Summary
Genetic recombination ensures proper chromosome segregation during meiosis and is essential for genome stability and tumor suppression. DNA synthesis after Rad51-mediated DNA strand invasion is a crucial step during recombination. PCNA is known as the processivity clamp for DNA polymerases. Here, we report the surprising observation that PCNA is specifically required to initiate recombination-associated DNA synthesis in the extension of the 3'-end of the invading strand in a D-loop. We show using a reconstituted system of yeast Rad51, Rad54, RPA, PCNA, RFC, and DNA polymerase δ that loading of PCNA by RFC targets DNA polymerase δ to the D-loop formed by Rad51 protein, allowing efficient utilization of the invading 3'-end and processive DNA synthesis. We conclude that PCNA has a specific role in the initiation of recombination-associated DNA synthesis, and that DNA polymerase δ promotes recombination-associated DNA synthesis.
Introduction
Homologous recombination is a key pathway to preserve genomic stability by repairing DNA double-stranded breaks, gaps, and interstrand crosslinks as well as mediating recovery of stalled or broken replication forks (Li and Heyer, 2008). Recombination also facilitates the pairing and segregation of homologous chromosomes during meiosis (Hunter, 2007). The signature reactions of recombination, homology search and DNA strand invasion, are carried out by the Rad51 protein, the eukaryotic homolog of the paradigmatic RecA protein (Krogh and Symington, 2004). Rad51 forms a right-handed helical filament on single-stranded DNA in a gap or resulting from the processing of a double-stranded DNA break. RPA, the eukaryotic single-stranded DNA binding protein, is involved in many DNA transactions and plays at least two distinct roles during recombination. First, RPA antagonizes secondary structure in the invading single-stranded DNA to favor formation of the active Rad51-ssDNA filament (Sugiyama et al., 1997). Second, RPA binds to the displaced strand in the D-loop to stabilize the invasion intermediate (Eggler et al., 2002). Binding of the displaced strand by RPA is likely occurring also during the DNA synthesis phase. Rad54 is a Snf2-family dsDNA motor protein with multiple functions during recombination in vitro. It stabilizes the Rad51 presynaptic filament and stimulates D-loop formation (Mazin et al., 2003; Mazin et al., 2000; Van Komen et al., 2000). Owing to its specific interaction with the Rad51-dsDNA filament, Rad54 promotes the dissociation of Rad51 from the heteroduplex DNA product after DNA strand invasion (Kiianitsa et al., 2006; Li et al., 2007; Solinger et al., 2002). This activity is required to allow access of DNA polymerases to the invading 3'-end, because Rad51, unlike bacterial RecA protein, does not readily release dsDNA after ATP hydrolysis (Li and Heyer, 2009; Li et al., 2007; Zaitseva et al., 1999).
Following DNA strand invasion, DNA synthesis is pivotal to restore the integrity of the chromosome. The identity of the DNA polymerases involved in homologous recombination remains to be defined. A biochemical screen in human cells for activities that could catalyze DNA synthesis primed at a synthetic D-loop identified DNA polymerase η (Pol η) and showed that DNA polymerase δ (Pol δ) was unable to do so (McIlwraith et al., 2005). However, genetic studies in budding yeast suggest an involvement of Pol δ in homologous recombination (Fabre et al., 1991; Giot et al., 1997; Lydeard et al., 2007; Maloisel et al., 2004; Maloisel et al., 2008; Paques and Haber, 1997; Wang et al., 2004). Single mutants displayed a rather modest reduction in recombination possibly owing to the use of conditional mutants necessitated by the essential function of Pol δ in DNA replication. In addition, the observed partial defects may indicate functional overlap with other DNA polymerases, such as Pol ε. While Pol η is required for DSB-induced gene conversion in chicken DT40 cells, it does not appear to be involved in recombination in yeast (Kawamoto et al., 2005; McDonald et al., 1997).
PCNA is involved with a host of DNA polymerases during DNA replication and other DNA metabolic processes (Majka and Burgers, 2004; Moldovan et al., 2007). Like its bacterial homolog the β-clamp, PCNA endows DNA polymerases with high processivity. PCNA was shown to be required for mating-type switching in budding yeast, a model double-strand break repair event by homologous recombination (Wang et al., 2004). Modification of PCNA modulates its protein interactions, regulating access to the primer-template (Moldovan et al., 2007). PCNA mono-ubiquitylation on K164 favors interaction with translesion DNA polymerases over replicative polymerases, whereas poly-ubiquitylation on the same residue may disengage polymerase interaction, favoring template switching by fork regression at stalled replication forks (Hoege et al., 2002; Stelter and Ulrich, 2003; Ulrich and Jentsch, 2000). In contrast, PCNA sumoylation on K164 (and K127) favors an interaction with Srs2, a helicase that dissociates Rad51 from ssDNA exerting an anti-recombinogenic effect (Papouli et al., 2005; Pfander et al., 2005). These results suggest that unmodified PCNA is the active form for recombination-associated DNA synthesis.
Using an in vitro system reconstituted with yeast proteins coupling Rad51-mediated DNA strand invasion in the presence of RPA and Rad54 with DNA synthesis, we show that DNA polymerase δ is efficiently targeted to the D-loop by PCNA, resulting in efficient usage of the invading 3'-end and processive DNA synthesis. This function of PCNA to specifically recruit DNA polymerases to a D-loop significantly extends the earlier biochemical work with human Pol δ in recombination (McIlwraith et al., 2005), resolving the apparent discrepancy between biochemical and genetic evidence suggesting a role of Pol δ in recombination.
