Abstract
Despite the growing number of atomic-resolution membrane protein structures, direct structural information about proteins in their native membrane environment is scarce. This problem is particularly relevant in the case of the highly-charged S1–S4 voltage-sensing domains responsible for nerve impulses, where interactions with the lipid bilayer are critical for the function of voltage-activated potassium channels. Here we use neutron diffraction, solid-state nuclear magnetic resonance spectroscopy, and molecular dynamics simulations to investigate the structure and hydration of bilayer membranes containing S1–S4 voltage-sensing domains. Our results show that voltage sensors adopt transmembrane orientations, cause a modest reshaping of the surrounding lipid bilayer, and that water molecules intimately interact with the protein within the membrane. These structural findings reveal that voltage sensors have evolved to interact with the lipid membrane while keeping the energetic and structural perturbations to a minimum, and that water penetrates into the membrane to hydrate charged residues and shape the transmembrane electric field.
Membrane-embedded S1–S4 voltage-sensing domains are used by membrane proteins to sense and react to changes in membrane voltage (Fig 1a). In the voltage-activated potassium (Kv), sodium and calcium channels, these S1–S4 domains drive opening and closing of an associated ion conducting pore domain (Fig 1a) to generate electrical signals1. In the Ciona intestinalis voltage-sensitive phosphatase, an S1–S4 domain controls the hydrolysis of phospholipids by an associated phosphatase2, and in voltage-activated proton channels the S1–S4 domain contains the permeation pathway for protons3. X-ray structures of S1–S4 domains show that the protein domain is comprised of four transmembrane α-helices (Fig 1a) and that its structure is well-conserved from archaebacteria to mammals1,4,5. A fundamental feature of S1–S4 domains is that they contain basic and acidic residues that enable the protein to change conformation in response to rapid fluctuations in membrane voltage1,6,7. In these voltage sensors, interactions with the surrounding lipid membrane play crucial roles. The S3b-S4 paddle motif within S1–S4 domains, for example, moves at the protein-lipid interface 5,8–13 and alterations in the composition of the lipid membrane alter voltage sensor activation14–17. The polar nature of voltage sensors and their intimate interactions with the bilayer, raise the possibility that these domains perturb the structure of the surrounding lipid bilayer. In addition, while spectroscopic and functional studies suggest that the electric field is focused across voltage sensors18–21, the structural basis for focusing is unclear. Might deformations of the membrane contribute to focusing the electric field, or is the shape and chemistry of the protein largely responsible? Crevices observed in X-ray structures of S1–S4 domains would be expected to reshape the electric field, but only if they persist and are filled with water when the domain is embedded in a lipid membrane. Although water penetration into the membrane has been inferred from accessibility studies9,13,19,22–26 and simulations12,27–29, hydration of voltage sensors has not been measured.
Figure 1. S1–S4 voltage-sensing domains and their biophysical properties in lipid bilayers.
a) Representation of membrane proteins containing S1–S4 voltage-sensing domains (red), embedded in the lipid bilayer (light grey). b) Circular Dichroism spectra of the S1–S4 voltage-sensing domain of KvAP in OG micelles (dotted red line) or reconstituted into POPC:POPG proteoliposomes at a protein/lipid molar ratio of 1:130 (solid red line). Mean residue elipticity (Θ) is given in units of deg cm2 dmol−1. CD spectra indicate high (~85 %) helical content (see Methods). c) Fluorescence emission spectra of Trp70 within the S1–S4 voltage-sensing domain after reconstitution in POPC:POPG (protein/lipid molar ratio of 1:100) in the absence (solid red line) and presence of 50 mM aqueous acrylamide (dotted red line). Emission spectra for the free Trp (25 µM) mixed with the POPC:POPG liposomes of identical lipid concentration in the absence (solid green line) and presence of 50 mM aqueous acrylamide (dotted green line). d) Stern-Volmer plots for acrylamide quenching of fluorescence emission for Trp70 within the S1–S4 voltage-sensing domain (red) or free Trp (green). Error bars are S.E.M (n=3). The Stern-Volmer constant for quenching was 0.4±0.02 M−1 for Trp70 and 26.2±0.2 M−1 for free Trp. e) Quenching of fluorescence emission of Trp70 within the S1–S4 voltage-sensing domain by Br atoms attached at different positions along the lipid hydrocarbon tail. The protein was reconstituted into the 1:1 mixture of POPG and either POPC or one of three dibrominated lipids (Br2(6,7)-PC, Br2(9,10)-PC, Br2(11,12)-PC (see Methods for nomenclature). Error bars are S.E.M (n=3).
Structure and hydration of membranes containing voltage sensors
To address these fundamental questions, we developed a homogeneous preparation of voltage-sensing domains incorporated into lipid membranes for use with neutron diffraction30–33. The neutron scattering length gives the relative amplitude of the de Broglie wave scattered from a nucleus and is analogous to the X-ray scattering length of an electron. We focused our efforts on the S1–S4 domain of KvAP, an archaebacterial channel from Aeropyrum pernix that can be robustly expressed, stably purified, and reconstituted into lipid membranes4,9,10,13,34. Following expression and purification of the S1–S4 domain of KvAP (see Methods), circular dichroism (CD) spectroscopy reveals that the domain has high α-helical content in detergent micelles or reconstituted into liposomes (Fig 1b), consistent with the X-ray structure of the domain4 and electron paramagnetic resonance studies 9,13,34. To investigate the topology of the S1–S4 domain in liposomes, we measured the fluorescence of the single Trp70 near the middle of the S2 helix. The emission spectrum of Trp70 is shifted towards shorter wavelengths compared to free Trp in aqueous solution (Fig 1c), indicating that Trp70 resides in a non-polar environment35. Moreover, its fluorescence is efficiently quenched by bromine atoms covalently bound to lipid hydrocarbon tails36 , but not by the aqueous quencher acrylamide (Fig 1c,d,e), consistent with a transmembrane topology of the S2 helix (see below).
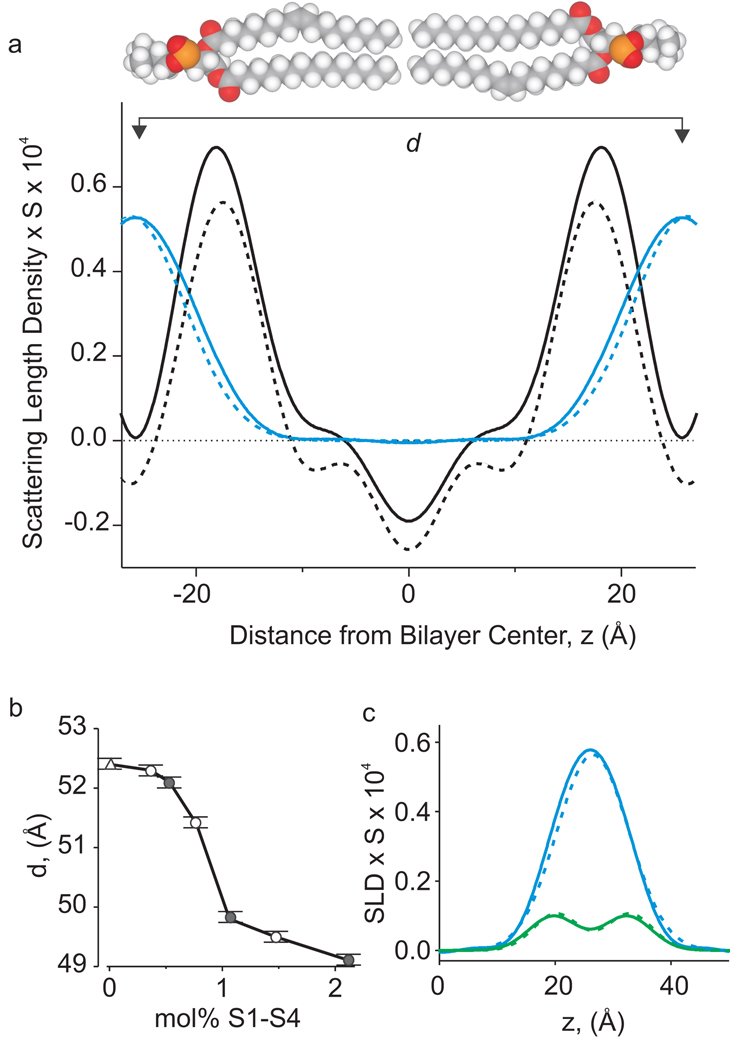
To determine the profile structure of bilayers containing S1–S4 domains, oriented lipid multilayers were produced by deposition of proteoliposomes or neat liposomes on glass substrates. When these multilayers were hydrated (86 or 93% relative humidity) and mounted in a cold neutron beam37, strong lamellar diffraction patterns with Bragg spacing (d) were observed (Supplementary Fig 1a,b). One dimensional, absolute-scale, scattering-length density profiles along the normal of the lipid bilayer plane were then computed from the observed structure factors (Fig 2a). The constructed neutron scattering-length density profiles for neat lipid bilayers show the distribution of lipid (black dashed line), with positive densities for the headgroup region, a trough for the hydrocarbon tails, and negative densities near the terminal methyl groups (Fig 2a). The scattering lengths for most of the relevant nuclear species (carbon, nitrogen, oxygen and phosphorous) have similarly positive values, with the notable exception of hydrogen, which has a negative scattering length. The average scattering-length density of the bilayer hydrocarbon core is close to zero because the scattering length of carbon is positive and that for hydrogen is negative. The headgroup peaks appear closer together than they would in an equivalent X-ray experiment33 because X-rays scatter most strongly from headgroup phosphates, while neutrons scatter most strongly from the carbonyl groups due to their relative lack of hydrogens. The overall scattering length density of the bilayer increases in the presence of the protein (solid black line), consistent with a transmembrane topology of the S1–S4 domain (see below). Comparison of the lipid bilayer profiles with and without S1–S4 domains reveals how the structure of the bilayer is influenced by the protein (Fig 2a). Although the S1–S4 domain does not radically alter the structure of the lipid bilayer, examination of the profiles shows that the voltage sensors produce a detectable thinning of the bilayer, as revealed by a decrease in d (Fig 2b). The thickness decrease depends upon the concentration of the protein in the membrane, with a maximal decrease of about 3 Å at protein to lipid ratios (molar) above 1 to 100 (Fig 2b). These results indicate that lipid molecules in the membrane maintain a bilayer-like arrangement around voltage sensors, consistent with the lipids resolved in the recent crystal structure of the Kv1.2/2.1 paddle chimaera5.
