Abstract
Previously, we have developed a unique in vitro LNCaP cell model, which includes androgen-dependent (LNCaP-C33), androgen-independent (LNCaP-C81) and an intermediate phenotype (LNCaP-C51) cell lines resembling the stages of prostate cancer progression to hormone independence. This model is advantageous in overcoming the heterogeneity associated with the prostate cancer up to a certain extent. We characterized and compared the gene expression profiles in LNCaP-C33 (androgen-dependent) and LNCaP-C81 (androgen-independent) cells using Affymetrix GeneChip array analyses. Multiple genes were identified exhibiting differential expression during androgen-independent progression. Among the important genes upregulated in androgen-independent cells were PCDH7, TPTE, TSPY, EPHA3, HGF, MET, EGF, TEM8, etc., whereas many candidate tumor suppressor genes (HTATIP2, CDKN2A, CDKN2B, CDKN1C, TP53, TP73, ICAM1, SOCS1/2, SPRY2, PPP2CA, PPP3CA, etc.) were decreased. Pathway prediction analysis identified important gene networks associated with growth-promoting and apoptotic signaling that were perturbed during androgen-independent progression. Further investigation of one of the genes, PPP2CA, which encodes the catalytic subunit of a serine phosphatase PP2A, a potent tumor suppressor, revealed that its expression was decreased in prostate cancer compared to adjacent normal/benign tissue. Furthermore, the downregulated expression of PPP2CA was significantly correlated with tumor stage and Gleason grade. Future studies on the identified differentially-expressed genes and signaling pathways may be helpful in understanding the biology of prostate cancer progression and prove useful in developing novel prognostic biomarkers and therapy for androgen-refractory prostate cancer.
Keywords: Prostate Cancer, Gene expression, Transcriptomic variation, Androgenin-independence, Gene-networks
INTRODUCTION
Prostate cancer is the most commonly diagnosed malignancy and second leading cause of cancer death in men in the United States [1]. It has been estimated that 218,890 new cases of prostate cancer will be diagnosed and 27,050 people will die due to prostate cancer in the year 2007 [1]. Clinical progression of prostate cancer is characterized by a transition from an androgen-dependent (AD) to an androgen-independent (AI) phenotype. Most patients treated by androgen deprivation therapy initially exhibit a dramatic regression of the androgen-dependent cancer cells; however, the tumor eventually progresses to an androgen-independent stage, resulting in a poor prognosis [2]. Therefore, an improved understanding of the molecular mechanisms underlying the AI progression of prostate cancer is required to develop novel effective therapies.
Initial insight into the molecular nature of prostate cancer progression can be gained by examining the gene expression patterns associated with AD and AI phenotypes. Prostate cancer, however, poses significant challenges to this approach due to the heterogeneity of primary and metastatic tumors [3]. To overcome this limitation, we have developed an in-vitro prostate cancer progression model consisting of early passage AD (LNCaP-C33), late passage AI (LNCaP-C81) and an intermediate-phenotype (LNCaP-C51) cell lines [4]. This model closely resembles with different progressive stages to hormone-independency and has been used previously to understand the disease mechanisms [4–6].
In this study, we have performed a genome-wide expression profiling and pathway prediction analyses in AD and AI prostate cancer cells to characterize the transcriptomic variation and identify the perturbed gene networks associated with the prostate cancer progression. Our study provides a list of candidate genes that could be useful for the development of new diagnostic/prognostic markers for human prostate cancer. Furthermore, it reveals that the androgen-independent progression of prostate cancer mainly involves a repression of cell signaling pathways. Functional studies on the identified differentially-expressed genes may be helpful in understanding the biology of prostate cancer progression and prove useful in developing novel treatment for androgen-refractory prostate cancer.
Materials and Methods
Cancer cell lines and tissue specimens
Human prostate cancer cell lines (LNCaP-C33, LNCaP-C81, LNCaP-C4-2, PC3 and DU145) were utilized in the study. LNCaP-C33 (androgen-sensitive) and LNCaP-C81 (androgen-independent) cell lines are of same genotypic lineage and serve as a good model for prostate cancer progression [4]. Furthermore, LNCaP cells express functional androgen receptors as is the case in majority of prostate carcinomas. All cell lines were maintained in the ATCC specified culture media supplemented with 10% FBS and 100µg/ml of penicillin-streptomycin (Gibco BRL, Grand Island, NY). Growth media were changed alternate days and the cells were trypsinized at near confluence. Prostate cancer tissue microarray containing 2 spots each from 35 cancer cases (formalin-fixed and paraffin-embedded) along with 1 spot from adjacent normal/benign tissue were obtained from a commercial source (Accumax™ Array, Petagen Inc., Seoul, Korea).
RNA isolation
Total RNA was extracted from cancer cell lines by using guanidine isothiocyanate-cesium chloride ultracentrifugation method and/or by using an RNeasy RNA isolation kit (Qiagen Inc., Valencia, CA). RNA concentration was measured spectrophotometrically, and its integrity was analyzed by electrophoresis on a formaldehyde agarose gel.
Affymetrix GeneChip array analysis
The mRNA expression profiles of LNCaP-C33 and -C81 cells were examined by Affymetrix Genechip microarray (Affymetrix, Santa Clara, CA, USA). Total RNA (5 µg) was reverse-transcribed, and biotin-labeled cRNA probes were generated using the Affymetrix labeling kit as per manufacturer’s instructions. Biotinylated fragmented cRNA probes were hybridized to the HGU133 plus2 Genechips (Affymetrix). Hybridization was performed at 45°C for 16 h in a hybridization oven (Affymetrix). The Genechips were then automatically washed and stained with streptavidin–phycoerythrin conjugate in an Affymetrix Genechip Fluidics Station. Fluorescence intensities were scanned using the Affymetrix GeneChip 3000 scanner in the UNMC microarray core facility. Quality metric parameters including noise level, background, and the efficiency of reverse transcription were ascertained for all hybridizations. The resultant microarray datasets were scaled to a target signal intensity of 500 using Affymetrix GCOS software. To identify differentially expressed genes and associated fold-change differences, the scaled intensities were compared to each other using Affymetrix comparison analysis software.
Pathway analysis
Pathway prediction analysis on the differentially expressed genes was performed using a web-based application ‘Ingenuity Pathway Analysis’ (Ingenuity Systems, Mountain View, CA). This web-delivered application searches its database to place differentially expressed genes in gene clusters linked to a molecular pathway(s) and is helpful in postulating the functional assumption from the large amount of gene expression data.
