SUMMARY
GATA factors interact with simple DNA motifs (WGATAR) to regulate critical processes, including hematopoiesis, but very few WGATAR motifs are occupied in genomes. Given the rudimentary knowledge of mechanisms underlying this restriction, and how GATA factors establish genetic networks, we used ChIP-seq to define GATA-1 and GATA-2 occupancy genome-wide in erythroid cells. Coupled with genetic complementation analysis and transcriptional profiling, these studies revealed a rich collection of targets containing a characteristic binding motif of greater complexity than WGATAR. GATA factors occupied loci encoding multiple components of the Scl/TAL1 complex, a master regulator of hematopoiesis and leukemogenic target. Mechanistic analyses provided evidence for cross-regulatory and autoregulatory interactions among components of this complex, including GATA-2 induction of the hematopoietic corepressor ETO-2 and an ETO-2 negative autoregulatory loop. These results establish fundamental principles underlying GATA factor mechanisms in chromatin and illustrate a complex network of considerable importance for the control of hematopoiesis.
INTRODUCTION
Master regulators of development are commonly transcription factors that instigate complex genetic networks. Mechanisms underlying the function of these regulators are highly stringent, as deviations in their expression, chromatin site selection and protein-protein interactions elicit catastrophic phenotypes. In the context of hematopoiesis, defective genetic networks cause anemias, leukemias and lymphomas. Given the essential role of GATA factors in controlling hematopoiesis (Cantor and Orkin, 2002) and mutations in human leukemias (Crispino, 2005), it is crucial to elucidate their mechanisms, with perhaps the most rudimentary goal to establish the ensemble of target genes genome-wide.
GATA-2 expression occurs early in hematopoiesis and is required for the maintenance and expansion of hematopoietic stem cells and/or multipotent progenitors (Tsai et al., 1994). GATA-1 expression is induced subsequent to GATA-2 and is essential for the development of erythrocytes (Pevny et al., 1991; Simon et al., 1992), megakaryocytes (Shivdasani et al., 1997), eosinophils (Yu et al., 2002) and mast cells (Migliaccio et al., 2003). Whereas GATA-1 and GATA-2 bind DNA with a similar specificity (Ko and Engel, 1993; Merika and Orkin, 1993), and function redundantly to promote primitive erythroblast development (Fujiwara et al., 2004), they also exert distinct functions. GATA factors activate and repress genes, with or without the coregulator Friend of GATA-1 (FOG-1) (Crispino et al., 1999; Johnson et al., 2007; Tsang et al., 1997). FOG-1-dependent activation involves facilitation of GATA-1 chromatin occupancy (Letting et al., 2004; Pal et al., 2004a) and GATA-2 displacement from target sites (Pal et al., 2004a). FOG-1-dependent repression can be accompanied by broad histone deacetylation (Grass et al., 2003), and FOG-1 binds two corepressors, NuRD (Hong et al., 2005) and CtBP (Turner and Crossley, 1998).
Analyses at several loci suggest that GATA-1 and GATA-2 occupy a small fraction of the abundant WGATAR motif (Grass et al., 2003; Grass et al., 2006; Im et al., 2005; Johnson et al., 2002; Pal et al., 2004b). Even the presence of a conserved motif appears to be insufficient for implicating a GATA factor in regulation. Given that GATA factor DNA binding specificities were defined with naked DNA, nucleotides flanking WGATAR or cis-elements near WGATAR may mediate occupancy in vivo, or WGATAR might not be critical in chromatin.
GATA-1 and GATA-2 function cooperatively with the master regulator of hematopoiesis Scl/TAL1 on E-box (CANNTG)-WGATAR-containing composite elements (Lahlil et al., 2004; Vyas et al., 1999; Wadman et al., 1997; Wozniak et al., 2007; Xu et al., 2003). GATA-1 (Tripic et al., 2008) and GATA-2 (Wozniak et al., 2008) co-localize on chromatin sites with Scl/TAL1. Scl/TAL1 assembles a multimeric complex containing E2A, LMO2, Ldb1 and GATA-1 (Gottgens et al., 2002; Lahlil et al., 2004; Lecuyer et al., 2002; Wadman et al., 1997; Xu et al., 2003). Another critical factor that binds the Scl/TAL1 complex is the corepressor ETO-2 (Amann et al., 2001; Schuh et al., 2005), which like Scl/TAL1 (Aplan et al., 1992) and LMO2 (Hacein-Bey-Abina et al., 2003), is disrupted in leukemia (Gamou et al., 1998). Targeted disruption of Cbfa2t3, which encodes ETO-2, revealed an ETO-2 requirement for hematopoietic progenitor fate decisions, proliferation, and stress-dependent hematopoiesis (Chyla et al., 2008). Although other studies implicated ETO-2 in controlling erythropoiesis (Goardon et al., 2006) and megakaryopoiesis (Hamlett et al., 2008), little is known about its function in GATA-2-expressing cells.
GATA-2 and Scl/TAL1 co-localize at chromatin sites containing E-box-WGATAR motifs (Wozniak et al., 2008). As only a small fraction of E-box-WGATAR motifs are occupied in chromatin, resembling that of WGATAR motifs (Wozniak et al., 2008), the composite motif does not appear to confer a major advantage for chromatin occupancy vs. WGATAR.
Since GATA factor DNA binding specificities have been deduced through in vitro analyses, it is critical to generate and validate datasets of GATA factor occupancy in vivo. The magnitude and qualitative features of GATA factor target gene ensembles is unclear. We describe ChIP-seq analysis, in conjunction with expression profiling, target validation in primary cells, and computational mining, which yielded principles governing GATA factor-chromatin interactions and a genetic network of considerable importance for controlling hematopoiesis.
RESULTS
ChIP-Seq and Expression Profiling Reveal a Rich Set of GATA Factor Targets
ChIP-seq was conducted with human K562 erythroleukemia cells that express GATA-1 (Tsai et al., 1989) and GATA-2 (Dorfman et al., 1992) and are studied intensively in the ENCODE project (http://genome.ucsc.edu/ENCODE). The ChIP assay was validated by measuring GATA-1 occupancy at β-globin LCR HS2 (Johnson et al., 2002). Immunoprecipitated DNA from two biological replicates was used to prepare libraries for deep-sequencing. Sequences were mapped to the UCSC Human Genome assembly. Replicates A and B yielded 4.4 million and 10.3 million uniquely mapped sequences, respectively. Using a false discovery rate of 0.001, we identified 1,536 and 6,104 GATA-1 binding sites (peaks) in replicates A and B respectively. To assess reproducibility, peak overlap was determined. 90% of the peaks (1,380) from replicate A were present in replicate B. All reads for both replicates were merged to yield 5,749 sites (Suppl. Table 1) corresponding to 4,061 genes. The peak heights ranged from 20-259 sequence reads, with an average height of 38 and width of 327 bp, and peak number did not correlate with chromosome size (Suppl. Table 2). The analysis revealed established and new GATA-1 targets, including hematopoietic transcription factors, signaling molecules, and red cell cytoskeletal components (Fig. 1).
Figure 1. Representative GATA-1 Targets Identified Through ChIP-seq Analysis.
GATA-1 signal maps are shown for representative hematopoietic transcription factors, signaling molecules and red cell cytoskeletal proteins. Arrows, ChIP-seq peak locations relative to the transcription start site of the respective GATA-1 target gene (kb).
K562 cells are transformed and have a primitive erythroid phenotype (Lozzio et al., 1979), whereas murine G1E cells are untransformed and resemble normal definitive proerythroblasts (Weiss et al., 1997; Welch et al., 2004). Given these differences, its target sites might differ greatly in the two cell types. Quantitative ChIP analysis was conducted with untreated and β-estradiol-treated G1E cells stably expressing ER-GATA-1 (G1E-ER-GATA-1) to test whether ER-GATA-1 and GATA-2 occupy sites containing conserved WGATAR motifs that were detected by ChIP-seq. Although the absolute levels of occupancy were higher for peaks with high vs. low peak values, ER-GATA-1 and GATA-2 occupancy were detected in 10/13 (Fig. 2A) and 8/13 (Fig. 2B) of the peaks, respectively.
Figure 2. Validation of Human ChIP-seq Results in Murine G1E-ER-GATA-1 cells.
Quantitative real-time ChIP analysis of ER-GATA-1 and GATA-2 occupancy at 13 high (A) and low (B) ChIP-seq hits containing conserved WGATAR motifs in both β-estradiol-untreated and -treated (1 μM, 24 h) G1E-ER-GATA-1 cells (mean +/− SE, 3 independent experiments).