Figure 2. Scattering length density profiles for bilayers containing S1-S4 voltage-sensing domains.
a) Scattering-length density profiles on an absolute scale32 of the S1–S4 voltage-sensing domain in lipid bilayers (black solid line) and the water distribution (blue solid line). Protein:lipid ratio is 1:130 (0.77 mol%) and relative humidity is 86%. Profiles for lipid in the absence of protein (dashed lines) are shown for comparison. The density profile amplitudes are presented in units of scattering-length per unit length, corresponding to the scattering-length density of a unit cell (0.4962:0.4962:0.0079 POPC:POPG:protein, plus 8.5 water molecules) multiplied by the area per lipid (S) (see Methods). The X axis shows the distance from the bilayer center (z) with 0 positioned in the middle of the bilayer. b) Effect of S1–S4 voltage sensing domains on the bilayer thickness (Bragg spacing, d) at different protein:lipid ratios (given as mol% protein). Triangle marks the value of d for neat lipid bilayers. Open circles are for voltage-sensing domains with His tag removed (see methods) and closed circles are for the His-tagged protein. c) Distribution of deuterium atoms in headgroup labeled phosphocholine (–C2H2-C2H2-; D4 lipid) in bilayers containing S1–S4 domains (solid green line) and comparison to the distribution of water (solid blue line). Dashed lines show distribution of D4 lipid and water in the absence of the protein. Protein:lipid ratio is as in b and 25 mol% D4-POPC is used in the mixture of POPC:POPG. Relative humidity is 93%.
Next we performed experiments to determine the distribution of water and to quantify the number of water molecules per lipid using contrast variation between water (1H2O) and deuterium oxide (2H2O), and comparing with lipids containing four deuterium atoms in the headgroup region (D4 lipids, Fig 2c, see Methods). This approach takes advantage of the fact that deuterated nuclei have a positive scattering length compared to a negative one for hydrogen, so that selective substitution of deuterium for hydrogen allows the deuterium atoms to be easily detected against the low scattering-length density of hydrocarbon core. Although the water distributions show that thinning of the bilayer brings water on the two sides of the bilayer closer together (Fig 2a; blue lines), we could not detect a change in the shape of the water distribution or the total water content. At 86% relative humidity, the unit cell of the membrane contains 8.1 ± 0.7 waters per lipid for neat lipid bilayers compared to 8.5 ± 0.5 in the presence of 0.77 mol% protein. At 93% relative humidity, water content per lipid was 10.6 ± 0.2 and 11.0 ± 0.2 in the absence and presence of 0.77 mol% protein (Supplementary Fig 2).
Distributions of S1–S4 voltage-sensing domains and water across membranes
To investigate the membrane topology and hydration of S1–S4 domains directly, we determined the protein distribution using contrast variation between protonated and deuterated S1–S4 domains. The S1–S4 domain of KvAP was uniformly deuterated to 74% (Fig 3a) and multilayers were formed with either protonated or deuterated protein at the same protein:lipid ratio and lipid composition. The two types of samples display similar diffraction patterns with the same number of observed diffraction orders and repeat spacing (Supplementary Fig 3). Subtraction of scaled profiles to obtain the protein density distribution reveals the distribution of the protein across the bilayer (Fig 3b; red line). Maxima in the density distribution are observed in the headgroup region of the bilayer and minima in the inter-bilayer space. This finding firmly establishes that S1–S4 domains adopt a transmembrane topology when embedded in a lipid membrane, with the four helices oriented roughly normal to the membrane plane. The significant protein density found in the interbilayer space is not surprising given that the dimensions of the S1–S4 helices4,5 are similar to the thickness of the bilayer (and may protrude somewhat outside the membrane).
Figure 3. Deuteration of S1–S4 voltage-sensing domains and distribution of the protein in lipid membranes.
a) Mass spectra (MS-MALDI) of the protonated S1–S4 domain of KvAP (1H S1–S4; purple) and uniformly deuterated S1–S4 domain (2H S1–S4; red). The difference in mass indicates that the protein is deuterated to 74%. b) Transbilayer distribution of the S1–S4 domain (white line surrounded by a broad red band) obtained in neutron diffraction experiments by the profile difference of deuterated and protonated S1–S4 domains. Profiles are shown on an absolute (per lipid) scale. Water distribution is shown in blue and lipid as a black line surrounded by a gray band. The broad bands represent estimates of experimental uncertainty computed using the methods of Wiener and White35. Protein:lipid ratio is 1:130 (0.77 mol%) and relative humidity is 93%. c) Neutron scattering length density profiles for the simulation system with 11 water molecules per lipid. Transbilayer distribution of the S1–S4 domain is shown as a white line surrounded by a broad red band (estimated experimental uncertainty), water in blue and lipid in black. d) Snapshots of the region in the vicinity of one of the two voltage-sensing domains from the MD simulation of a stack of two bilayers with 11 water molecules per lipid (left) and excess water (right). Waters within 6 Å of protein are shown as red/white spheres while all other waters are colored purple. Phosphocholine headgroups are colored yellow and the acyl chains are colored light green. Ribbon diagram of the S1–S4 domain is colored red with the outer four arginines in S4 shown as blue CPK models.
Having determined the distribution of protein in the bilayer (Fig 3b; red), we then compared it to that of water (Fig 3b; blue) to ascertain whether S1–S4 domains are hydrated. Strikingly, the two distributions exhibit extensive overlap within the confines of the lipid membrane, in particular for the outer halves of the bilayer. Because the voltage sensor does not detectably alter the shape of the water distribution or water content, the hydration detected in these neutron diffraction experiments is largely from water that is already present in the bilayer. The voltage sensors may bring additional water molecules into the bilayer, but it is unlikely that we would detect those in the experiments thus far. For example, molecular dynamics simulations predict that 45 to 47 water molecules intimately associate with each voltage sensor (see below), which would not detectably alter the shape of the water distribution determined in neutron diffraction experiments because that represents less than 4% of the water molecules in the system for each voltage sensor (at 0.77 mol% protein and 93% humidity, the protein:lipid:water ratio is 1:130:1430).
Predicted distributions of water, lipid and protein with varying hydration
To explore whether the distributions of water, lipid and protein observed in neutron diffraction experiments are compatible with those predicted from molecular dynamics simulations, we calculated neutron diffraction structure factors from molecular dynamics simulation trajectories for S1–S4 domains of KvAP embedded in a lipid bilayer in a transmembrane orientation (see Methods). The resulting Bragg spacing is in excellent agreement with the experimental results, and the overall bilayer scattering length density profile and water distributions, determined by applying the same procedures as in the experimental data reduction, are in good agreement with the experimental results (Fig 3c). Both the simulation and experiment show comparable overall protein density distributions in the membrane interior, as well as overlap between the distributions of protein and water, suggesting a similar disposition of protein and water in the lipid bilayer. The protein studied experimentally contains 18 additional residues on the N-terminus compared with that used for simulations (see Methods), precluding a quantitative comparison of the experimental and simulated protein profiles. Based on the location of the corresponding segment in the structure of the Kv1.2/2.1 paddle chimaera5, the excess scattering length density in the experimental data compared to the simulation near the membrane-water interface (|z| > 10 Å) can be reasonably attributed to the 18-residue amphipathic segment absent in the simulation.