Quantitative reverse transcription-polymerase chain reaction (Q-RT-PCR)
Total RNA (2µg) from each of the prostate cancer cell line was reverse transcribed using the first-strand cDNA synthesis kit (Perkin Elmer, Branchburg, NJ) and oligo-d(T) primers according to the manufacturer’s instructions. Real-time PCR amplifications were carried out with 100 ng of first strand cDNA in 10-µl reaction volumes. The reaction mixture was subjected to a two step cyclic program (95°C for 10 min. followed by 40 cycles of 95°C for 15 sec. and 60°C for 1 min.) as per manufacturer’s protocol on ABI 7500 sequence detection system (Applied Biosystems, Foster City, CA) with SYBR chemistry. Pre-designed PCR primers for SPRY2, ALCAM, TPTE, HGF, MET, PTK6, PCDH7 and GAPDH were purchased from a commercial source (Superarray Biosciences Corporation, Frederick, MD). The relative fold difference in gene expression was calculated by ΔΔCT method [7] using GAPDH as normalization control.
Western-blot analysis
Total protein was extracted from prostate cancer cell lines, resolved and analyzed by western blotting as previously described [8]. In brief, cells were washed with cold-phosphate buffered-saline (PBS), scraped in RIPA buffer (100 mM Tris, 5 mM EDTA, 5% NP40, pH-8.0) containing protease inhibitors cocktail (Roche diagnostics, Mannheim, Germany) and allowed for lysis for at least 30 min on ice with intermittent vortexing. Cells were subjected to further lysis by one freeze-thaw cycle and centrifuged at 14,000×g for 30 min at 4°C. Supernatants were carefully removed and protein concentrations were determined by Bio-Rad-DC protein estimation kit. Electrophoresis was performed on polyacrylamide gel (10%) using equal amounts of protein samples under reducing conditions. Resolved proteins were transferred to the PVDF membranes and probed with specific primary antibodies [anti-PP2A rabbit Mab, Clone Y119 (Epitomics, Inc., Burlingame, California); anti-PGK rabbit polyclonal antibodies (a gift from Dr. J.K. Vishwanatha, UNMC)] followed by incubation with corresponding horseradish peroxidase-conjugated secondary antibodies (Amarsham Biosciences Buckinghamshire, UK). Signal was detected with ECL electro-chemiluminescence (ECL) Kit (Amarsham Biosciences).
Immunohistochemistry
Tissue microarray sections were deparaffinized using EZ-DeWax™ (Bio Genex, San Ramon, CA) and then rehydrated with graded alcohols. Heat-induced antigen retrieval was performed in citrate buffer (pH 6.0) by heating slides in a microwave oven at 700 W for 15 min. and the sections were processed for immunohistochemistry as described previously [8]. In brief, the tissue sections were washed three times with phosphate buffered saline (PBS) and incubated with 0.3% H2O2 in methanol: PBS (1:1) solution for 30 min to quench the endogenous peroxidase activity. Slides were then washed with PBS and incubated with normal serum (Vectastain ABC kit, Vector Laboratories, Burlingame, CA) for 30 min. for blocking the non-specific immunostaining. The sections were then incubated with a 1:100 dilution of anti-PP2A antibody at room temperature for one hour. Slides were washed (3 × 5 min) with PBS containing 0.05% Tween-20 (PBS-T) and incubated with biotinylated- secondary antibody for 30 min. After washing (3 × 5 min) with PBS-T, the slides were incubated with ABC solution (Vector Laboratories) at room temperature. The reaction color was developed by treating the tissue sections with 3, 3 –diaminobenzidine (DAB) substrate (DAB substrate kit, Vector Laboratories) as per the manufacturer's instructions. A reddish-brown precipitate indicated positive immunoreactivity. The slides were washed with water, counter-stained with hematoxylin, dehydrated and mounted with Vectamount permanent mounting media (Vector Laboratories).
Assessment of antigen staining
All slides were analyzed using a Nikon Eclipse E400 Microscope (Japan) and the intensity of immunoreactivity was scored. Staining intensity was graded on a 0 to 3 scale (0 for no staining, + for weak immunoreactivity; ++ for moderate immunoreactivity; and +++ for strong immunoreactivity). The percentage of cells that showed positive immunoreactivity within the normal epithelial/cancerous region of the section was scored as follows: 1 for 0–25%; 2 for 26–50%; 3 for 51–75%; and 4 for 76–100%. The values of the staining intensity and the percent of immunoreactive cells were multiplied to obtain a composite score ranging from 0–12
Statistical analysis
Paired t-tests were used to see if there was significant difference in the composite score values of PP2A, when comparing benign and prostate cancer samples.
RESULTS
A genome-wide survey of differentially-expressed genes in androgen dependent versus androgen-independent prostate cancer cells
Progression of prostate cancer from an AD phenotype to AI nature may involve alterations in the expression of multiple genes. To identify these genes, we aimed at characterizing and comparing the genome-wide expression profiles in LNCaP-C33 (AD) and LNCaP-C81 (AI) prostate cancer cells. LNCaP human prostate cancer cell model was employed because this model recapitulate many characteristics associated with progression of prostate cancer cells from AD to androgen-refractory phenotype [4].
Total RNA was extracted from the cell lines and analyzed using Affymetrix Genechips (Human Genome U133plus2 GeneChip) to determine the global gene expression patterns. The Human Genome U133plus2 GeneChip contains greater than 54,000 probe sets and can be used to analyze the expression level of more than 47,000 transcripts and variants, including approximately 38,500 well-characterized human genes. Over 2,000 probe sets were determined to be differentially expressed (with equal to and/or more than two-fold difference) in LNCaP-C33 vs. LNCaP-C81 cells (data not shown). Notably, in our study, more differentially-expressed genes were found to be under-expressed (n=1460) than upregulated (n=876) in AI prostate cancer cells. Table 1 and Table 2 present the list of some of the selected genes over-expressed and under-expressed, respectively, during the AI growth of LNCaP prostate cancer cells. We have selected these genes based on their fold-change in expression, comparison with the Oncomine cancer profiling database (www.oncomine.org) and established pathological significance in prostate and/or other cancers.
Table 1.
