SUMMARY
Formins stimulate actin filament assembly for fundamental cellular processes including division, adhesion, establishing polarity and motility. A formin inhibitor would be useful because most cells express multiple formins whose functions are not known, and because metastatic tumor formation depends upon the deregulation of formin-dependent processes. We identified a general small molecule inhibitor of formin homology 2 domains (SMIFH2) by screening compounds for the ability to prevent formin-mediated actin assembly in vitro. SMIFH2 targets formins from evolutionarily diverse organisms including yeast, nematode worm and mice, with a half-maximal inhibitor concentration of ~5 to 15 μM. SMIFH2 prevents both formin nucleation and processive barbed-end elongation, and decreases formin’s affinity for the barbed end. Furthermore, low micromolar concentrations of SMIFH2 disrupt formin-dependent, but not Arp2/3 complex-dependent, actin cytoskeletal structures in fission yeast and mammalian NIH 3T3 fibroblasts.
INTRODUCTION
Cells assemble diverse actin-dependent structures for a variety of fundamental processes, each of which is thought to be reliant upon specific actin nucleation factors such as the Arp2/3 complex, spire and formin (Chhabra and Higgs, 2007). Determining which factors drive actin filament assembly for particular cellular functions is daunting. Formins are evolutionarily conserved proteins that stimulate actin assembly for a variety of processes including division, motility, establishing polarity, stress fiber formation, focal adhesions and cell-to-cell adhesions (Faix and Grosse, 2006; Goode and Eck, 2007; Higgs, 2005), which are often deregulated during tumor cell transformation and metastasis (Sahai, 2005). Not surprising then, formins are involved in malignant tumor function and are highly overexpressed in different cancer cell types including colorectal, carcinoma, leukemia, melanoma, and lymphoid (Favaro et al., 2003; Favaro et al., 2006; Kitzing et al., 2007; Sarmiento et al., 2008; Schuster et al., 2007; Zhu et al., 2008). Determining formin’s numerous roles is particularly complicated because most organisms express multiple isoforms (Goode and Eck, 2007). For example, there are at least 20 formin genes in plants, 18 in mammals, six each in Drosophila melanogaster and Caenaorhabditis elegans, three in fission yeast and two in budding yeast (Higgs, 2005). A major challenge is to elucidate all formin-dependent cellular functions in both healthy and diseased tissues.
Formins are multi-domain proteins that contain a highly conserved actin assembly formin homology 2 FH2 domain, and its associated profilin-binding FH1 domain, which are flanked on either side by regulatory domains that coordinate localization and activation (Goode and Eck, 2007; Higgs, 2005). Some formins are auto-inhibited by association of their N- and C-terminal regulatory domains, and activated by GTP-bound small Rho-GTPases (Goode and Eck, 2007; Wallar and Alberts, 2003). Upon activation the FH2 domain either nucleates actin filament assembly, or associates with pre-existing filaments, and remains continuously associated with the elongating actin filament barbed end (Higashida et al., 2004; Kovar and Pollard, 2004). Processive association allows formins to protect barbed ends from capping proteins (Harris et al., 2004; Kovar et al., 2005; Moseley et al., 2004; Zigmond et al., 2003), and to stimulate the addition of FH1 domain bound profilin-actin (Kovar et al., 2006; Kovar and Pollard, 2004; Neidt et al., 2008; Paul and Pollard, 2008; Romero et al., 2004; Vavylonis et al., 2006). As a result, formins produce long-straight filaments that are often bundled in cells and pulled by myosin motors to produce contractile forces, such as the cytokinetic contractile ring and stress fibers (Glotzer, 2005; Naumanen et al., 2007).
Given that the roles of most formins in different cell types are not known, and the potential for formin as an anti-cancer target, inhibitors of formin-mediated actin assembly would be extremely useful. We report here a simple fluorescence based in vitro screen, and identification of the first general small molecule inhibitor of formin-mediated actin assembly that disrupts formin-dependent processes from yeast to mammals.
RESULTS AND DISCUSSION
Identification of a Small Molecule that Inhibits Formin-Mediated Actin Assembly
We screened ~10,000 commercially available small molecules (ChemBridge) for the ability to prevent mouse formins mDia1(FH1FH2) and mDia2(FH1FH2) from stimulating actin assembly in vitro (see Experimental Procedures). We identified a compound that we named SMIFH2 for small molecule inhibitor of formin FH2 domain-mediated actin assembly (Figure 5A). This 2-thiooxodihydropyrimidine-4,6-dione derivative inhibits mouse formin mDia1(FH1FH2)-mediated assembly of profilin-actin in a concentration dependent manner (Figures 1A and 1B). Half-maximal inhibition occurs at ~15.0 μM SMIFH2, and at saturating concentrations of SMIFH2 the rate of assembly is identical to the rate without formin. Control experiments demonstrate that SMIFH2 is specific for formin-mediated actin assembly. High concentrations of SMIFH2 did not eliminate the spontaneous assembly of actin monomers without formin (Figure 1C), the addition of actin to the barbed end of pre-assembled filaments without formin (Figures 1G and 1I, and Supplemental Figures 1C and1E), or actin polymerization stimulated with Arp2/3 complex (Figures 1E and 1F).
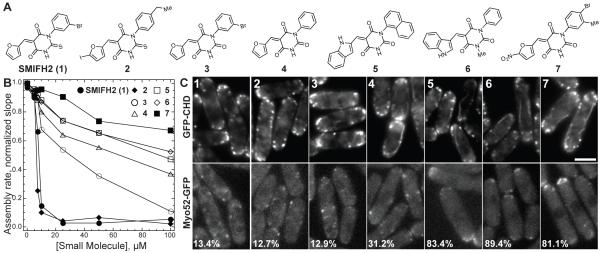
Figure 5. Structure and Activity of SMIFH2 Analog Molecules.
(A) Structure of SMIFH2 (1) and analog molecules 2 – 7.
(B) Plot of the dependence of the assembly rate of 2.5 μM Mg-ATP actin monomers (20% pyrene-labeled) in the presence of 25 nM mouse formin mDia1 on the concentration of SMIFH2 (1) (● ) and analog molecules 2 (◆ ), 3 (○ ), 4 (△ ), 5 (□ ), 6 (◇ ) and 7 (■ ). Conditions were the same as in Figure 1.
(C) Fission yeast cells expressing either GFP-CHD (upper panels) to label the entire actin cytoskeleton, or type V myosin Myo52-GFP (lower panels), following treatment for 30 minutes at 25° C with DMSO or 10 μM of the indicated analog. Numbers in the left corner of lower panels represent the percent of cells in which Myo52-GFP is localized specifically to cell tips via formin-dependent actin cables.
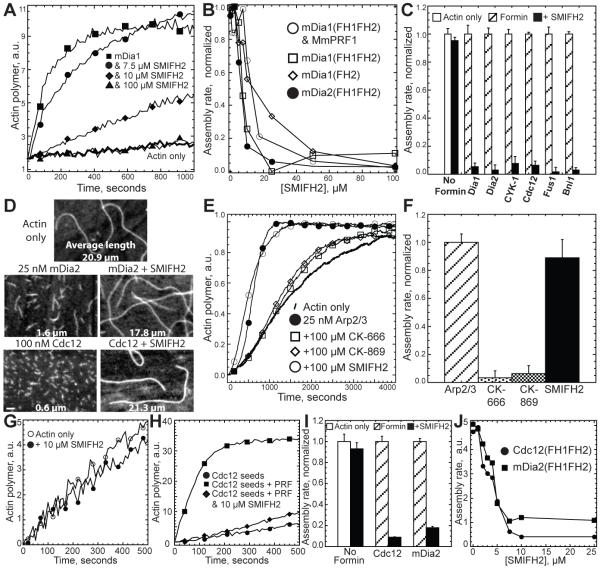
Figure 1. SMIFH2 Inhibits Formin-Mediated Actin Assembly In Vitro.
