/ja903404v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xi Z, Goldberg IH. In: Comprehensive Natural Products Chemistry. Kool ET, editor. Vol 7. Amsterdam: Elsevier; 1999. pp. 553–592. [Google Scholar]
- 14.Goldberg IH. Acc. Chem. Res. 1991;24:191–198. [Google Scholar]
- 15.Chen B, Bohnert T, Zhou X, Dedon PC. Chem. Res. Toxicol. 2004;17:1406–1413. doi: 10.1021/tx049818e. [DOI] [PubMed] [Google Scholar]
- 16.Chin DH, Kappen LS, Goldberg IH. Proc. Nat. Acad. Sci. USA. 1987;84:7070–7074. doi: 10.1073/pnas.84.20.7070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Georgakilas A. Mol. BioSyst. 2008;4:30–35. doi: 10.1039/b713178j. [DOI] [PubMed] [Google Scholar]
- 18.Kozmin SG, Sedletska Y, Reynaud-Angelin A, Gasparutto D, Sage E. Nucl. Acids Res. 2009;37:1767–1777. doi: 10.1093/nar/gkp010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paap B, Wilson I, D M, Sutherland BM. Nucl. Acids Res. 2008;36:2717–2727. doi: 10.1093/nar/gkn118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lomax ME, Gulston MK, O'Neill P. Radiat. Prot. Dosim. 2002;99:63–68. doi: 10.1093/oxfordjournals.rpd.a006840. [DOI] [PubMed] [Google Scholar]
- 21.Blaisdell JO, Wallace SS. Proc. Natl. Acad. Sci. USA. 2001;98:7426–7430. doi: 10.1073/pnas.131077798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gulston M, de Lara C, Jenner T, Davis E, O'Neil P. Nucleic Acids Res. 2004;32:1602–1609. doi: 10.1093/nar/gkh306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sczepanski J, Jacobs AC, Van Houten B, Greenberg MM. Biochemistry. 2009:7565–7567. doi: 10.1021/bi901006b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen B, Vu CC, Byrns MC, Dedon PC, Peterson LA. Chem. Res. Toxicol. 2006;19:982–985. doi: 10.1021/tx0601197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gingipalli L, Dedon PC. J. Am. Chem. Soc. 2001;123:2664–2665. doi: 10.1021/ja0056421. [DOI] [PubMed] [Google Scholar]
- 26.Byrns MC, Vu CC, Neidigh JW, Abad J-L, Jones RA, Peterson LA. Chem. Res. Toxicol. 2006;19:414–420. doi: 10.1021/tx050302k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Byrns MC, Predecki DP, Peterson LA. Chem. Res. Toxicol. 2002;15:373–379. doi: 10.1021/tx0101402. [DOI] [PubMed] [Google Scholar]
- 28.Kodama T, Greenberg MM. J. Org. Chem. 2005;70:9916–9924. doi: 10.1021/jo051666k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dhar S, Kodama T, Greenberg MM. J. Am. Chem. Soc. 2007;129:8702–8703. doi: 10.1021/ja073014e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aso M, Usui K, Fukuda M, Kakihara Y, Goromaru T, Suemune H. Org. Lett. 2006;15:3183–3186. doi: 10.1021/ol060987v. [DOI] [PubMed] [Google Scholar]
- 31.Usiv K, Aso M, Fukuda M, Suemune H. J. Org. Chem. 2008;73:241–248. doi: 10.1021/jo702080r. [DOI] [PubMed] [Google Scholar]
- 32.See Supporting Information.
- 33.Millard JT, Weidner MF, Kirchner JJ, Ribeiro S, Hopkins PB. Nucleic Acids Res. 1991;19:1885–1892. doi: 10.1093/nar/19.8.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mueller R, et al. J. Med. Chem. 2004;47:5183–5197. doi: 10.1021/jm0400395. [DOI] [PubMed] [Google Scholar]
- 35.Bae S, Lakshman MK. J. Org. Chem. 2008;73:3707–3713. doi: 10.1021/jo702558n. [DOI] [PubMed] [Google Scholar]
- 36.Shishkina IG, Johnson F. Chem. Res. Toxicol. 2000;13:907–912. doi: 10.1021/tx000108s. [DOI] [PubMed] [Google Scholar]
- 37.Huang H, Greenberg MM. J. Org. Chem. 2008;73:2695–2703. doi: 10.1021/jo702614p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mulzer J, Angermann A, Münch W. Liebigs Ann. Chem. 1986:825–838. [Google Scholar]
- 39.Abushanab E, Vemishetti P, Leiby RW, Singh HK, Mikkilineni AB, Wu DCJ, Saibaba R, Panzica RP. J. Org. Chem. 1988;53:2598–2602. [Google Scholar]
- 40.Yadav JS, Nirranjan Kumar N, Sridhar Reddy M, Prasad AR. Tetrahedron. 2007;63:2689–2694. [Google Scholar]
- 41.Byrns MC, Vu CC, Peterson LA. Chem. Res. Toxicol. 2004;17:1607–1613. doi: 10.1021/tx049866z. [DOI] [PubMed] [Google Scholar]
- 42.Bamford CH, Tipper CFH. The Theory of Kinetics. Vol. 2. Amsterdam: Elsevier; 1969. [Google Scholar]
- 43.Stone MP, Cho Y-J, Huang H, Kim H-Y, Kozekov ID, Kozekova A, Wang H, Minko IG, Lloyd RS, Harris TM, Rizzo CJ. Acc. Chem. Res. 2008;41:793–804. doi: 10.1021/ar700246x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cho YJ, Wang H, Kozekov ID, Kozekova A, Kurtz AJ, Jacob J, Voehler M, Smith J, Harris TM, Rizzo CJ, Lloyd RS, Stone MP. Chem. Res. Toxicol. 2006;19:1019–1029. doi: 10.1021/tx0600604. [DOI] [PubMed] [Google Scholar]
- 45.Wang H, Kozekov ID, Kozekova A, Tamura PJ, Marnett LJ, Harris TM, Rizzo CJ. Chem. Res. Toxicol. 2006;19:1467–1474. doi: 10.1021/tx060137o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sodum RS, Shapiro R. Bioorg. Chem. 1988;16:272–282. [Google Scholar]
- 47.Chen H-JC, Chung F-L. Chem. Res. Toxicol. 1994;7:857–860. doi: 10.1021/tx00042a021. [DOI] [PubMed] [Google Scholar]
- 48.Weinert EE, Frankenfield KN, Rokita SE. Chem. Res. Toxicol. 2005;18:1364–1370. doi: 10.1021/tx0501583. [DOI] [PubMed] [Google Scholar]
- 49.Veldhuyzen WF, Shallop AJ, Jones RA, Rokita SE. J. Am. Chem. Soc. 2001;123:11126–11132. doi: 10.1021/ja011686d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.