List of selected genes that are overexpressed in androgen-independent (LNCaP-C81) compared to androgen-dependent (LNCaP-33) prostate cancer cells.
| Gene | Product | Fold-change (Logbase2) |
Function | Genomic location |
|---|---|---|---|---|
| PCDH7 | Protocadherin 7 | 7.9 | Cell adhesion | 4p15 |
| NRXN3 | Neurexin 3 | 7.7 | " | 14q31 |
| TPTE/ PTEN2 |
Transmembrane phosphatase with tensin homology |
5.4 | Signal transduction | 21p11 |
| EPHA3 | EPH receptor A3 | 5.3 | " | 3p11.2 |
| TSPY | Testis specific protein, Y-linked | 4.5 | Hormone-response | Yp11.2 |
| HGF | Hepatocyte growth factor | 4.4 | Signal transduction | 7q21.1 |
| PKIA | Protein kinase inhibitor alpha | 4.4 | " | 8q21.12 |
| PRLR | Prolactin receptor | 3.8 | " | 5p14-p13 |
| NR2F1 | Nuclear receptor subfamily 2, group f, member 1 |
3.6 | Hormone-response | 5q14 |
| TNS | Tensin | 3.6 | Signal transduction, cell migration | 2q35-q36 |
| MET | Hepatocyte growth factor receptor | 3.2 | Signal transduction | 7q31 |
| CALN1 | Calneuron 1 | 3.2 | Calcium-binding protein | 7q11 |
| ADAM-TS3 | A disintegrin and metalloprotease with thrombospondin motifs-3 |
3.1 | propeptidase | 4q13.3 |
| NPL4 | Nuclear protein localization 4 | 3 | Protein trafficking | 17qter |
| MYO3A | Myosin IIIA | 3 | Actin-dependent motor protein | 10p11.1 |
| GNA11 | Guanine nucleotide binding protein (G protein), alpha 11 |
2.8 | Signal transduction | 19p13.3 |
| GAGEB1/PA GE1 |
P antigen family, member 1 (prostate associated) |
2.7 | Tumor-linked protein, function unknown |
Xp11.23 |
| AKAP9 | A kinase anchor protein 1 | 2.6 | Signal transduction | 17q21-q23 |
| TCF4 | Transcription factor 4 | 2.6 | " | 18q21.1 |
| MYLK | Myosin, light polypeptide kinase | 2.6 | cell migration | 3q21 |
| KLK2 | Kallikrein 2 | 2.4 | peptidase | 19q13.41 |
| HOXA10 | Homeo box A10 | 2.3 | Signal transduction | 7p15-p14 |
| DDR2 | Discoidin domain receptor family, member 2 | 2.2 | " | 1q12-q23 |
| EGF | Epidermal growth factor | 2.2 | Cell growth | 4q25 |
| TEM8 | Tumor endothelial marker 8 | 2.1 | angiogenesis | 2p13.1 |
| XAGE-1 | X antigen family, member 1 | 2 | Cancer/testis-associated gene, unknown function |
Xp11.22-p11.21 |
| GAS2 | Growth arrest-specific 2 | 2 | Regulation of cell shape, apoptosis | 11p14.3-p15.2 |
Table 2.
List of selected genes that are under-expressed in androgen-independent (LNCaP-C81) compared to androgen-dependent (LNCaP-33) prostate cancer cells.
| Gene | Product | Fold- change (Logbase2) |
Function | Genomic location |
|---|---|---|---|---|
| ANGPT2 | Angiopoietin 2 | 7.8 | Context-dependent vascular regression and angiogenesis | 8p23.1 |
| THBS1 | Thrombospondin 1 | 7.3 | Cell-cell and cell-extracellular matrix interaction | 15q15 |
| CCL5 | Chemokine (c-c motif) ligand 5 | 6.2 | Chemokine, immunoregulatory | 17q11.2-12 |
| PTK6 | Protein tyrosine kinase 6 | 6.1 | Signal transduction | 20q13.3 |
| CXCL11 | Chemokine (c-x-c motif) ligand 11 | 5.8 | Chemokine, immunoregulatory | 4q21.2 |
| S100P | S100 calcium binding protein p | 5.1 | Hormone response | 4p16 |
| PSMD5 | Proteasome 26s non-ATPase subunit, 5 | 4.9 | Multi-subunit protease | 9q33.2 |
| CDKN2B | Cyclin-dependent kinase inhibitor 2b | 4.7 | tumor suppressor | 9p21 |
| PEG3 | Paternally expressed 3 | 4.5 | " | 19q13.4 |
| HTATIP2 | Hiv-1 tat interactive protein 2 | 4.4 | " | 11p15.1 |
| CXCL2 | Chemokine (c-x-c motif) ligand 2 | 4.4 | Chemokine, immunoregulatory | 4q21 |
| RND1 | Rho family gtpase 1 | 4.2 | Cell motility | 12q12-q13 |
| NK4 | Natural killer4/Interleukin 32 | 4.1 | cytokine | 16p13.3 |
| ACPP | Prostatic acid phosphotase | 4.1 | Hormone response, growth signaling | 3q21-q23 |
| ICAM1 | Intercellular adhesion molecule 1 | 4 | tumor suppressor, cell migration | 19p13.3-13.2 |
| FOSL2 | Fos-like antigen 2 | 3.9 | Cell proliferation/differentiation | 2p23.3 |
| SFN | Stratifin | 3.7 | Tumor suppressor | 1p36.11 |
| PLAT | Plasminogen activator, tissue | 3.5 | Cell migration, tissue remodeling | 8p12 |
| INHBE | Inhibin, beta E | 3 | tumor suppressor | 12q13.3 |
| CDKN1C | Cyclin-dependent kinase inhibitor 1c | 2.9 | tumor suppressor | 11p15.5 |
| TP73 | Tumor protein p73 | 2.7 | tumor suppressor | 1p36.3 |
| SOCS2 | Suppressor of cytokine signaling 2 | 2.7 | growth suppressor | 12q |