(A) Time course of the polymerization of 2.5 μM Mg-ATP actin monomers (20% pyrene-labeled) with 2.5 μM profilin MmPRF1 in either the absence (thick curve) or presence of 25 nM mDia1(FH1FH2) and 0.0 (■ ), 7.5 (● ), 10 (◆ ), or 100 μM SMIFH2 (▲ ). Conditions: 10 mM imidazole, pH 7.0, 50 mM KCl, 1 mM MgCl2, 1 mM DGTA, 0.5 mM DTT, 0.2 mM ATP, 90 μM CaCl2.
(B) Plot of the dependence of the maximum polymerization rate of 2.5 μM Mg-ATP actin on the concentration of SMIFH2 in the presence of the indicated formin constructs.
(C) Bar graph of the effect of 50 μM SMIFH2 on the maximum polymerization rate of 2.5 μM actin monomer in the absence (No Formin) or presence of diverse formin(FH1FH2) constructs: 25 nM mouse mDia1, 25 nM mouse mDia2, 100 nM nematode worm CYK-1, 100 nM fission yeast Cdc12, 10 nM fission yeast Fus1 and 75 nM budding yeast Bni1. (Error bars, s.d.; n = 3).
(D) Fluorescent micrographs of the products of actin polymerization assays from (C) stained with rhodamine-phalloidin. Bar, 1.0 μm.
(E and F) Effect of 100 μM SMIFH2 or Arp2/3 complex inhibitors CK-666 and CK-869 (Nolen et al., 2009), on the polymerization of 2.5 μM actin monomers with 25 nM Arp2/3 complex and 100 nM GST-WASP-VCA. (Error bars, s.d.; n = 3).
(G-J) Effect of SMIFH2 on the elongation of filaments pre-assembled by formin. 2.5 μM unlabeled actin was pre-assembled alone or in the presence of 50 nM Cdc12(FH1FH2) or mDia2(FH1FH2), treated with a range of concentrations of SMIFH2, and diluted 15-fold into new reactions with 0.5 μM Mg-ATP-actin (10% pyrene-labeled) and 5.0 μM profilin. (G) Time-course of the elongation of control filaments pre-assembled without formin in the absence (○ ) or presence of 10 μM SMIFH2 (● ). (H) Time-course of the elongation of Cdc12-assembled filaments alone (● ), with profilin (■ ) and with profilin and 10 μM SMIFH2 (◆ ). (I) Bar graph of the effect of 10 μM SMIFH2 on the maximum elongation rate of control and formin-assembled filaments. (Error bars, s.d.; n = 3). (J) Plot of the dependence of the polymerization rate on the concentration of SMIFH2 for filaments pre-assembled by formin.
With similar potency SMIFH2 also inhibits mDia1(FH1FH2) without profilin, as well as the construct mDia1(FH2) lacking the profilin binding FH1 domain (Figure 1B). Thus, the molecular target of SMIFH2 is likely the highly conserved FH2 domain. SMIFH2 also inhibits actin assembly by evolutionarily diverse formin FH1FH2 constructs including C. elegans CYK-1, S. pombe Cdc12, S. pombe Fus1, S. cerevisae Bni1 and M. musculus mDia2 (Figures 1B and 1C). Therefore SMIFH2 is a general inhibitor of actin assembly mediated by formin FH2 domains.
SMIFH2 inhibits both formin-mediated nucleation and elongation
Formins nucleate actin assembly and drive rapid elongation of profilin-actin by remaining continually associated with the elongating barbed end (Kovar, 2006). We determined that SMIFH2 inhibits formin-mediated nucleation by visualizing the products of spontaneous assembly reactions upon reaching plateau (Figure 1D). Filament length is proportional to the number of filaments. Control reactions without formin produce long filaments (~21 μm) compared to reactions with formins (~1.5 and ~0.5 μm for mouse mDia2 and fission yeast Cdc12). SMIFH2 inhibits formin nucleation, resulting in filament lengths similar to controls without formin (~18 and ~21 μm for mDia2 and Cdc12).
We determined that SMIFH2 inhibits formin-mediated elongation by measuring the addition of profilin-actin to pre-assembled formin-associated filaments (Figures 1G-1J). In the absence of SMIFH2, formin-associated filaments rapidly assemble upon addition of profilin-actin. Conversely, SMIFH2 severely blocked the elongation of both Cdc12- and mDia2-associated filaments. Half-maximal inhibition occurs at ~4.0 μM SMIFH2 (Figure 1J). SMIFH2 did not inhibit the barbed end elongation of filaments without formin (Figures 1G and 1I).
SMIFH2 decreases the affinity of formin for the actin filament barbed end
We determined that SMIFH2 decreases the affinity of formin for the barbed end by three assays. First, high concentrations of SMIFH2 allow barbed ends to depolymerize in the presence of formin (Supplemental Figures 1A and 1B). In the absence of SMIFH2, formin significantly inhibits the depolymerization of pre-assembled filaments upon dilution below the barbed end critical concentration. Concentrations of SMIFH2 above 25 to 50 μM allow rapid barbed end depolymerization in the presence of both Cdc12 and mDia2. Conversely, SMIFH2 had no effect on the barbed end depolymerization rate without formin (Supplemental Figure 1B).
Second, we determined the effect of SMIFH2 on the ability of formin to bind to the barbed end and reduce the rate of monomer addition (Supplemental Figures 1C-1E). Cdc12 binds to pre-assembled barbed ends and severely inhibits their elongation in the absence of profilin (Supplemental Figures 1D and 1E) (Kovar et al., 2003). In a concentration dependent manner SMIFH2 prevents Cdc12 from binding to the barbed end, allowing filaments to elongate near the control rate (Supplemental Figures 1D and 1E). Conversely, SMIFH2 had no effect on the rate of barbed end elongation in the absence of formin (Supplemental Figures 1C and 1E).
Third, we determined that SMIFH2 does not allow formin to inhibit the end-to-end annealing of actin filaments (Supplemental Figure 2). Formin-associated filaments do not anneal (Kovar et al., 2003). SMIFH2 allowed pre-assembled filaments severed in the presence of Cdc12 (Supplemental Figures 2A and 2B), or Cdc12- and mDia2-nucleated filament seeds (Supplemental Figures 2C and 2D) to anneal. Conversely, SMIFH2 had no effect on annealing in the absence of formin (Supplemental Figures 2A and 2B).
Direct Observation of Formin Inhibition by Total Internal Reflection Fluorescence (TIRF) Microscopy
To verify that SMIFH2 specifically inhibits formin-mediated actin assembly, we utilized TIRF microscopy to observe individual filaments assembled from formin in solution (Figure 2) or from formin immobilized on beads (Figure 3). When 1.5 μM Mg-ATP actin monomers (33% labeled with Oregon green for visualization) are assembled in the presence of 5.0 nM mDia1(FH1FH2) and 2.5 μM profilin, two filament populations are present that differ by both elongation rate and brightness (Figure 2A) (Kovar et al., 2006). The first population consists of ‘bright’ control filaments whose barbed ends are not associated with formin and elongate at ~14 subunits/second (sub/s) (Figures 2A and 2B). The second population consists of ‘dim’ formin-associated filaments which elongate their barbed ends significantly faster at ~60 sub/s. 450 seconds after the reaction was initiated, just over 80% of the filaments were associated with formin (Figure 2G).
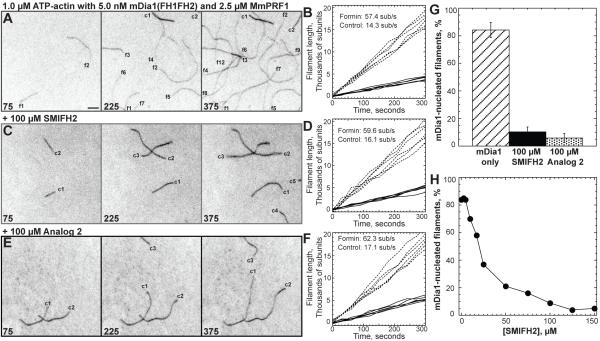
Figure 2. Evanescent Wave Fluorescent Microscopy of the Effect of SMIFH2 on Spontaneous Actin Assembly.