To explore whether hydration of the preparation influences these distributions, we compared a simulation with 11 waters per lipid (corresponding to 93 % relative humidity) to a previously reported simulation in a lipid bilayer in excess water 27 (Fig 3d). Similar scattering length density profiles are observed for simulations at 11 waters per lipid when compared to excess water (Supplementary Fig 4). The structure of the S1–S4 domain was also relatively insensitive to hydration level (Supplementary Fig 5), and in each case a similar number of water molecules (45–47 at 11 waters per lipid compared to 48–49 in excess water) are intimately associated with the protein domain within the hydrophobic core of the bilayer (Supplementary Fig 6). In addition, solvation of crucial S4 Arg residues by both phosphate headgroups and water is similarly observed at varying hydration levels (Supplementary Fig 7). Together, these observations suggest that the extent to which hydration of the preparation influences the structure of lipid membranes containing voltage sensors is minor, and would not be discernible in the neutron scattering profiles at the protein concentrations studied.
Interaction between water and S1–S4 voltage-sensing domains
While the neutron diffraction experiments indicate that the distribution of water and protein in the bilayer overlap, they do not directly address whether water is intimately associated with voltage sensors (Fig 4a). To explore this possibility, we used solid-state nuclear magnetic resonance (NMR) spectroscopy to measure magnetization transfer from water to lipid via intermolecular 1H dipole-dipole interactions in the presence of the voltage sensor. Well-resolved lipid resonances (Fig 4b; black spectra) were observed when rapidly spinning the sample (10 kHz) at the magic angle (54.7°) to the magnetic field (B0), a procedure that averages out anisotropic dipolar interactions that broaden resonance lines. We performed saturation transfer difference experiments 38,39 by comparing lipid spectra before (Fig 4b; black spectra) and after (Fig 4b; blue spectra) applying saturating radio frequency pulses at the 1H2O resonance frequency (4.79 ppm). Magnetization transfer to lipid would cause a decrease in lipid resonances, which can be quantified as an attenuation factor for different saturating field strengths (Fig 4c). Control experiments in which neat lipid membranes were studied show that magnetization transfer from 1H2O to lipid is inefficient when radio frequency pulses are applied to 1H2O (Fig 4c; blue triangles). In the presence of the protein, magnetization transfer is similar in 1H2O and 2H2O when radio frequency pulses are applied directly to the protein amide resonance frequency (8.5 ppm; Fig 4c; red filled and open circles, respectively). In contrast, magnetization transfer from water to lipid is very efficient in membranes containing S1–S4 voltage-sensing domains in the presence of 1H2O (Fig 4c; blue filled circles). Much weaker transfer is observed when 1H2O is substituted with 2H2O (Fig 4c; blue open circles), demonstrating that most of the attenuation results from magnetization transfer originating from 1H2O rather than from aliphatic 1H which overlap 4.79 ppm. (The attenuation observed in 2H2O at a saturating field >200 Hz likely originate from aliphatic 1H.) When considered together with the overlap in water and protein distributions observed in neutron diffraction experiments (Fig 3b), these NMR results indicate that water intimately associated with the protein within the bilayer.
Figure 4. Interaction of water and S1–S4 voltage-sensing domains within lipid membranes.
a) Schematic representation of crevices in S1–S4 voltage-sensing domains (red) filled with water (blue) and an experiment in which selective, saturating radio frequency (RF) pulses were applied at the water resonance (4.792 ppm). Magnetization transfer from water through protein to the surrounding lipid results in the attenuation of the lipid 1H NMR signals. b) Aliphatic region of MAS 1H NMR spectra of a lipid sample containing S1–S4 voltage-sensing domains in the presence of 1H2O (left spectrum) or 2H2O (right spectrum). Lipid resonances for both POPC and POPG (present in a 1:1 mix) are indicated on the spectra, with peaks corresponding to underlined 1H atoms. Attenuation of the methylene resonance (1.3 ppm) is observed (blue traces) when saturating RF pulses (field strength 232 Hz) are applied at 4.792 ppm to a sample containing 1H2O (left spectrum), but not to one containing 2H2O (right spectrum). Attenuation is defined as signal intensity recorded without saturation divided by signal intensity with saturation. c) Attenuation factors plotted as a function of RF field strength (power, kHz). The carrier frequency was placed on either the water resonance (4.792 ppm) (blue) or protein amide region 8.5 ppm (red). All circles are for S1–S4 domains in lipid bilayers where protein: lipid ratio is 1:100. Triangles are for samples containing lipid alone. Samples containing 1H2O are shown as filled symbols, whereas those in 2H2O are shown as open symbols.
Discussion
The objective of the present study was to investigate the structure and hydration of lipid membranes containing S1–S4 voltage-sensing domains. Previously, only computational approaches have been used to explore how different types of membrane proteins influence the structure of the bilayer 12,27–29,40,41. We adapted neutron diffraction techniques to determine how voltage sensors influence membrane structure; these protein domains are highly polar and exhibit important interactions with the lipid membrane, making them a particularly interesting test case. We succeeded in reconstructing the bilayer profile in the presence of the S1–S4 domain of KvAP, which shows that the structure of the lipid bilayer remains largely intact around the protein. The most notable change is that the protein causes a thinning of the bilayer by about 3 Å. Neutron diffraction measurements reflect changes in the structure of the bilayer, averaged over the entire membrane plane. Molecular dynamics simulations yielding a value of the Bragg spacing consistent with the diffraction experiments, predict that the distortion of the lipid bilayer by the protein is restricted to the lipids immediately surrounding the voltage sensors (Supplementary Fig 8). Taken together, the modest membrane-averaged thinning and local adaptation of the lipid bilayer to the presence of the voltage sensor suggest that the protein has evolved to interact with lipid molecules while minimizing the energetic and structural perturbations of the bilayer
Our neutron diffraction, solid-state NMR, and simulation results indicate that S1–S4 voltage-sensing domains are hydrated in the bilayer and that water interacts intimately with the protein. The observed hydration can explain the accessibility of water-soluble reagents to residues in S1–S49,13,19,22–26 and suggests that the crevices seen in X-ray structures4,5, which house the Arg residues that carry gating charge6,7, actually contain water when the protein is embedded in a lipid membrane, as illustrated in Fig 3d. Hydration of these critical residues will raise the local dielectric within the bilayer, ensuring that the Arg residues remain charged and thereby move in response to changes in membrane voltage. Consideration of the observed average water distributions in the presence of S1–S4 domains indicates that the membrane electric field drops over a distance of no more than about 25Å, the hydrophobic thickness of the bilayer in the presence of the voltage-sensing domain. The water-filled crevices in the structure of the S1–S4 domain would be expected to focus the electric field further, which is in agreement with our simulations showing the transmembrane potential is contoured by the structure of the protein and drops over a distance of about 20Å (Fig 5). These simulations do show significant distortions of the lipid bilayer in the local vicinity of the protein, but these do not have pronounced effects on the transmembrane voltage.
Figure 5. Effects of the voltage-sensing domain on a lipid bilayer as revealed by molecular dynamics simulation.
a) The lipid bilayer interface, represented as a 2D Delaunay triangulation for the average positions of lipid carbonyl carbon atoms, reveals local distortions around the voltage-sensing domain. b) The transmembrane potential on a slice passing through the system center (the R133-D62 salt-bridge) shows focusing features in the voltage-sensing domain cavities. Contributions to the molecular surface by aliphatic chains (green); polar groups (yellow); and protein (white, transparent) of the corresponding cutaway view are shown as background. The dashed box indicates the region of the system considered atomistic in the calculation of the transmembrane potential (see Methods). In both panels the voltage-sensing domain is in ribbon representation with the outer four arginines in S4, and their salt-bridge partners, shown in CPK representation and colored by atom.
Although the effects of S1–S4 voltage-sensing domains on the physical thickness of the bilayer are rather modest, the bilayer thinning we observe indicates that the protein and bilayer do interact, thereby providing a basis for understanding how lipid modification can influence voltage sensor function14–17. For example, based on theoretical considerations and studies on gramicidin channels42–44, the thinning we observed would be expected to have profound effects on the mechanical properties of channels containing S1–S4 domains and may help to explain the sensitivity of voltage-activated ion channels to alterations in the mechanical properties of the lipid bilayer41,45–47.