Results
DNA polymerase δ requires PCNA for recombination-associated DNA synthesis
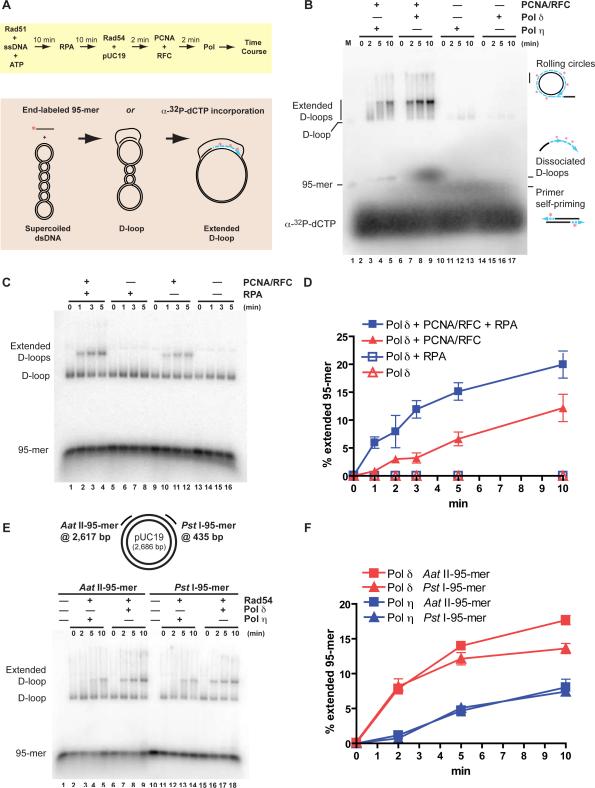
To study the DNA synthesis step during homologous recombination we reconstituted recombination in vitro using the D-loop reaction, in which a 95-mer single-stranded DNA (ssDNA) invades a homologous supercoiled duplex DNA by the combined action of yeast Rad51, Rad54, and RPA (Fig. 1A). Rad54 is a double-stranded DNA (dsDNA) motor protein that enhances D-loop formation and dissociates Rad51 from the product heteroduplex DNA after DNA strand exchange. We have previously shown that this activity allows access to the invading 3'-end by DNA polymerase (Li and Heyer, 2009). We compared recombination-associated DNA synthesis by yeast Pol δ with that by yeast Pol η by measuring the incorporation of α-32P-dCTP (Fig. 1B). Consistent with previous results using D-loops that were pre-assembled from oligonucleotides (McIlwraith et al., 2005), Pol η but not Pol δ could extend the 3'-end in a D-loop, albeit very inefficiently (Fig. 1B, lanes 10–17). Both polymerases were equally efficient in extending standard primed templates under these reactions conditions (Supplemental Figure 1). Both Pol δ and Pol η require PCNA for processive DNA synthesis (Haracska et al., 2001; Prelich et al., 1987). Addition of the processivity clamp PCNA and its clamp loader RFC resulted in strong stimulation of DNA synthesis by Pol η (Fig. 1B, lanes 2–5). Strikingly, recombination-associated DNA synthesis by Pol δ was completely dependent on PCNA. These results were confirmed for Pol δ in experiments with an end-labeled 95-mer, which allows quantitation of both the D-loop (no DNA synthesis) and the D-loop with an extended 3'-end (extended D-loop) (Fig. 1C–F). Extension of the invading 95-mer by Pol δ depended on the combined presence of both PCNA and RFC, consistent with the model that PCNA requires appropriate loading by its clamp loader RFC (Fig. 1C, D; Supplemental Figure 2). The presence of PCNA not only allowed DNA synthesis by Pol δ, but also led to increased total product formation (D-loops + extended D-loops) (Supplemental Figure 3). This suggests that PCNA not only acts as a processivity factor, its known function in DNA replication (Majka and Burgers, 2004), but also recruits Pol δ to the 3'-end of the D-loop.
Figure 1. PCNA/RFC stimulate Pol η in recombination-associated DNA synthesis and are necessary for Pol δ-dependent DNA synthesis during D-loop extension.
(A) Schematic representation of the reconstituted recombination reaction (top) and the assays (bottom) using either a 5'-end-labeled invading 95-mer or α-32P-dCTP incorporation to identify DNA synthesis products.
(B) α-32P-dCTP incorporation assay. Reactions were either complete (Pol η, lanes 2–5; Pol δ, lanes 6–9) or lacked PCNA/RFC (Pol η, lanes 10–13; Pol δ, lanes 14–17) using the AatII-95-mer. Lane 1 contains a D-loop reaction with 5'-end-labeled AatII-95-mer as a marker (M), and the positions of the 95-mer and D-loop are indicated. The signal above the extended D-loops in lanes 3–5 and 7–9 represents products of rolling circle replication with greater than full-length synthesis products (see Fig. 3D) that also depend on DNA strand invasion (Fig. 2B). The signal running just above the 95-mer marker in lanes 3–5 and 7–9 represents dissociated D-loops, which form during sample processing and electrophoresis (see Supplemental Figure 6). The signal co-migrating with the 95-mer in lanes 10–17 is due to self-priming of the 95-mer, and was particularly evident with Pol η (see also Fig. 2B, C).
(C) D-loop extension by Pol δ with 5'-end-labeled AatII-95-mer. Reactions were either complete (lanes 1–4), lacking PCNA/RFC (lanes 5–8), lacking RPA (lanes 9–12), or lacking PCNA/RFC and RPA (lanes 13–16).
(D) Quantitation of the results in (C) and additional experiments. Shown are means from at least three independent experiments and one standard deviation, except for the reactions containing Pol δ only, which were repeated twice.
(E) Efficiency of D-loop extension is independent of sequence context. The relative position of the homologies on the duplex DNA target for the AatII- and PstI-95-mers is schematically indicated. Reactions were complete with either Pol η (lanes 2–5, 11–14) or Pol δ (lanes 6–9, 15–18) with 5'-end-labeled invading ssDNA, either the AatII-95-mer (lanes 1–9) or the PstI-95-mer (lanes 10–18). Lanes 1 and 10 were controls with complete reactions lacking only Rad54 and polymerase.