Assembly of 1.0 μM ATP-actin with 0.5 μM Oregon green labeled ATP-actin in the presence of 5.0 nM mDia1 and 2.5 μM MmPRF1 on slides coated with NEM-myosin II. Conditions: 10 mM imidazole, pH 7.0, 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 50 mM DTT, 0.2 mM ATP, 50 μM CaCl2, 15 mM glucose, 20 μg/mL catalase, 100 μg/mL glucose oxidase, 0.5% (500 centipoise) methylcellulose, 1.7% DMSO at 25° C. Bar = 5 μm. Movies of time lapses are published as supplemental data on the Chemistry and Biology web site.
(A, C and E) Time-lapse micrographs of representative 45 × 45 μm areas, with time in seconds indicated at the bottom. Labels indicate control (c) and formin-associated (f) filaments.
(B, D and F) Plots of the length of six individual filaments versus time for control (solid lines) and formin-associated filaments (dashed lines). The average barbed end elongation rates are indicated.
(A-B) Control reaction without SMIFH2.
(C-D) Reaction with 100 μM SMIFH2.
(E-F) Reaction with 100 μM Analog 2.
(G) Bar graph of the average percent of formin-nucleated filaments that appear by 450 seconds in the entire 133 × 133 μm field in both the absence and presence of either 100 μM SMIFH2 or 100 μM Analog 2. (Error bars, s.d.; n = 3).
(H) Dependence of the percent of formin-nucleated filaments on the concentration of SMIFH2.
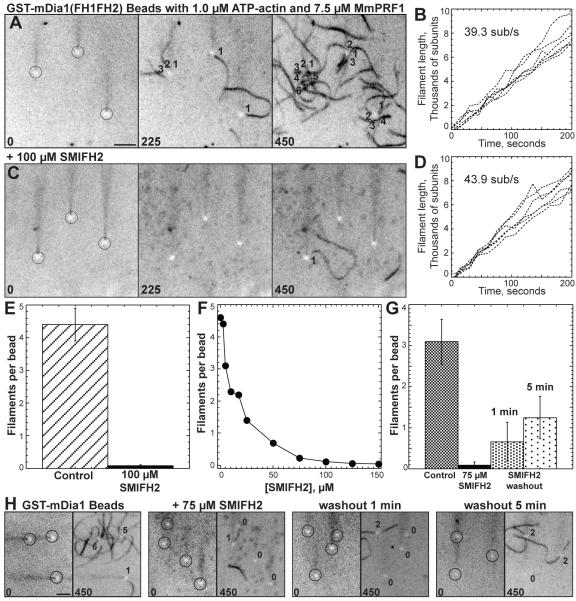
Figure 3. Evanescent Wave Fluorescent Microscopy of the Effect of SMIFH2 on Formin-Mediated Actin Assembly From Beads.
Assembly of 1.0 μM ATP-actin with 0.5 μM Oregon green labeled ATP-actin on beads coated with GST-mDia1(FH1FH2) in the presence of 7.5 μM MmPRF1. Conditions were the same as in Figure 2. Movies of time lapses shown in (A) and (C) are published as supplemental data on the Chemistry and Biology web site.
(A and C) Time-lapse micrographs of representative areas containing three beads (circled), with time in seconds indicated at the bottom. Numbers indicate individual actin filaments assembled from each bead. Bar = 5 μm.
(B and D) Plots of the growth of six individual filament barbed ends versus time for filaments assembled from beads. The average barbed end elongation rate of 10 filaments is indicated.
(A-B) Control reaction without SMIFH2.
(C-D) Reaction with 100 μM SMIFH2.
(E) Bar graph of the average number of filaments per bead that appear by 375 seconds in both the absence and presence of 100 μM SMIFH2. (Error bars, s.d.; n = 3).
(F) Dependence of the average number of filaments per bead that appear by 375 seconds on the concentration of SMIFH2.
(G and H) Effect of washing beads following treatment with 75 μM SMIFH2. (G) Bar graph of the average number of filaments per bead that appear by 375 seconds in reactions without SMIFH2, with SMIFH2, one minute after SMIFH2 was washed out, and five minutes after SMIFH2 was washed out. (Error bars, s.d.; n = 3). (H) Representative regions before and after 450 seconds. The number of filaments polymerized from each bead (circled) are indicated. Bar = 5 μm.
In reactions containing 100 μM SMIFH2 control filaments were unaffected, whereas the average number of dim formin-associated filaments was significantly reduced to ~10% of the total filament population (Figures 2C-2D, and 2G). The few formin-associated filaments present in reactions containing 100 μM SMIFH2 elongate their barbed ends at the same rate as formin-associated filaments in control reactions without SMIFH2 (Figure 2D; ~60 sub/s), suggesting that SMIFH2 inhibits formin-mediated actin assembly by targeting formin not actin. A plot of the dependence of the percent of formin-nucleated filaments on the concentration of SMIFH2 revealed half-maximal inhibition at ~25 μM SMIFH2 (Figure 2H).
When 1.5 μM Mg-ATP actin monomers (33% labeled with Oregon green) and 7.5 μM profilin is flowed into a chamber containing 2.0 μm carboxylate beads pre-absorbed with GST-mDia1(FH1FH2), actin filaments are nucleated from and remain processively associated with formin immobilized on the beads (Figure 3A) (Michelot et al., 2007; Romero et al., 2004). Each formin-absorbed bead averaged slightly more than four filaments by 375 seconds (Figure 3E), which elongated at a rate of ~40 sub/s (Figure 3B). Filaments processively associated with mDia1 elongate their barbed ends 33% slower in the presence of 7.5 μM profilin than in the presence of 2.5 μM profilin (Figures 2B and 3B) (Kovar et al., 2006). When incubated with 100 μM SMIFH2 each formin-absorbed bead averaged less than 0.05 filaments (Figures 3C and 3E), which elongated at ~44 sub/s (Figure 3D). A plot of the dependence of the average number of filaments per bead on the concentration of SMIFH2 revealed half-maximal inhibition at ~15 μM SMIFH2 (Figure 3F).
Although formin is likely the molecular target, it is theoretically possible that SMIFH2 binds to actin in a manner that inhibits formin-mediated assembly but not (1) spontaneous actin assembly without formin (Figure 1C), (2) barbed end elongation without formin (Figures 1G and 1I, and Supplemental Figures 1C and 1E), (3) barbed end depolymerization without formin (Supplemental Figure 1B) and (4) Arp2/3 complex-mediated actin assembly (Figures 1E and 1F). To establish that the molecular target of SMIFH2 is formin, we tested the effect of washing out SMIFH2 from formin-associated beads before the addition of actin monomers (Figures 3G and 3H). In this case formin-associated beads, but not actin, are exclusively treated with SMIFH2. We found that extensively washing beads one and five minutes before the addition of actin monomers only partly recovered the nucleation activity of formin-associated beads to ~0.5 and ~1.25 filaments per bead (Figure 3G). Increased nucleation activity after five minutes likely reflects the dissociation rate of SMIFH2 from formin.
Inhibition of the Fission Yeast Formin-Dependent Actin Cytoskeleton
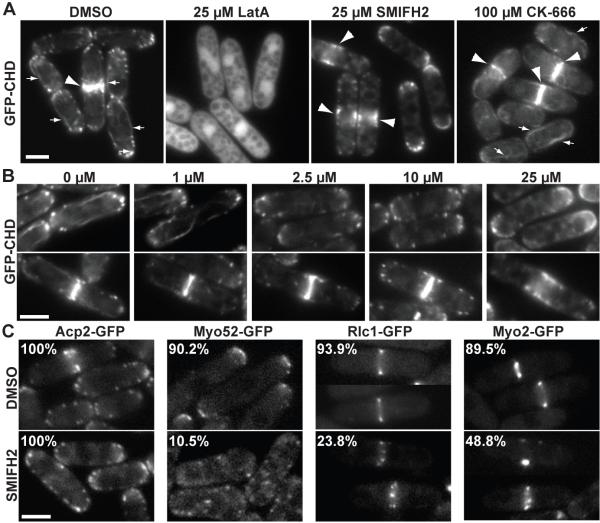
We examined the effect of SMIFH2 on fission yeast to determine whether the compound inhibits formin-mediated processes in cells. Fission yeast assembles four distinct filamentous actin structures that depend upon different actin nucleation factors (Figure 4A) (La Carbona et al., 2006; Marks and Hyams, 1985). The Arp2/3 complex is required for endocytic actin patches at sites of polarized growth (Balasubramanian et al., 1996; McCollum et al., 1996). Three formin isoforms are specifically required for polarized actin cables (For3p; Feierbach and Chang, 2001; Nakano et al., 2002), the cytokinetic contractile ring (Cdc12p; Chang et al., 1997) and the mating projection tip (Fus1p; Petersen et al., 1998). We imaged fission yeast cells expressing GFP-CHD (actin binding domain of Rng2) to label the entire filamentous actin cytoskeleton (Figure 4A) (Martin and Chang, 2006). An asynchronous population of control cells treated with DMSO contained Arp2/3 complex-dependent patches as well as formin-dependent actin cables and contractile rings. The non-specific actin depolymerizing drug LatA completely disassembled all filamentous actin structures (Figure 4A). The Arp2/3 complex inhibitor CK-666 specifically disassembled Arp2/3 complex-dependent actin patches (Figure 4A) (Nolen et al., 2009). On the other hand, 25 μM SMIFH2 disassembled formin-dependent actin cables and contractile rings, but not actin patches (Figure 4A). The concentration of SMIFH2 required to disassemble actin cables and contractile rings varied (Figure 4B). Whereas actin cables were lost in the presence of only 2.5 μM SMIFH2, disassembly of contractile rings required 25 μM SMIFH2. It is possible that contractile rings are more robust than actin cables, or that For3 is more sensitive to SMIFH2 than is Cdc12.