| BMPR1B | Bone morphogenetic protein receptor, type 1B | 2.7 | Signal transduction | 4q22-q24 |
| CCL2 | Chemokine (c-c motif) ligand 2 | 2.6 | chemokine | 17q11.2-q21.1 |
| VIM | Vimentin | 2.6 | Mesenchymal cytoskeletal protein | 10p13 |
| SPRY2 | Sprouty 2 | 2.6 | tumor suppressor | 13q31.1 |
| CXCL10 | Chemokine (c-x-c motif) ligand 10 | 2.6 | Chemokine, angiostatic | 4q21 |
| LCN2 | Lipocalin 2 | 2.5 | Tumor suppressor/promoter | 9q34 |
| AKAP12 | A kinase anchor protein 12 | 2.4 | tumor/metastasis suppressor | 6q24-q25 |
| KLF4 | Kruppel-like factor 4 | 2.4 | Transcriptional activator | 9q31 |
| ATF3 | Activating transcription factor 3 | 2.4 | apoptosis, cell migration | 1q32.3 |
| FBN1 | Fibrillin 1 | 2.4 | ECM protein, cell attachment | 15q21.1 |
| TWIST1 | Twist homolog 1 | 2.4 | Invasion, metastasis | 7p21.2 |
| ALCAM | Activated leukocyte cell adhesion molecule | 2.3 | Cell adhesion | 3q13.1 |
| MSX2 | Msh homeo box homolog 2 | 2.2 | tumor suppressor | 5q34-q35 |
| LOX | Lysyl oxidase | 2.2 | Cross-linking of ECM proteins | 5q23.2 |
| BTG1 | B-cell translocation gene 1 protein | 2.2 | Hormone response, anti-proliferative | 12q22 |
| SOCS1 | Suppressor of cytokine signaling 1 | 2.2 | tumor suppressor | 16p13.13 |
| PPP2CA | Protein phosphatase 2 (formerly 2a), catalytic subunit, alpha isoform |
1.5 | Serine phosphatase, control of cell division |
5q23-q31 |
| PPP3CA | Protein phosphatase 3 (formerly 2b), catalytic subunit, alpha isoform |
1.3 | Serine phosphatase, apoptosis | 4q21-q24 |
Validation of Affymetrix data by quantitative reverse transcriptase–polymerase chain reaction
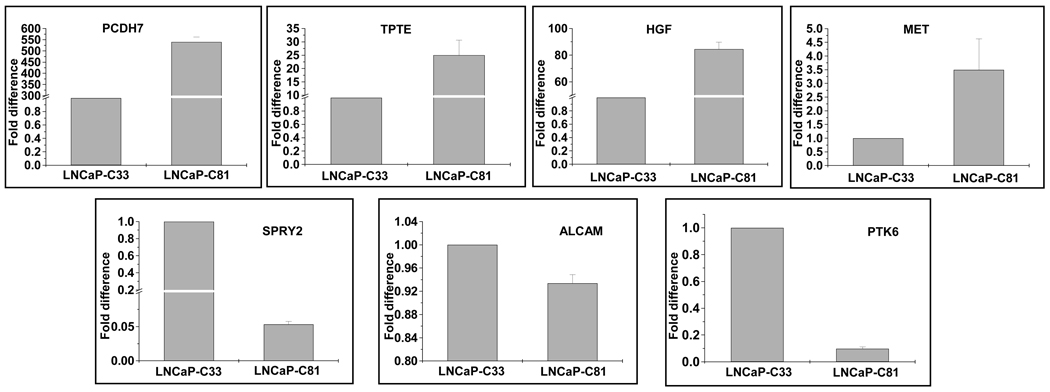
To confirm that gene expression changes observed in microarray analysis were indeed associated with androgen-independent phenotype, we examined the expression of a few select genes by quantitative real-time reverse transcription–polymerase chain reaction (RT–PCR) (Figure 1). Consistent with the GeneChip data, Q-RT–PCR confirmed an increased expression of PCDH7, HGF, Met, and TPTE and a decreased expression of ALCAM, SPRY2 and PTK6 in AI (LNCaP-C81) cells after normalizing the data with that of a housekeeping gene, glyceraldehyde-3-phophate dehydrogenase (GAPDH).
Figure 1.
Expression analysis of differential gene expression by real-time RT-PCR. Total RNA was reverse-transcribed and subsequently amplified by a real-time PCR using gene-specific primers. A fold-change in expression was calculated relative to LNCaP-C33 cells using GAPDH as an internal control by ΔΔCT method. Each experiment was repeated at least three times and data presented as mean ± standard deviation.
Identification of signaling pathways associated with the progression of prostate cancer cells toward an androgen-refractory phenotype
To identify the genetic networks altered during the androgen-refractory progression of prostate cancer cells, we utilized microarray data to perform a pathway prediction analysis using web-based application ‘Ingenuity pathway analysis’ (Ingenuity Systems, Mountain View, CA). This web-delivered soft-ware searches its database to place differentially expressed genes in gene clusters linked to a molecular pathway(s). We identified several genetic networks to be perturbed during androgen-independent progression (Table 3). Most of the signaling pathways central to the identified gene networks were found to be suppressed, except EGF, HGF and E2F4 (Table 3). Thus, our data suggest that the prostate cancer mainly involves a repression of cell signaling pathways to attain an androgen-refractory phenotype.
Table 3.
Top gene networks identified to be perturbed during androgenindependent progression by Ingenuity pathway analysis
| Gene network(s) |
Score | Focus genes | Signaling Pathway(s) |
Biological function(s) | Status |
|---|---|---|---|---|---|
| 1 | 36 | 35 | TP53 | Negative regulation of growth | repressed |
| 2 | 36 | 35 | HGF | Growth signaling | activated |
| 3,4 | 36,36 | 35,35 | AP-1 (Jun/Fos) | Oncogenesis | repressed |
| 5 | 36 | 35 | VEGF | Angiogenesis | repressed |
| 6 | 36 | 35 | IL8, CCL2 | Inflammatory response, angiogenesis | repressed |
| 7 | 36 | 35 | EGF | Cell proliferation and differentiation | activated |
| 8 | 36 | 35 | TRAIL | Apoptosis | repressed |
| 9 | 36 | 35 | PLCB1 | Cell cycle regulation | repressed |