Computational Mining of ChIP-seq and Expression Profiling Datasets
Prior work defined GATA-1 and GATA-2 occupancy at dispersed regions of several loci (Grass et al., 2003; Grass et al., 2006; Im et al., 2005; Johnson et al., 2007; Johnson et al., 2002; Martowicz et al., 2005; Munugalavadla et al., 2005; Pal et al., 2004b; Scherzer et al., 2008; Wozniak et al., 2007). At the β-globin locus, GATA-1 instigates chromatin looping (Vakoc et al., 2005), which brings dispersed complexes in close proximity to each other. Since GATA-1 occupancy at very few loci were analyzed, it is unclear whether occupancy occurs mainly at promoters or other regions. Location analysis with the 5,749 peaks, using the Cis-regulatory Element Annotation System (http://ceas.cbi.pku.edu.cn/), revealed occupancy predominantly within introns (37%) and >1 kb away from RefSeq genes (“enhancer”) (47%) (Fig. 3A). The highest frequency of peaks was at −10 to −100 kb and +10 kb to +100 kb (Suppl. Fig. 1). Only 10% of the sites reside in proximal promoters (<1 kb upstream of RefSeq 5′ start).
Figure 3. Computational/Statistical Mining of ChIP-seq Data.
(A, B) The 5,749 ChIP-seq peaks were classified by (A) locations relative to nearest-neighbor genes and (B) GATA motif pattern within each peak. (C) Sequence logos from 5051 WGATAR-containing (left) and 301 E-box-WGATAR-containing (right) peaks. The logo on the left was obtained by de novo motif finding using MEME, while the logo on the right was generated by aligning E-box-WGATAR sequences within peaks. Information content was measured in bits (ranging from 0 to 2 for a given position of sequence). A position in the motif at which all nucleotides occur with equal probability has a value of 0, while a position at which only a single nucleotide can occur has a value of 2. (D) Merged ChIP-seq and Illumina expression profiling results. ChIP-seq peaks were merged with array data profiling expression in untreated and β-estradiol-treated (1 μM, 24 h) G1E-ER-GATA-1 cells (2 independent experiments). Among 296 genes shared by both datasets, the top 60 activated and repressed genes are shown. Asterisk, gene demonstrated previously to be differentially expressed in G1E-ER-GATA-1 cells (Welch et al., 2004). (E) Quantitative ChIP analysis of GATA-1 occupancy in primary murine Ter119+ bone marrow cells (mean +/− SE, 2 independent experiments). Asterisk, significantly greater GATA-1 occupancy compared to the Ey promoter (p < 0.05). Preimmune signals were analyzed with all primer sets and did not exceed 0.0025.
GATA-1 preferentially binds WGATAR-containing DNA (Evans et al., 1988). In a site selection analysis, recombinant chicken GATA-1 preferentially bound NNAGATAANN (Ko and Engel, 1993). Site selection with recombinant mouse GATA-1 (Merika and Orkin, 1993) and Mouse Erythroleukemia Cell (MEL) extracts (Wadman et al., 1997) yielded consensus sequences (G/C/A)NGAT(A/G/T)G(GCT) and CGATAA, respectively.
Based on the highly restricted occupancy of WGATAR-containing sites in cells, sequences preferred in vitro might not be an obligate requirement in chromatin. DNA binding-defective mutants of Scl/TAL1 (Porcher et al., 1999) and the glucocorticoid receptor (Reichardt et al., 1998) can function in vivo, presumably through binding DNA-bound activators. Thus, GATA-1 tethered to DNA-bound factors might crosslink to sites lacking WGATAR motifs.
De novo motif finding with Cosmo (Bembom et al., 2007) and Meme (Bailey and Elkan, 1995) was used to ascertain the percent of targets containing specific cis-elements. Of the 5,749 ChIP-seq peaks, 5,051 (88%), 155 (3%), 193 (3%), 79 (1%) contained WGATAR, WGATA, GATAR, and WGATA + GATAR motifs (Fig. 3B), respectively, which were conserved similarly from mouse to man. Of the 6,976,111 WGATAR motifs in the human genome, 9,741 (0.14%) reside within the 5,051 peaks (0.07-0.14% occupancy). A small subset of these motifs (6.0%) reside in E-box-WGATAR motifs. De novo motif finding using peaks containing at least one WGATAR identified a highly significant (E-value = 2.8e−3685) position weight matrix with the consensus (C/G)(A/T)GATAA(G/A/C)(G/A/C) (Fig. 3C, left), which occurs 297,124 times in the human genome. Of the 5,051 WGATAR-containing peaks, 3,165 (63%) contain this consensus. Based on 3,165 peaks containing ≥1 copy of the consensus, 0.71% were occupied by GATA-1, an order of magnitude higher than the 0.07% occupancy of WGATAR (p < 0.0001). As the three extended positions (2, 9, 10) exhibit compositions that deviate greatly from the equal probability of bases and the [(A, T): 0.3] and [(C, G):0.2] configuration (p < 1e−100), we further evaluated the significance of this composition by randomly drawing the same number of WGATAR occurrences from the genome and constructing a position weight matrix with WGATAR and its first left and two right flanking positions. We repeated this process 1000 times, and the flanking region information contents were significantly smaller than that of the position weight matrix constructed from WGATAR-containing peaks (p = 0 based on 1000 randomization experiments).
Of the 62,412 E-box-WGATAR composite elements in the human genome, 307 reside within 304 peaks (0.49% occupancy). Analysis of the 301 peaks containing one E-boxWGATAR element revealed the logo in Fig. 3C, right. Positions 16, 23, and 24 resemble the respective positions of the more complex WGATAR motif (Fig. 3C) and deviate from random nucleotides (p < 1e−90). The analysis also revealed unexpected information content in the NN residues of CANNTG (Fig. 3C, right). GATA-1-occupied composite elements had a similar probability of having G or T in the 1st N position and C, A, or G in the 2nd N position (Fig. 3C). Site-selection analysis with recombinant Scl/TAL1-E2A heterodimers identified the consensus AACAGATGGT, and 86-100% of bound sequences had GA in the NN positions (Hsu et al., 1994). The in vitro preference for AA and GT at the 5′ and 3' ends deviated from occupied chromatin sites, which had no sequence preference at the 5′ end and either C, G, or T following TG. Site-selection studies with MEL cell extracts identified E-box-GATA elements with the consensus CAGGTG(N)9GATA (Wadman et al., 1997). The GG sequence in the NN positions differed from GATA-1-occupied E-box-WGATAR elements in chromatin.
De novo motif finding on the 698 peaks lacking WGATAR identified GGAATGGAATG as overrepresented in this group. This sequence appears 3 and 64 times in WGATAR-containing and -lacking peaks, respectively (p < 2.2e−16). GGAATGGAATG resides in microsatellite repeats (Gangwal et al., 2008) and contains the Ets binding motif GGAA (Sharrocks, 2001). The oncogenic Ewings Sarcoma protein fusion to FLI functions through this sequence (Gangwal et al., 2008). GAATGGAATGGAAT-containing GATA-1 occupancy sites defined by ChIP-seq lack WGATAR motifs. As GATA factors physically associate with Ets factors (Rekhtman et al., 1999), a DNA-bound Ets factor might tether GATA-1 to chromatin at this class of sites.
To assess whether the ChIP-seq peaks pinpoint GATA-1-regulated genes, gene expression was profiled in untreated and β-estradiol-treated G1E-ER-GATA-1 cells (Suppl. Table 3). ER-GATA-1 induced and repressed 1,166 and 1,010 genes, respectively, >1.5 fold. Merging ChIP-seq and profiling datasets revealed 142 and 154 activated and repressed genes, respectively, which were GATA-1-occupied (the top 60 are shown in Fig. 3D). These genes include known GATA-1 targets, such as Slc4a1 and Epb4.9 (Kim et al., 2007) encoding red cell cytoskeletal proteins (Mohandas and Gallagher, 2008), yet many had not been implicated in GATA-1 function or hematopoiesis. 44% of the genes (Fig. 3D) were not described in a prior profiling analysis in this system (Welch et al., 2004). In primary Ter119+ bone marrow erythroblasts, GATA-1 occupied 32/36 of the sites significantly higher than the negative control Ey promoter and 14/36 higher than the positive control β-globin HS2, respectively (p < 0.05) (Fig. 3E). Using tiled microarrays containing sequences from 120 genes of Fig. 3D (with 120,000 bp of upstream and downstream sequence), ChIP-chip analysis in primary Ter119+ bone marrow erythroblasts revealed significant GATA-1 occupancy at 90% of the targets (98% and 82% of activated and repressed targets, respectively) (Fig. 3F, Suppl. Table 4).
