Abstract
The tumour microenvironment can be a potent carcinogen, not only by facilitating cancer progression and activating dormant cancer cells, but also by stimulating tumour formation1. We have previously investigated stromelysin-1/matrix metalloproteinase-3 (MMP-3), a stromal enzyme upregulated in many breast tumours2, and found that MMP-3 can cause epithelial–mesenchymal transition (EMT) and malignant transformation in cultured cells3–5, and genomically unstable mammary carcinomas in transgenic mice3. Here we explain the molecular pathways by which MMP-3 exerts these effects: exposure of mouse mammary epithelial cells to MMP-3 induces the expression of an alternatively spliced form of Rac1, which causes an increase in cellular reactive oxygen species (ROS). The ROS stimulate the expression of the transcription factor Snail and EMT, and cause oxidative damage to DNA and genomic instability. These findings identify a previously undescribed pathway in which a component of the breast tumour microenvironment alters cellular structure in culture and tissue structure in vivo, leading to malignant transformation.
Cancer is characterized by a progressive series of alterations that disrupt cell and tissue homeostasis. Whereas many of these alterations can be induced by specific mutations, faulty signals from the microenvironment also can act as inducers of tumour development and progression1. MMPs are prominent contributors to such micro-environmental signals, because these proteolytic enzymes degrade structural components of the extracellular matrix (ECM), permitting tumour invasion and metastasis. Additionally, MMPs can release cell-bound inactive precursor forms of growth factors, degrade cell–cell and cell–ECM adhesion molecules, activate precursor zymogen forms of other MMPs, and inactivate inhibitors of MMPs and other proteases6. Our observations that MMP-3 can induce transformation in mammary epithelial cells in culture4,5,7 and in transgenic mice3 prompted investigations into the underlying molecular mechanisms. Here we show that MMP-3 can lead directly to genomic instability and EMT, and identify the pathway that mediates these events.
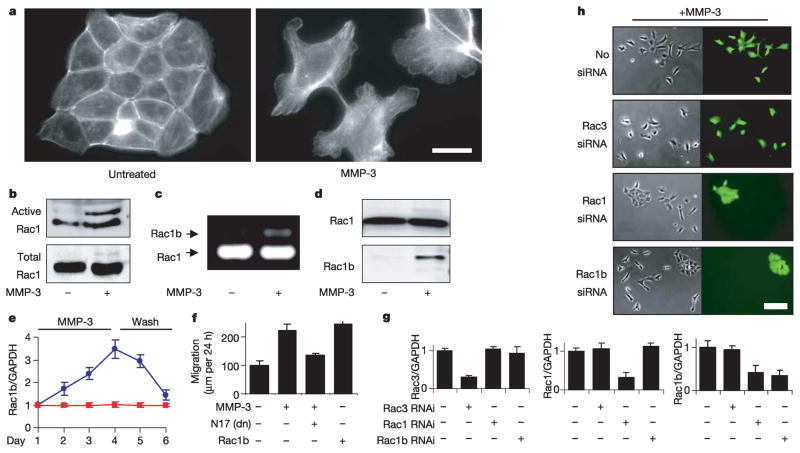
Induction of EMT by treatment of SCp2 mouse mammary epithelial cells with MMP-3 is associated with a loss of intact E-cadherin, increased motility and invasiveness, downmodulation of epithelial markers, and upregulation of mesenchymal markers (refs 4, 5; Supplementary Fig. 1a,b), through a process that is initially reversible (refs 4, 5; Supplementary Fig. 1c). The MMP-3-induced alteration of the F-actin cytoskeleton (Fig. 1a) indicated the possible involvement of members of the Rho GTPase family, and although the activities of RhoA and Cdc42 were unchanged (not shown), we were intrigued by an additional band in the Rac activity assay of MMP-3-treated cells (Fig. 1b). A highly activated splice isoform of Rac1, designated Rac1b, containing 57 additional nucleotides that result in an in-frame insertion of 19 additional amino acid residues, was discovered recently in breast and colorectal tumours8,9 and has transforming characteristics when exogenously expressed in cultured cells10. We identified the additional Rac band induced by MMP-3 as Rac1b by polymerase chain reaction with reverse transcription (RT–PCR) (Fig. 1c) and through the use of an antibody raised against the mouse Rac1b insertion sequence (Fig. 1d); we also found that induction of Rac1b by treatment with MMP-3 was initially reversible (Fig. 1e). We determined that the activity of Rac1b was required for the MMP-3-induced alterations in vimentin expression (Supplementary Fig. 2) and for MMP-3-induced motility (Fig. 1f), because dominant-negative Rac1N17 attenuated the effects of MMP-3, and expression of Rac1b could substitute for MMP-3 (Fig. 1f).