(F) Quantitation of the results in (E) and additional experiments. Shown are means from at least three independent experiments and one standard deviation.
RPA is the eukaryotic, heterotrimeric ssDNA binding protein that enhances D-loop formation by binding to the displaced strand (Eggler et al., 2002). As expected, RPA stimulated D-loop formation (Supplemental Figure 4), leading to a corresponding increase in extension products (Fig. 1D).
PCNA-stimulated DNA synthesis by Pol δ appeared more robust than that by Pol η (Fig. 1B, lanes 2–5 vs. 6–9). This inference was confirmed in experiments using 5'-end-labeled 95-mers (Fig. 1E, F; Supplemental Figure 5). We tested two different homology regions in the duplex DNA target, designated AatII-95-mer and PstI-95-mer, with virtually identical results, indicating that the apparent preference for extension by Pol δ is not dependent on the sequence context.
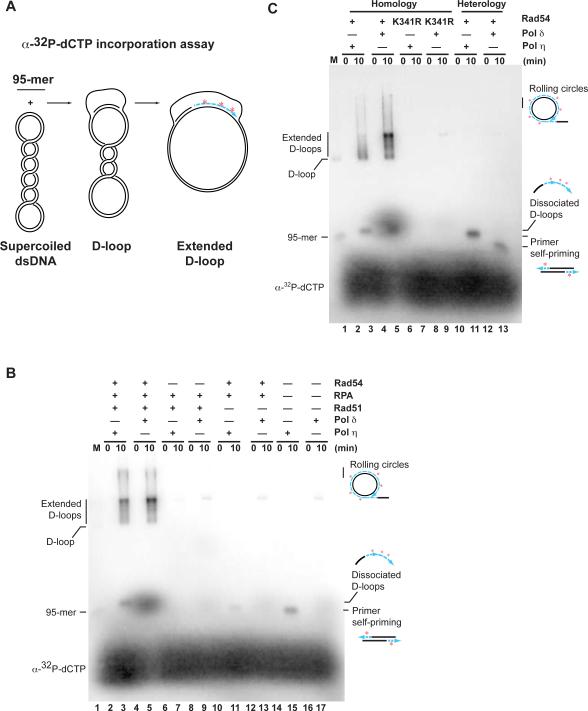
Control experiments omitting Rad51 demonstrated that the observed DNA synthesis depended on recombination (Fig. 2B, lanes 10–13). Also the Rad54 protein (lanes 6–9) was essential for recombination-associated DNA synthesis. Substituting with the Rad54-K341R mutant protein, which eliminates Rad54 ATPase and motor activities (Amitani et al., 2006), abrogated recombination-associated DNA synthesis consistent with previous studies (Li and Heyer, 2009). In the reactions with Pol δ, we observed low, but reproducible incorporation of α-32P-dCTP in the absence of Rad51 or Rad54 (Fig. 2B, lanes 9, 13, 17) or with Rad54-K341R protein (Fig. 2C, lane 9). This signal is due to Pol δ priming after spontaneous annealing of the 95-mer to the homologous sequence in the negatively supercoiled DNA. Accordingly, in reactions using a heterologous 95-mer, this low level incorporation was essentially lost (Fig. 2C, lane 13).
Figure 2. Recombination-associated DNA synthesis depends on Rad51, Rad54, and homology.
(A) Scheme of the α-32P-dCTP incorporation assay.
(B) D-loop extension requires Rad51 and Rad54. α-32P-dCTP incorporation assays were complete with either Pol η or Pol δ (lanes 2–5), omitted Rad54 (lanes 6–9), Rad51 (lanes 10–13), or RPA, Rad51 and Rad54 (lanes 14–17). Lane 1 contains a D-loop reaction with 5'-end-labeled AatII-95-mer as a marker (M), and the positions of the 95-mer and D-loop are indicated.
(C) D-loop extension requires the Rad54 ATPase activity and homology. Reactions were complete with either Pol η or Pol δ and wild type Rad54 protein (lanes 2–5) or the ATPase-deficient Rad54-K341R protein (lanes 6–9). In lanes 10–13, reactions using a heterologous 95-mer (olWDH640) were analyzed. Lane 1 contains size markers as described in (B). The signals representing rolling-circles, dissociated D-loops and primer self-priming were described in the legend to Figure 1B.
D-loop extension is limited by topological constraint
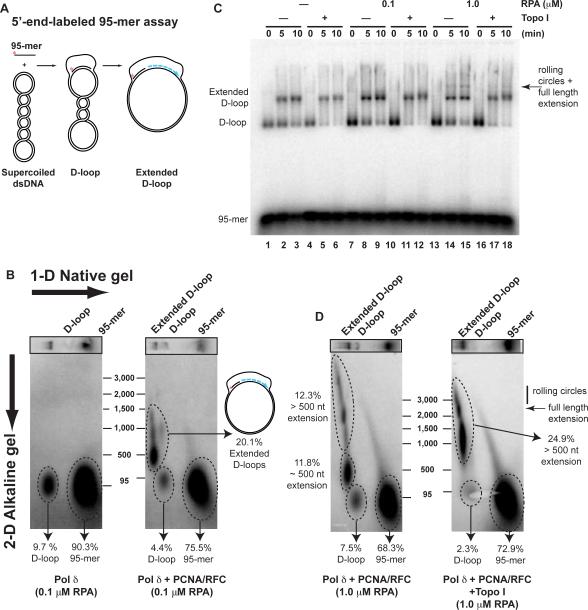
To determine the length of DNA synthesis in individual reaction products, the extended D-loops were analyzed by native/denaturing 2-dimensional gel electrophoresis. In both assays (α-32P-dCTP incorporation, 5'-end-labeled 95-mer), the products of both polymerases center around a length of 500 nt with evidence of some full-length (2.7 kb) products that have replicated around the entire plasmid (Fig. 3, Supplemental Figure 6). In accordance with the data from the incorporation assay (Fig. 1B), absence of PCNA prevented detectable extension of the 5'-end-labeled 95-mer by Pol δ (Fig. 3B, left panel). In titrating the ssDNA binding protein RPA, we showed that the efficiency of extension was stimulated by the presence of RPA but not limited by the RPA concentration used (Supplemental Figure 7).