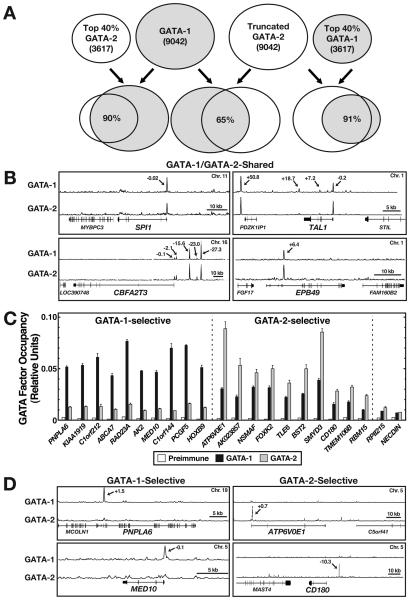
Our previous comparison of GATA-1 and GATA-2 occupancy at several loci in G1E and G1E-ER-GATA-1 cells revealed that almost all of the GATA-1-occupied sites were occupied by GATA-2 in uninduced G1E-ER-GATA-1 cells and G1E cells. To assess the extent of overlap in a cell expressing endogenous GATA-1 and GATA-2, we analyzed GATA-2 occupancy by ChIP-seq in K562 cells. Analysis of two biological replicates (21,167 peaks) (Suppl. Table 5) revealed major overlap (Fig. 4A, B). Since the GATA-1 and GATA-2 samples analyzed by ChIP-seq were isolated on different days, we conducted quantitative ChIP analysis for GATA-1 and GATA-2 at the same time. Sampling representative GATA-1- and GATA-2-unique peaks identified GATA-1- and GATA-2-selective targets (Fig. 4C, D). The extensive sharing of sites by GATA-1 and GATA-2 provides insights into the finding that GATA-1 and GATA-2 function redundantly to generate primitive erythroblasts (Fujiwara et al., 2004). Our analysis also revealed GATA factor-selective targets, including: a kinase critical for controlling hematopoiesis (AK2) (Lagresle-Peyrou et al., 2009); a cell type-specific component of the Mediator complex (MED10) that regulates Wnt and Nodal signaling (Lin et al., 2007b); a Hox gene (HOXB9) induced by Wnt signaling (Nguyen et al., 2009); a Forkhead transcription factor (FOXK2); a factor (BST2) that suppresses HIV-1 release from the cell surface (Goffinet et al., 2009) and is downregulated by Kaposi's sarcoma herpesvirus (Mansouri et al., 2009); a Set domain-containing histone H3K4 methyltransferase (SMYD3) (Nguyen et al., 2009); and a putative RNA binding protein (RBM15) that regulates Notch signaling (Ma et al., 2007), controls hematopoiesis (Raffel et al., 2007) and is implicated in acute megakaryoblastic leukemia (Ma et al., 2001). As GATA-1 mutations are linked to acute megakaryoblastic leukemia (Wechsler et al., 2002), and GATA-1 represses GATA2 transcription (Grass et al., 2003), our discovery of RBM15 as a GATA-2-selective target is intriguing.
Figure 4. Comparison of GATA-1 and GATA-2 Chromatin Occupancy Genome-Wide by ChIP-seq.
(A) Peaks were called on the merged GATA-1 and merged GATA-2 replicate datasets (9,042 and 21,167 for GATA-1 and GATA-2, respectively). GATA-2 peaks were ranked and truncated to the size of the GATA-1 peak list. A comparison of the 9,042 GATA-1 and GATA-2 peaks revealed 65% overlap. Using the ENCODE overlap rule, the top 40% of each peak file was compared to the entire set of 9,042 peaks for the other factor. A 90-91% overlap was observed, indicating that the majority of the highest ranked peaks for each factor are contained within the peak set for the other factor. When the ENCODE overlap rule is applied to replicate datasets of the same factor, high quality datasets often overlap by 80-90%. (B) ChIP-seq signal maps for GATA-1/GATA-2-shared targets. Arrows, ChIP-seq peak locations relative to the transcription start site (kb). (C) Quantitative ChIP analysis of GATA factor occupancy at GATA-1- and GATA-2-selective targets in K562 cells (mean +/− SE, 3 independent experiments). (D) ChIP-seq signal maps for GATA-1- and GATA-2-selective targets. Arrows, ChIP-seq peak locations relative to the transcription start site (kb).
Linking GATA-2 and Scl/TAL1 to Regulation of the Hematopoietic Corepressor ETO-2, a Component of GATA Factor and Scl/TAL1 Complexes
ChIP-seq identified five GATA factor-bound sites at CBFA2T3 (Fig. 4B), which encodes ETO-2, a co-repressor that controls hematopoiesis. We tested whether GATA factors occupy all or a subset of conserved WGATAR motifs at and near Cbfa2t3. Conserved WGATAR motifs reside at −37.3, −35.7, −21.7, −21.6, −21.6, −21.5, −13.3, −1.9, −0.2, +13.3 and +15.1 kb relative to the start site (Fig. 5A). Conservation of the immediate region was high (>75%) at −35.7, −21.7, −21.6, −21.6, −21.5, −13.3, −1.9 and −0.2 kb, intermediate (>50%) at +13.3 kb, and low (<50%) at −37.3 and +15.1 kb (Fig. 5A). Among five ChIP-seq peaks at human CBFA2T3 (Fig. 4B), −27.3, −15.6, −2.1, and −0.1 kb peaks corresponded to −21.6, −13.3, −1.9 kb, and the promoter of murine Cbfa2t3 (Fig. 5A). GATA-2 occupied −37.3, −21.6, −13.3, −1.9, −0.2, and +13.3 kb sites (Suppl. Fig. 2A).
Figure 5. ETO-2 Negative Autoregulatory Loop.
(A) Cbfa2t3 locus organization (UCSC genome assembly: uc009ntn.1). Open and filled boxes, noncoding and coding exons, respectively. Asterisk, locations of conserved E-boxWGATAR motif; downward pointing arrows, WGATAR motifs conserved from mouse to human. Four WGATAR motifs located at −21.7, −21.6, −21.6, and −21.5 kb were analyzed as a cluster. The VISTA plot depicts sequence identity between mouse and human, using mouse as a reference. (B) Quantitative RT-PCR analysis of Cbfa2t3 mRNA (left) (mean +/− SE, 4 independent experiments) and anti-ETO-2 Western blot of whole cell extracts (right) from G1E cells transfected with siRNA against mouse Cbfa2t3 or control siRNA. Asterisk, cross-reactive band. (C) Analysis of ETO-2 occupancy at the Cbfa2t3 locus (−21.6 kb) in ETO-2-knockdown and control G1E cells (mean +/− SE, 2 independent experiments). (D) Cbfa2t3 primary transcripts were quantitated by real-time RT-PCR analysis in control and ETO-2-knockdown G1E cells (mean +/− SE, 4 independent experiments). Two primer sets (Intron1/Intron1 and Intron11/Exon12) were used. Gapdh mRNA was quantitated as a control. (E) Analysis of Pol II and P-Ser5-Pol II occupancy at Cbfa2t3 (left and middle) and RPII215 (right) promoters in ETO-2-knockdown and control G1E cells (mean +/− SE, 2 independent experiments). Two primer sets were used to analyze the Cbfa2t3 promoter. (F) Analysis of AcH3 at the Cbfa2t3 −21.6 kb (left), Cbfa2t3 promoter-B (middle), and RPII215 promoter (right) in ETO-2-knockdown and control G1E cells (mean +/− SE, 2 independent experiments).
Scl/TAL1 forms a complex with E2A, LMO2, and Ldb1 (Lahlil et al, 2004; Wadman et al., 1997), which co-localizes with GATA-2 (Wozniak et al., 2008) and GATA-1 (Anguita et al., 2004; Lahlil et al., 2004; Tripic et al., 2008; Xu et al., 2003) in chromatin. We tested whether this complex resides at GATA-2-occupied regions of Cbfa2t3. Scl/TAL1 occupied all except the −37.3, −35.7, and +15.1 kb sites (Suppl. Fig. 2B). The Scl/TAL1-interacting factor ETO-2 (Schuh et al., 2005) occupied only the Scl/TAL1-bound sites (Suppl. Fig. 2C). The occupancy of each factor correlated with the others (Suppl. Fig. 2D).
Regulatory Interactions Among Components of Complexes Containing Master Regulators of Hematopoiesis and Instigators of Leukemogenesis
To test whether ETO-2 occupancy at Cbfa2t3 (Fig. 5A, Suppl. Fig. 2C) reflects negative or positive autoregulation, ETO-2 was knocked-down in G1E cells with Cbfa2t3 siRNA. Cbfa2t3 primary transcripts were quantitated as a metric of transcription. Western blotting and real-time RT-PCR revealed a knockdown of ETO-2 protein and Cbfa2t3 mRNA, respectively (Fig. 5B), and ETO-2 occupancy at −21.6 kb was reduced by 74% (p = 0.002) (Fig. 5C). The knockdown increased Cbfa2t3 primary transcripts (58%, p = 0.01; 71%; p = 0.00009) (Fig. 5D), suggesting that ETO-2 represses Cbfa2t3. To further evaluate negative autoregulation, we measured Pol II and Ser 5-phosphorylated Pol II (P-Ser5-Pol II) at the Cbfa2t3 promoter. The knockdown increased Pol II and P-Ser5-Pol II occupancy at two sites (~2.5 and ~2 fold, respectively, p < 0.05), without affecting occupancy at the RPII215 promoter (Fig. 5E). As ETO-2 interacts with class I HDACs (Amann et al., 2001), we asked whether knocking-down ETO-2 affects histone acetylation at Cbfa2t3. Acetylated histone H3 increased at −21.6 kb (p = 0.015) and the promoter (p = 0.016), but not at the RPII215 promoter (Fig. 5F). These results establish an ETO-2 negative autoregulatory loop.