Figure 1. MMP-3 induces EMT through Rac1b.
a, MMP-3-induced alterations in actin cytoskeleton. Scale bar, 25μm. b, Analysis of active and total levels of Rac. c, RT–PCR of Rac1 and Rac1b. d, Rac1b protein expression. e, Rac1b transcript levels in response to MMP-3 treatment (days 1–4) and washout (days 5–6); blue circles, treated; red squares, untreated. f, Cell motility assessed by scratch assay. dn, dominant-negative. g, Quantification of knockdown of endogenous gene expression. h, Selective knockdown of Rac1b inhibits MMP-3-induced cell scattering. Scale bar, 25μm. For all graphs, error bars represent s.e.m.
We also evaluated the relationship between the induction of Rac1b and downstream EMT by specific transcript knockdown with short interfering RNA (siRNA). SCp2 cells were co-transfected transiently with yellow fluorescent protein (YFP), YFP-Rac1 or YFP-Rac1b, and either no siRNA, siRNA targeting Rac3 (as negative control), siRNA targeting Rac1 (which also targets Rac1b) or siRNA selectively targeting the splice insertion sequence in Rac1b. We found that Rac1 siRNA blocked the expression of co-transfected YFP-Rac1b, and that the specific Rac1b siRNA blocked the expression of co-transfected YFP-Rac1b only, not YFP-Rac1 (Supplementary Fig. 3); none of the siRNAs affected the expression of co-transfected YFP. The effect on endogenous gene expression levels was consistent with effective knockdown of all transiently transfected cells (about 70% transfection efficiency) and showed the following: Rac1 siRNA inhibits the expression of both Rac1 and Rac1b but does not affect Rac3; Rac1b siRNA selectively targets Rac1b and does not affect Rac1 or Rac3; and Rac3 siRNA selectively targets Rac3 and does not affect expression of Rac1 or Rac1b (Fig. 1g). When SCp2 cells were transiently co-transfected with YFP and either no siRNA or siRNA targeting Rac3, Rac1/Rac1b or Rac1b, and then treated with MMP-3 for 4 days, we observed that siRNA for Rac1/Rac1b or Rac1b inhibited MMP-3-induced cell motility in the co-transfected colonies, whereas siRNA targeting Rac3 had no effect (Fig. 1h).
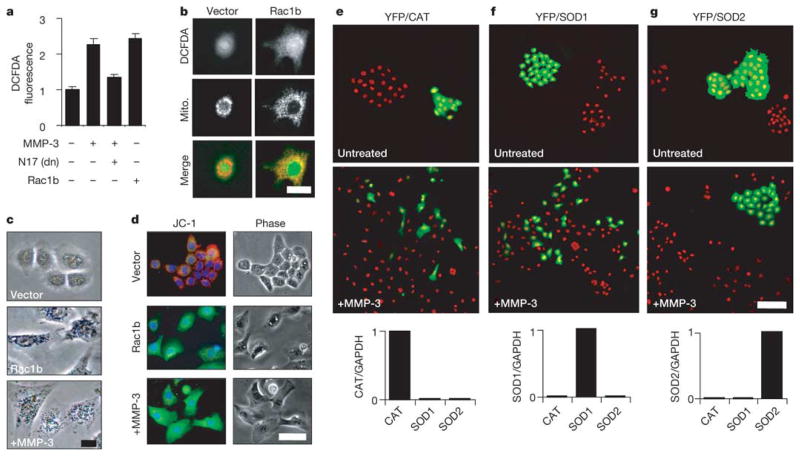
How can increased Rac activity lead to the diverse alterations induced by MMP-3? Previous studies11,12 showed that active Rac can stimulate the production and release of mitochondrial superoxide into the cytoplasm. The production of excess superoxide can cause oxidative DNA damage and genomic instability13, transform cells in culture14 and potentiate tumour progression15; in addition, super-oxide is readily converted to other forms of ROS that stimulate further tumorigenic processes15–17. We found that treatment with MMP-3 or the expression of Rac1b produced increases in cellular ROS, as assessed by the fluorophore dichlorodihydrofluorescein diacetate (DCFDA), and that the expression of Rac1N17 attenuated the induction of ROS by MMP-3 (Fig. 2a). The DCFDA fluorescence partly colocalized with a mitochondrial marker protein (Fig. 2b), and the identity of the induced ROS as mitochondrial superoxide was indicated by the staining pattern of nitroblue tetrazolium (Fig. 2c), which forms an insoluble blue formazan in the presence of super-oxide12, and by the altered fluorescence pattern of cells