| 10 | 36 | 35 | JAK/Stat | Cell proliferation, apoptosis and differentiation | repressed |
| 11 | 36 | 35 | TP73, NCAM1 | Growth suppression, cell adhesion | repressed |
| 12 | 24 | 29 | p16, E2F4 | Negative regulation of cell cycle | repressed, activated |
Immunohistochemical analyses of protein phosphatase 2, catalytic subunit, (PPP2CA) in prostate cancer
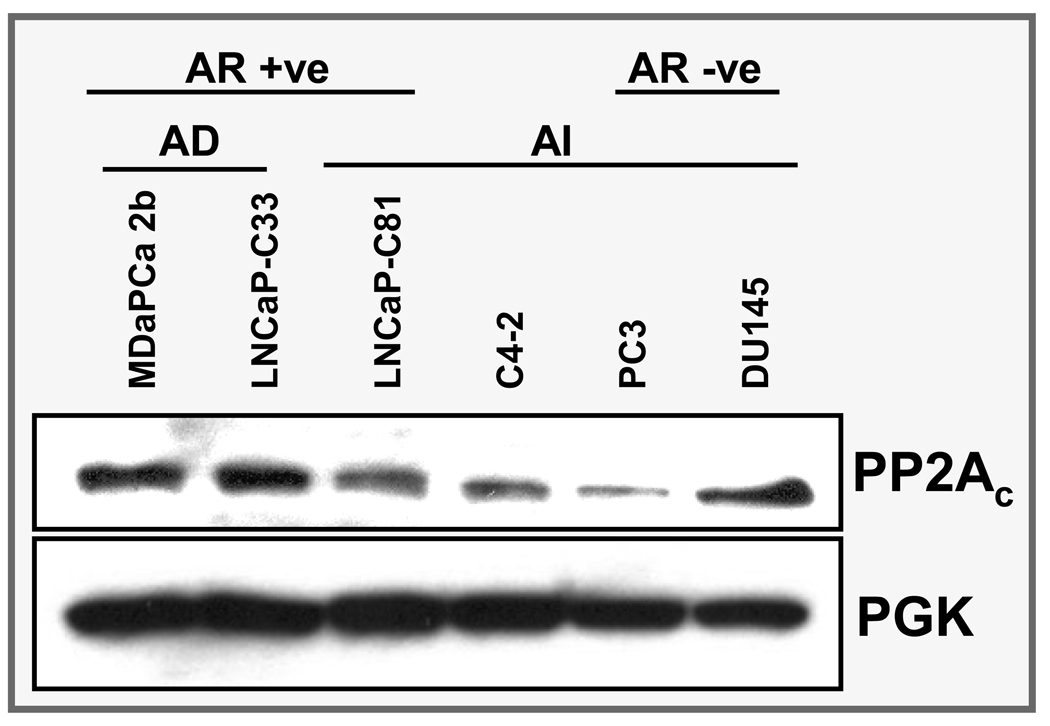
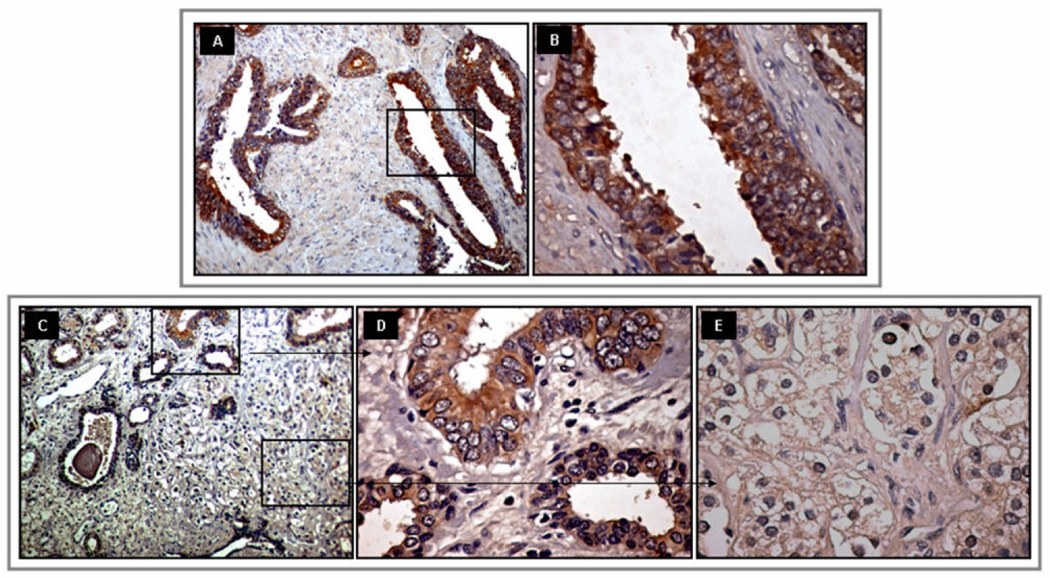
Genome-wide survey of the differential gene expression resulted in the identification of multiple genes with potential pathobiological importance in androgen-refractory progression of prostate cancer. From the long list of genes, we chose PPP2CA for further analysis at protein level in human prostate cancer tissues to predict its prospective significance. PPP2CA encodes the catalytic subunit (alpha-isoform) of protein phosphatase 2A (PP2A), which is a serine phosphatase implicated in the negative control of the cell division and survival. Immunoblot analysis demonstrated its variable expression pattern in AD and AI prostate cancer cell lines (Figure 2). Consistent with the microarray data, PP2A was downregulated in LNCaP-C81 (AI) cells compared to the LNCaP-C33 (AD) cells. Furthermore, data from immunohistochemical analysis also showed an overall downregulation of PP2A in cancer lesions compared to the adjacent normal/benign tissue (Figure 3). Out of the 35 human tissue samples, 22 (62.9%) showed positive reactivity in normal/benign region, whereas only 20 (57.0%) cancer cases were positive for PP2A (Table 4). No intense immunoreactivity was observed in either normal/benign or cancer cells. Composite score values for normal/benign cells ranged from 0 to 6 with a mean composite score of 1.6. In cancer cells, however, the composite score values ranged from 0 to 3 (mean composite score=0.7) due to less number of immunoreactive cells (% positivity). The difference between mean composite scores of benign and cancer samples was statistically significant (P=0.0073). Interestingly, when we compared the composite score differences in the stage and Gleason grade subgroups, the composite score were significantly greater in late stage [stages 3 & 4] (P=0.015) and high Gleason grade [Gleason score 8–10] (P=0.042) subgroups as compared to early stage and low Gleason grade subgroups, respectively.
Figure 2.
Immunoblot analysis of PPP2CA (alpha isoform of catalytic subunit of protein phosphatase 2A). Consistent with the microarray analysis, a reduced expression of PPP2CA is observed in androgen-independent (AI) LNCaP-C81 cells compared to androgen-dependent (AD) LNCaP-C33 cells. Except DU145, a reduced expression of PPP2CA is also evident in other AI cell lines (C4-2 and PC3). Phospho-glycerol kinase (PGK) was used as an internal control.
Figure 3.
Immunohistochemical analysis of PPP2CA expression on a prostate cancer tissue array. A and B, Normal/benign tissue at 10X and 40 × magnifications, respectively. C, Prostate cancer section with various grade lesions (10× magnification), D and E, magnified (40X) view of section depicting lesions with Gleason grade 6 (3+3) and 8 (4+4), respectively. A decreased expression of PPP2CA is observed in prostate cancer, which further correlates with Gleason grade.
Table 4.
Expression of PP2A antigen in Benign Prostatic Hyperplasia (BPH) and Prostate Cancer (PCa) lesions, n= 35
| Immunostaining Grade | BPH | PCa | |||||
|---|---|---|---|---|---|---|---|
| −ve (%) | 0 (n) | 37.1% | 13 (37.1%) | Mean composite score ± SEM |
43.0% | 15 (42.8%) | Mean composite score ± SEM |
| +ve (%) | +1 (n) | 62.9% | 20 (57.2%) | 57.0% | 19 (54.3%) | ||
| +2 (n) | 02 (05.7%) | 1.63 ± .25 | 01 (02.9%) | 0.70 ± .13 | |||
| +3 (n) | 00 (00.0%) | 00 (00.0%) | |||||
DISCUSSION
The present study examined the differential gene expression in androgen-dependent (AD) LNCaP-C33 and androgen-independent (AI) LNCaP-C81 cells via genome-wide expression profiling. Our study resulted in the identification of many key genes that could be of clinical importance and future targets for prostate cancer therapy. Furthermore, our data indicated that the androgen-refractory progression of prostate cancer mainly involves a repression mechanism as a majority of the differentially-expressed genes and signalling networks were downregulated in AI prostate cancer cells.