Figure 3. Topological constraint limits DNA synthesis.
(A) Scheme of the assay with the 5'-end-labeled 95-mer.
(B) Extension of the invading 3'-end in a D-loop by Pol δ requires PCNA. 2-dimensional product analysis assays with 5'-end-labeled invading ssDNA (AatII-95-mer). D-loop extension products in complete reactions (10 min time point) with Pol δ (left) or omitting PCNA/RFC (right) were separated in the first dimension by native electrophoresis and in the second dimension under denaturing, alkaline conditions. Quantitation of the substrate and product species is provided in % of total.
(C) Product length is limited by topological constraint. Complete reactions with Pol δ were carried out with a titration of RPA (0–1 μM) using end-labeled AatII-95-mer. Eukaryotic topoisomerase I (lanes 4–6, 10–12, 16–18) or buffer control (lanes 1–3, 7–9, 13–15) was added at 2.5 min after addition of Pol δ and products were analyzed on native gels.
(D) 2-dimensional product analysis. Products of the 10 min time points of reactions containing 1 μM RPA (see lanes 15, 18 in C) supplemented (right) or not (left) with topoisomerase I were analyzed by 2-dimensional gel electrophoresis as in B. Quantitation of the different substrate and product species is provided as % of total. The positions of full-length extension products (2.7 kb) and rolling circle products (>2.7 kb) are indicated.
In the D-loop assay, extension of the 3'-end can lead to topological constraints, as with each 10.5 nucleotides synthesized one positive supercoil is induced in the closed circular template DNA. Experiments with the bacterial RecA protein showed that D-loop extension by E. coli Pol III holoenzyme to full-length products required gyrase to relieve this topological stress (Xu and Marians, 2002). In contrast, studies with the phage T4 recombination system have shown that DNA synthesis was unobstructed by topological problems because the D-loop migrated along with DNA synthesis. Such D-loop migration (bubble migration) relieves the topological stress by dissociating the D-loop at one end as it is extended at the other end, thereby keeping it at a constant size (Formosa and Alberts, 1986). To distinguish between these possibilities, we supplemented the reconstituted recombination reactions with eukaryotic topoisomerase I (Topo I), which relieves both positive and negative supercoils. Topo I was added 2.5 min after DNA synthesis was initiated, when D-loops have formed and synthesis is ongoing. Addition of Topo I stimulated the extent of DNA synthesis by Pol δ. In the absence of Topo I ~50% of the extension products (11.8% of total label; Fig. 3D left) had an average length of 500 nt. After Topo I addition, this entire population shifted to a size of about 1,500 nts; Fig. 3D, right). As expected, addition of Topo I led to the disappearance of D-loops (Fig. 3C, Supplemental Figure 8), because D-loop stability depends on the negative superhelicity of the duplex DNA (Van Komen et al., 2000). Addition of saturating amounts of RPA (1 μM, enough to cover a full-length displaced strand) led to the accumulation of a new, slower migrating species (Fig. 3C arrow). This band represents full-length extension of the 95-mer (Fig. 3D arrow) and accumulated to 2% of the input DNA corresponding to more than 10% of the extended D-loops. These results suggest that DNA synthesis initiated at the D-loop is limited by topological constraints.
Loading of PCNA at the D-loop targets DNA polymerase δ
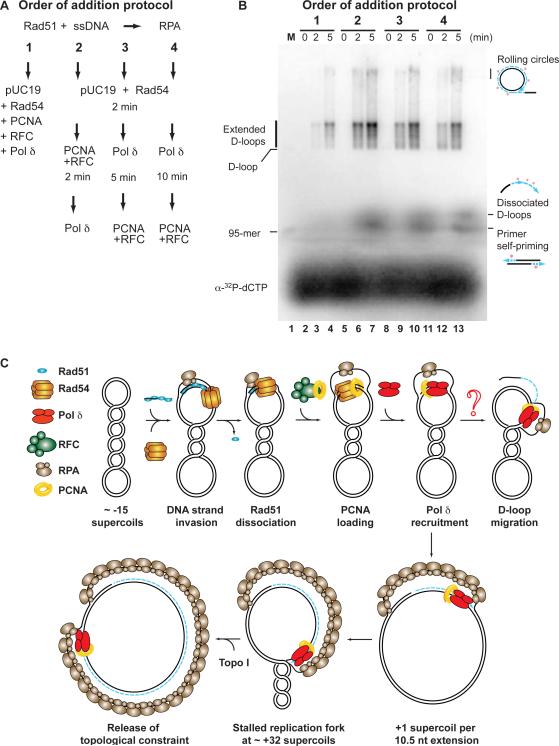
In DNA replication, RFC loads PCNA at the primer-template to act as a processivity clamp for the DNA polymerase. Control experiments showed that RFC was required for recombination-associated DNA synthesis by Pol δ and PCNA (Supplemental Figure 2). To corroborate the sequence of events, D-loop formation -> PCNA loading -> polymerase recruitment, we performed order-of addition experiments, where we changed the relative order of the addition of PCNA, RFC and Pol δ. As schematically diagrammed in Figure 4A, dsDNA, Rad54, PCNA, RFC, and Pol δ were either added at the same time (order #1), in sequence (dsDNA + Rad54 -> PCNA/RFC -> Pol δ = order #2), or in a scrambled sequence (dsDNA + Rad54 -> Pol δ -> PCNA/RFC after 5 min (order #3) or 10 min (order #4)). The results show that recombination-associated DNA synthesis has faster kinetics and higher yield when PCNA/RFC is supplied after D-loop formation and Pol δ is added last (Fig. 4B, lanes 5–7; order #2). This order is consistent with a model that loading of PCNA by RFC at the D-loop recruits Pol δ to the 3'-end of the invading strand for processive DNA synthesis.