stained with JC-1, in which the punctate red mitochondrial staining of the J-aggregate of JC-1 was replaced by diffuse cytoplasmic green staining of the monomeric form (Fig. 2d), consistent with the dissipation of membrane potential after the mitochondrial production of super-oxide12,18. To determine whether the induction of mitochondrial superoxide by MMP-3/Rac1b was essential for the induction of EMT, we co-transfected cells with expression plasmids encoding YFP and either catalase, superoxide dismutase 1 (SOD1) or SOD2. Catalase stimulates the decomposition of H2O2 into water and molecular oxygen, whereas SOD1 and SOD2 convert superoxide into H2O2 and molecular oxygen; catalase and SOD1 are cytoplasmic enzymes, whereas SOD2 is localized to the mitochondria. These experiments demonstrated that YFP/SOD2 cells were resistant to MMP-3-induced scattering (Fig. 2g), whereas YFP/catalase and YFP/SOD1 cells responded in a similar fashion to adjacent untransfected cells (Fig. 2e, f).
Figure 2. MMP-3/Rac1b stimulate mitochondrial production of ROS.
a, Cellular ROS levels assessed by DCFDA. dn, dominant-negative. Error bars represent s.e.m. b, Mitochondrial pattern of DCFDA fluorescence. Scale bar, 25μm. c, Precipitation of nitroblue tetrazolium. Scale bar, 15μm. d, Mitochondrial depolarization assessed with JC-1. Scale bar, 50μm. e–g, Cells co-transfected with EYFP and catalase (CAT; e), superoxide dismutase 1 (SOD1; f) or superoxide dismutase 2 (SOD2; g) and then cultured in the absence (top) or presence (middle) of MMP-3 for 6 days. Green, EYFP fluorescence; red, nuclei. Graphs at bottom show gene transcript levels in transfected cell populations. Scale bar, 100μm.
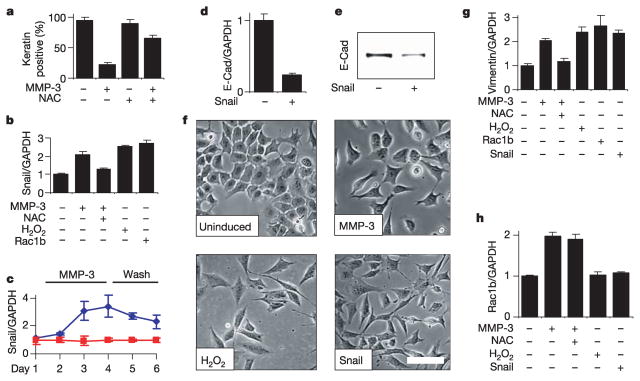
ROS can alter gene expression15–17 and stimulate cell invasiveness19, and we found that the ROS-quenching agent N-acetyl cysteine (NAC) effectively inhibited the MMP-3-induced downregulation of epithelial cytokeratins (Fig. 3a) and the upregulation of mesenchymal vimentin (Fig. 3g). NAC also inhibited MMP-3-induced cell motility, invasion and morphological alterations (not shown). The induction of EMT involves the coordinated regulation of many genes20; here we focused on MMP-3/ROS-dependent alterations in the expression levels of transcriptional regulatory proteins that mediate EMT. We determined that MMP-3 enhances expression of the transcription factor Snail21,22 (Fig. 3c), and that this effect could be blocked by treatment with NAC, or induced in the absence of MMP-3 by elevating ROS levels with H2O2 or by the expression of Rac1b (Fig. 3b). Expression of Snail in SCp2 cells was sufficient to induce EMT: induction caused the downmodulation of E-cadherin transcript and protein levels (Fig. 3d, e) and led to cell scattering comparable to that induced by MMP-3 or H2O2 (Fig. 3f). We also found that whereas MMP-3, Rac1b, H2O2 or Snail can stimulate the expression of mesenchymal vimentin (Fig. 3g), only MMP-3 could stimulate the expression of Rac1b (Fig. 3h). When combined with the data presented in Figs 1b–f and 2a–d, these results show that treatment with MMP-3 stimulates the expression of Rac1b, which causes increases in cellular ROS, leading in turn to an increased expression of Snail and to EMT.