Figure 4. SMIFH2 Inhibits Formin-Mediated Actin Assembly in Fission Yeast.
(A and B) Representative micrographs demonstrate that SMIFH2 disrupts the formin-dependent actin cytoskeleton in fission yeast. Scale bars = 5 μm. Fission yeast cells expressing GFP-CHD to label the entire actin cytoskeleton: Arp2/3-dependent actin patches, and formin-dependent actin cables (small arrows) and contractile rings (large arrowheads). (A) Cells treated for 30 minutes at 25° C with either DMSO, 25 μM actin monomer binding drug Latrunculin A, 25 μM SMIFH2 or 100 μM Arp2/3 complex inhibitor CK-666. (B) Cells treated with a range of SMIFH2 concentrations for 30 minutes at 25° C. Upper panels show actin cables, and lower panels show contractile rings. Cables are lost with 2.5 μM SMIFH2, whereas contractile rings are present until 25 μM SMIFH2.
(C) Representative micrographs of fission yeast cells expressing various GFP fusions that label specific actin cytoskeleton structures. Cells were treated for 30 minutes at 25° C with either DMSO or 10 μM SMIFH2. Numbers in the top left corner indicate the percent of cells containing Arp2/3 complex-dependent actin capping protein patches (Acp2-GFP), type V myosin localized to the cell tip via formin-dependent actin cables (Myo52-GFP) (Win et al., 2000), or normal (not punctate) localization of regulatory light chain (Rlc1-GFP) and type II myosin (Myo2-GFP) to the formin-dependent contractile ring.
To quantify the inhibitory effect of SMIFH2 on formin-dependent actin cables and contractile rings, we imaged fission yeast cells expressing integrated GFP fusions to proteins that localize to particular actin structures (Figure 4C). As expected, actin capping protein-labeled actin patches (Acp2-GFP) were observable in 100% of cells treated with 10 μM SMIFH2. However, the localization of GFP-tagged proteins to formin-dependent actin cables and the contractile ring was abrogated. Although type V myosin (Myo52-GFP) normally localizes to the tips of interphase cells via actin cables, 10 μM SMIFH2 caused Myo52-GFP to become distributed throughout the cell. Myo52-GFP also distributes throughout the cell in formin For3 null cells that completely lack actin cables (Feierbach and Chang, 2001). Additionally, both the regulatory light chain (Rlc1-GFP) and type II myosin (Myo2-GFP) appeared as punctate spots rather than continuous smooth formin-dependent contractile rings at the division site in 10 μM SMIFH2 treated cells.
Structure-Activity Relationship of SMIFH2
To gain insight into the structure-function profile of SMIFH2 (1), we characterized the ability of six other compounds (2–7), including both pyrimidine-2,4,6-triones and 2-thiooxodihydropyrimidine-4,6-diones, to inhibit formin-mediated actin assembly in vitro and disrupt the fission yeast actin cytoskeleton (Figure 5). We found that the ability of analogs to inhibit formin-mediated actin assembly in vitro (Figures 5B and 2E-2G), correlated closely with their ability to disrupt the fission yeast formin-dependent actin cytoskeleton (Figure 5C). None of the analogs removed Arp2/3 complex-dependent actin patches. Analogs 2 and 3 were the most potent both in vitro and in fission yeast. Similar to SMIFH2 (1), cells treated with 10 μM of analogs 2 and 3 for 30 minutes lost actin cables resulting in mislocalization of the type V myosin Myo52-GFP from cell tips to throughout the cell. Analog 4 had lower potency, whereas analogs 5, 6 and 7 had little effect on both formin-mediated actin assembly in vitro and actin cables in fission yeast cells. Our results strongly suggested that the thiourea moiety of SMIFH2 is an important pharmacophoric element of this compound since a substitution of this unit by the urea fragment resulted in substantial loss in activity. A series of structurally related pyrimidine-2,4,6-triones have been previously reported to inhibit cell migration by modulating the activity of mucosal addressin cell adhesion molecule-1 with compounds 5 and 6 being the most potent in the series (Harriman et al., 2008). Despite notable structural homology between 5, 6 and SMIFH2 (1), there is a substantial difference in their ability to inhibit formin-mediated actin assembly. This data suggests that the cell-based activity of SMIFH2 is due to selective modulation of formin and that the activity profile is uniquely linked to the thiourea-containing structure of this compound.
SMIFH2-Induced Cytotoxicity and Growth Inhibition of NIH 3T3 Fibroblasts
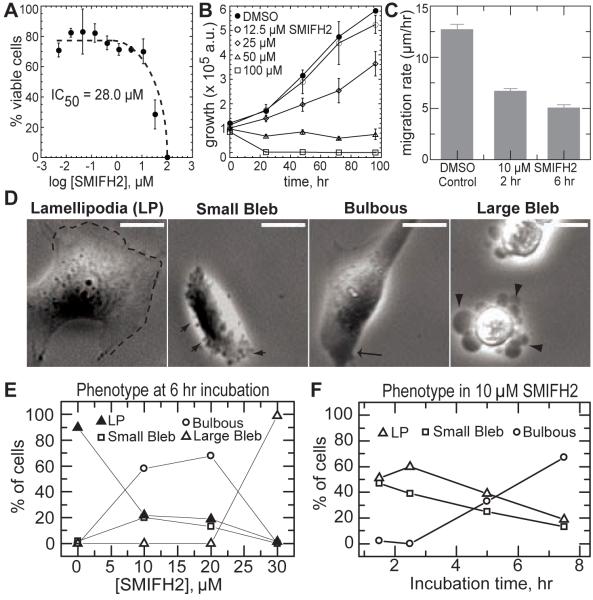
NIH 3T3 fibroblasts were incubated with a range of concentrations of SMIFH2 to determine its effects on adherent mammalian cells. Cell viability was assessed after 24 hours. SMIFH2 elicited cytotoxicity with an IC50 of 28 μM (Figure 6A). We also observed dose-dependent growth inhibition of NIH 3T3 fibroblasts within a period of 96 hours (Fig. 6B). In addition, cells cultured in SMIFH2 with concentrations as low as 2.5 μM were observed to have cytokinesis defects, as measured by the percentage of multi-nucleated cells. After 4 days, 8.6 ± 2% of cells cultured in 2.5 μM SMIFH2 had multiple nuclei as compared to 1.1 ± 0 .7% of cells treated with a DMSO control. This cytokinesis defect is consistent with recent data showing that siRNA depletion of mDia2 in NIH3T3 fibroblasts increases the percent of binucleate cells to ~35% (Watanabe et al., 2008).
Figure 6. Effect of SMIFH2 on Death, Growth and Migration of NIH 3T3 Fibroblasts.