The hydration and reshaping of the lipid membrane that we observe for voltage sensors will likely be relevant for other classes of membrane proteins. For example, the presence of binding sites for water soluble ligands deep within G protein-coupled receptors48 and transporters49, implies that these proteins are hydrated within the membrane. In the case of intramembrane enzymes, such as the rhomboid protease50, hydrolysis of the peptide bond requires the presence of water molecules in an active site located in the hydrophobic interior of the membrane. In each of these instances, little is known about the structure of the surrounding lipid membrane or whether adaptations may play important roles in the mechanism of the protein.
METHODS SUMMARY
The S1–S4 domain of KvAP was expressed and purified as previously described4 and reconstituted into a 1:1 mixture of POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine) and POPG (1-palmitoyl-2-oleoyl-sn-glycero-3-[phospho-rac-(1-glycerol)]) using rapid dilution. Lipid multilayer samples for neutron diffraction were prepared by deposition of aqueous dispersions of liposomes on glass slides. A solution containing ~4 mg of lipid was applied to the glass surface (15 mm diameter), dried under vacuum and rehydrated from water vapor in a sealed chamber containing saturated salt solutions. Three types of samples were prepared using: 1) protonated voltage-sensing domain and protonated lipid, 2) deuterated voltage-sensing domain and protonated lipid, and 3) protonated voltage-sensing domain and headgroup deuterated POPC lipid (D4 lipid: (CH3)3-N-C2H2-C2H2-). Oriented multilayers (~ 10 µm thick, corresponding to 2000–3000 bilayers) were transferred into the sample chamber of the neutron diffractometer, and hydrated through the vapor phase at a temperature of 298K. Neutron diffraction measurements were performed at the Advanced Neutron Diffractometer/Reflectometer (AND/R)37, located at the NIST Center for Neutron Research. Monochromatic cold neutrons of wavelength λ=5 Å and a wavelength spread Δλ/λ=1% were diffracted by the sample and counted with a pencil-type 3He detector. Specular (Θ − 2Θ) scans were taken to assure that the momentum transfer (Qz; typically 0–1.2 Å−1) between the incident and the diffracted neutron wavevector was always perpendicular to the multilayer plane, thus probing the bilayer normal (Supplementary Fig 1). Solid-state NMR experiments were performed on proteoliposome pellets in 1H2O or 2H2O at a water:lipid ratio of 30:1. Lipid spectra were recorded on a 800 MHz Bruker AV800 spectrometer equipped with a 4 mm 1H/13C/2H CP-MAS probe (Bruker Biospin Corp., Billerica, MA). All-atom MD simulations of the S1–S4 domain of KvAP were carried out in POPC bilayers hydrated with 9 and 11 water molecules/lipid (corresponding to experiments at 86% and 93% relative humidity). Each system consisted of two stacked lipid bilayers, each containing a single S1–S4 domain, arranged to form a single pseudo centrosymmetric unit cell. The simulations were run at a constant temperature of 295 K and a constant pressure of 1 bar.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
METHODS
Voltage-sensing domain expression, solubilization and purification
The KvAP gene was amplified from Aeropyrum pernix genomic DNA using PCR and cloned into the pQE-60 vector (Qiagen). A pQE-60 plasmid containing the S1–S4 voltage-sensing domain of KvAP (amino acid residues Met1-Lys147) was obtained by deletion of the C-terminal region of KvAP using PCR and its sequence was confirmed by DNA sequencing. The recombinant voltage-sensing domain was expressed in XL1-Blue strain of E.coli as previously described 4. The plasmid was transformed into the chemically competent E.coli cells (subcloning grade) (Stratagene). One colony was inoculated into 100 ml LB broth supplemented with Ampicillin (100 µg/ml) and grown overnight at 37°C with continuous shaking at 200 rpm. One liter of LB/Ampicillin broth was inoculated with 10 ml of the starting culture and the protein expression was induced with 1 mM isopropyl-β-D-thiogalactopyranoside (Calbiochem) when the optical density of the cells at 600 nm reached 0.6. After 3 hours induction, cells were harvested by centrifugation at 5000xg for 20 min. Cells were re-suspended in 10 mM EDTA solution and collected by centrifugation, and then twice re-suspended in 20 mM Tris/HCl, 100 mM KCl, pH 7.8 and collected by centrifugation. Cells were then re-suspended in 40 ml of 20 mM Tris/HCl, 100 mM KCl, pH 7.8 buffer, supplemented with 100 µL of protease inhibitor cocktail (Sigma) and 100 µL of 26 mg/ml PMSF in isopropanol. Cells were sonicated for 5 minutes on ice, 50 µL of protease inhibitor cocktail was added, and the S1–S4 domain extracted by solubilization of the homogenate in 2.5 % (w/v) decylmaltoside (Anatrace) in 20 mM Tris/HCl, 100 mM KCl, pH 7.8. Lysate was then centrifuged at 100,000g for 1 hour at 4°C and the supernatant collected. Supernatant was mixed with Co-Talon resin (Clontech) and mixture was transferred to a chromatography column (Bio-Rad) and the solution was allowed to pass through. The resin with bound protein was washed with 0.25% (w/v) decylmaltoside, 10 mM imidazole in 20 mM Tris/HCl, 100 mM KCl, pH 7.8. Buffer was then exchanged to 3 % (w/v) n-octyl-β-D-glucopyranoside (OG) in 20 mM Tris/HCl, 100 mM KCl, pH 7.8 and the protein eluted with 400 mM imidazole in the same buffer. One unit of thrombin (Sigma) was added per mg of protein and the mixture was dialyzed against 3 % (w/v) OG in 20 mM Tris/HCl, 100 mM KCl, pH 7.8 overnight at 4°C in a dialysis cassette with MW cutoff of 10 kDa (Pierce). The protein was analyzed by SDS-PAGE electrophoresis, followed by Coomassie staining (Invitrogen) and by MALDI-MS using the Invitrosol MALDI protein solubilizer kit (Invitrogen). The efficiency of the thrombin cleavage and the His-tag removal during the dialysis was confirmed by His-Probe HRP reagent kit (Pierce) and by MALDI-MS. S1–S4 KvAP protein identity was confirmed by mass spectrometric (MS) analysis of protein digest fragments and N-terminal Edman sequencing. N-terminal sequencing revealed that the first 5 amino acids are removed during protein expression, consistent with the earlier observations 4. The uniformly deuterated protein was obtained by expression in Bioexpress media (Cambridge Isotope Laboratories) supplemented with 80 % 2H2O and the molecular weight of the purified protein was determined by MS-MALDI using the Invitrosol MALDI protein solubilizer kit (Invitrogen). The concentration of the protein was determined spectrophotometrically using an extinction coefficient (ε 280 nm = 17,210 M−1cm−1) calculated from the deduced protein composition 51.
Lipid reconstitution of the voltage-sensing domain of KvAP
The S1–S4 voltage-sensing domain of KvAP was reconstituted to different molar ratios of protein to lipid as previously described 14,52–54 using a 1:1 mixture of POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine) and POPG (1-palmitoyl-2-oleoyl-sn-glycero-3-[phospho-rac-(1-glycerol)]). All lipids were purchased from Avanti Polar Lipids and mixtures dried from solution in chloroform under a stream of nitrogen gas and desiccated under vacuum overnight. Lipid films were solubilized in 20 mM Tris/HCl, 100 mM KCl buffer, pH 7.8 with 3% (w/v) OG and protein was added to the lipid to form mixed detergent-lipid micelles. Proteoliposomes were formed by rapid dilution of the protein-detergent-lipid mixed micelles well below the critical micelle concentration of the OG detergent. Proteoliposome pellets were collected by ultracentrifugation at 200,000 g at 4°C using an Optima TL 100 TLX ultracentrifuge and 100.3 TLA rotor (Beckman). Proteoliposome pellets were resuspended in H2O and sedimented by ultracentrifugation, resuspended and mildly sonicated for 1 minute in water bath sonicator. The resultant proteoliposomes were analyzed for the lipid content using the modified method of Bartlett 55 and residual detergent contents determined using the modified phenol-sulfuric acid assay 56 and by dissolving aliquots of the sample in deuterated MeOH and analysis of the components of the mixture by 1H NMR.