Figure 4. Order-of-addition experiments.
(A) Scheme of the order-of-addition experiments.
(B) Complete α-32P-dCTP incorporation assays with the AatII-95-mer containing Pol δ were staged by the different protocols schematically indicated on the left hand side. Lane 1 contains a D-loop reaction with end-labeled AatII-95-mer as size markers (M), and the positions of the 95-mer and D-loop are indicated. The signals representing rolling-circles, dissociated D-loops and primer self-priming were described in the legend to Figure 1B.
(C) PCNA targets DNA polymerase δ to the 3'-OH end of the invading strand in a recombination-mediated D-loop. Working model depicting the sequence of DNA strand invasion and D-loop extension by Pol δ in the presence of PCNA/RFC. The topological constraint limiting the extent of DNA synthesis can be relieved by the addition of Topo I. The question mark indicates a possible pathway leading to D-loop migration, which would require additional activities. For more discussion see text.
Discussion
DNA synthesis plays a key role in the restoration of the genetic information during recombinational DNA repair. The identities of the DNA polymerases involved in homologous recombination remained rather uncertain, in particular the involvement of DNA polymerase δ, because of conflicting genetic and biochemical results. Genetic experiments in S. cerevisiae suggested a role of Pol δ and demonstrated a requirement for PCNA (and by implication RFC) (Fabre et al., 1991; Giot et al., 1997; Lydeard et al., 2007; Maloisel et al., 2004; Maloisel et al., 2008; Paques and Haber, 1997; Wang et al., 2004). Break-induced replication, a recombination-mediated pathway, depends on the Pol δ holoenzyme (the complex that includes the non-essential Pol32 subunit) (Jain et al., 2009; Lydeard et al., 2007). Moreover, a small C-terminal truncation of the Pol δ catalytic subunit affects conversion tract length in mitotic and meiotic recombination (Maloisel et al., 2004; Maloisel et al., 2008). Finally, the proof-reading exonuclease activity of Pol δ was shown to process terminal heterology of the invading 3'-end, suggesting that Pol δ has access to the nascent (unextended) D-loop (Fig. 4C) (Paques and Haber, 1997). These genetic data are consistent with a central role for Pol δ in homologous recombination, and our data now provide biochemical evidence for this role. The previous biochemical experiments with human Pol δ omitted PCNA/RFC (McIlwraith et al., 2005) and found that Pol δ alone could not access the 3'-end of the invading strand, consistent with our data.
Using an in vitro reconstituted recombination reaction, we coupled DNA strand invasion and D-loop formation by Rad51, Rad54 and RPA with the ensuing DNA synthesis catalyzed by Pol δ and its accessory factors PCNA and RFC. This system allowed us to identify a role for PCNA in the initiation of DNA synthesis at a recombination-mediated D-loop. This surprising finding consolidates the genetic evidence in yeast that suggested a role for Pol δ in homologous recombination with biochemical data. Taken together, our studies suggest the following model (Fig. 4C). First, the Rad51-ssDNA filament performs homology search and DNA strand invasion. Next, D-loops are stabilized by RPA binding to the displaced strand. Rad54 dissociates Rad51 from the duplex DNA after DNA strand exchange to allow access to the 3'-end of the invading strand (Li and Heyer, 2009). This is in agreement with the need for E. coli RecA protein to hydrolyze ATP and dissociate from the heteroduplex DNA before Pol III holoenzyme can access the invading 3'-end (Xu and Marians, 2002). Finally, loading of PCNA by RFC targets DNA Pol δ to the D-loop, leading to efficient and processive DNA synthesis from the invading 3'-end.
The unmodified form of PCNA used here is likely the relevant form for recombination, as mono- and poly-ubiquitylation of PCNA favor alternative pathways and sumoylation antagonizes recombination (Moldovan et al., 2007). The in vitro system identified a preference of Pol δ over Pol η in D-loop extension, consistent with unmodified PCNA favoring binding to Pol δ over the translesion polymerase η (Garg and Burgers, 2005). It is unclear, how DNA polymerases are selected for access at the D-loop beyond the inherent preference identified here. This may involve post-translational modification of the polymerases themselves or other mechanisms. While an involvement of Pol α - primase had been excluded in genetic experiments (Wang et al., 2004), clearly further studies are needed to address the potential for functional overlap between the other five nuclear polymerases (δ, ε, η, ζ, γ) in recombination
The in vitro system demonstrates that Pol δ is capable of efficient displacement synthesis in the context of a D-loop, where the displaced strand is contiguous, unlike in lagging strand replication, where the displaced strand is discontiguous (Jin et al., 2003). The D-loop appears to grow in size during recombination-associated DNA synthesis, to about 500 nt, before stalling due to topological constraints estimated to be about 32 positive supercoils (Fig. 4C). The topological stalling can be relieved by the addition of eukaryotic Topo I, but it is unclear whether this is a physiological role of type I topoisomerases. It is unlikely that topological constraint limits the length of conversion tracts to 500 nts in vivo, because mitotic gene conversion tracts are significantly longer (Lee et al., 2009). However, it is possible that topological constraint leads to polymerase stalling and D-loop dissociation followed by reinvasion, which might explain the observed template-switching during break-induced replication (BIR) (Smith et al., 2007). In this scenario, recombination events would involve multiple rounds of invasion and DNA synthesis. Alternatively, our reconstituted system might be lacking an activity that is required for D-loop migration (see Fig. 4C). It is possible that Sgs1/BLM (Bachrati et al., 2006; Gangloff et al., 2000; Lo et al., 2006; van Brabant et al., 2000), members of the RecQ 3'–5' DNA helicase family, or other helicases/motor proteins (Srs2 ? (Dupaigne et al., 2008)) are involved. Biochemical experiments have also suggested a potential role of the Rad54 motor protein in D-loop migration (Bugreev et al., 2007), however, this functions of Rad54 appears to be limited in our reconstituted system.