(A) Cytotoxicity of SMIFH2 in 3T3 fibroblasts after 24-hour incubation, determined by measuring ATP content as described in Materials and Methods. The percentages of viable cells are relative to cells treated with DMSO only.
(B) Inhibition of the growth of 3T3 fibroblasts by various concentrations of SMIFH2 over 96 hours. Cell viability was measured by ATP content, and the luminescence outputs were plotted against incubation time.
(C) Migration rate of NIH 3T3 fibroblasts treated with DMSO control or 10 μM SMIFH2 for 2 and 6 hours.
(D) Fibroblast morphologies assessed by time-lapse phase contrast imaging of cells treated with SMIFH2. Lamellipodia (LP) are thin, sheet like protrusions in spread cells characterized by periods of extension and retraction (indicated by dashed line). Small Bleb phenotype is characterized by phase-dark blebs (~0.5 mm in diameter) that extend and retract along the cell periphery in poorly spread cells (indicated by small arrow). Bulbous phenotype is characterized by poorly spread cells with phase dark regions near the periphery (arrow) and minimal protrusive activity. Large Bleb phenotype is characterized by blebs (>2 mm in diameter) that are static (indicated by large arrowheads). Scale bar is 10 mm. Movies of time lapses are published as supporting data on the Chemistry and Biology web site.
(E) Quantification of the percent of fibroblasts with the indicated cellular morphologies as a function of SMIFH2 concentration.
(F) Distribution of cellular morphologies observed as a function of incubation time for fibroblasts treated with 10 μM SMIFH2 (n > 100 cells for each measurement).
Cellular Morphology and Protrusion Phenotypes Observed in NIH 3T3 Fibroblasts
We took time-lapse images of NIH 3T3 fibroblasts to visualize SMIFH2-induced phenotypes in mammalian cells. The migration rate of cells treated with 10 μM SMIFH2 for >2 hrs was reduced two-fold compared to control cells (Figure 6C), which is consistent with previous studies implicating mDia2 in cell migration (Gupton et al., 2007). Control cells are characterized by a spread morphology and thin sheet-like protrusions termed lamellipodium that requires Arp2/3 complex-mediated actin assembly. Over 90% of control cells have lamellipodium (Figures 6D and 6E). Despite a less spread morphology, approximately 20% of cells treated with less than 20 μM SMIFH2 for six hours still extended lamellipodial protrusions (Figures 6D and 6E). Mouse formin mDia2 has been implicated in the assembly of cortical actin required for maintaining plasma membrane integrity, and inhibition of mDia2 led to abundant nonapoptotic membrane blebbing (Eisenmann et al., 2007). Consistent with this result, we observed that 20% of cells treated with 10–20 μM SMIFH2 for six hours exhibit constitutive, dynamic, small (< 1 μm) blebs (Figures 6D and 6E). At 30 μM SMIFH2 nearly all the cells exhibited large blebs (Figures 6D and 6E), which is consistent with an IC50 of 28 μM SMIFH2 for NIH 3T3 fibroblasts (Figure 6A). Effects of SMIFH2 perfusion were reversible on the time scale of 5 to 10 minutes (data not shown).
The observed phenotype changes slowly over time when cells are incubated with 10 μM SMIFH2 (Figure 6F). After two hours 60% still retain the lamellipodial protrusive phenotype, whereas 40% protrude by means of small dynamic blebs (Figure 6F). After seven hours the percentage of cells exhibiting either lamellipodial protrusions or constitutive small blebbing decreases dramatically, while the percentage of cells that lack protrusive activity and appear phase dark near the cell periphery increases (Figure 6F). Because of the static, round shape of these cells, we term this phenotype “bulbous” (Figure 6D). The phase dark appearance near the cell periphery suggests an increase in cell thickness in this region, which may be due to decreased stability of the filamentous actin cortex.
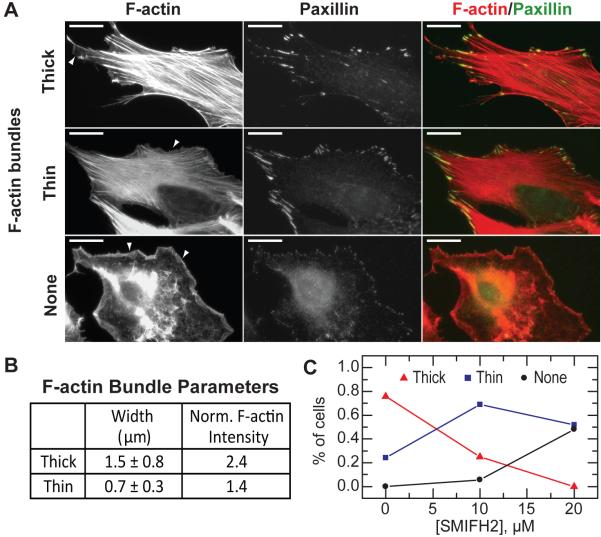
F-Actin Cytoskeleton in SMIFH2-Treated 3T3 Fibroblasts
We imaged filamentous actin and the focal adhesion marker paxillin to assess the organization of the actin cytoskeleton in cells treated with SMIFH2 (Figure 7A). In fibroblasts the actin cytoskeleton is typically organized into prominent type II myosin-dependent contractile bundles. The width and actin filament density of these bundles were characterized by quantifying the variation in intensity of fluorescent phalloidin across the bundles. ‘Thick’ F-actin bundles have a mean width of 1.5 μm and a 2.4-fold higher local actin filament density compared with the surrounding region (Figures 7A and 7B). ‘Thin’ F-actin bundles have a mean width of 0.7 μm and a 1.5-fold enhanced actin filament density. Interestingly, focal adhesions are similar in cells with thick or thin F-actin bundles (Figures 7A). In control cells, over 80% of the fibroblasts exhibited predominately thick F-actin bundles, with only 20% exhibiting thin bundles (Figure 7C). However, 60% of cells incubated with 10 μM of SMIFH2 for 2 hours have thin F-actin bundles, and only 20% have thick F-actin bundles (Figure 7C). Moreover, approximately 5% of the spread cells have a disorganized lamellar network and no F-actin bundles (Figure 7A and 7C). In cells lacking F-actin bundles, focal adhesion plaques were significantly reduced in size, reminiscent of nascent focal complexes that are observed to form in the absences of myosin-II driven tension (Figure 7A) (Choi et al., 2008). As the concentration of SMIFH2 was increased to 20 μM, the percentage of cells that lacked F-actin bundles increases to 50%, with the rest of the population showing only thin bundles (Figure 7C). Importantly, all spread cells have a F-actin-rich lamellipodia near the cell periphery, which is dependent upon Arp2/3 complex-mediated actin assembly (arrows, Figure 7A). Thus, at intermediate SMIFH2 concentrations the filamentous actin organization of cells that remain spread is in the form of thin F-actin bundles or an isotropic lamellar meshwork. This is consistent with previous reports that mDia1, in addition to the Rho effector Rho kinase, is essential to building contractile stress fibers and the growth of focal complexes (Hotulainen and Lappalainen, 2006; Riveline et al., 2001; Watanabe et al., 1999). Changes in F-actin organization and adhesion were not observed with analog compounds (data not shown).
Figure 7. Effect of SMIFH2 on the Actin Cytoskeleton in NIH 3T3 Fibroblasts.
(A) Immunofluorescence of filamentous actin and paxillin in NIH 3T3 fibroblasts. Cells that remained spread after SMIFH2 treatment exhibited three distinct actin cytoskeleton organizations: thick F-actin bundles, thin F-actin bundles and no F-actin bundles. Lamellipodial actin is observed in all phenotypes, and indicated by arrows. Scale bar is 10 mm.
(B) F-actin bundles observed in (A) are characterized by their mean width and the maximum filamentous actin intensity normalized to the local background.
(C) Percentage of spread cells exhibiting different actin cytoskeleton phenotypes after 2 hr incubation with indicated concentrations of SMIFH2 (n > 75 cells for each concentration).
Impact of SMIFH2 on a Carcinoma Cell Line A549
We treated the carcinoma cell line A549 with a range of SMIFH2 concentrations to determine the effect of SMIFH2 on diverse cells. A549 cells are exceptionally more tolerant, with an IC50 near 75 μM SMFH2 and a higher dose of SMIFH2 was required to inhibit growth (Supplemental Figures 3A and 3B). Treatment of A549 cells for four days with 30 μM SMIFH2 increases the percentage of multi-nucleated cells by four-fold, suggesting a SMIFH2-induced cytokinesis defect.