Over the last decade, DNA array technique has contributed significantly in identifying novel molecules that play potential role in the disease progression as well as in the classification of various cancers [9–11]. Moreover, recent technological advances have enabled the comprehensive surveying of a cell’s transcriptional landscape. Genome-wide analysis allows the simultaneous monitoring of whole transcriptome, and is helpful in identifying the functional and biochemical pathway(s) that are perturbed during the disease progression and targeted by the therapeutic drugs [12;13]. In prostate cancer, several studies have been conducted on different types of DNA arrays using tumor biopsy specimens, tumor cell lines, and more recently tissues obtained from rapid autopsies [14;15]. What we have learnt thus far is that prostate cancer is a heterogeneous group of diseases [3] and therefore a crucial examination of current or newly identified biomarkers is needed, while considering their potential as diagnostic, prognostic and therapeutic targets.
In this study, we utilized a unique in vitro LNCaP cell model, which includes different LNCaP cells (AD: C33 and AI: C81), closely resembling early and late stages of tumor progression [4]. One advantage of using this model is that it overcomes the heterogeneity, associated with the prostate cancer [3] to a certain extent. Furthermore, LNCaP cells express functional androgen receptors as is the case in majority of prostate carcinomas. We hypothesized that identification of differentially regulated genes between AD (C33) and AI (C81) prostate cancer cells may provide insights into its clinical course of progression and may be exploited for a variety of clinical applications. Genome-wide expression analysis generated a large amount of interesting data and identified many genes that were differentially regulated in AI prostate cancer cells. Genes identified in our study include those involved in cell proliferation, apoptosis, angiogenesis, hormone response, etc. These genes are of potential interest for follow-up studies. While some have already been reported to have potential as clinical biomarkers [15–17], many others seem to be novel in context to their association with AI phenotype. Among the selected overexpressed genes in androgen-independent (C81) cells, two (TPTE and TSPY) have previously been shown to exhibit a testis-specific restricted pattern of expression in normal conditions [18;19]. TPTE (transmembrane phosphatase with tensin homology) encodes a PTEN-related tyrosine phosphatase (also referred as PTEN2) with four potential transmembrane domains [19]. PTEN (phosphatase and tensin homolog) acts as a tumor suppressor and its multiple implications in prostate cancer progression have also been reported [20]. No clear biological function has yet been assigned to TPTE and future studies are warranted to understand its role in tumor progression. In contrast to our finding, another study has reported recurrent loss of a gene cluster on Y-chromosome involving TSPY (tumor-specific protein-Y linked) in prostate cancer [21]. This further supports the heterogeneous nature of AI prostate cancer as previously described [3] and suggests the existence of different signaling mechanisms among subtypes of metastatic prostate cancer. Nonetheless, a recent study has shown the oncogenic potential of TSPY in Hela and NIH3T3 cells [22]. Other important overexpressed genes were HGF (hepatocyte growth factor) and MET/HGFR (hepatocyte growth factor receptor). HGF and its receptor, c-Met, have been demonstrated to elicit a number of key functions in numerous tissues that are important in the progression, invasion and metastasis of cancer [23]. Involvement of HGF-Met signalling in metastatic prostate cancer development has also been reported [24].
It is interesting to note that relatively more differentially-expressed genes were found to be under-expressed (n=1460) than overexpressed (n=876) in AI prostate cancer cells (data not shown). In our study, Angiopoietin 2 (ANGP2), whose expression has been shown to correlate with increasing grade of prostate cancer [25], was found to be downregulated in AI prostate cancer cells. Thrombospondin 1 (THBS1) is another important gene that was highly repressed in AI prostate cancer cells. In previous studies, it has been shown to possess anti-angiogenic activity [26–28]. The intratumoral administration of a THBS1 expression vector into androgen-independent DU-145 prostate cancer xenografts was shown to inhibit tumor growth in vivo through inhibition of tumor angiogenesis [28]. Interestingly, androgens have been shown to repress the expression of thrombospondin 1 in normal and neoplastic prostate tissues [27]. HTATIP2, another tumor and metastasis suppressor gene [29], was also repressed in androgen-refractory PC cells. Ectopic expression of HTATIP2 in v-SCLC cells induced expression of a number of genes including the apoptosis-related genes Bad and Siva, as well as metastasis suppressor NM23-H2 [30]. SPRY2, an inhibitor of receptor tyrosine kinase (RTK) signaling, found to be under-expressed in our study has been shown to be epigenetically inactivated in prostate cancer [31]. Progressive loss of ALCAM, which is downregulated in AI prostate cancer cells, has been reported in high-grade prostate carcinomas [32]. Among other genes of importance exhibiting downregulated expression in AI prostate cancer cells were CDKN2B/TP53, CDKN1C/TP57, TP73. These genes are known tumor suppressors and are frequently silenced in cancers [33–35].
To better understand the functional premise related to androgen-independent progression, we searched for patterns of gene expression using Ingenuity pathway analysis. Many signaling networks (Table 3) were identified, suggesting that perturbation of these molecular pathways is associated with the gain of androgen-refractory phenotype. In the literature, these pathways have been shown to be linked with various aspects of tumor progression [36–40]. Of particular interest is the finding that most of the pathways identified in our analysis were repressed in androgen-refractory prostate cancer cells. Importantly, many (TP53, TP73, p16, and TRAIL) among these downregulated pathways are associated with growth inhibitory cell signaling.
PPP2CA, which encodes for the catalytic-subunit (alpha-isoform) of the protein phosphatase 2A (PP2AC), was one of several genes of interest that exhibited a downregulated expression in our analysis in AI prostate cancer cells. PP2A is a potent tumor suppressor and is involved in broad cellular functions [41]. In light of recent observations, it seems to be of potential interest in prostate cancer [42–45]. Data mining analysis using Oncomine cancer profiling database (www.oncomine.org) demonstrate that PPP2CA is significantly downregulated in prostate cancer compared to normal/benign prostate (4 studies) and in metastatic prostate cancer compared to primary prostate carcinoma (3 studies). Interestingly, in a recent report the expression of the beta-isoform of catalytic-subunit of PP2A (PPP2CB) is also shown to be downregulated in prostate cancer [43]. Our immunohistochemical analysis in normal/benign and matched prostate cancer tissues corroborated the downregulated expression of PP2A in prostate cancer. However, we were not able to obtain patient-matched, androgen-dependent and androgen-independent specimens to examine the expression of PP2A during progression to AI phenotype.
In summary, our study offers a significant contribution to our knowledge of the molecular characteristics of AI prostate cancer. Overall comparison of the expression data between AD and AI prostate cancer cells is intriguing demonstrating the decreased expression of several tumor suppressor genes in the AI tumor cells, while many genes that may potentially promote growth and metastasis were upregulated. Protein phosphatase 2A was identified as differentially-expressed protein and its downregulated expression may be of potential importance during the prostate cancer development and further during AI progression of the disease. Many of these differentially-expressed genes may be important as potential diagnostic/prognostic and therapeutic targets for the prostate cancer.