Circular Dichroism Spectroscopy
Circular Dichroism spectra were recorded in 20 mM Tris/HCl, 100 mM KCl buffer, pH 7.8, using a JASCO J-815 spectropolarimeter equipped with thermally controlled cuvette holder. Spectra were recorded on voltage-sensing domain samples in 0.1–1 mm quartz cuvettes, from 180 nm to 250 nm with 1 nm step resolution and 4 sec integration time. The helix content of the protein sample was calculated according to ref 57, which indicated high (~ 85%) helical content of the protein both in OG micelles and when reconstituted in lipid, consistent with the X-ray structure of the S1–S4 domain of KvAP4, and EPR results on the S1–S4 domain and full length KvAP channel 9,13.
Fluorescence Spectroscopy
Fluorescent emission spectra for Trp-70 within the S2 helix of the voltage-sensing domain of KvAP were recorded for the protein in either OG micelles or reconstituted into lipid. (Trp-70 is the only Trp residue within S1–S4.) Fluorescence spectra were recorded in 20 mM Tris/Cl, 100 mM KCl, pH 7.4 at 25°C with stirring in a total volume of 2 ml using the SPEX FluoroMax 3 spectrofluorometer. Quartz 1 cm × 1 cm curettes were used for all fluorescence measurements. An excitation wavelength of 295 nm (5 nm band pass) was used and the emission was scanned between 300 and 400 nm (5 nm band pass) with an increment of 0.5 nm. The polarizer was configured to excitation-90°, emission-0° 58 and emission spectra of OG buffer or lipid alone were subtracted. Quenching of Trp-70 fluorescence was examined by titration with acrylamide (Q), an aqueous quencher of Trp fluorescence. Stern-Volmer quenching constants, KSV, were calculated from the best fits of equation 1, where F0 and F are fluorescence of the Trp-70 in the absence and presence of a quencher, respectively, and [Q] is the concentration of the quencher:
| Equation 1 |
To determine the disposition of the Trp-70 in model membranes, we compared quenching by bromine atoms attached to different positions on the hydrocarbon tail35,36,59. For these experiments, protein was reconstituted into proteoliposomes using a 1:1 mixture of POPG and either Br2(6,7))-PC (1-palmitoyl-2-stearoyl(6–7)dibromo-sn-glycero-3-phosphatidylcholine, C16:0,C18:0), Br2(9,10))-PC (1-palmitoyl-2-stearoyl(9–10)dibromosn-sn-glycero-3-phosphatidylcholine, C16:0,C18:0), or Br2(11,12))-PC (1-palmitoyl-2-stearoyl(11–12)dibromo-sn-glycero-3-phosphatidylcholine, C16:0,C18:0).
Determining structure from neutron diffraction data and deuterium contrast variation
Lamellar diffraction patterns yield transbilayer distributions of scattering-length projected onto the bilayer normal (z-axis), called bilayer profiles written as ρ(z). The profiles presented in this paper have been placed on the absolute per-lipid scale. The simplest profiles are obtained by Fourier transformation of the measured structure factors FM(h) = √Ih, where Ih is the corrected intensity (below) of the hth diffracted intensity. In this case, ρ(z) varies along the bilayer normal and has an average value of 0 when integrated over the unit cell defined by the Bragg spacing d. The amplitude of ρ(z) is arbitrary, determined only by the units used to measure intensities, such as neutron counts observed in a given time period. This simple approach provides limited information about the disposition of molecules dissolved within the bilayer.
Useful information can be obtained only when the profiles are placed on an absolute scale, meaning that (1) the average value of ρ(z), ρ0, corresponds to the total scattering length of the unit cell and (2) the variation of ρ(z) around ρ0 shows absolute changes in scattering-length density. To determine ρ0, the contents of the unit cell (lipid, water, and protein) must be known. To calibrate the variation of ρ(z), an isomorphous substitution of atoms of known scattering-length bsub must be introduced into the sample. In this case, the integral over the unit cell of the so-called difference profile, Δρ(z) ≡ ρsub(z) − ρ(z), must equal bsub. This procedure, described in detail by White and colleagues 32,60,61 yields instrumental constants k(h) that lead to absolute-scale structure factors F(h) = k(h)FM(h). For a centrosymmetric bilayer containing two lipids per unit cell, the average scattering-length density is given by ρ0 = (2/Sd)∑bi, where S is the area per lipid and ∑bi is the sum of the scattering-lengths of all of the atoms in the unit cell. One rarely knows the value of S. To circumvent this problem, Jacobs and White 32 introduced what they called the relative absolute scale in which ρ*(z) = Sρ(z). A better and more descriptive term is the per-lipid scale, because the scattering-length density describes the scattering-length per lipid rather than per unit volume. To use this scale, one needs only the average numbers per lipid of waters and other components in the unit cell. The profiles presented in this paper have placed on the absolute per-lipid scale.
The absolute per-lipid scattering-length density is given by
| Equation 2 |
where ϕ(h) is the sign of the absolute-scale structure factor F(h) whose absolute value is |F(h)| and hmax is the index highest-angle observable structure factor. For centrosymmetric unit cells, which prevail for the data presented here, ϕ(h) = ±1 (i.e. cos(0) or cos(π). Methods for determining the signs have been discussed in detail elsewhere 30,31.
Treatment of diffraction data
In the kinematical approximation, the structure factors, F(h), at all observed diffraction orders, are determined as the square root of the integrated peak intensities, after background correction, absorption correction (B), and Lorentz factor correction (sin(2θh)), where θh represents the angle of incidence corresponding to the hth order of diffraction. The absorption correction was calculated as: B(h)= sin(θh)/(2αt)*[1-exp(-2αt/sin(θh)], where t=sample thickness and α=linear absorption coefficient30. We determined phases by measuring a sample in a series of different 1H2O-2H2O contrasts (i.e. using different mole fractions of 2H2O in the salt solutions for hydrating the sample). Assuming a Gaussian distribution of water hydrating the lipid headgroups 60, the difference structure factors corresponding to a lipid bilayer hydrated with 2H2O (FD) and 1H2O (FH), respectively, can be modeled according to Equation 3.
| Equation 3 |
where A and Z denote the 1/e half-width and the mean position of the Gaussian describing the labeled component distribution (water, in this case). The prefactor xD scales with the amount of deuterium per lipid: xD = 2(bD-bH)fDnD,, where bH and bD are the scattering lengths of deuterium and hydrogen, respectively, fD is the fraction of deuterated component and nD is the number of deuterium atoms per molecule in the labeled component. The cosine factor in Equation 3 determines the slope of the linear dependence ΔF(h)=f(xD), and thus the phases ϕ(h).
To determine the transbilayer distributions of water, -CH2-CH2- group of the phosphorylcholine (PC), and the S1–S4 domain of KvAP, we substituted those molecular components by their deuterated counterparts, for both protein-containing or pure lipid samples, and compared their density profiles with those of protonated samples using the absolute per-lipid scale.
Scaling of the neutron data and determination of the amount of water per lipid
Because the raw diffraction intensities collected are not normalized, the absolute (per lipid) scale is determined based on the sample composition (e.g. protein concentration and amount of water). While the protein concentration was determined by UV absorbance at 280 nm, the amount of water accumulated in the protein-containing membranes at the relative humidity of our experiments is unknown, and has to be determined by additional experiments. We used a lipid deuteration scheme (manuscript in preparation), which includes deuteration of the water of hydration (2H2O) and the PC -C2H2-C2H2- group (D4 lipid), respectively, to resolve the absolute scale and the number of waters per lipid. The two homologous samples (protonated and D4-lipid) were each measured under at least two different 1H2O-2H2O contrast conditions by exchanging 1H2O with 2H2O in different proportions (e.g. 1H2O:2H2O=100:0; 50:50; 80:20) in the saturated salt solutions used in the chamber. Both the water and the D4 label are components that can be described by Gaussian distributions 33,60. In Equation 3 we compare the data from the protonated and D4 lipid homologous samples, measured in either 1H2O or 20% 2H2O, to determine the parameters describing the D4 distribution. Knowing the prefactor xD, we determine the position Z and width A, as well as the scale factors kH (protonated lipid) and kD (D4-lipid), by a least-squares minimization procedure. Once scaled, the data collected in 1H2O and 20% 2H2O, for a given sample, are compared in Equation 3 to determine the water distribution parameters and the number of waters per lipid contained in the prefactor xD. The water content of neat POPC multilayers determined using this approach is indistinguishable from that determined by independent methods 62.