Figure 3. MMP-3-induced EMT is dependent on ROS.
a, NAC inhibits MMP-3-induced downregulation of epithelial cytokeratin protein levels. b, Induction of Snail by MMP-3, and dependence on ROS. c, Snail transcript levels in response to MMP-3 treatment (days 1–4) and washout (days 5–6); blue diamonds, treated; red squares, untreated. d, e, Exogenous expression of Snail in SCp2 cells reduces E-cadherin transcript (d) and protein levels (e). f, Cell scattering induced by treatment with MMP-3 or H2O2, or by exogenous expression of Snail. Scale bar, 50μm. g, h, ROS and Snail dependence of vimentin (g) and Rac1b (h) expression. For all graphs, error bars represent s.e.m.
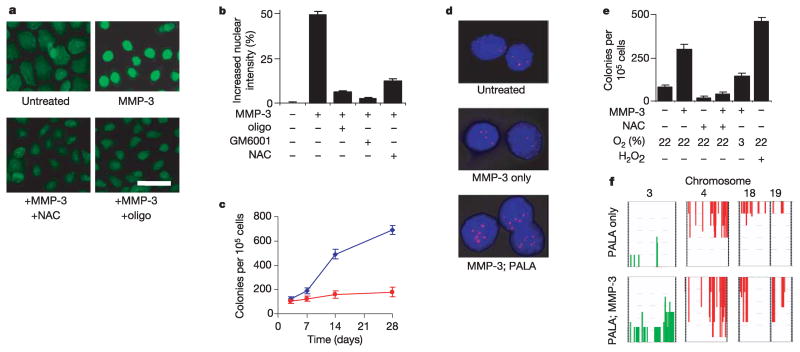
We had previously found that tumours in the MMP-3-expressing transgenic mice showed common patterns of genomic rearrangements3, indicating that MMP-3 might lead to genomic instability in target epithelial cells in vivo. Given the known genotoxic effects of ROS, we investigated the effects of MMP-3-induced ROS on the integrity of the genome under defined conditions in culture. To test for DNA damage, we used fluorescein isothiocyanate (FITC)-conjugated avidin, because this reagent binds to 8-oxodeoxyguanosine, an oxidative DNA lesion with a structural similarity to biotin23. Cells treated with MMP-3 showed significantly increased FITC-avidin nuclear staining (Fig. 4a) that was blocked by preincubation with an oligonucleotide containing 8-oxodeoxyguanosine (but not with a control oligonucleotide; not shown), by inhibiting the proteolytic activity of MMP-3 with GM6001, or by treatment with NAC (Fig. 4b). To test for induction of genomic instability, we assayed for increased resistance of MMP-3-treated SCp2 mouse mammary epithelial cells to N-(phosphonacetyl)-L-aspartate (PALA)24, because resistance to PALA is acquired through amplification of the CAD gene, which encodes carbamoylphosphate synthetase/aspartate carbamyltransferase/dihydroorotase25. Exposure to MMP-3 led to a progressive increase in the fraction of cells that had acquired PALA resistance (Fig. 4c), an increase that was due to amplification of the CAD locus (Fig. 4d). This effect could also be inhibited by treatment with NAC or by culturing under reduced oxygen tension, and could be reproduced in the absence of MMP-3 by treatment with H2O2 (Fig. 4e). That the genomic instability induced by MMP-3 was not limited to the CAD locus was shown by comparative genomic hybridization analysis, because many additional genomic amplifications and deletions were found in MMP-3-treated cells (Fig. 4f), including characteristic alterations previously observed in tumours derived from the MMP-3 transgenic mice3.