Acknowledgements
This work was supported by the U.S. Department of Defense- Idea Award (PC04502). The UNMC Microarray Core Facility receives partial support from the INBRE Program of the National Center for Research Resources (NIH grant number P20 RR016469). We thank the Molecular Biology Core Facility, UNMC, for oligonucleotide synthesis, Ms. Fen F. Lin for technical help in cell culture and Kristi L.W. Berger (Eppley Institute) for editorial assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J.Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 2.Feldman BJ, Feldman D. The development of androgen-independent prostate cancer. Nat.Rev.Cancer. 2001;1:34–45. doi: 10.1038/35094009. [DOI] [PubMed] [Google Scholar]
- 3.Shah RB, Mehra R, Chinnaiyan AM, Shen R, Ghosh D, Zhou M, Macvicar GR, Varambally S, Harwood J, Bismar TA, Kim R, Rubin MA, Pienta KJ. Androgen-independent prostate cancer is a heterogeneous group of diseases: lessons from a rapid autopsy program. Cancer Res. 2004;64:9209–9216. doi: 10.1158/0008-5472.CAN-04-2442. [DOI] [PubMed] [Google Scholar]
- 4.Igawa T, Lin FF, Lee MS, Karan D, Batra SK, Lin MF. Establishment and characterization of androgen-independent human prostate cancer LNCaP cell model. Prostate. 2002;50:222–235. doi: 10.1002/pros.10054. [DOI] [PubMed] [Google Scholar]
- 5.Karan D, Schmied BM, Dave BJ, Wittel UA, Lin MF, Batra SK. Decreased androgen-responsive growth of human prostate cancer is associated with increased genetic alterations. Clin.Cancer Res. 2001;7:3472–3480. [PubMed] [Google Scholar]
- 6.Veeramani S, Igawa T, Yuan TC, Lin FF, Lee MS, Lin JS, Johansson SL, Lin MF. Expression of p66(Shc) protein correlates with proliferation of human prostate cancer cells. Oncogene. 2005;24:7203–7212. doi: 10.1038/sj.onc.1208852. [DOI] [PubMed] [Google Scholar]
- 7.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 8.Singh AP, Chauhan SC, Bafna S, Johansson SL, Smith LM, Moniaux N, Lin MF, Batra SK. Aberrant expression of transmembrane mucins, MUC1 and MUC4, in human prostate carcinomas. Prostate. 2006;66:421–429. doi: 10.1002/pros.20372. [DOI] [PubMed] [Google Scholar]
- 9.Hayes DN, Monti S, Parmigiani G, Gilks CB, Naoki K, Bhattacharjee A, Socinski MA, Perou C, Meyerson M. Gene expression profiling reveals reproducible human lung adenocarcinoma subtypes in multiple independent patient cohorts. J.Clin.Oncol. 2006;24:5079–5090. doi: 10.1200/JCO.2005.05.1748. [DOI] [PubMed] [Google Scholar]
- 10.Pusztai L, Mazouni C, Anderson K, Wu Y, Symmans WF. Molecular classification of breast cancer: limitations and potential. Oncologist. 2006;11:868–877. doi: 10.1634/theoncologist.11-8-868. [DOI] [PubMed] [Google Scholar]
- 11.Sorlie T. Molecular portraits of breast cancer: tumour subtypes as distinct disease entities. Eur.J.Cancer. 2004;40:2667–2675. doi: 10.1016/j.ejca.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 12.Bono H, Okazaki Y. Functional transcriptomes: comparative analysis of biological pathways and processes in eukaryotes to infer genetic networks among transcripts. Curr.Opin.Struct.Biol. 2002;12:355–361. doi: 10.1016/s0959-440x(02)00335-4. [DOI] [PubMed] [Google Scholar]
- 13.Liu S, Seidel-Dugan C. In search of p53 target genes for the therapeutic manipulation of cancer. Curr.Opin.Drug Discov.Devel. 2006;9:176–183. [PubMed] [Google Scholar]
- 14.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, Rubin MA, Chinnaiyan AM. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 15.Varambally S, Yu J, Laxman B, Rhodes DR, Mehra R, Tomlins SA, Shah RB, Chandran U, Monzon FA, Becich MJ, Wei JT, Pienta KJ, Ghosh D, Rubin MA, Chinnaiyan AM. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell. 2005;8:393–406. doi: 10.1016/j.ccr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 16.Knudsen BS, Gmyrek GA, Inra J, Scherr DS, Vaughan ED, Nanus DM, Kattan MW, Gerald WL, Vande Woude GF. High expression of the Met receptor in prostate cancer metastasis to bone. Urology. 2002;60:1113–1117. doi: 10.1016/s0090-4295(02)01954-4. [DOI] [PubMed] [Google Scholar]
- 17.Naughton M, Picus J, Zhu X, Catalona WJ, Vollmer RT, Humphrey PA. Scatter factor-hepatocyte growth factor elevation in the serum of patients with prostate cancer. J.Urol. 2001;165:1325–1328. [PubMed] [Google Scholar]
- 18.Krick R, Jakubiczka S, Arnemann J. Expression, alternative splicing and haplotype analysis of transcribed testis specific protein (TSPY) genes. Gene. 2003;302:11–19. doi: 10.1016/s0378-1119(02)01104-6. [DOI] [PubMed] [Google Scholar]
- 19.Wu Y, Dowbenko D, Pisabarro MT, llard-Telm L, Koeppen H, Lasky LA. PTEN 2, a Golgi-associated testis-specific homologue of the PTEN tumor suppressor lipid phosphatase. J.Biol.Chem. 2001;276:21745–21753. doi: 10.1074/jbc.M101480200. [DOI] [PubMed] [Google Scholar]
- 20.Mulholland DJ, Dedhar S, Wu H, Nelson CC. PTEN and GSK3beta: key regulators of progression to androgen-independent prostate cancer. Oncogene. 2006;25:329–337. doi: 10.1038/sj.onc.1209020. [DOI] [PubMed] [Google Scholar]
- 21.Vijayakumar S, Hall DC, Reveles XT, Troyer DA, Thompson IM, Garcia D, Xiang R, Leach RJ, Johnson-Pais TL, Naylor SL. Detection of recurrent copy number loss at Yp11.2 involving TSPY gene cluster in prostate cancer using array-based comparative genomic hybridization. Cancer Res. 2006;66:4055–4064. doi: 10.1158/0008-5472.CAN-05-3822. [DOI] [PubMed] [Google Scholar]
- 22.Oram SW, Liu XX, Lee TL, Chan WY, Lau YF. TSPY potentiates cell proliferation and tumorigenesis by promoting cell cycle progression in HeLa and NIH3T3 cells. BMC.Cancer. 2006;6(154):154. doi: 10.1186/1471-2407-6-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hurle RA, Davies G, Parr C, Mason MD, Jenkins SA, Kynaston HG, Jiang WG. Hepatocyte growth factor/scatter factor and prostate cancer: a review. Histol.Histopathol. 2005;20:1339–1349. doi: 10.14670/HH-20.1339. [DOI] [PubMed] [Google Scholar]