Molecular dynamics simulations
Two simulation systems with 9 and 11 waters per lipid and 130 lipid molecules per protein (corresponding to the conditions of 86% and 93% relative humidity, and a protein to lipid ratio of 0.77%) were prepared from the end configuration of a simulation trajectory of the S1–S4 domain of KvAP in a POPC bilayer with excess water reported in reference (27). The initial atomistic model in the excess water simulation corresponded to residues 24 to 147 in the model of the KvAP full channel proposed by MacKinnon and collaborators 63. The pore domain of the full channel model provides an unambiguous constraint for the orientation of the S1–S4 domain in the lipid bilayer. The final placement of the protein in the lipid bilayer along the transmembrane direction was determined by assuring that the 5 Tyr side chains in the S2–S3 connecting turn and the S3–S4 end were, simultaneously, in contact with the headgroup region on opposite sides of the lipid bilayer. Further details of excess water simulation system set up and MD trajectory generation can be found in reference (27). The low-hydration simulation systems consisted of two stacked lipid bilayers, each containing a single S1–S4 domain, arranged to form a single pseudo centrosymmetric unit cell. The system with 9 waters per lipid was prepared by removing the necessary water and lipid molecules from the end configuration of the excess water simulation. The initial equilibration consisted of 1000 steps of energy minimization, followed by 1 ns molecular dynamics run at constant volume and constant temperature (295 K) with the protein backbone held fixed. The full simulation was then carried out at a constant temperature of 295 K and constant pressure of 1 atm. The protein was progressively released from its initial configuration over the first 5.5 ns using harmonic restraints. The simulation was carried out in the absence of restraints for 37.5 ns. The system with 11 waters per lipid was prepared from the end configuration of 9 waters per lipid system by adding the necessary number of water molecules. The initial equilibration consisted of 1000 steps of energy minimization, followed by a 20 ps run at constant volume and constant temperature (295 K) over the newly added waters, and 80 ps over the whole system. The full simulation was then carried out at a constant temperature of 295 K and constant pressure of 1 atm for 25.2 ns. All of the molecular dynamics trajectories were generated with the NAMD 2.6 software package 64. The CHARMM22 and the revised CHARMM27 force fields 65–67 were used for the peptide and the lipids, respectively, and the TIP3P model was used for water 68. The smooth particle mesh Ewald (PME) method 69,70 was used to calculate electrostatic interactions, and the short-range real-space interactions were cut off at 11 Å, employing a switching function. A reversible multiple time–step algorithm 71 was employed to integrate the equations of motion with a time step of 4 fs for electrostatic forces, 2 fs for short-range non-bonded forces, and 1 fs for bonded forces. All bond lengths involving hydrogen atoms were held fixed using the SHAKE and SETTLE algorithms. A Langevin dynamics scheme was used for thermostating. Nose-Hoover-Langevin pistons were used for pressure control 72,73. Molecular graphics and simulation analyses were performed with the VMD 1.8.6 software package 74 over the last 10 ns of each simulation. To compare simulations with 11 waters per lipid with the experimental data directly, neutron diffraction structure factors75 for the nth order of diffraction F(n) were computed from the MD trajectory according to Equation 4,
| Equation 4 |
where the sum is over all the atoms in the simulation unit cell, bj and zj are the neutron scattering length and z-coordinate of the jth atom, respectively, d half of the simulation cell length along the transmembrane direction and n is an integer. The scaling factor of half the simulation cell length for the spatial coordinates corresponds to the repeat distance of an oriented stack of bilayers. The oriented bilayers diffract as centrosymmetrical objects independent of the presence of the protein. The purpose of the double-bilayer simulation system is to model the two equally probable orientations of the protein in the lipid bilayer. Therefore, since the total scattering length of a single simulation cell is twice that of a single repeat unit in the diffraction experiment, each atom in the simulation cell is considered to have an occupation factor of one-half. Structure factors were averaged over 10 system configurations (one per ns of trajectory), and the total scattering density profiles were constructed from the structure factors exactly as in the analysis of the experimental diffraction data (see Eq. 2 in neutron diffraction methods section). Component densities were computed following the experimental protocol (see Eq 3 in neutron diffraction methods section) assuming uniform labeling at the same mole fraction as in the neutron diffraction experiments. The average length of the simulation cell in the dimension perpendicular to the membranes was 104 Å, corresponding to a d-spacing of 51.8 Å for a single bilayer containing the S1–S4 voltage-sensing domain of KvAP, in excellent agreement with the experimental value of 52 Å obtained at 93% relative humidity (corresponding to 11 waters per lipid).
The electrostatic potential in the excess hydration simulation was calculated using the linearized Poisson-Boltzmann theory, treating all the system components as linear, isotropic dielectrics under an applied potential difference across the membrane, as previously described 76–78. For a given configuration along the simulation trajectory, the electrostatic potential was calculated over a composite system consisting of a cuboid region of space (dashed rectangle in Figure 5b), containing the atomistic configurations of the protein and most of the lipids. This region is embedded in a continuum composed of a semi-infinite planar slab, representing the membrane, between two half-spaces that represent the electrolyte solution 77. The calculations were performed over the last 16 ns of the simulation trajectory, taking one configuration per ns, using the PBEQ module of the CHARMM 32a2 software package 79. The linearized Poisson-Boltzmann equation was solved by finite differences, using the successive over-relaxation method, over a cubic grid of 161 nodes with a grid spacing of 1 Å. The continuum slab thickness was set to the separation between carbonyl distributions in the atomistic system. A dielectric constant of 2 was assigned to lipids and protein. The solvent dielectric constant was set to 80 and the salt concentration was set to 150 mM. The molecular surface was used to define the atomistic dielectric boundaries using the CHARMM force field 65 van der Waals radii.
NMR Saturation Transfer Difference using MAS
Saturation Transfer Difference (STD) experiments 80 were implemented to study hydration of the voltage-sensing domains in membranes. Magic Angle Spinning (MAS) conditions were used as previously described 38,81 to resolve lipid resonances. 1H NMR spectra of lipids were recorded and resonance attenuation measured in response to radio frequency (RF) pulses. The saturating RF pulses (field strength 0–1.2 kHz) consisted of twenty Gaussian-shaped 50-msec pulses. The saturation frequency was set to the amide region of the protein (8.5 ppm) or water resonance (4.792 ppm). The attenuation of the lipid methylene signal defined as resonance amplitude recorded without saturation divided by the amplitude with saturation was followed as indicator of magnetization transfer to lipid. The proteoliposomes were packed into 4 mm MAS rotors (Bruker) and hydrated with either 2H2O or 1H2O to the final water to lipid ratio of 30:1. 16 scans with a recycle delay of 10 s were acquired at 295.1°K. All spectra were recorded on a 800 MHz Bruker AV800 spectrometer equipped with a 4 mm 1H/13C/2H CP-MAS probe (Bruker Biospin Corp., Billerica, MA) at a MAS frequency of 10 kHz.
Supplementary Material
Acknowledgements
We thank the NINDS DNA sequencing facility for DNA sequencing, Howard Jaffe in the NINDS protein sequencing facility for mass spectrometry and peptide sequencing, and Tetsuya Kitaguchi for cloning KvAP. We also thank Tomohiro Kimura, Mark Mayer, Mirela Milescu, Joe Mindell and Shai Silberberg for helpful discussions. This work was supported by the Intramural Research Programs of the NINDS, NIH (K.J.S.) and NIAAA, NIH (K.G.), NIH grants GM74737 (S.H.W.) and Program Project GM86685 from NINDS and NIGMS (S.H.W.; D.J.T.), and NSF grant CHE-0750175 (D.J.T.). We are grateful for allocation of computer time on the NSF-supported Teragrid resources provided by the Texas Advanced Computing Center, and the support of the National Institute of Standards and Technology, U.S. Department of Commerce, in providing the neutron research facilities used for neutron diffraction experiments. The identification of any commercial product or trade name does not imply endorsement or recommendation by the National Institute of Standards and Technology.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions D.K. performed the biochemistry experiments, M.M., D.K. and D.L.W performed neutron diffraction experiments, D.K. and K.G. performed the solid-state NMR experiments and J.A.F. and E.V.S. performed MD simulations. All authors contributed to the study design and to writing the manuscript.
Author Information Reprints and permissions information is available at npg.nature.com/reprints. The authors declare no competing financial interests.