ETO-2-mediated negative autoregulation might reflect a non-redundant repressor function at all of its target genes. We asked whether ETO-2 occupied and regulated other GATA factor targets (Fig. 6A), including Scl/TAL1 (Lugus et al., 2007) and Lmo2 (Landry et al., 2009). ETO-2 occupied these sites with a level comparable to that at Cbfa2t3 in G1E cells (Fig. 6B, Suppl. Fig. 2C). ETO-2 knockdown induced expression of certain GATA factor targets (Icam4, Epb4.9, Slc4a1, μmajor, Eraf, and Alas2), while others were unaffected (Fig. 6C). Similarly, ETO-2 knockdown facilitated ER-GATA-1-mediated activation of certain, but not all, targets (Fig. 6D). The knockdown might not reduce ETO-2 below a threshold at which occupancy at all targets would be impaired. If ETO-2 interacts with targets in different chromatin environments with distinct affinities, this could explain the differential sensitivities. Thus, knocking-down ETO-2 would reduce its concentration sufficiently to impair occupancy and regulation at sites with the lowest apparent affinities. However, the knockdown significantly decreased ETO-2 occupancy at sensitive and resistant targets (Fig. 6B), inconsistent with this possibility.
Figure 6. Context-Dependent ETO-2 Corepressor Function at Endogenous Loci.
(A) The diagram depicts the location of the ChIP-seq peaks analyzed. Asterisk, location of the corresponding peak in mouse; shaded boxes, coding regions. (B) Analysis of ETO-2 occupancy at GATA target genes in G1E cells transfected with control or Cbfa2t3 siRNA (mean +/− SE, 2 independent experiments). Asterisk, p < 0.05. (C, D) Real-time RT-PCR analysis of GATA target genes in G1E cells (C) and G1E-ER-GATA1 cells (D) transfected with control or Cbfa2t3 siRNA. For G1E-ER-GATA1 cells, β-estradiol was added 24 h after the initial siRNA transfection and cells were cultured for 24 h. mRNA levels were normalized to Gapdh mRNA (mean +/− SE, 3 and 4 independent experiments for Fig. 6C and 6D, respectively). The relative transcript level for control G1E-ER-GATA-1 cells was designated as 1. Asterisk, p < 0.05. (E) Models for ETO-2-dependent repression of Slc4a1. GATA-2 occupies the repressed Slc4a1 promoter (left), and ETO-2 loss suffices for induction (right, upper) or is coupled to GATA-2 loss (right, lower). (F) Analysis of GATA-2 occupancy at the Slc4a1 promoter in G1E cells transfected with control or Cbfa2t3 siRNA (mean +/− SE, 2 independent experiments).
Since Slc4a1, which is strongly induced by the ETO-2 knockdown, is bound by GATA-2 in the repressed state, ETO-2 loss might suffice for induction, or might be inextricably coupled to GATA-2 loss (Fig. 6E). The ETO-2 knockdown induced ETO-2, but not GATA-2, loss from the Slc4a1 promoter (Fig. 6F), indicating that transcriptional activation solely requires ETO-2 eviction.
Despite the lack of GATA-1 in G1E cells, knocking-down ETO-2 induced certain GATA-1 targets. GATA-1-mediated activation might therefore require ETO-2 displacement. However, GATA-1 activation of Slc4a1 in G1E-ER-GATA-1 cells is associated with increased ETO-2 occupancy at its promoter (data not shown). Thus, GATA-1-associated coactivators dominate over ETO-2 corepressor activity, negating the need to evict ETO-2, or ETO-2 has dual corepressor/coactivator activities.
GATA-1, GATA-2, and Scl/TAL1 occupancy at Cbfa2t3 suggested that these factors regulate Cbfa2t3 transcription. Interactions among components of GATA factor – Scl/TAL1 complexes include GATA-1 repression of Gata2 (Grass et al., 2003), Scl/TAL1 induction of Gata2 (Lugus et al., 2007), and GATA-2 induction of Scl/TAL1 (Chan et al., 2007; Lugus et al., 2007; Wozniak et al., 2008). We used knockout and conditional overexpression approaches in ES cells to examine all possible regulatory influences of GATA-1, GATA-2 and Scl/TAL1 on Gata1, Gata2, Scl/TAL1, Lmo2, Ldb1 and Cbfa2t3 expression. Gene expression was analyzed upon conditional expression of GATA-2 and Scl/TAL1 during ES cell differentiation (Lugus et al., 2007) and in Gata2−/− and Scl/TAL1−/− vs. wild-type EBs. Dox-mediated induction of GATA-2 induced Gata1, Cbfa2t3, and Lmo2 (383, 6.2, and 5.1-fold; p = 0.003, 0.03, and 0.002, respectively) (Fig. 7A). Previously, we demonstrated that GATA-2 increases Scl/TAL1 expression in this system 12-fold (Wozniak et al., 2008). Gata1, Cbfa2t3, Scl/TAL1, and Lmo2 were weakly to modestly downregulated in Gata2-null EBs (p = 0.0055, 0.09, 0.20 and 0.58, respectively) (Fig. 7A). As GATA-3 can compensate for GATA-2 (Kobayashi-Osaki et al., 2005), this might limit the downregulation. In Gata2-null cells, reduced Cbfa2t3 expression would abrogate the negative autoregulatory loop. The altered Cbfa2t3 expression in GATA-2-expressed and Gata2-null ES cells, as well as in G1E-ERGATA-1 cells, establishes Cbfa2t3 as a GATA factor target. Dox-mediated induction of Scl/TAL1 increased Gata2 (p < 0.001) and Lmo2 expression (p < 0.0001), and their expression, as well as that of Gata1, was downregulated in Scl/TAL1-null EBs (p = 0.0044, 0.0089, and 0.04 respectively). Cbfa2t3 was repressed in Scl-null EBs (p = 0.025), and Scl/TAL1 induction in iSCL EBs repressed its expression (-2.2 fold, p < 0.0001) (Fig. 7A).
Figure 7. Regulatory Interactions Among Components of Complexes Containing Master Regulators of Hematopoiesis and Leukemogenic Factors.
(A) The table summarizes changes in the expression of genes in day 3/4, 6 and 8 EBs derived from mouse ES cells following Dox-mediated GATA-2 induction (mean +/− SE, 9 independent experiments for day 3/4 and day 6 EBs; 6 independent experiments for day 8 EBs), day 4 EBs from mouse ES cells following Dox-mediated Scl/TAL1 induction (mean +/− SE, 3 independent experiments), and G1E-ER-GATA1 cells after β-estradiol-treatment (1 μM for 3, 8 and 24 h; mean +/− SE, 2 independent experiments). For EBs derived from Gata2−/− and Scl/TAL1−/− ES cells, values are expressed relative to that of wild-type EBs, which has a value of 1.0. mRNA levels were quantitated by real-time RT-PCR, and the expression level was divided by that of corresponding control cells. (B) The model summarizes cross-regulatory interactions among Scl/TAL1 complex components. (C) Model demonstrating GATA-2 activation of Cbfa2t3 transcription, ETO-2 negative autoregulatory loop, and GATA switch-mediated Cbfa2t3 repression.
DISCUSSION
GATA Factor Chromatin Occupancy Rules
The genome-wide analysis of GATA-1 chromatin occupancy identified a consensus element as a hallmark of the repertoire of GATA-1 chromatin occupancy sites, which changes the paradigm of how GATA-1 selects sequences at target genes. Contrasting with naked DNA binding in which WGATAR is believed to be sufficient, specific nucleotides at the 5′ and 3′ flanks of WGATAR were preferred, and A dominated in the R position, both with WGATAR alone and E-box-WGATAR elements, yielding the chromatin occupancy consensus (C/G)(A/T)GATAA(G/A/C)(G/A/C) (Fig. 3C). Within GATA-1-occupied E-box-WGATAR elements, the NN residues of the CANNTG E-box consensus exhibited significant sequence preferences. As FOG-1 facilitates GATA-1 chromatin occupancy (Letting et al., 2004; Pal et al., 2004a), and FoxA1 stabilizes GATA-4 chromatin complexes (Sekiya et al., 2009), protein-protein interactions are also important determinants. An obvious candidate for controlling GATA factor occupancy at E-box-WGATAR composite elements is the E-box, although GATA-1 occupancy was only slightly greater at composite elements vs. WGATAR motifs.