The identification of Pol δ as a DNA polymerase involved in D-loop extension is a critical step in developing a complete mechanistic understanding of homologous recombination and will help to unravel the mechanisms that govern the differences between various recombination modes (synthesis-dependent strand annealing, gap repair, double Holliday junction formation, break-induced replication) that are distinguished by their DNA synthesis reactions (Jain et al., 2009).
EXPERIMENTAL PROCEDURES
Proteins and DNA Substrates
Proteins were purified as described (Fortune et al., 2006; Garg et al., 2005; Gomes et al., 2000; Solinger et al., 2002). Oligonucleotide sequences are available upon request. pUC19 DNA (2,686 bps) was prepared by Triton/SDS lysis and purified by isopycnic density centrifugation on CsCl/Ethidium bromide gradients. T4 polynucleotide kinase and DNA polymerase I Klenow fragment were from New England Biolabs, wheat germ topoisomerase I from Promega.
D-loop extension assay
The reaction was carried out at 30°C in SEB buffer (30 mM Tris-acetate pH 7.5, 1 mM DTT, 50 μg/ml BSA, 4 mM ATP, 5 mM Mg2+-acetate, 20 mM phosphocreatine, 20 U/ml creatine kinase) supplemented with three dNTPs (dATP, dGTP, dTTP at 11.1 μM each) and α-32P-dCTP (0.22 μM; 6,000 μCi/mmol, Perkin Elmer). 95-mer (2 μΜ nt) was incubated with Rad51 (0.67 μM) for 10 min to assemble nucleoprotein filaments. RPA (0.1 μM) was then added to the reaction for an additional 10 min. The formation of D-loops was catalyzed by addition of Rad54 (72 nM) and pUC19 plasmid (56.6 μM bp, 20 nM molecule) to the reaction for 2 min. PCNA (20 nM) and RFC-1D complex (20 nM) were added to the D-loop reaction for an additional 2 min. The initiation of D-loop extension was started by addition of either Pol δ (20 nM) or Pol η (20 nM). At each time point, aliquots were mixed with stop buffer (1% SDS, 0.1 M EDTA, 4 mg/ml Proteinase K) and incubated for 10 min at 30°C. The DNA was separated on 1% agarose gels at 6 V/cm for 150 min. The gels were dried and analyzed by a phosphoImager. For the assays with 5'-end labeled 95-mers, the experimental procedure was as described except all four unlabeled dNTPs were present at 100 μM each. To release the topological constraint introduced by DNA synthesis, at 2.5 min after the addition of DNA polymerase, wheat germ topoisomerase I (0.42 U/μl) was added to the reaction, and the reactions were incubated another 7.5 min before analysis on gels.
Two-dimensional gel electrophoresis
For analysis of the extension products, the D-loop extension assays were carried out with either Pol δ or Pol η, and the reactions were stopped at 10 min. The sample of each reaction was split into two halves and separated in two lanes on a 0.8% native agarose gel at 5.5 V/cm for 70 min (α-32P-dCTP incorporation assay) or for 90 min (assays with 5'-end-labeled 95-mer). After electrophoresis, both lanes were excised and one lane was dried and served as one-dimensional reference. The other lane was soaked into the alkaline gel running buffer (50 mM NaOH, 1 mM EDTA) for 40 min, and then inserted into a 1% alkaline agarose gel. Electrophoresis was carried out at 2 V/cm for 11 hr. The gel was dried and analyzed by a phosphoImager.