References
- 1.Swartz KJ. Sensing voltage across lipid membranes. Nature. 2008;456:891–897. doi: 10.1038/nature07620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murata Y, Iwasaki H, Sasaki M, Inaba K, Okamura Y. Phosphoinositide phosphatase activity coupled to an intrinsic voltage sensor. Nature. 2005;435:1239–1243. doi: 10.1038/nature03650. [DOI] [PubMed] [Google Scholar]
- 3.Lee SY, Letts JA, MacKinnon R. Functional reconstitution of purified human Hv1 H+ channels. J Mol Biol. 2009;387:1055–1060. doi: 10.1016/j.jmb.2009.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang Y, et al. X-ray structure of a voltage-dependent K+ channel. Nature. 2003;423:33–41. doi: 10.1038/nature01580. [DOI] [PubMed] [Google Scholar]
- 5.Long SB, Tao X, Campbell EB, MacKinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007;450:376–382. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- 6.Seoh SA, Sigg D, Papazian DM, Bezanilla F. Voltage-sensing residues in the S2 and S4 segments of the Shaker K+ channel. Neuron. 1996;16:1159–1167. doi: 10.1016/s0896-6273(00)80142-7. [DOI] [PubMed] [Google Scholar]
- 7.Aggarwal SK, MacKinnon R. Contribution of the S4 segment to gating charge in the Shaker K+ channel. Neuron. 1996;16:1169–1177. doi: 10.1016/s0896-6273(00)80143-9. [DOI] [PubMed] [Google Scholar]
- 8.Jiang Y, Ruta V, Chen J, Lee A, MacKinnon R. The principle of gating charge movement in a voltage-dependent K+ channel. Nature. 2003;423:42–48. doi: 10.1038/nature01581. [DOI] [PubMed] [Google Scholar]
- 9.Cuello LG, Cortes DM, Perozo E. Molecular architecture of the KvAP voltage-dependent K+ channel in a lipid bilayer. Science. 2004;306:491–495. doi: 10.1126/science.1101373. [DOI] [PubMed] [Google Scholar]
- 10.Ruta V, Chen J, MacKinnon R. Calibrated measurement of gating-charge arginine displacement in the KvAP voltage-dependent K+ channel. Cell. 2005;123:463–475. doi: 10.1016/j.cell.2005.08.041. [DOI] [PubMed] [Google Scholar]
- 11.Alabi AA, Bahamonde MI, Jung HJ, Kim JI, Swartz KJ. Portability of paddle motif function and pharmacology in voltage sensors. Nature. 2007;450:370–375. doi: 10.1038/nature06266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sands ZA, Sansom MS. How does a voltage sensor interact with a lipid bilayer? Simulations of a potassium channel domain. Structure. 2007;15:235–244. doi: 10.1016/j.str.2007.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chakrapani S, Cuello LG, Cortes DM, Perozo E. Structural dynamics of an isolated voltage-sensor domain in a lipid bilayer. Structure. 2008;16:398–409. doi: 10.1016/j.str.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmidt D, Jiang QX, MacKinnon R. Phospholipids and the origin of cationic gating charges in voltage sensors. Nature. 2006;444:775–779. doi: 10.1038/nature05416. [DOI] [PubMed] [Google Scholar]
- 15.Ramu Y, Xu Y, Lu Z. Enzymatic activation of voltage-gated potassium channels. Nature. 2006;442:696–699. doi: 10.1038/nature04880. [DOI] [PubMed] [Google Scholar]
- 16.Milescu M, et al. Interaction between lipids and voltage-sensor paddles detected with tarantula toxins. Nature Structural and Molecular Biology. doi: 10.1038/nsmb.1679. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu Y, Ramu Y, Lu Z. Removal of phospho-head groups of membrane lipids immobilizes voltage sensors of K+ channels. Nature. 2008;451:826–829. doi: 10.1038/nature06618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Asamoah OK, Wuskell JP, Loew LM, Bezanilla F. A fluorometric approach to local electric field measurements in a voltage-gated ion channel. Neuron. 2003;37:85–97. doi: 10.1016/s0896-6273(02)01126-1. [DOI] [PubMed] [Google Scholar]
- 19.Starace DM, Bezanilla F. A proton pore in a potassium channel voltage sensor reveals a focused electric field. Nature. 2004;427:548–553. doi: 10.1038/nature02270. [DOI] [PubMed] [Google Scholar]
- 20.Chanda B, Asamoah OK, Blunck R, Roux B, Bezanilla F. Gating charge displacement in voltage-gated ion channels involves limited transmembrane movement. Nature. 2005;436:852–856. doi: 10.1038/nature03888. [DOI] [PubMed] [Google Scholar]
- 21.Ahern CA, Horn R. Focused electric field across the voltage sensor of potassium channels. Neuron. 2005;48:25–29. doi: 10.1016/j.neuron.2005.08.020. [DOI] [PubMed] [Google Scholar]
- 22.Yang N, Horn R. Evidence for voltage-dependent S4 movement in sodium channels. Neuron. 1995;15:213–218. doi: 10.1016/0896-6273(95)90078-0. [DOI] [PubMed] [Google Scholar]
- 23.Larsson HP, Baker OS, Dhillon DS, Isacoff EY. Transmembrane movement of the shaker K+ channel S4. Neuron. 1996;16:387–397. doi: 10.1016/s0896-6273(00)80056-2. [DOI] [PubMed] [Google Scholar]
- 24.Starace DM, Bezanilla F. Histidine scanning mutagenesis of basic residues of the S4 segment of the shaker k+ channel. J Gen Physiol. 2001;117:469–490. doi: 10.1085/jgp.117.5.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neale EJ, Rong H, Cockcroft CJ, Sivaprasadarao A. Mapping the membrane-aqueous border for the voltage-sensing domain of a potassium channel. J Biol Chem. 2007;282:37597–37604. doi: 10.1074/jbc.M706437200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tombola F, Pathak MM, Isacoff EY. Voltage-sensing arginines in a potassium channel permeate and occlude cation-selective pores. Neuron. 2005;45:379–388. doi: 10.1016/j.neuron.2004.12.047. [DOI] [PubMed] [Google Scholar]
- 27.Freites JA, Tobias DJ, White SH. A Voltage-Sensor Water Pore. Biophys J. 2006 doi: 10.1529/biophysj.106.096065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jogini V, Roux B. Dynamics of the Kv1.2 voltage-gated K+ channel in a membrane environment. Biophys J. 2007;93:3070–3082. doi: 10.1529/biophysj.107.112540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Treptow W, Tarek M. Environment of the gating charges in the Kv1.2 Shaker potassium channel. Biophys J. 2006;90:L64–L66. doi: 10.1529/biophysj.106.080754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Worcester DL, Franks NP. Structural analysis of hydrated egg lecithin and cholesterol bilayers. II. Neutrol diffraction. J Mol Biol. 1976;100:359–378. doi: 10.1016/s0022-2836(76)80068-x. [DOI] [PubMed] [Google Scholar]
- 31.Blasie JK, Schoenborn BP, Zaccai G. Direct methods for the analysis of lamellar neutron diffraction from oriented multilayers: a difference Patterson deconvolution approach. Brookhaven Symp Biol. 1976:III58–III67. [PubMed] [Google Scholar]
- 32.Jacobs RE, White SH. The nature of the hydrophobic binding of small peptides at the bilayer interface: implications for the insertion of transbilayer helices. Biochemistry. 1989;28:3421–3437. doi: 10.1021/bi00434a042. [DOI] [PubMed] [Google Scholar]
- 33.Wiener MC, White SH. Structure of a fluid dioleoylphosphatidylcholine bilayer determined by joint refinement of x-ray and neutron diffraction data. III. Complete structure. Biophys J. 1992;61:434–447. doi: 10.1016/S0006-3495(92)81849-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vamvouka M, Cieslak J, Van Eps N, Hubbell W, Gross A. The structure of the lipid-embedded potassium channel voltage sensor determined by double-electron-electron resonance spectroscopy. Protein Sci. 2008;17:506–517. doi: 10.1110/ps.073310008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee AG. Measurement of lipid-protein interactions in reconstituted membrane vesicles using fluorescence spectroscopy. Methods Mol Biol. 1994;27:101–107. doi: 10.1385/0-89603-250-7:101. [DOI] [PubMed] [Google Scholar]
- 36.McIntosh TJ, Holloway PW. Determination of the depth of bromine atoms in bilayers formed from bromolipid probes. Biochemistry. 1987;26:1783–1788. doi: 10.1021/bi00380a042. [DOI] [PubMed] [Google Scholar]
- 37.Dura JA, et al. AND/R: A neutron diffractometer/reflectometer for investigation of thin films and multilayers for the life sciences. Review Scientific Instruments. 2006;77:074301. doi: 10.1063/1.2219744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grossfield A, Pitman MC, Feller SE, Soubias O, Gawrisch K. Internal hydration increases during activation of the G-protein-coupled receptor rhodopsin. J Mol Biol. 2008;381:478–486. doi: 10.1016/j.jmb.2008.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ader C, et al. Structural rearrangements of membrane proteins probed by water-edited solid-state NMR spectroscopy. J Am Chem Soc. 2009;131:170–176. doi: 10.1021/ja806306e. [DOI] [PubMed] [Google Scholar]