The localization of 90% of the occupied sites away from promoters (Fig. 3A) indicates that a canonical mode of GATA factor function involves long-range control. GATA-1 induces looping at the β-globin locus (Kim et al., 2007; Kim et al., 2009; Vakoc et al., 2005) and alters a pre-existing loop at c-kit (Jing et al., 2008), but whether looping is common or infrequent for GATA factors was unknown. Estrogen receptor-α induces looping (Carroll et al., 2005), and genome-wide analyses of estrogen receptor-α chromatin occupancy (Lin et al., 2007a) revealed abundant non-promoter sites. By contrast, >80% of E2F1 targets are promoters (Bieda et al., 2006). While GATA-1 (Blobel et al., 1998) and E2F1 (Fry et al., 1999; Trouche and Kouzarides, 1996) utilize CBP/p300 to regulate transcription, GATA-1 uniquely utilizes FOG-1 (Crispino et al., 1999) and MED1 (Stumpf et al., 2006). The primary sequence determinants and topographic constraints constitute chromatin occupancy rules that govern how GATA-1 establishes genetic networks that control critical processes.
Analysis of histone modification patterns in K562 cells generated by the ENCODE project with the identical line of K562 cells used in our ChIP-seq analysis (UCSC Genome Browser) revealed GATA-1 occupancy in introns that was often mutually exclusive with histone H3K36 trimethylation (Suppl. Fig. 3). Trimethylated H3K36 marks open reading frames, facilitates HDAC recruitment, and prevents aberrant transcription initiation (Carrozza et al., 2005; Li et al., 2007). Acetylated H3K9, dimethylated H3K4, and monomethylated K3K4 were commonly enriched at GATA-1 occupancy sites (Suppl. Fig. 3). This result suggests that H3K36 trimethylation is incompatible with GATA-1 function at intronic sites. Since H3K36 trimethylation mediates HDAC recruitment (Li et al., 2007), H3K36 trimethylation-dependent deacetylation at GATA-1-bound introns might oppose GATA-1-induced looping and/or intergenic transcription (Kim et al., 2009). Alternatively, H3K36 trimethylation or the associated HDAC recruitment might disfavor the assembly of functional GATA factor complexes within introns. As mutually exclusive factor occupancy and H3K36 trimethylation had not been described, it will be important to assess whether this is unique to GATA factors or can be applied in a broader context.
Biological Insights Derived from the Genomic Screen
Mining the consolidated ChIP-seq and transcriptional profiling dataset revealed a rich set of GATA factor-shared and -selective targets, which were validated with high fidelity in primary erythroblasts. Many of these targets had not been implicated in GATA factor function or hematopoiesis, while others, such as RBM15, had been implicated in hematopoiesis and leukemogenesis, but were not known to function in GATA factor pathways. Our GATA-1 and GATA-2 ChIP-seq data provides a important resource for elucidating hematopoietic regulatory mechanisms.
Using rigorous computational and mechanistic analyses, our results establish a conceptual framework for understanding how the actions of GATA factors, Scl/TAL1, and ETO-2 are integrated to establish a genetic network that controls hematopoiesis and instigates leukemogenesis. GATA-1 directly represses Gata2 by displacing GATA-2 from this locus (Grass et al., 2003; Grass et al., 2006; Martowicz et al., 2005). Given the short t1/2 of GATA-2 (Lurie et al., 2008), GATA switches rapidly yield cells solely expressing GATA-1. By contrast to GATA-2 and GATA-1, which are expressed early and late in hematopoiesis, respectively, Scl/TAL1 is expressed in both stages (Begley et al., 1989; Lecuyer et al., 2002; Porcher et al., 1996; Schuh et al., 2005; Shivdasani et al., 1995). Though the mechanism by which LDB1 and Lmo2 mediate Scl/TAL1 function is unclear, ETO-2 is an attractive candidate for differentially controlling Scl/TAL1 activity during hematopoiesis. Reduced ETO-2 expression during erythropoiesis (Goardon et al., 2006) can be explained by our discovery that GATA-2 directly induces ETO-2, which is followed by ETO-2 negative autoregulation and GATA switch-mediated ETO-2 repression (Fig. 7B,C).
Our loss-of-function and gain-of-function studies define fundamental insights into how cell type-specific trans-acting factors function combinatorially to control a complex developmental process. The specific interactions include: GATA-2 induction of interacting trans-acting factors (Scl/TAL1, GATA-1, ETO-2) and an interacting co-repressor (LMO2); GATA-1 repression of GATA-2; GATA-1 repression of ETO-2, and ETO-2 negative autoregulation (Fig. 7B,C). Since GATA-2 and Scl/TAL1 co-localize (Wozniak et al., 2008) on chromatin, and ETO-2 antagonizes Scl/TAL1 (Goardon et al., 2006; Schuh et al., 2005), ETO-2 indirectly inhibits GATA-2. Accordingly, ETO-2 counteracts GATA-2-mediated induction of GATA-1, and given that GATA-1 represses ETO-2 expression, the balance between these positive and negative interactions must be strictly managed and dynamically regulated to ensure high fidelity of hematopoiesis. The elaborate integration of the activities of GATA factors, Scl/TAL1, and ETO-2, ensures the efficient and rapid transition from a GATA-2- to a GATA-1-driven genetic network. The differential co-repressor function of ETO-2 at endogenous targets, including key regulators of erythropoiesis and erythroid cell function, suggests that minimizing ETO-2 level/activity during later stages of hematopoiesis enables GATA-1 and Scl/TAL1 to efficiently establish the genetic network of the developing red blood cell. Further mining of our highly validated resource is expected to reveal additional fundamental insights into hematopoiesis and a broader spectrum of important biological processes.
EXPERIMENTAL PROCEDURES
Cell Culture
GATA-1-null G1E (Weiss et al., 1997) and G1E-ER-GATA-1 cells (Gregory et al., 1999; Johnson et al., 2002) were maintained as described (Gregory et al., 1999; Johnson et al., 2002) and in Supplementary Experimental Procedures. ES cells were cultured and differentiated as described (Park et al., 2004) and in Supplemental Experimental Procedures.
Antibodies
Antibodies are described in Supplementary Experimental Procedures.
Quantitative ChIP Assay
Quantitative chromatin immunoprecipitation (ChIP) analysis was conducted and validated as described (Im et al., 2004).
ChIP-Seq Cell and Data Processing
ChIP-Seq analysis was conducted as described in Supplementary Experimental Procedures.
siRNA-mediated Knockdown
The knockdown was conducted as described in Supplementary Experimental Procedures.
Quantitative Analysis of RNA and Protein
Quantitative RT-PCR analysis was conducted as described in Supplementary Experimental Procedures
Isolation of Primary Murine Bone Marrow Erythroid Precursors
Murine bone marrow erythroblasts were separated by magnetic cell sorting system (Miltenyi Biotec) using anti-Ter119 microbeads (Miltenyi Biotec).
ChIP-chip Analysis
A tiled microarray was constructed by Nimblegen, which contains sequences from 120 GATA-1 target genes (Fig. 3D), including 120,000 bp of upstream and downstream sequence, and ChIP-chip was conducted as described in Supplementary Experimental Procedures.