Figure 4. MMP-3-induced ROS cause DNA damage and genomic instability.
a, b, 8-Oxoguanosine-induced treatment with MMP-3 (a; scale bar, 50μm) and quantification of increased nuclear staining relative to untreated (b; error bars, 95% confidence intervals). c, Induction of PALA resistance by MMP-3 (blue diamonds, MMP-3; red squares, untreated). d, Fluorescence in situ hybridization of the CAD gene locus (red). e, ROS and oxygen dependence of PALA resistance induced by 14 days of treatment with MMP-3. f, Frequency plots of comparative genomic hybridization analyses of cells grown in the absence (top) or presence (bottom) of MMP-3, and then selected with PALA. Green, amplifications; red, deletions. For all graphs, error bars represent s.e.m.
Our results show that a key event in the MMP-3-induced malignant transformation of SCp2 cells is the induction of Rac1b, an alternative splice isoform of Rac1 that was initially identified in breast and colon cancers8,9. Many oncogenic splice isoforms are induced in cancers26, and although most of these produce proteins that lack key functional domains and act by sequestering factors involved in tumour suppressive pathways into nonfunctional complexes, Rac1b is unusual in that it becomes more highly activated27,28. The fact that Rac1b is the only apparent splice isoform of Rac1 found in MMP-3-treated cells is also notable, because Rac1b is also the only apparent splice isoform in breast cancer cells9.
How does treatment with MMP-3 lead to alternative splicing of Rac1b? It is clear that the extracellular proteolytic activity of MMP-3 is essential (Supplementary Fig. 4). We have shown that MMP-3 effectively cleaves E-cadherin, resulting in a loss of cell–cell adhesions and the relocalization of transcriptionally active β-catenin to the nucleus (refs 4, 5; not shown). It is important to note that MMP-3 is not the only protease capable of initiating this pathway, as we have found that MMP-9 (but not MMP-2) can substitute for MMP-3 in our experimental system (not shown), and MMP-7 and MMP-14 are also known to induce tumours when expressed in transgenic mice2. Furthermore, MMPs are not the only microenvironmental components implicated in tumour induction or progression. Oncogenic properties have also been attributed to transforming growth factor-β, growth factors, and hormones, and the tumour-promoting activities of chronic inflammation are well known1. Our investigations of MMP-3 show how this factor can directly stimulate phenotypic and genotypic malignant transformation in normally functioning cells. We expect that similar or parallel pathways may be induced by other elements of the tumour microenvironment, and we suspect that such mechanisms may be much more relevant for the generation of genomic instability than predicted by current models of tumour progression.