In contrast to the NIH 3T3 fibroblasts, a majority of A549 cells treated with 30 μM SMIFH2 remained spread over the course of an 8 hr time-lapse movie. However, ~40% have spherical bleb-like protrusions which rapidly extended and retracted at the cell periphery (Supplemental Figures 3C and 3D). In 10% of cells, these protrusions were stabilized by adhesion to the external matrix and resulted in changes in cell shape (Supplemental Figures 3C and 3D). Immunofluorescence images revealed regions near the cell periphery with high filamentous actin intensity and numerous small focal complexes that existed beyond the zone of large focal adhesions (Supplemental Figure 3E). Thus, it appears that SMIFH2 treatment induces bleb-like protrusions near the cell periphery without significantly altering the formation of focal complexes. This is consistent with reports suggesting that the RhoA pathway and its effector, mDia1, are instrumental in maintaining cortical integrity to limit protrusive activity (Worthylake and Burridge, 2003). Such bleb-like protrusions have been observed in several cell types, but the relative contributions of actin polymerization and local cortical tension in regulating cellular protrusions has not been well established (Fackler and Grosse, 2008). These results suggest that reduction in formin-mediated actin polymerization at the plasma membrane results in local cortical weakening. In this scenario, the forces for protrusion might originate from osmotic pressure driven changes in membrane shape rather than from actin polymerization.
Additional work is required to elucidate the mechanism of differences in drug toxicity between these two cell lines, possible reasons are changes in expression of different isoforms of formins or different formin activity due to different levels of the formin activator RhoA.
Future Considerations
Analysis of SMIFH2 and the Arp2/3 complex inhibitor CK-666 demonstrates the usefulness of fission yeast to determine the specificity of drugs on actin cytoskeleton processes. Extensive work is required to determine which specific actin-modulating proteins are required for the at least 15 processes in vertebrates (Chhabra and Higgs, 2007). Fission yeast assembles actin into just four structures (La Carbona et al., 2006; Marks and Hyams, 1985). The ease of genetic manipulation and visualization of functional GFP-tagged proteins expressed from their endogenous locus has provided a nearly complete profile of which evolutionarily conserved actin-binding proteins are required for each structure in budding and fission yeast (La Carbona et al., 2006; Moseley and Goode, 2006). Inhibitor specificity can be investigated in yeast, and then utilized to determine which processes in animals depend upon the drug’s target. Yeast has also been useful for testing the specificity of drugs targeting other fundamental cellular processes (Lehar et al., 2008; Lopez et al., 2008).
As a general inhibitor, which targets diverse formin isoforms with similar potency, SMIFH2 may be a useful drug for identifying cellular processes dependent upon formin-mediated actin assembly in a broad range of experimental systems. However, it will be important to isolate formin isoform specific inhibitors to determine which formin is required for a particular cellular process. Futhermore, particular formin isoforms are involved in malignant tumor formation and are highly overexpressed in a range of cancer cell types (Favaro et al., 2003; Kitzing et al., 2007; Sarmiento et al., 2008; Zhu et al., 2008). Isoform specific inhibitors are likely to be better anti-metastatic tumor drugs because they may target tumor cells while having less impact on surrounding healthy tissues.
SIGNIFICANCE
Formins are required to stimulate actin filament assembly at the correct time and place to drive fundamental cellular processes such as division, adhesion and establishing polarity. Formin inhibitors would be an extremely useful reagent because most eukaryotic organisms express multiple formins whose cellular roles have not been determined, and because formins are required for processes deregulated during metastic tumor formation.
We established a simple fluorescent-based in vitro screen to discover for the first time a general small molecular formin inhibitor that we named SMIFH2 (small molecule inhibitor of formin FH2 domains). SMIFH2 is a general formin inhibitor that targets evolutionarily diverse formin isoforms from budding yeast, fission yeast, nematode worm and mouse, with similar ~5 to 15 μM half-maximal potency. SMIFH2 inhibits formin at multiple points from preventing both nucleation and processive barbed-end elongation, and decreases formin’s affinity for the barbed end. We verified that SMIFH2 abolishes formin-dependent actin cytoskeletal structures in both fission yeast and animal cells. SMIFH2 disrupts formin-dependent actin cables and contractile rings in fission yeast, but does not target endocytic actin patches, which are dependent upon the other major actin nucleation factor Arp2/3 complex. Although it is possible that SMIFH2 has additional cellular targets, the potency of analog compounds on formin-mediated actin assembly in vitro directly correlates with their potency on the fission yeast formin actin cytoskeleton. In animal cells, SMIFH2 severely disrupts formin-dependent stress fibers and perturbs cell division and motility, but does not disrupt Arp2/3 complex-dependent lamellipodia near the cell periphery.
SMIFH2 may be a useful drug to elucidate formin-dependent processes in a wide-range of organisms and cell types. Future work will focus on elucidating where SMIFH2 binds formin, optimization of SMIFH2 activity, and expansion of the in vitro screen to identify formin isoform specific inhibitors.
EXPERIMENTAL PROCEDURES
Compounds and Protein Purification
Compounds were purchased from ChemBridge Corporation: SMIFH2 (catalog number 5992446), analog 2 (6062767), analog 3 (5991944), analog 4 (5174640), analog 5 (5240678), analog 6 (5241190), and analog 7 (6062214). Procedures for protein purification can be found in supplemental data on the Chemistry and Biology website.
Formin Inhibitor Screen
Small molecules were initially screened by a single-point pyrene-fluorescence assay. In 96-well plates, 1.5 μM Mg-ATP-actin (20% pyrene-labeled) and either 10 nM mDia1(FH1FH2) or 20 nM mDia2(FH1FH2) with 1.5 μM mouse profilin MmPRF1 was assembled in the presence of 960 pools of 8 small molecules (Chembridge; 12.5 μM final concentration of each small molecule) for 30 minutes and read on a Safire2 fluorescent plate reader (Tecan). Positive hits had 50% or lower fluorescence than control reactions without small molecules. 61 positive pools were re-screened kinetically by following the time course of pyrene-fluorescence (actin assembly) of 1.5 μM Mg-ATP-actin with 10 nM mDia1(FH1FH2) or 20 nM mDia1(FH1FH2) and 1.5 μM MmPRF1. 46 positive pools assembled at least two-fold slower than control reactions without small molecules. The 368 individual compounds were subsequently screened against both mDia1 and mDia2 by the single-point assay (42 compounds inhibited pyrene fluorescence by at least 50%), and by the kinetic assay (34 compounds slowed the rate of assembly by at least two-fold). 30 of the 34 compounds did not inhibit the rate of 1.5 μM actin assembly in the absence of formin. The final 30 compounds were screened kinetically over a range of concentrations. Two small molecules, SMIFH2 and analog 2, inhibited formin-mediated actin assembly by at least 50% at a concentration of 10 μM and were characterized further.
Actin Biochemistry
Procedures for fluorescence spectroscopy, microscopy of fluorescently labeled actin filaments, and TIRF microscopy can be found in supplemental data on the Chemistry and Biology website.
Fission Yeast
Fission yeast strains and microscopy procedures can be found in supplemental data on the Chemistry and Biology website.
Tissue Culture
NIH 3T3 Fibroblasts (ATCC) were cultured in high-glucose DMEM (CellGro), supplemented with 10% FBS (Hyclone), L-Glutamine (Gibco) and Penicillin/Streptomycin (Gibco). A549 carcinomic human alveolar basal epithelial cells were cultured in F12K medium (ATCC), supplemented with 10% FBS (ATCC) and Penicillin-streptomycin-glutamine solution (ATCC).