Supplementary Material
ACKNOWLEDGEMENTS
This work was funded by NIH grants DK50107 (EHB), DK68634 (EHB), HG003747 (SK), HL55337 (KC) and 1U54HG004558 (PJF). We thank Stuart Orkin for providing Gata2−/− and Scl−/− ES cells.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Amann JM, Nip J, Strom DK, Lutterbach B, Harada H, Lenny N, Downing JR, Meyers S, Hiebert SW. ETO, a target of t(8:21) in acute leukemia, makes distinct contacts with multiple histone deacetylases and binds Sin3A through its oligomerization domain. Mol Cell Biol. 2001;21:6470–6483. doi: 10.1128/MCB.21.19.6470-6483.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anguita E, Hughes J, Heyworth C, Blobel GA, Wood WG, Higgs DR. Globin gene activation during hematopoiesis is driven by protein complexes nucleated by GATA-1 and GATA-2. EMBO J. 2004;23:2841–2852. doi: 10.1038/sj.emboj.7600274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aplan PD, Lombardi DP, Reaman GH, Sather HN, Hammond GD, Kirsch IR. Involvement of the putative hematopoietic transcription factor SCL in T- cell acute lymphoblastic leukemia. Blood. 1992;79:1327–1333. [PubMed] [Google Scholar]
- Bailey TL, Elkan C. Unsupervised learning of multiple motifs in biopolymers using EM. Machine Learning. 1995;21:51–80. [Google Scholar]
- Begley CG, Aplan PD, Denning SM, Haynes BF, Waldmann TA, Kirsch IR. The gene SCL is expressed during early hematopoiesis and encodes a differentiation-related DNA-binding motif. Proc Natl Acad Sci U S A. 1989;86:10128–10132. doi: 10.1073/pnas.86.24.10128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bembom O, Keles S, van der Laan MJ. Supervised detection of conserved motifs in DNA sequences with cosmo. Statistical Applications in Genetics and Molecular Biology. 2007;6 doi: 10.2202/1544-6115.1260. Article 8. [DOI] [PubMed] [Google Scholar]
- Bieda M, Xu X, Singer MA, Green R, Farnham PJ. Unbiased location analysis of E2F1-binding sites suggests a widespread role for E2F1 in the human genome. Genome Res. 2006;16:595–605. doi: 10.1101/gr.4887606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel GA, Nakajima T, Eckner R, Montminy M, Orkin SH. CREB-binding protein cooperates with transcription factor GATA-1 and is required for erythroid differentiation. Proc Natl Acad Sci USA. 1998;95:2061–2066. doi: 10.1073/pnas.95.5.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor AB, Orkin SH. Transcriptional regulation of erythropoiesis: an affair involving multiple partners. Oncogene. 2002;21:3368–3376. doi: 10.1038/sj.onc.1205326. [DOI] [PubMed] [Google Scholar]
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, et al. Chromosome mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Fiorens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- Chan WY, Follows GA, Lacaud G, Pimanda JE, Landry JR, Kinston S, Knezevic K, Piltz S, Donaldson IJ, Gambardella L, et al. The paralogous hematopoietic regulators Lyl1 and SCL are co-regulated by Ets and GATA factors yet Lyl1 cannot rescue the early SCL−/− phenotype. Blood. 2007;109:1908–1916. doi: 10.1182/blood-2006-05-023226. [DOI] [PubMed] [Google Scholar]
- Chyla BJ, Moreno-Miralles I, Steapleton MA, Thompson MA, Bhaskara S, Engel M, Hiebert SW. Deletion of Mtg16, a target of t(16:21), alters hematopoietic progenitor cell proliferation and lineage allocation. Mol Cell Biol. 2008;28:6234–6247. doi: 10.1128/MCB.00404-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crispino JD. GATA-1 in normal and malignant hematopoiesis. Semin Cell Dev Biol. 2005;16:137–147. doi: 10.1016/j.semcdb.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Crispino JD, Lodish MB, MacKay JP, Orkin SH. Use of altered specificity mutants to probe a specific protein-protein interaction in differentiation: the GATA-1:FOG complex. Mol Cell. 1999;3:219–228. doi: 10.1016/s1097-2765(00)80312-3. [DOI] [PubMed] [Google Scholar]
- Dorfman DM, Wilson DB, Bruns GA, Orkin SH. Human transcription factor GATA-2. Evidence for regulation of preproendothelin-1 gene expression in endothelial cells. J Biol Chem. 1992;267:1279–1285. [PubMed] [Google Scholar]
- Evans T, Reitman M, Felsenfeld G. An erythrocyte-specific DNA-binding factor recognizes a regulatory sequence common to all chicken globin genes. Proc Natl Acad Sci U S A. 1988;85:5976–5980. doi: 10.1073/pnas.85.16.5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry CJ, Pearson A, Malinowski E, Bartley SM, Greenblatt J, Farnham PJ. Activation of the murine dihydrofolate reductase promoter by E2F1. A requirement for CBP recruitment. J Biol Chem. 1999;274:15883–15891. doi: 10.1074/jbc.274.22.15883. [DOI] [PubMed] [Google Scholar]
- Fujiwara Y, Chang AN, Williams AM, Orkin SH. Functional overlap of GATA-1 and GATA-2 in primitive hematopoietic development. Blood. 2004;103:583–585. doi: 10.1182/blood-2003-08-2870. [DOI] [PubMed] [Google Scholar]
- Gamou T, Kitamura E, Hosoda F, Shimizu K, Shinsohara K, Hayashi Y, Nagese T, Yokoyama Y, Ohki M. The partner gene of AML1 in t(16:21) myeloid malignancies is a novel member of the MTG8(ETO) family. Blood. 1998;91:4028–4037. [PubMed] [Google Scholar]
- Gangwal K, Sankar S, Hollenhorst PC, Kinsey M, Haroldsen SC, Shah AA, Boucher KM, Watkins WS, Jorde LB, Graves BJ, Lessnick SL. Microsatellites as EWS/FLI response elements in Ewing's sarcoma. Proc Natl Acad Sci U S A. 2008;105:10149–10154. doi: 10.1073/pnas.0801073105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goardon N, Lambert JA, Rodriguez P, Nissaire P, Herblot S, Thibault P, Dumenil D, Strouboulis J, Romeo PH, Hoang T. ETO2 coordinates cellular proliferation and differentiation during erythropoiesis. EMBO J. 2006;25:357–366. doi: 10.1038/sj.emboj.7600934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffinet C, Allespach I, Homann S, Tervo HM, Habermann A, Rupp D, Oberbremer L, Kern C, Tibroni N, Welsch S, et al. HIV-1 antagonism of CD317 is species specific and involves Vpu-mediated proteasomal degradation of the restriction factor. Cell Host Microbe. 2009;5:285–297. doi: 10.1016/j.chom.2009.01.009. [DOI] [PubMed] [Google Scholar]
- Gottgens B, Nastos A, Kinston S, Piltz S, Delabesse EC, Stanley M, Sanchez MJ, Ciau-Uitz A, Patient RK, Green AR. Establishing the transcriptional programme for blood: the SCL stem cell enhancer is regulated by a multiprotein complex containing Ets and GATA factors. EMBO J. 2002;21:3039–3050. doi: 10.1093/emboj/cdf286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grass JA, Boyer ME, Pal S, Wu J, Weiss MJ, Bresnick EH. GATA-1-dependent transcriptional repression of GATA-2 via disruption of positive autoregulation and domain-wide chromatin remodeling. Proc Natl Acad Sci U S A. 2003;100:8811–8816. doi: 10.1073/pnas.1432147100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grass JA, Jing H, Kim S-I, Martowicz ML, Pal S, Blobel GA, Bresnick EH. Distinct functions of dispersed GATA factor complexes at an endogenous gene locus. Mol Cell Biol. 2006;26:7056–7067. doi: 10.1128/MCB.01033-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory T, Yu C, Ma A, Orkin SH, Blobel GA, Weiss MJ. GATA-1 and erythropoietin cooperate to promoter erythroid cell survival by regulating bcl-xl expression. Blood. 1999;94:87–96. [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, Lim A, Osborne CS, Pawliuk R, Morillon E, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- Hamlett I, Draper J, Strouboulis J, Iborra F, Porcher C, Vyas P. Characterization of megakaryocyte GATA1-interacting proteins: the corepressor ETO2 and GATA1 interact to regulate terminal megakaryocyte maturation. Blood. 2008;112:2738–2749. doi: 10.1182/blood-2008-03-146605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong W, Nakazawa M, Chen YY, Kori R, Vakoc CR, Rakowski C, Blobel GA. FOG-1 recruits the NuRD repressor complex to mediate transcriptional repression by GATA-1. EMBO J. 2005;24:67–78. doi: 10.1038/sj.emboj.7600703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu HL, Huang L, Tsan JT, Funk W, Wright WE, Hu JS, Kingston RE, Baer R. Preferred sequences for DNA recognition by the TAL1 helix-loop-helix proteins. Mol Cell Biol. 1994;14:1256–1265. doi: 10.1128/mcb.14.2.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im H, Grass JA, Johnson KD, Boyer ME, Wu J, Bresnick EH. Measurement of protein-DNA interactions in vivo by chromatin immunoprecipitation. Methods Mol Biol. 2004;284:129–146. doi: 10.1385/1-59259-816-1:129. [DOI] [PubMed] [Google Scholar]