- 24.MacDougall CA, Vargas M, Soares CR, Holzer RG, Ide AE, Jorcyk CL. Involvement of HGF/SF-Met signaling in prostate adenocarcinoma cells: evidence for 21 alternative mechanisms leading to a metastatic phenotype in Pr-14c. Prostate. 2005;64:139–148. doi: 10.1002/pros.20226. [DOI] [PubMed] [Google Scholar]
- 25.Lind AJ, Wikstrom P, Granfors T, Egevad L, Stattin P, Bergh A. Angiopoietin 2 expression is related to histological grade, vascular density, metastases, and outcome in prostate cancer. Prostate. 2005;62:394–399. doi: 10.1002/pros.20163. [DOI] [PubMed] [Google Scholar]
- 26.Bocci G, Francia G, Man S, Lawler J, Kerbel RS. Thrombospondin 1, a mediator of the antiangiogenic effects of low-dose metronomic chemotherapy. Proc.Natl.Acad.Sci.U.S.A. 2003;100:12917–12922. doi: 10.1073/pnas.2135406100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Colombel M, Filleur S, Fournier P, Merle C, Guglielmi J, Courtin A, Degeorges A, Serre CM, Bouvier R, Clezardin P, Cabon F. Androgens repress the expression of the angiogenesis inhibitor thrombospondin-1 in normal and neoplastic prostate. Cancer Res. 2005;65:300–308. [PubMed] [Google Scholar]
- 28.Jin RJ, Kwak C, Lee SG, Lee CH, Soo CG, Park MS, Lee E, Lee SE. The application of an anti-angiogenic gene (thrombospondin-1) in the treatment of human prostate cancer xenografts. Cancer Gene Ther. 2000;7:1537–1542. doi: 10.1038/sj.cgt.7700266. [DOI] [PubMed] [Google Scholar]
- 29.Ito M, Jiang C, Krumm K, Zhang X, Pecha J, Zhao J, Guo Y, Roeder RG, Xiao H. TIP30 deficiency increases susceptibility to tumorigenesis. Cancer Res. 2003;63:8763–8767. [PubMed] [Google Scholar]
- 30.Xiao H, Palhan V, Yang Y, Roeder RG. TIP30 has an intrinsic kinase activity required for up-regulation of a subset of apoptotic genes. EMBO J. 2000;19:956–963. doi: 10.1093/emboj/19.5.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McKie AB, Douglas DA, Olijslagers S, Graham J, Omar MM, Heer R, Gnanapragasam VJ, Robson CN, Leung HY. Epigenetic inactivation of the human sprouty2 (hSPRY2) homologue in prostate cancer. Oncogene. 2005;24:2166–2174. doi: 10.1038/sj.onc.1208371. [DOI] [PubMed] [Google Scholar]
- 32.Kristiansen G, Pilarsky C, Wissmann C, Stephan C, Weissbach L, Loy V, Loening S, Dietel M, Rosenthal A. ALCAM/CD166 is up-regulated in low-grade prostate cancer and progressively lost in high-grade lesions. Prostate. 2003;54:34–43. doi: 10.1002/pros.10161. [DOI] [PubMed] [Google Scholar]
- 33.Herman JG, Jen J, Merlo A, Baylin SB. Hypermethylation-associated inactivation indicates a tumor suppressor role for p15INK4B. Cancer Res. 1996;56:722–727. [PubMed] [Google Scholar]
- 34.Lodygin D, Epanchintsev A, Menssen A, Diebold J, Hermeking H. Functional epigenomics identifies genes frequently silenced in prostate cancer. Cancer Res. 2005;65:4218–4227. doi: 10.1158/0008-5472.CAN-04-4407. [DOI] [PubMed] [Google Scholar]
- 35.Yang B, House MG, Guo M, Herman JG, Clark DP. Promoter methylation profiles of tumor suppressor genes in intrahepatic and extrahepatic cholangiocarcinoma. Mod.Pathol. 2005;18:412–420. doi: 10.1038/modpathol.3800287. [DOI] [PubMed] [Google Scholar]
- 36.Crosby ME, Almasan A. Opposing roles of E2Fs in cell proliferation and death. Cancer Biol.Ther. 2004;3:1208–1211. doi: 10.4161/cbt.3.12.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eferl R, Wagner EF. AP-1: a double-edged sword in tumorigenesis. Nat.Rev.Cancer. 2003;3:859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- 38.Igney FH, Krammer PH. Death and anti-death: tumour resistance to apoptosis. Nat.Rev.Cancer. 2002;2:277–288. doi: 10.1038/nrc776. [DOI] [PubMed] [Google Scholar]
- 39.Kim WY, Sharpless NE. The regulation of INK4/ARF in cancer and aging. Cell. 2006;127:265–275. doi: 10.1016/j.cell.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 40.Melino G, De L, V, Vousden KH. p73: Friend or foe in tumorigenesis. Nat.Rev.Cancer. 2002;2:605–615. doi: 10.1038/nrc861. [DOI] [PubMed] [Google Scholar]
- 41.Janssens V, Goris J, Van HC. PP2A: the expected tumor suppressor. Curr.Opin.Genet.Dev. 2005;15:34–41. doi: 10.1016/j.gde.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 42.Li L, Ren CH, Tahir SA, Ren C, Thompson TC. Caveolin-1 maintains activated Akt in prostate cancer cells through scaffolding domain binding site interactions with and inhibition of serine/threonine protein phosphatases PP1 and PP2A. Mol.Cell Biol. 2003;23:9389–9404. doi: 10.1128/MCB.23.24.9389-9404.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prowatke I, Devens F, Benner A, Grone EF, Mertens D, Grone HJ, Lichter P, Joos S. Expression analysis of imbalanced genes in prostate carcinoma using tissue microarrays. Br.J.Cancer. 2007;96:82–88. doi: 10.1038/sj.bjc.6603490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang X, Chen MW, Terry S, Vacherot F, Bemis DL, Capodice J, Kitajewski J, de la TA, Benson MC, Guo Y, Buttyan R. Complex regulation of human androgen receptor expression by Wnt signaling in prostate cancer cells. Oncogene. 2006;25:3436–3444. doi: 10.1038/sj.onc.1209366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang CS, Vitto MJ, Busby SA, Garcia BA, Kesler CT, Gioeli D, Shabanowitz J, Hunt DF, Rundell K, Brautigan DL, Paschal BM. Simian virus 40 small t antigen mediates conformation-dependent transfer of protein phosphatase 2A onto the androgen receptor. Mol.Cell Biol. 2005;25:1298–1308. doi: 10.1128/MCB.25.4.1298-1308.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]