Cell Viability
All assays were performed using at least three replicate wells for each compound concentration tested. Three-fold or two-fold serial dilutions in DMSO were performed using a stock solution of SMIFH2 in DMSO. We seeded cells in 96-well white plates at the density of 1,000 cells/well (A549 cell line) or 2,000 cells/well (3T3 cell line) in 100 μl of the appropriate cell culture media. The cells were allowed to attach and grow for 24 hours before treatment with serially diluted SMIFH2 solutions pre-diluted in 25 μL growth media for each data point. After indicated incubation periods, cell viability was determined with an ATPlite 1step luminescence assay kit (PerkinElmer) according to the manufacturer’s instructions. Luminescence was analyzed using a Wallac Victor 3 plate reader (PerkinElmer).
Immunofluorescence
NIH 3T3 Fibroblasts and A549 carcinomic human alveolar basal epithelial cells were plated onto glass coverslips, and allowed to spread for 24 hrs. Cells were fixed at room temperature for 15 min in 4% paraformaldehyde, 0.5% Triton X-100, and 1.5% bovine serum albumin as a blocking agent. Paxillin antibody (1:400, Santa Cruz Biotechnology) and Alex Fluor 488 phalloidin (1:400, Molecular Probes) were diluted in 1.5% BSA in phosphate buffered saline (PBS), and incubated for 1 hr at room temperature. After three washes in PBS, coverslips were incubated overnight at 4° in Alexa 568 conjugated anti-rabbit (1:400, Molecular Probes), and were subsequently washed three times in PBS, and mounted in Prolong Gold Antifade reagent (Molecular Probes). Images were acquired with an inverted Nikon Ti microscope equipped with a cooled CCD camera (Photometrics, Coolsnap HQ2).
Time-Lapse Imaging
NIH 3T3 Fibroblasts or A549 carcinomic human alveolar basal epithelial cells were plated onto glass coverslips, and allowed to spread for 24 hrs. Coverslips were placed in a sealed chamber with the designated quantity of drug and 20 μM HEPES. Images were collected with a CCD Photometric CoolSnap HQ Camera at 5-minute intervals over 8–12 hrs with a 20X lens on an automated (Metamorph 7.5.4.0; Universal Imaging Corporation) microscope (Nikon Eclipse Ti) with a heated and motorized stage. For each experiment, 10 individual cell fields were recorded. Phenotypic frequency was measured by scoring incidence of occurrence at each indicated time point. Cells that did not spread, or left the field of view during the length of the experiment were not scored.
Supplementary Material
ACKNOWLEDGMENTS
We thank Geoff Greene and Matt Walker (The University of Chicago) for access to the small molecule library, Fred Chang (Columbia University) for fission yeast strains, Brad Nolen (University of Oregon) for Arp2/3 inhibitors, and members of the Gardel, Kovar and Kozmin labs for reagents, technical expertise and helpful discussions. This work was supported by a National Institutes of Health grant RO1GM079265 (to D.R.K.) and 5DP1OD003354 (to M.L.G.), and American Cancer Society grant RSG-04-017-CDD (to S.A.K.). M.L.G. acknowledges support from a Burroughs Wellcome Career Award at the Scientific Interface.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Balasubramanian MK, Feoktistova A, McCollum D, Gould KL. Fission yeast Sop2p: a novel and evolutionarily conserved protein that interacts with Arp3p and modulates profilin function. Embo J. 1996;15:6426–6437. [PMC free article] [PubMed] [Google Scholar]
- Chang F, Drubin D, Nurse P. cdc12p, a protein required for cytokinesis in fission yeast, is a component of the cell division ring and interacts with profilin. J Cell Biol. 1997;137:169–182. doi: 10.1083/jcb.137.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhabra ES, Higgs HN. The many faces of actin: matching assembly factors with cellular structures. Nat Cell Biol. 2007;9:1110–1121. doi: 10.1038/ncb1007-1110. [DOI] [PubMed] [Google Scholar]
- Choi CK, Vicente-Manzanares M, Zareno J, Whitmore LA, Mogilner A, Horwitz AR. Actin and alpha-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner. Nat Cell Biol. 2008;10:1039–1050. doi: 10.1038/ncb1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenmann KM, West RA, Hildebrand D, Kitchen SM, Peng J, Sigler R, Zhang J, Siminovitch KA, Alberts AS. T cell responses in mammalian diaphanous-related formin mDia1 knock-out mice. J Biol chem. 2007;282:25152–25158. doi: 10.1074/jbc.M703243200. [DOI] [PubMed] [Google Scholar]
- Fackler OT, Grosse R. Cell motility through plasma membrane blegging. J Cell Biol. 2008;181:879–884. doi: 10.1083/jcb.200802081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faix J, Grosse R. Staying in shape with formins. Dev Cell. 2006;10:693–706. doi: 10.1016/j.devcel.2006.05.001. [DOI] [PubMed] [Google Scholar]
- Favaro PM, de Souza Medina S, Traina F, Basseres DS, Costa FF, Saad ST. Human leukocyte formin: a novel protein expressed in lymphoid malignancies and associated with Akt. Biochem Biophys Res Commun. 2003;311:365–371. doi: 10.1016/j.bbrc.2003.10.012. [DOI] [PubMed] [Google Scholar]
- Feierbach B, Chang F. Roles of the fission yeast formin for3p in cell polarity, actin cable formation and symmetric cell division. Curr Biol. 2001;11:1656–1665. doi: 10.1016/s0960-9822(01)00525-5. [DOI] [PubMed] [Google Scholar]
- Glotzer M. The molecular requirements for cytokinesis. Science. 2005;307:1735–1739. doi: 10.1126/science.1096896. [DOI] [PubMed] [Google Scholar]
- Goode BL, Eck MJ. Mechanism and function of formins in the control of actin assembly. Annu Rev Biochem. 2007;76:593–627. doi: 10.1146/annurev.biochem.75.103004.142647. [DOI] [PubMed] [Google Scholar]
- Gupton SL, Eisenmann K, Alberts AS, Waterman-Storer CM. mDia2 regulates actin and focal adhesion dynamics and organization in the lamella for efficient epithelial cell migration. J Cell Sci. 2007;120:3475–3487. doi: 10.1242/jcs.006049. [DOI] [PubMed] [Google Scholar]
- Harriman GC, Brewer M, Bennett R, Kuhn C, Bazin M, Larosa G, Skerker P, Cochran N, Gallant D, Baxter D, et al. Selective cell adhesion inhibitors: Barbituric acid based alpha4beta7--MAdCAM inhibitors. Bioorg Med Chem Lett. 2008;18:2509–2512. doi: 10.1016/j.bmcl.2007.07.068. [DOI] [PubMed] [Google Scholar]
- Harris ES, Li F, Higgs HN. The mouse formin, FRLalpha, slows actin filament barbed end elongation, competes with capping protein, accelerates polymerization from monomers, and severs filaments. J Biol Chem. 2004 doi: 10.1074/jbc.M312718200. [DOI] [PubMed] [Google Scholar]
- Higashida C, Miyoshi T, Fujita A, Oceguera-Yanez F, Monypenny J, Andou Y, Narumiya S, Watanabe N. Actin polymerization-driven molecular movement of mDia1 in living cells. Science. 2004;303:2007–2010. doi: 10.1126/science.1093923. [DOI] [PubMed] [Google Scholar]
- Higgs HN. Formin proteins: a domain-based approach. Trends Biochem Sci. 2005;30:342–353. doi: 10.1016/j.tibs.2005.04.014. [DOI] [PubMed] [Google Scholar]
- Hotulainen P, Lappalainen P. Stress fibers are generated by two distinct actin assembly mechanisms in motile cells. J Cell Biol. 2006;173:383–394. doi: 10.1083/jcb.200511093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitzing TM, Sahadevan AS, Brandt DT, Knieling H, Hannemann S, Fackler OT, Grosshans J, Grosse R. Positive feedback between Dia1, LARG, and RhoA regulates cell morphology and invasion. Genes Dev. 2007;21:1478–1483. doi: 10.1101/gad.424807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar DR. Molecular details of formin-mediated actin assembly. Curr Opin Cell Biol. 2006;18:11–17. doi: 10.1016/j.ceb.2005.12.011. [DOI] [PubMed] [Google Scholar]