METHODS
Cell culture, antibodies and plasmids
Cell culture was performed as described previously4,5; for gene repression, a 5 mg ml−1 stock solution of tetracycline in 100% ethanol was diluted 1:1,000 into culture medium and changed daily. To stimulate cells with MMP-3, we used medium that had been conditioned by SCp2 cells containing the tetracycline-regulated, autoactivated MMP-3 construct4,5 with expression induced by growth in the absence of tetracycline; conditioned medium from cells repressed by treatment with tetracycline was used for controls. This conditioned medium was analysed by zymography to verify that MMP-3 was the only MMP being expressed, and that EMT was induced by extracellular proteolytic activity (Supplementary Fig. 4). Except as otherwise indicated, cells were incubated for 4 days in the presence of conditioned medium containing MMP-3 and for 7 days with 25μM H2O2. NAC was used at a concentration of 10 mM. Antibodies against cytokeratin and vimentin were described previously4,5. Rac antibody was from Upstate, and the Rac1b antibody was raised against the peptide Ac-CGKDRPSRGKDKPIA-amide (antibody validation in Supplementary Fig. 5). Human catalase complementary DNA was obtained from R. Arnold, and human SOD1 and SOD2 cDNAwere obtained from T.-T. Huang. SOD1, SOD2 and catalase were cloned into pcDNA3.1 expression vectors; all other constructs were subcloned into the tetracycline-repressible expression system used previously for the expression of MMP-3 (described in refs 4, 5). Rac1 and Rac1b were cloned from SCp2 cDNA and expressed as unmodified proteins or as fused with YFP. Rac1V12 and Rac11N17 mutants of Rac1 (Supplementary Figs 6 and 7), and aslo the catalytically inactive E217A mutant of MMP-3, were generated with the Quickchange mutagenesis kit (Stratagene); the sequences of all modified plasmids were verified. Transcript levels were assessed with RT–PCR by isolating RNA (Tripure; Roche Diagnostics), synthesizing cDNA and performing quantitative, real-time PCR (Lightcycler, Roche Diagnostics); all these experiments were normalized to GAPDH. For analysis of Rac1 and Rac1b (Fig. 1c), oligonucleotide primers that hybridize to sequences flanking the splice insertion site were used; for specific analysis of Rac1b (Figs 1d and 3h), oligonucleotide primers specific for the Rac1b splice isoform were used.
Rho GTPase assays
Cells were lysed in glutathione S-transferase (GST)-Fish buffer (10% glycerol, 50 mM Tris-HCl pH 7.4, 100 mM NaCl, 1% Nonidet P40, 2 mM MgCl2, 10 μg ml−1 leupeptin, 10 μg ml−1 pepstatin, 10μg ml−1 aprotinin, 10μg ml−1 E64 and 1 mM Pefabloc). Equal amounts of protein supernatants were incubated on ice for 45 min with GST-PAK-CD (Rac-binding domain and Cdc42-binding domain) or GST-C21 (Rho-binding domain) fusion protein-coated Sepharose beads. The beads were washed, eluted in sample buffer, and then analysed by SDS–polyacrylamidegel electrophoresis and western blotting with antibodies against Rac, Cdc42 and Rho. Dominant-negative and consitutively active Rac1 expression constructs were provided by D. Kalman. Rac1 and Rac3 siRNA were SMARTpool reagents (Dharmacon), whereas Rac1b siRNA used the sequence 5′-UGGAGACACAUGUGGUAAAGAUAGA-3′; siRNAs were transfected into SCp2 cells with Lipofectamine 2000 (Gibco) in accordance with the manufacturer’s protocols. For analysis of endogenous gene knockdown, RNA was harvested after 24 h and analysed by RT–PCR with primer pairs selective for Rac1, Rac1b or Rac3. For MMP-3-induced EMT, siRNA mixtures were co-transfected with YFP-C1 and then treated with MMP-3 for 4 days; they were then evaluated for scatter of fluorescent (co-transfected with YFP and siRNA) and nonfluorescent (non-transfected control) colonies.
8-Oxodeoxyguanosine analyses and genomic instability assays
To measure ROS concentrations, cells were incubated in the dark with 50 mM DCFDA (Molecular Probes) for 30 min in serum-free and phenol-red-free medium. For 8-oxodeoxyguanosine analysis, a modification of published techniques23,29 was used: cells were fixed in methanol (20 min, −20 °C), permeabilized with TBST (Tris-buffered saline containing 0.1% Triton X-100; 15 min, 25 °C), blocked for non-specific binding (TBST containing 15% fetal calf serum; 2 h, 25 °C), and stained with 15μg ml−1 FITC-conjugated avidin (Sigma; 1 h, 37 °C). To verify the specificity of staining, FITC-avidin was preincubated with a tenfold excess of either the blocking oligonucleotide 5′-GAA CTA GT°G ATC CCC CGG GTC GC-3′ (where °G is 8-oxodeoxyguanosine) or the control oligonucleotide 5′-GAA CTA GTG ATC CCC CGG GTC GC-3′. Images were captured with a Nikon Diaphot 300 microscope and Spot RT camera and software (Technical Instruments). Fluorescence was quantified with IMAGEJ (http://rsb.info.nih.gov/ij/index.html). For DCFDA staining, cellular fluorescence was quantified; for FITC-avidin staining, nuclear fluorescence was measured (with a 4,6-diamidino-2-phenylindole image mask). More than 250 measurements were made for each data point. JC-1 and nitroblue tetrazolium labelling were performed essentially as in ref. 12. The PALA assay and the comparative genomic hybridization analyses were performed using modifications of previously published protocols24,25,30, as described in the Supplementary Information.