- Im H, Grass JA, Johnson KD, Kim S-I, Boyer ME, Imbalzano AN, Bieker JJ, Bresnick EH. Chromatin domain activation via GATA-1 utilization of a small subset of dispersed GATA motifs within a broad chromosomal region. Proc Natl Acad Sci U S A. 2005;102:17065–17070. doi: 10.1073/pnas.0506164102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing H, Vakoc CR, Ying L, Mandat S, Wang H, Zheng X, Blobel GA. Exchange of GATA factors mediates transitions in looped chromatin organization at a developmentally regulated gene locus. Mol Cell. 2008;29:232–242. doi: 10.1016/j.molcel.2007.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KD, Boyer ME, Kim S-I, Kang SY, Wickrema A, Cantor AB, Bresnick EH. Friend of GATA-1-independent transcriptional repression: a novel mode of GATA-1 function. Blood. 2007;109:5230–5233. doi: 10.1182/blood-2007-02-072983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KD, Grass JD, Boyer ME, Kiekhaefer CM, Blobel GA, Weiss MJ, Bresnick EH. Cooperative activities of hematopoietic regulators recruit RNA polymerase II to a tissue-specific chromatin domain. Proc Natl Acad Sci USA. 2002;99:11760–11765. doi: 10.1073/pnas.192285999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S-I, Bultman SJ, Jing H, Blobel GA, Bresnick EB. Dissecting molecular steps in chromatin domain activation during hematopoietic differentiation. Mol Cell Biol. 2007;27:4551–4565. doi: 10.1128/MCB.00235-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S-I, Bultman SJ, Kiefer CM, Dean A, Bresnick EH. BRG1 requirement for long-range interaction of a locus control region with a downstream promoter. Proc Natl Acad Sci U S A. 2009;106:2259–2264. doi: 10.1073/pnas.0806420106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko LJ, Engel JD. DNA-binding specificities of the GATA transcription factor family. Mol Cell Biol. 1993;13:4011–4022. doi: 10.1128/mcb.13.7.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi-Osaki M, Ohneda O, Suzuki N, Minegishi N, Yokomizo T, Takahashi S, Lim KC, Engel JD, Yamamoto M. GATA motifs regulate early hematopoietic lineage-specific expression of the Gata2 gene. Mol Cell Biol. 2005;25:7005–7020. doi: 10.1128/MCB.25.16.7005-7020.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagresle-Peyrou C, Six EM, Picard C, Rieux-Laucat F, Michel V, Ditadi A, Demerens-de Chappedelaine C, Morillon E, Valensi F, Simon-Stoos KL, et al. Human adenylate kinase deficiency causes a profound hematopoietic defect associated with sensorineural deafness. Nat Genet. 2009;41:106–111. doi: 10.1038/ng.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahlil R, Lecuyer E, Herblot S, Hoang T. SCL assembles a multifactorial complex that determines glycophorin A expression. Mol Cell Biol. 2004;24:1439–1452. doi: 10.1128/MCB.24.4.1439-1452.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry JR, Bonadies N, Kinston S, Knezevic K, Wilson NK, Oram SH, Janes M, Piltz S, Hammett M, Carter J, et al. Expression of the leukaemia oncogene Lmo2 is controlled by an array of tissue specific elements dispersed over 100 kb and bound by Tal1/Lmo2, Ets, and GATA factors. Blood. 2009;113:5783–5792. doi: 10.1182/blood-2008-11-187757. [DOI] [PubMed] [Google Scholar]
- Lecuyer E, Herblot S, Saint-Denis M, Martin R, Begley CG, Porcher C, Orkin SH, Hoang T. The SCL complex regulates c-kit expression in hematopoietic cells through functional interaction with Sp1. Blood. 2002;100:2430–2440. doi: 10.1182/blood-2002-02-0568. [DOI] [PubMed] [Google Scholar]
- Letting DL, Chen YY, Rakowski C, Reedy S, Blobel GA. Context-dependent regulation of GATA-1 by friend of GATA-1. Proc Natl Acad Sci U S A. 2004;101:476–481. doi: 10.1073/pnas.0306315101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Gogol M, Carey M, Lee D, Seidel C, Workman JL. Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science. 2007;316:1050–1054. doi: 10.1126/science.1139004. [DOI] [PubMed] [Google Scholar]
- Lin C-Y, Vega VB, Thomsen JS, Zhang T, Kong SL, Xie M, Chiu KP, Lipovich L, Barnett DH, Stossi F, et al. While-genome cartography of estrogen receptor alpha binding sites. PLoS Genet. 2007a;3:867–885. doi: 10.1371/journal.pgen.0030087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Rinaldo L, Fazly AF, Xu X. Depletion of Med10 enhances Wnt and suppresses Nodal signaling during zebrafish embryogenesis. Dev Biol. 2007b;303:536–548. doi: 10.1016/j.ydbio.2006.11.034. [DOI] [PubMed] [Google Scholar]
- Lozzio CB, Lozzio BB, Machado EA, Fuhr JE, Lair SV, Bamberger EG. Effects of sodium butyrate on human chronic myelogenous leukemia cell line K562. Nature. 1979;281:709–710. doi: 10.1038/281709b0. [DOI] [PubMed] [Google Scholar]
- Lugus JJ, Chung YS, Mills JC, Kim SI, Grass JA, Kyba M, Doherty JM, Bresnick EH, Choi K. GATA2 functions at multiple steps in hemangioblast development and differentiation. Development. 2007;134:393–405. doi: 10.1242/dev.02731. [DOI] [PubMed] [Google Scholar]
- Lurie LJ, Boyer ME, Grass JA, Bresnick EH. Differential GATA factor stabilities: implications for chromatin occupancy by structurally similar transcription factors. Biochem. 2008;47:859–869. doi: 10.1021/bi701692p. [DOI] [PubMed] [Google Scholar]
- Ma X, Renda MJ, Wang L, Cheng EC, Niu C, Morris SW, Chi AS, Krause DS. Rbm15 modulates Notch-induced transcriptional activation and affects myeloid differentiation. Mol Cell Biol. 2007;27:3056–3064. doi: 10.1128/MCB.01339-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Morris SW, Valentine V, Li M, Herbrick JA, Cui X, Bouman D, Li Y, Mehta PK, Nizetic D, et al. Fusion of two novel genes, RBM15 and MKL1 in the t(1:22)(p13:q13) of acute megakaryoblastic leukemia. Nat Genet. 2001;28:220–221. doi: 10.1038/90054. [DOI] [PubMed] [Google Scholar]
- Mansouri M, Viswanathan K, Douglas JL, Hines J, Gustin J, Moses AV, Fruh K. Molecular mechanism of BST2/tetherin downregulation by K5/MIR2 of Kaposi's sarcoma herpesvirus. J Virol July 15. 2009 doi: 10.1128/JVI.00597-09. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martowicz ML, Grass JA, Boyer ME, Guend H, Bresnick EH. Dynamic GATA factor interplay at a multi-component regulatory region of the GATA-2 locus. J Biol Chem. 2005;280:1724–1732. doi: 10.1074/jbc.M406038200. [DOI] [PubMed] [Google Scholar]
- Merika M, Orkin SH. DNA-binding specificity of GATA family transcription factors. Mol Cell Biol. 1993;13:3999–4010. doi: 10.1128/mcb.13.7.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migliaccio AR, Rana RA, Sanchez M, Lorenzini R, Centurione L, Bianchi L, Vannucchi AM, Migliaccio G, Orkin SH. GATA-1 as a regulator of mast cell differentiation revealed by the phenotype of the GATA-1low mouse mutant. J Exp Med. 2003;197:281–296. doi: 10.1084/jem.20021149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohandas N, Gallagher PG. Red cell membrae: past, present, and future. Blood. 2008;112:3939–3948. doi: 10.1182/blood-2008-07-161166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munugalavadla V, Dore LC, Tan BL, Hong L, Vishnu M, Weiss MJ, Kapur R. Repression of c-kit and its downstream substrates by GATA-1 inhibits cell proliferation during erythroid maturation. Mol Cell Biol. 2005;25:6747–6759. doi: 10.1128/MCB.25.15.6747-6759.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DX, Chiang AC, Zhang XH, Kim JY, Kris MG, Ladanyi M, Gerald WL, Massague J. WNT/TCF signaling through LEF1 and HOXB9 mediates lung adenocarcinoma metastasis. Cell. 2009;138:51–62. doi: 10.1016/j.cell.2009.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S, Cantor AB, Johnson KD, Moran T, Boyer ME, Orkin SH, Bresnick EH. Coregulator-dependent facilitation of chromatin occupancy by GATA-1. Proc Natl Acad Sci U S A. 2004a;101:980–985. doi: 10.1073/pnas.0307612100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S, Nemeth MJ, Bodine DM, Miller JL, Svaren J, Thein SL, Lowry PJ, Bresnick EH. Neurokinin-B transcription in erythroid cells: direct activation by the hematopoietic transcription factor GATA-1. J Biol Chem. 2004b;279:31348–31356. doi: 10.1074/jbc.M403475200. [DOI] [PubMed] [Google Scholar]