Supplementary Material
Acknowledgements
We thank Steve Kowalczykowski, Neil Hunter, and Amitabh Nimonkar as well as members of the Heyer laboratory (Shannon Ceballos, Kirk Ehmsen, Clare Fasching, Ryan Janke, Jie Liu, Damon Meyer, Erin Schwartz, Jessica Sneeden, William Wright, Xiao-Ping Zhang) for discussion and comments on the manuscript. We thank Rita Alexeeva for help with DNA preparations, as well as Jie Liu, Kirk Ehmsen, and Xiao-Ping Zhang for help with protein preparations. This study was supported by National Institutes of Health grant GM32431 awarded to P.M.B. as well as CA92276 and GM58015 to W.D.H.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amitani I, Baskin RJ, Kowalczykowski SC. Visualization of Rad54, a chromatin remodeling protein, translocating on single DNA molecules. Mol Cell. 2006;23:143–148. doi: 10.1016/j.molcel.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Bachrati CZ, Borts RH, Hickson ID. Mobile D-loops are a preferred substrate for the Bloom's syndrome helicase. Nucleic Acids Res. 2006;34:2269–2279. doi: 10.1093/nar/gkl258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugreev DV, Hanaoka F, Mazin AV. Rad54 dissociates homologous recombination intermediates by branch migration. Nature Struct Mol Biol. 2007;14:746–753. doi: 10.1038/nsmb1268. [DOI] [PubMed] [Google Scholar]
- Dupaigne P, Le Breton C, Fabre F, Giangloff S, Le Cam E, Veaute X. The Srs2 helicase activity is stimulated by Rad51 filaments on dsDNA: Implications for crossover incidence during mitotic recombination. Mol. Cell. 2008;29:243–254. doi: 10.1016/j.molcel.2007.11.033. [DOI] [PubMed] [Google Scholar]
- Eggler AL, Inman RB, Cox MM. The Rad51-dependent pairing of long DNA substrates is stabilized by replication protein. A. J. Biol. Chem. 2002;277:39280–39288. doi: 10.1074/jbc.M204328200. [DOI] [PubMed] [Google Scholar]
- Fabre F, Boulet A, Faye G. Possible involvement of the yeast POLIII DNA polymerase in induced gene conversion. Mol. Gen Genet. 1991;229:353–356. doi: 10.1007/BF00267455. [DOI] [PubMed] [Google Scholar]
- Formosa T, Alberts BM. DNA synthesis dependent on genetic recombination: Characterization of a reaction catalyzed by purified bacteriophage T4 proteins. Cell. 1986;47:793–806. doi: 10.1016/0092-8674(86)90522-2. [DOI] [PubMed] [Google Scholar]
- Fortune JM, Stith CM, Kissling GE, Burgers PM, Kunkel TA. RPA and PCNA suppress formation of large deletion errors by yeast DNA polymerase delta. Nucleic Acids Res. 2006;34:4335–4341. doi: 10.1093/nar/gkl403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff S, Soustelle C, Fabre F. Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nature Genet. 2000;25:192–194. doi: 10.1038/76055. [DOI] [PubMed] [Google Scholar]
- Garg P, Burgers PM. Ubiquitinated proliferating cell nuclear antigen activates translesion DNA polymerases eta and REV1. Proc Natl Acad Sci U S A. 2005;102:18361–18366. doi: 10.1073/pnas.0505949102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg P, Stith CM, Majka J, Burgers PM. Proliferating cell nuclear antigen promotes translesion synthesis by DNA polymerase zeta. J Biol Chem. 2005;280:23446–23450. doi: 10.1074/jbc.C500173200. [DOI] [PubMed] [Google Scholar]
- Giot L, Chanet R, Simon M, Facca C, Faye G. Involvement of the yeast DNA polymerase delta in DNA repair in vivo. Genetics. 1997;146:1239–1251. doi: 10.1093/genetics/146.4.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes XV, Gary SL, Burgers PM. Overproduction in Escherichia coli and characterization of yeast replication factor C lacking the ligase homology domain. J Biol Chem. 2000;275:14541–14549. doi: 10.1074/jbc.275.19.14541. [DOI] [PubMed] [Google Scholar]
- Haracska L, Kondratick CM, Unk I, Prakash S, Prakash L. Interaction with PCNA is essential for yeast DNA polymerase eta function. Mol. Cell. 2001;8:407–415. doi: 10.1016/s1097-2765(01)00319-7. [DOI] [PubMed] [Google Scholar]
- Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- Hunter N. Meiotic recombination. In: Aguilera A, Rothstein R, editors. Homologous Recombination. Springer-Verlag; Berlin-Heidelberg: 2007. pp. 381–441. [Google Scholar]
- Jain S, Sugawara N, Lydeard J, Vaze M, Tanguy Le Gac N, Haber JE. A recombination execution checkpoint regulates the choice of homologous recombination pathway during DNA double-strand break repair. Genes Dev. 2009;23:291–303. doi: 10.1101/gad.1751209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin YH, Ayyagari R, Resnick MA, Gordenin DA, Burgers PMJ. Okazaki fragment maturation in yeast - II. Cooperation between the polymerase and 3'-5'-exonuclease activities of Pol delta in the creation of a ligatable nick. J. Biol. Chem. 2003;278:1626–1633. doi: 10.1074/jbc.M209803200. [DOI] [PubMed] [Google Scholar]
- Kawamoto T, Araki K, Sonoda E, Yamashita YM, Harada K, Kikuchi K, Masutani C, Hanaoka F, Nozaki K, Hashimoto N, Takeda S. Dual roles for DNA polymerase eta in homologous DNA recombination and translesion DNA synthesis. Mol. Cell. 2005;20:793–799. doi: 10.1016/j.molcel.2005.10.016. [DOI] [PubMed] [Google Scholar]