Supplementary Material
Acknowledgments
We thank G. Stark for advice with the PALA assay; M. LaBarge, M. Adriance and other members of the Bissell laboratory for discussions; J. Campisi for critical reading of the manuscript; and C. Chen for technical assistance. This work was supported by grants from the OBER office of the Department of Energy and an Innovator award from the Department of Defense (to M.J.B.) and from the National Institutes of Health (to M.J.B. and Z.W.), and by fellowships from the American Cancer Society (D.C.R.), the National Cancer Institute (L.E.L.), the Department of Defense (H.L. and C.M.N.) and the California Breast Cancer Research Program (J.E.F).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Information Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests.
References
- 1.Bissell MJ, Radisky D. Putting tumours in context. Nature Rev Cancer. 2001;1:46–54. doi: 10.1038/35094059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sternlicht MD, Werb Z. How matrix metalloproteinases regulate cell behaviour. Annu Rev Cell Dev Biol. 2001;17:463–516. doi: 10.1146/annurev.cellbio.17.1.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sternlicht MD, et al. The stromal proteinase MMP3/stromelysin-1 promotes mammary carcinogenesis. Cell. 1999;98:137–146. doi: 10.1016/s0092-8674(00)81009-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lochter A, et al. Misregulation of stromelysin-1 expression in mouse mammary tumour cells accompanies acquisition of stromelysin-1-dependent invasive properties. J Biol Chem. 1997;272:5007–5015. doi: 10.1074/jbc.272.8.5007. [DOI] [PubMed] [Google Scholar]
- 5.Lochter A, et al. Matrix metalloproteinase stromelysin-1 triggers a cascade of molecular alterations that leads to stable epithelial-to-mesenchymal conversion and a premalignant phenotype in mammary epithelial cells. J Cell Biol. 1997;139:1861–1872. doi: 10.1083/jcb.139.7.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nature Rev Cancer. 2002;2:161–174. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- 7.Boudreau N, Sympson CJ, Werb Z, Bissell MJ. Suppression of ICE and apoptosis in mammary epithelial cells by extracellular matrix. Science. 1995;267:891–893. doi: 10.1126/science.7531366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jordan P, Brazao R, Boavida MG, Gespach C, Chastre E. Cloning of a novel human Rac1b splice variant with increased expression in colorectal tumors. Oncogene. 1999;18:6835–6839. doi: 10.1038/sj.onc.1203233. [DOI] [PubMed] [Google Scholar]
- 9.Schnelzer A, et al. Rac1 in human breast cancer: overexpression, mutation analysis, and characterization of a new isoform, Rac1b. Oncogene. 2000;19:3013–3020. doi: 10.1038/sj.onc.1203621. [DOI] [PubMed] [Google Scholar]
- 10.Singh A, et al. Rac1b, a tumour associated, constitutively active Rac1 splice variant, promotes cellular transformation. Oncogene. 2004;23:9369–9380. doi: 10.1038/sj.onc.1208182. [DOI] [PubMed] [Google Scholar]
- 11.Kheradmand F, Werner E, Tremble P, Symons M, Werb Z. Role of Rac1 and oxygen radicals in collagenase-1 expression induced by cell shape change. Science. 1998;280:898–902. doi: 10.1126/science.280.5365.898. [DOI] [PubMed] [Google Scholar]
- 12.Werner E, Werb Z. Integrins engage mitochondrial function for signal transduction by a mechanism dependent on Rho GTPases. J Cell Biol. 2002;158:357–368. doi: 10.1083/jcb.200111028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Samper E, Nicholls DG, Melov S. Mitochondrial oxidative stress causes chromosomal instability of mouse embryonic fibroblasts. Aging Cell. 2003;2:277–285. doi: 10.1046/j.1474-9728.2003.00062.x. [DOI] [PubMed] [Google Scholar]