- Park C, Afrikanova I, Chung YS, Zhang WJ, Arentson E, Fong GH, Rosendahl A, Choi K. A hierarchical order of factors in the generation of FLK1- and SCL-expressing hematopoietic and endothelial progenitors from embryonic stem cells. Development. 2004;131:2749–2762. doi: 10.1242/dev.01130. [DOI] [PubMed] [Google Scholar]
- Pevny L, Simon MC, Robertson E, Klein WH, Tsai SF, D'Agati V, Orkin SH, Costantini F. Erythroid differentiation in chimaeric mice blocked by a targeted mutation in the gene for transcription factor GATA-1. Nature. 1991;349:257–260. doi: 10.1038/349257a0. [DOI] [PubMed] [Google Scholar]
- Porcher C, Liao EC, Fujiwara Y, Zon LI, Orkin SH. Specification of hematopoietic and vascular development by the bHLH transcription factor SCL without direct DNA binding. Development. 1999;126:4603–4615. doi: 10.1242/dev.126.20.4603. [DOI] [PubMed] [Google Scholar]
- Porcher C, Swat W, Rockwell K, Fujiwara Y, Alt FW, Orkin SH. The T cell leukemia oncoprotein SCL/tal-1 is essential for development of all hematopoietic lineages. Cell. 1996;86:47–57. doi: 10.1016/s0092-8674(00)80076-8. [DOI] [PubMed] [Google Scholar]
- Raffel GD, Mercher T, Shigematsu H, Williams IR, Cullen DE, Akashi K, Bernard OA, Gilliland DG. Ott1 (Rbm15) has pleitropic roles in hematopoietic development. Proc Natl Acad Sci U S A. 2007;104:6001–6006. doi: 10.1073/pnas.0609041104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichardt HM, Kaestner KH, Tuckermann J, Kretz O, Wessely O, Bock R, Gass P, Schmid W, Herrlich P, Angel P, Schutz G. DNA binding of the glucocorticoid receptor is not essential for survival. Cell. 1998;93:531–541. doi: 10.1016/s0092-8674(00)81183-6. [DOI] [PubMed] [Google Scholar]
- Rekhtman N, Radparvar F, Evans T, Skoultchi AI. Direct interaction of hematopoietic transcription factors PU.1 and GATA-1: functional antagonism in erythroid cells. Genes Dev. 1999;13:1398–1411. doi: 10.1101/gad.13.11.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherzer CR, Grass JA, Liao Z, Pepivani I, Zheng B, Eklund AC, Ney PA, J N, McGoldrick M, Mollenhauer B, et al. GATA transcription factors directly regulate the Parkinson's disease-linked gene alpha-synuclein. Proc Natl Acad Sci USA. 2008;105:10907–10912. doi: 10.1073/pnas.0802437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh AH, Tipping AJ, Clark AJ, Hamlett I, Guyot B, Iborra FJ, Rodriguez P, Strouboulis J, Enver T, Vyas P, Porcher C. ETO-2 associates with SCL in erythroid cells and megakaryocytes and provides repressor functions in erythropoiesis. Mol Cell Biol. 2005;25:10235–10250. doi: 10.1128/MCB.25.23.10235-10250.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya T, Muthurajan UM, Luger K, Tulin AV, Zaret KS. Nucleosome-binding affinity as a primary determinant of the nuclear mobility of the pioneer transcription factor FoxA. Genes Dev. 2009;23:804–809. doi: 10.1101/gad.1775509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrocks AD. The ETS-domain transcription factor family. Nat Rev Mol Cell Biol. 2001;2:827–837. doi: 10.1038/35099076. [DOI] [PubMed] [Google Scholar]
- Shivdasani RA, Fujiwara Y, McDevitt MA, Orkin SH. A lineage-selective knockout establishes the critical role of transcription factor GATA-1 in mekagaryocyte growth and platelet development. EMBO J. 1997;16:3965–3973. doi: 10.1093/emboj/16.13.3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivdasani RA, Mayer EL, Orkin SH. Absence of blood formation in mice lacking the T-cell leukaemia oncoprotein tal-1/SCL. Nature. 1995;373:432–434. doi: 10.1038/373432a0. [DOI] [PubMed] [Google Scholar]
- Simon MC, Pevny L, Wiles MV, Keller G, Costantini F, Orkin SH. Rescue of erythroid development in gene targeted GATA-1- mouse embryonic stem cells. Nat Genet. 1992;1:92–98. doi: 10.1038/ng0592-92. [DOI] [PubMed] [Google Scholar]
- Stumpf M, Waskow C, Krotschel M, van Essen D, Rodriguez P, Zhang X, Guyot B, Roeder RG, Borggrefe T. The mediator complex functions as a coactivator for GATA-1 in erythropoiesis via subunit Med1/TRAP220. Proc Natl Acad Sci U S A. 2006;103:18504–18509. doi: 10.1073/pnas.0604494103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripic T, Deng W, Cheng Y, Vakoc CR, Gregory GD, Hardison RC, Blobel GA. SCL and associated protein distinguish active from repressive GATA transcription factor complexes. Blood. 2008;113:2191–2201. doi: 10.1182/blood-2008-07-169417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trouche D, Kouzarides T. E2F1 and E1A(12S) have a homologous activation domain regulated by RB and CBP. Proc Natl Acad Sci U S A. 1996;93:1439–1442. doi: 10.1073/pnas.93.4.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai FY, Keller G, Kuo FC, Weiss M, Chen J, Rosenblatt M, Alt FW, Orkin SH. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature. 1994;371:221–226. doi: 10.1038/371221a0. [DOI] [PubMed] [Google Scholar]
- Tsai SF, Martin DI, Zon LI, D'Andrea AD, Wong GG, Orkin SH. Cloning of cDNA for the major DNA-binding protein of the erythroid lineage through expression in mammalian cells. Nature. 1989;339:446–451. doi: 10.1038/339446a0. [DOI] [PubMed] [Google Scholar]
- Tsang AP, Visvader JE, Turner CA, Fujuwara Y, Yu C, Weiss MJ, Crossley M, Orkin SH. FOG, a multitype zinc finger protein as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell. 1997;90:109–119. doi: 10.1016/s0092-8674(00)80318-9. [DOI] [PubMed] [Google Scholar]
- Turner J, Crossley M. Cloning and characterization of mCtBP2, a co-repressor that associates with basic Kruppel-like factor and other mammalian transcriptional regulators. EMBO J. 1998;17:5129–5140. doi: 10.1093/emboj/17.17.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakoc CR, Letting DL, Gheldof N, Sawado T, Bender MA, Groudine M, Weiss MJ, Dekker J, Blobel GA. Proximity among distant regulatory elements at the beta globin locus requires GATA-1 and FOG-1. Mol Cell. 2005;17:453–462. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- Vyas P, McDevitt MA, Cantor AB, Katz SG, Fujiwara Y, Orkin SH. Different sequence requirements for expression in erythroid and megakaryocytic cells within a regulatory element upstream of the GATA-1 gene. Development. 1999;126:2799–2811. doi: 10.1242/dev.126.12.2799. [DOI] [PubMed] [Google Scholar]
- Wadman IA, Osada H, Grutz GG, Agulnick AD, Westphal H, Forster A, Rabbitts TH. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 1997;16:3145–3157. doi: 10.1093/emboj/16.11.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler J, Greene M, McDevitt MA, Anastasi J, J.E K, LeBeau MM, Crispino JD. Acquired mutations in GATA-1 in the megakaryoblastic leukemia of Down syndrome. Nat Genet. 2002;32:148–152. doi: 10.1038/ng955. [DOI] [PubMed] [Google Scholar]
- Weiss MJ, Yu C, Orkin SH. Erythroid-cell-specific properties of transcription factor GATA-1 revealed by phenotypic rescue of a gene-targeted cell line. Mol Cell Biol. 1997;17:1642–1651. doi: 10.1128/mcb.17.3.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch JJ, Watts JA, Vakoc CR, Yao Y, Wang H, Hardison RC, Blobel GA, Chodosh LA, Weiss MJ. Global regulation of erythroid gene expression by transcription factor GATA-1. Blood. 2004;104:3136–3147. doi: 10.1182/blood-2004-04-1603. [DOI] [PubMed] [Google Scholar]
- Wozniak RJ, Boyer ME, Grass JA, Lee Y-S, Bresnick EH. Context-dependent GATA factor function: combinatorial requirements for transcriptional control in hematopoietic and endothelial cells. J Biol Chem. 2007;282:14665–14674. doi: 10.1074/jbc.M700792200. [DOI] [PubMed] [Google Scholar]
- Wozniak RJ, Keles S, Lugus JJ, Young K, Boyer ME, Tran TT, Choi K, Bresnick EH. Molecular hallmarks of endogenous chromatin complexes containing master regulators of hematopoiesis. Mol Cell Biol. 2008;28:6681–6694. doi: 10.1128/MCB.01061-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Huang S, Chang LS, Agulnick AD, Brandt SJ. Identification of a TAL1 target gene reveals a positive role for the LIM domain-binding protein (Ldb1) in erythroid gene expression and differentiation. Mol Cell Biol. 2003;23:7585–7599. doi: 10.1128/MCB.23.21.7585-7599.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Cantor AB, Yang H, Browne C, Wells RA, Fukiwara Y, Orkin SH. Targeted deletion of a high-affinity GATA-binding site in the GATA-1 promoter leads to selective loss of the eosinophil lineage in vivo. J Exp Med. 2002;195:1387–1395. doi: 10.1084/jem.20020656. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.