- 40.Lindahl E, Sansom MS. Membrane proteins: molecular dynamics simulations. Curr Opin Struct Biol. 2008;18:425–431. doi: 10.1016/j.sbi.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 41.Phillips R, Ursell T, Wiggins P, Sens P. Emerging roles for lipids in shaping membrane-protein function. Nature. 2009;459:379–385. doi: 10.1038/nature08147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang HW. Deformation free energy of bilayer membrane and its effect on gramicidin channel lifetime. Biophys J. 1986;50:1061–1070. doi: 10.1016/S0006-3495(86)83550-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nielsen C, Goulian M, Andersen OS. Energetics of inclusion-induced bilayer deformations. Biophys J. 1998;74:1966–1983. doi: 10.1016/S0006-3495(98)77904-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goulian M, et al. Gramicidin channel kinetics under tension. Biophys J. 1998;74:328–337. doi: 10.1016/S0006-3495(98)77790-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tabarean IV, Juranka P, Morris CE. Membrane stretch affects gating modes of a skeletal muscle sodium channel. Biophys J. 1999;77:758–774. doi: 10.1016/S0006-3495(99)76930-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Laitko U, Juranka PF, Morris CE. Membrane stretch slows the concerted step prior to opening in a Kv channel. J Gen Physiol. 2006;127:687–701. doi: 10.1085/jgp.200509394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmidt D, Mackinnon R. Voltage-dependent K+ channel gating and voltage sensor toxin sensitivity depend on the mechanical state of the lipid membrane. Proc Natl Acad Sci U S A. 2008;105:19276–19281. doi: 10.1073/pnas.0810187105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rosenbaum DM, Rasmussen SG, Kobilka BK. The structure and function of G-protein-coupled receptors. Nature. 2009;459:356–363. doi: 10.1038/nature08144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krishnamurthy H, Piscitelli CL, Gouaux E. Unlocking the molecular secrets of sodium-coupled transporters. Nature. 2009;459:347–355. doi: 10.1038/nature08143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Erez E, Fass D, Bibi E. How intramembrane proteases bury hydrolytic reactions in the membrane. Nature. 2009;459:371–378. doi: 10.1038/nature08146. [DOI] [PubMed] [Google Scholar]
- 51.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 52.Heginbotham L, Kolmakova-Partensky L, Miller C. Functional reconstitution of a prokaryotic K+ channel. J Gen Physiol. 1998;111:741–749. doi: 10.1085/jgp.111.6.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Perozo E, Cortes DM, Cuello LG. Three-dimensional architecture and gating mechanism of a K+ channel studied by EPR spectroscopy [see comments] Nat Struct Biol. 1998;5:459–469. doi: 10.1038/nsb0698-459. [DOI] [PubMed] [Google Scholar]
- 54.Cuello LG, Romero JG, Cortes DM, Perozo E. pH-dependent gating in the Streptomyces lividans K+ channel. Biochemistry. 1998;37:3229–3236. doi: 10.1021/bi972997x. [DOI] [PubMed] [Google Scholar]
- 55.Hurst RO. The Determination of Nucleotide Phosphorus with a Stannous Chloride-Hydrazine Sulphate Reagent. Can J Biochem Physiol. 1964;42:287–292. doi: 10.1139/o64-033. [DOI] [PubMed] [Google Scholar]
- 56.Dubois M, Gilles K, Hamilton JK, Rebers PA, Smith FA. colorimetric method for the determination of sugars. Nature. 1951;168:167. doi: 10.1038/168167a0. [DOI] [PubMed] [Google Scholar]
- 57.Chen YH, Yang JT, Chau KH. Determination of the helix and beta form of proteins in aqueous solution by circular dichroism. Biochemistry. 1974;13:3350–3359. doi: 10.1021/bi00713a027. [DOI] [PubMed] [Google Scholar]
- 58.Ladokhin AS, Jayasinghe S, White SH. How to measure and analyze tryptophan fluorescence in membranes properly, and why bother? Anal Biochem. 2000;285:235–245. doi: 10.1006/abio.2000.4773. [DOI] [PubMed] [Google Scholar]
- 59.Abrams FS, London E. Extension of the parallax analysis of membrane penetration depth to the polar region of model membranes: use of fluorescence quenching by a spin-label attached to the phospholipid polar headgroup. Biochemistry. 1993;32:10826–10831. doi: 10.1021/bi00091a038. [DOI] [PubMed] [Google Scholar]
- 60.Wiener MC, King GI, White SH. Structure of a fluid dioleoylphosphatidylcholine bilayer determined by joint refinement of x-ray and neutron diffraction data. I. Scaling of neutron data and the distributions of double bonds and water. Biophys J. 1991;60:568–576. doi: 10.1016/S0006-3495(91)82086-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wiener MC, White SH. Transbilayer distribution of bromine in fluid bilayers containing a specifically brominated analogue of dioleoylphosphatidylcholine. Biochemistry. 1991;30:6997–7008. doi: 10.1021/bi00242a027. [DOI] [PubMed] [Google Scholar]
- 62.Hristova K, White SH. Determination of the hydrocarbon core structure of fluid dioleoylphosphocholine (DOPC) bilayers by x-ray diffraction using specific bromination of the double-bonds: effect of hydration. Biophys J. 1998;74:2419–2433. doi: 10.1016/S0006-3495(98)77950-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee SY, Lee A, Chen J, Mackinnon R. Structure of the KvAP voltage-dependent K+ channel and its dependence on the lipid membrane. Proc Natl Acad Sci U S A. 2005 doi: 10.1073/pnas.0507651102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Phillips JC, et al. Scalable molecular dynamics with NAMD. Journal of Computational Chemistry. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.MacKerell AD, Jr, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J.Phys.Chem.B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 66.Feller SE, MacKerell AD., Jr An improved empirical potential energy function for molecular simulations of phospholipids. J.Phys.Chem.B. 2000;104:7510–7515. [Google Scholar]
- 67.Klauda JB, Brooks BR, MacKerell AD, Jr, Venable RM, Pastor RW. An ab initio study on the torsional surface of alkanes and its effect on molecular simulations of alkanes and a DPPC bilayer. J Phys Chem B. 2005;109:5300–5311. doi: 10.1021/jp0468096. [DOI] [PubMed] [Google Scholar]
- 68.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J.Chem.Phys. 1983;79:926–935. [Google Scholar]
- 69.Darden T, York D, Pedersen L. Particle mesh Ewald: An N•log(N) method for Ewald sums in large systems. J.Chem.Phys. 1993;98:10089–10092. [Google Scholar]
- 70.Essmann U, et al. A smooth particle mesh Ewald method. J.Chem.Phys. 1995;103:8577–8593. [Google Scholar]
- 71.Grubmüller H, Heller H, Windemuth A, Schulten K. Generalized Verlet algorithm for efficient molecular dynamics simulations with long-range interactions. Molecular Simulation. 1991;6:121–142. [Google Scholar]
- 72.Martyna GJ, Tobias DJ, Klein ML. Constant-pressure molecular-dynamics algorithms. J.Chem.Phys. 1994;101:4177–4189. [Google Scholar]
- 73.Feller SE, Zhang Y, Pastor RW, Brooks BR. Constant pressure molecular dynamics simulation: The Langevin piston method. J.Chem.Phys. 1995;103:4613–4621. [Google Scholar]
- 74.Humphrey W, Dalke W, Schulten K. VMD: Visual molecular dynamics. J.Mol.Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 75.Benz RW, Castro-Román F, Tobias DJ, White SH. Experimental validation of molecular dynamics simulations of lipid bilayers: A new approach. Biophys.J. 2005;88:805–817. doi: 10.1529/biophysj.104.046821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Roux B. Influence of the membrane potential on the free energy of an intrinsic protein. Biophys J. 1997;73:2980–2989. doi: 10.1016/S0006-3495(97)78327-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Roux B. The membrane potential and its representation by a constant electric field in computer simulations. Biophys J. 2008;95:4205–4216. doi: 10.1529/biophysj.108.136499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Grabe M, Lecar H, Jan YN, Jan LY. A quantitative assessment of models for voltage-dependent gating of ion channels. Proc Natl Acad Sci U S A. 2004;101:17640–17645. doi: 10.1073/pnas.0408116101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Brooks BR, et al. CHARMM: the biomolecular simulation program. J Comput Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Forsen SH. A. Study of moderately rapid chemical exchange reactions by means of nuclear magnetic double resonance. J. Chem. Physics. 1963;39:2892–2901. [Google Scholar]
- 81.Soubias O, Gawrisch K. Probing specific lipid-protein interaction by saturation transfer difference NMR spectroscopy. J Am Chem Soc. 2005;127:13110–13111. doi: 10.1021/ja0538942. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.