- Kovar DR, Harris ES, Mahaffy R, Higgs HN, Pollard TD. Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell. 2006;124:423–435. doi: 10.1016/j.cell.2005.11.038. [DOI] [PubMed] [Google Scholar]
- Kovar DR, Kuhn JR, Tichy AL, Pollard TD. The fission yeast cytokinesis formin Cdc12p is a barbed end actin filament capping protein gated by profilin. J Cell Biol. 2003;161:875–887. doi: 10.1083/jcb.200211078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar DR, Pollard TD. Insertional assembly of actin filament barbed ends in association with formins produces piconewton forces. Proc Natl Acad Sci U S A. 2004;101:14725–14730. doi: 10.1073/pnas.0405902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar DR, Wu JQ, Pollard TD. Profilin-mediated Competition between Capping Protein and Formin Cdc12p during Cytokinesis in Fission Yeast. Mol Biol Cell. 2005;16:2313–2324. doi: 10.1091/mbc.E04-09-0781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Carbona S, Le Goff C, Le Goff X. Fission yeast cytoskeletons and cell polarity factors: connecting at the cortex. Biol Cell. 2006;98:619–631. doi: 10.1042/BC20060048. [DOI] [PubMed] [Google Scholar]
- Lehar J, Stockwell BR, Giaever G, Nislow C. Combination chemical genetics. Nat Chem Biol. 2008;4:674–681. doi: 10.1038/nchembio.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez A, Parsons AB, Nislow C, Giaever G, Boone C. Chemical-genetic approaches for exploring the mode of action of natural products. Prog Drug Res. 2008;66:239–271. doi: 10.1007/978-3-7643-8595-8_5. [DOI] [PubMed] [Google Scholar]
- Marks J, Hyams JS. Localization of F-actin through the cell division cycle of Schizosaccharomyces pombe. Eur J Cell Biol. 1985;39:27–32. [Google Scholar]
- Martin SG, Chang F. Dynamics of the formin for3p in actin cable assembly. Curr Biol. 2006;16:1161–1170. doi: 10.1016/j.cub.2006.04.040. [DOI] [PubMed] [Google Scholar]
- McCollum D, Feoktistova A, Morphew M, Balasubramanian M, Gould KL. The Schizosaccharomyces pombe actin-related protein, Arp3, is a component of the cortical actin cytoskeleton and interacts with profilin. Embo J. 1996;15:6438–6446. [PMC free article] [PubMed] [Google Scholar]
- Michelot A, Berro J, Guerin C, Boujemaa-Paterski R, Staiger CJ, Martiel JL, Blanchoin L. Actin-filament stochastic dynamics mediated by ADF/cofilin. Curr Biol. 2007;17:825–833. doi: 10.1016/j.cub.2007.04.037. [DOI] [PubMed] [Google Scholar]
- Moseley JB, Goode BL. The yeast actin cytoskeleton: from cellular function to biochemical mechanism. Microbiol Mol Biol Rev. 2006;70:605–645. doi: 10.1128/MMBR.00013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley JB, Sagot I, Manning AL, Xu Y, Eck MJ, Pellman D, Goode BL. A conserved mechanism for Bni1- and mDia1-induced actin assembly and dual regulation of Bni1 by Bud6 and profilin. Mol Biol Cell. 2004;15:896–907. doi: 10.1091/mbc.E03-08-0621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano K, Imai J, Arai R, Toh EA, Matsui Y, Mabuchi I. The small GTPase Rho3 and the diaphanous/formin For3 function in polarized cell growth in fission yeast. J Cell Sci. 2002;115:4629–4639. doi: 10.1242/jcs.00150. [DOI] [PubMed] [Google Scholar]
- Naumanen P, Lappalainen P, Hotulainen P. Mechanisms of actin stress fibre assembly. J Microsc. 2007;231:446–454. doi: 10.1111/j.1365-2818.2008.02057.x. [DOI] [PubMed] [Google Scholar]
- Neidt EM, Skau CT, Kovar DR. The cytokinesis formins from the nematode worm and fission yeast differentially mediate actin filament assembly. J Biol Chem. 2008;283:23872–23883. doi: 10.1074/jbc.M803734200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolen BJ, Tomasevic N, Russell A, Pierce DW, Jia Z, McCormick CD, Hartman J, Sakowicz R, Pollard TD. Characterization of two classes of small molecule inhibitors of Arp2/3 complex. Nature. 2009;460:1031–1035. doi: 10.1038/nature08231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul AS, Pollard TD. The role of the FH1 domain and profilin in formin-mediated actin-filament elongation and nucleation. Curr Biol. 2008;18:9–19. doi: 10.1016/j.cub.2007.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen J, Nielsen O, Egel R, Hagan IM. FH3, a domain found in formins, targets the fission yeast formin Fus1 to the projection tip during conjugation. J Cell Biol. 1998;141:1217–1228. doi: 10.1083/jcb.141.5.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riveline D, Zamir E, Balaban NQ, Schwarz US, Ishizaki T, Narumiya S, Kam S, Geiger B, Bershadsky AD. Focal contacts as mechanosensors: externally applied local mechanical force induces growth of focal contacts by an mDia1-dependent and ROCK-independent mechanism. J Cell Biol. 2001;153:1175–1186. doi: 10.1083/jcb.153.6.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero S, Le Clainche C, Didry D, Egile C, Pantaloni D, Carlier MF. Formin is a processive motor that requires profilin to accelerate actin assembly and associated ATP hydrolysis. Cell. 2004;119:419–429. doi: 10.1016/j.cell.2004.09.039. [DOI] [PubMed] [Google Scholar]
- Sahai E. Mechanisms of cancer cell invasion. Curr Opin Gen Dev. 2005;15:87–96. doi: 10.1016/j.gde.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Sarmiento C, Wang W, Dovas A, Yamaguchi H, Sidani M, El-Sabai M, DesMarais V, Holman HA, Kitchen S, Backer JM, et al. WASP family members and formin proteins coordinate regulation of cell protrusions in carcinoma cells. J Cell Biol. 2008;180:1245–1260. doi: 10.1083/jcb.200708123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavylonis D, Kovar DR, O’Shaughnessy B, Pollard TD. Model of formin-associated actin filament elongation. Mol Cell. 2006;21:455–466. doi: 10.1016/j.molcel.2006.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallar BJ, Alberts AS. The formins: active scaffolds that remodel the cytoskeleton. Trends Cell Biol. 2003;13:435–446. doi: 10.1016/s0962-8924(03)00153-3. [DOI] [PubMed] [Google Scholar]
- Watanabe N, Kato T, Fujita A, Ishizaki T, Narumiya S. Cooperation between mDia1 and ROCK in Rho-induced actin reorganization. Nat Cell Biol. 1999;1:136–143. doi: 10.1038/11056. [DOI] [PubMed] [Google Scholar]
- Watanabe S, Ando Y, Yasuda S, Hosoya H, Watanabe N, Ishizaki T, Narumiya S. mDia2 induces the actin scaffold for the contractile ring and stabilizes its position during cytokinesis in NIH 3T3 cells. Mol Biol Cell. 2008;19:2328–2338. doi: 10.1091/mbc.E07-10-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Win TZ, Gachet Y, Mulvihill DP, May KM, Hyams JS. Two type V myosins with non-overlapping functions in the fission yeast Schizosaccharomyces pombe: Myo52 is concerned with groth polarity and cytokinesis, Myo51 is a component of th cytokinetic actin ring. J Cell Sci. 2000;114:69–79. doi: 10.1242/jcs.114.1.69. [DOI] [PubMed] [Google Scholar]
- Worthylake RA, Burridge K. RhoA and ROCK promote migration by limiting membrane protrusions. J Biol Chem. 2003;278:13578–13584. doi: 10.1074/jbc.M211584200. [DOI] [PubMed] [Google Scholar]
- Zhu X-L, Liang L, Ding Y-Q. Overexpression of FMNL2 is closely related to metastasis of colorectal cancer. In J Colorectal Dis. 2008;23:1041–1047. doi: 10.1007/s00384-008-0520-2. [DOI] [PubMed] [Google Scholar]
- Zigmond SH, Evangelista M, Boone C, Yang C, Dar AC, Sicheri F, Forkey J, Pring M. Formin leaky cap allows elongation in the presence of tight capping proteins. Curr Biol. 2003;13:1820–1823. doi: 10.1016/j.cub.2003.09.057. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.