- 14.Suh YA, et al. Cell transformation by the superoxide-generating oxidase Mox1. Nature. 1999;401:79–82. doi: 10.1038/43459. [DOI] [PubMed] [Google Scholar]
- 15.Droge W. Free radicals in the physiological control of cell function. Physiol Rev. 2002;82:47–95. doi: 10.1152/physrev.00018.2001. [DOI] [PubMed] [Google Scholar]
- 16.Puri PL, et al. A myogenic differentiation checkpoint activated by genotoxic stress. Nature Genet. 2002;32:585–593. doi: 10.1038/ng1023. [DOI] [PubMed] [Google Scholar]
- 17.Finkel T. Oxidant signals and oxidative stress. Curr Opin Cell Biol. 2003;15:247–254. doi: 10.1016/s0955-0674(03)00002-4. [DOI] [PubMed] [Google Scholar]
- 18.Madesh M, Hajnoczky G. VDAC-dependent permeabilization of the outer mitochondrial membrane by superoxide induces rapid and massive cytochrome c release. J Cell Biol. 2001;155:1003–1015. doi: 10.1083/jcb.200105057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mori K, Shibanuma M, Nose K. Invasive potential induced under long-term oxidative stress in mammary epithelial cells. Cancer Res. 2004;64:7464–7472. doi: 10.1158/0008-5472.CAN-04-1725. [DOI] [PubMed] [Google Scholar]
- 20.Kalluri R, Neilson EG. Epithelial-mesenchymal transition and its implications for fibrosis. J Clin Invest. 2003;112:1776–1784. doi: 10.1172/JCI20530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nieto MA. The snail superfamily of zinc-finger transcription factors. Nature Rev Mol Cell Biol. 2002;3:155–166. doi: 10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- 22.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nature Rev Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 23.Struthers L, Patel R, Clark J, Thomas S. Direct detection of 8-oxodeoxyguanosine and 8-oxoguanine by avidin and its analogues. Anal Biochem. 1998;255:20–31. doi: 10.1006/abio.1997.2354. [DOI] [PubMed] [Google Scholar]
- 24.Johnson RK, Inouye T, Goldin A, Stark GR. Antitumor activity of N-(phosphonacetyl)-L-aspartic acid, a transition-state inhibitor of aspartate transcarbamylase. Cancer Res. 1976;36:2720–2725. [PubMed] [Google Scholar]
- 25.Wahl GM, Padgett RA, Stark GR. Gene amplification causes overproduction of the first three enzymes of UMP synthesis in N-(phosphonacetyl)-L-aspartate-resistant hamster cells. J Biol Chem. 1979;254:8679–8689. [PubMed] [Google Scholar]
- 26.Shin C, Manley JL. Cell signalling and the control of pre-mRNA splicing. Nature Rev Mol Cell Biol. 2004;5:727–738. doi: 10.1038/nrm1467. [DOI] [PubMed] [Google Scholar]
- 27.Matos P, Collard JG, Jordan P. Tumor-related alternatively spliced Rac1b is not regulated by Rho-GDP dissociation inhibitors and exhibits selective downstream signalling. J Biol Chem. 2003;278:50442–50448. doi: 10.1074/jbc.M308215200. [DOI] [PubMed] [Google Scholar]
- 28.Fiegen D, et al. Alternative splicing of Rac1 generates Rac1b, a self-activating GTPase. J Biol Chem. 2004;279:4743–4749. doi: 10.1074/jbc.M310281200. [DOI] [PubMed] [Google Scholar]
- 29.Neumann CA, et al. Essential role for the peroxiredoxin Prdx1 in erythrocyte antioxidant defence and tumour suppression. Nature. 2003;424:561–565. doi: 10.1038/nature01819. [DOI] [PubMed] [Google Scholar]
- 30.Snijders AM, et al. Shaping of tumour and drug-resistant genomes by instability and selection. Oncogene. 2003;22:4370–4379. doi: 10.1038/sj.onc.1206482. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.