- Kiianitsa K, Solinger JA, Heyer WD. Terminal association of Rad54 protein with the Rad51-dsDNA filament. Proc. Natl. Acad. Sci. USA. 2006;103:9767–9772. doi: 10.1073/pnas.0604240103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh BO, Symington LS. Recombination proteins in yeast. Annu. Rev. Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- Lee PS, Greenwell PW, Dominska M, Gawel M, Hamilton M, Petes TD. A fine-structure map of spontaneous mitotic crossovers in the yeast Saccharomyces cerevisiae. PLoS Genet. 2009;5:e1000410. doi: 10.1371/journal.pgen.1000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Heyer WD. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 2008;18:99–113. doi: 10.1038/cr.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Heyer WD. RAD54 controls access to the invading 3-OH end after RAD51-mediated DNA strand invasion in homologous recombination in Saccharomyces cerevisiae. Nucleic Acids Res. 2009;37:638–646. doi: 10.1093/nar/gkn980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhang XP, Solinger JA, Kiianitsa K, Yu X, Egelman EH, Heyer WD. Rad51 and Rad54 ATPase activities are both required to modulate Rad51-dsDNA filament dynamics. Nucleic Acids Res. 2007;35:4124–4140. doi: 10.1093/nar/gkm412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo YC, Paffett KS, Amit O, Clikernan JA, Sterk R, Brenneman MA, Nickoloff JA. Sgs1 regulates gene conversion tract lengths and crossovers independently of its helicase activity. Mol. Cell. Biol. 2006;26:4086–4094. doi: 10.1128/MCB.00136-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lydeard JR, Jain S, Yamaguchi M, Haber JE. Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature. 2007;448:820–U810. doi: 10.1038/nature06047. [DOI] [PubMed] [Google Scholar]
- Majka J, Burgers PMJ. The PCNA-RFC families of DNA clamps and clamp loaders. Prog.Nucl. Acid Res. Mol. Biol. 2004;78:227–260. doi: 10.1016/S0079-6603(04)78006-X. [DOI] [PubMed] [Google Scholar]
- Maloisel L, Bhargava J, Roeder GS. A role for DNA polymerase delta in gene conversion and crossing over during meiosis in Saccharomyces cerevisiae. Genetics. 2004;167:1133–1142. doi: 10.1534/genetics.104.026260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloisel L, Fabre F, Gangloff S. DNA polymerase delta is preferentially recruited during homologous recombination to promote heteroduplex DNA extension. Mol. Cell. Biol. 2008;28:1373–1382. doi: 10.1128/MCB.01651-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazin AV, Alexeev AA, Kowalczykowski SC. A novel function of Rad54 protein - Stabilization of the Rad51 nucleoprotein filament. J. Biol. Chem. 2003;278:14029–14036. doi: 10.1074/jbc.M212779200. [DOI] [PubMed] [Google Scholar]
- Mazin AV, Bornarth CJ, Solinger JA, Heyer W-D, Kowalczykowski SC. Rad54 protein is targeted to pairing loci by the Rad51 nucleoprotein filament. Mol. Cell. 2000;6:583–592. doi: 10.1016/s1097-2765(00)00057-5. [DOI] [PubMed] [Google Scholar]
- McDonald JP, Levine AS, Woodgate R. The Saccharomyces cerevisiae RAD30 gene, a homologue of Escherichia coli dinB and umuC, is DNA damage inducible and functions in a novel error-free postreplication repair mechanism. Genetics. 1997;147:1557–1568. doi: 10.1093/genetics/147.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIlwraith MJ, Vaisman A, Liu YL, Fanning E, Woodgate R, West SC. Human DNA polymerase eta promotes DNA synthesis from strand invasion intermediates of homologous recombination. Mol. Cell. 2005;20:783–792. doi: 10.1016/j.molcel.2005.10.001. [DOI] [PubMed] [Google Scholar]
- Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Papouli E, Chen SH, Davies AA, Huttner D, Krejci L, Sung P, Ulrich HD. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol. Cell. 2005;19:123–133. doi: 10.1016/j.molcel.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Paques F, Haber JE. Two pathways for removal of nonhomologous DNA ends during double-strand break repair in Saccharomyces cerevisiae. Mol. Cell. Biol. 1997;17:6765–6771. doi: 10.1128/mcb.17.11.6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature. 2005;436:428–433. doi: 10.1038/nature03665. [DOI] [PubMed] [Google Scholar]
- Prelich G, Tan CK, Kostura M, Mathews MB, So AG, Downey KM, Stillman B. Functional identity of proliferating cell nuclear antigen and a DNA polymerase-delta auxiliary protein. Nature. 1987;326:517–520. doi: 10.1038/326517a0. [DOI] [PubMed] [Google Scholar]
- Smith CE, Llorente B, Symington LS. Template switching during break-induced replication. Nature. 2007;447:102–105. doi: 10.1038/nature05723. [DOI] [PubMed] [Google Scholar]
- Solinger JA, Kiianitsa K, Heyer W-D. Rad54, a Swi2/Snf2-like recombinational repair protein, disassembles Rad51:dsDNA filaments. Mol. Cell. 2002;10:1175–1188. doi: 10.1016/s1097-2765(02)00743-8. [DOI] [PubMed] [Google Scholar]
- Stelter P, Ulrich HD. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature. 2003;425:188–191. doi: 10.1038/nature01965. [DOI] [PubMed] [Google Scholar]
- Sugiyama T, Zaitseva EM, Kowalczykowski SC. A single-stranded DNA-binding protein is needed for efficient presynaptic complex formation by the Saccharomyces cerevisiae Rad51 protein. J. Biol. Chem. 1997;272:7940–7945. doi: 10.1074/jbc.272.12.7940. [DOI] [PubMed] [Google Scholar]
- Ulrich HD, Jentsch S. Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. Embo J. 2000;19:3388–3397. doi: 10.1093/emboj/19.13.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Brabant AJ, Ye T, Sanz M, German JL, Ellis NA, Holloman WK. Binding and melting of D-loops by the Bloom syndrome helicase. Biochemistry. 2000;39:14617–14625. doi: 10.1021/bi0018640. [DOI] [PubMed] [Google Scholar]
- Van Komen S, Petukhova G, Sigurdsson S, Stratton S, Sung P. Superhelicity-driven homologous DNA pairing by yeast recombination factors Rad51 and Rad54. Mol. Cell. 2000;6:563–572. doi: 10.1016/s1097-2765(00)00055-1. [DOI] [PubMed] [Google Scholar]
- Wang XA, Ira G, Tercero JA, Holmes AM, Diffley JFX, Haber JE. Role of DNA replication proteins in double-strand break-induced recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 2004;24:6891–6899. doi: 10.1128/MCB.24.16.6891-6899.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Marians KJ. A dynamic RecA filament permits DNA polymerasecatalyzed extension of the invading strand in recombination intermediates. J. Biol. Chem. 2002;277:14321–14328. doi: 10.1074/jbc.M112418200. [DOI] [PubMed] [Google Scholar]
- Zaitseva EM, Zaitsev EN, Kowalczykowski SC. The DNA binding properties of Saccharomyces cerevisiae Rad51 protein. J. Biol. Chem. 1999;274:2907–2915. doi: 10.1074/jbc.274.5.2907. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.