Abstract
Advantageous organismal and technical attributes of the zebrafish are being increasingly applied to study cancer biology. Along with other tumor models, zebrafish that develop melanomas have been generated. In both genetics and phenotype, zebrafish melanomas are strikingly similar to their human counterparts. For this reason, studies in the zebrafish are poised to make significant contributions to melanoma biology. In this review, we summarize important features of human melanoma and discuss how the zebrafish can be used to address many questions that remain unanswered about this devastating disease.
Introduction
Melanoma is the most aggressive and lethal form of skin cancer. Annually, melanoma is responsible for an estimated 8110 deaths in the United States and at least 48,000 deaths worldwide.1,2 Whereas advances in detection and treatment have reduced rates of many cancers, the incidence of melanoma has risen sharply in recent decades.3,4 There is a high mortality rate for metastatic melanoma, in part because these tumors are radioresistant and refractory to available chemotherapies.5 However, early stage melanomas, those primary tumors that have not spread to lymph nodes, can be excised with little risk of recurrence. For these reasons it is important to identify and monitor individuals genetically at risk for melanoma and better recognize premetastatic lesions in these and other patients. More effective treatments of metastatic disease are also necessary.
Melanoma arises from melanocytes, which are the pigment-producing cells of human skin. Melanocytes can proliferate and give rise to various types of benign nevi, which are commonly referred to as moles. Transformed melanocytes can yield melanomas that generally begin growth radially in the epidermis. Radial growth then transitions to a vertical growth phase that involves invasion through the basement membrane into the underlying dermis. Metastasis follows thereafter.
There are many areas of melanoma biology that are underexplored. For example, the steps by which a normal melanocyte becomes or generates a melanoma cell are largely unknown. How the disease subverts properties of normal melanocytes and their neural crest progenitors has been a topic of recent investigations, and this will be a fertile area of research in the future. Lastly, while great strides have been made in identifying genetic defects that contribute to melanoma,6 there are clearly many more genes that remain to be tied to this disease.
The zebrafish is becoming increasingly utilized to address some of these issues. Certain attributes of zebrafish make it useful for the study of melanocyte biology and melanoma. Zebrafish melanocytes (also referred to as melanophores) are externally visible, and single cells can be visualized in a living animal. In mammals, melanin pigment–containing melanosomes are transported to neighboring keratinocytes. However, zebrafish melanocytes retain melanin, which consequently serves as a reliable and useful cell-type marker. The development of melanocytes from the embryonic neural crest is well characterized (Fig. 1). Further, a wealth of zebrafish pigmentation mutants that affect melanocyte specification, differentiation, and function are available, and the genes defective in these mutants have conserved roles in mammals.7,8 The goal of this review is to highlight important findings in melanoma research and discuss issues that may be tractably addressed using zebrafish.
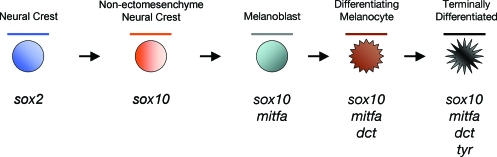
FIG. 1.
Factors involved in the development of the melanocyte lineage in zebrafish. Embryonic precursors of the melanocyte lineage can be identified by specific markers. sox2 is expressed in the developing ectoderm, and becomes down-regulated at the edge of the neural plate from where the crest cells emerge. sox10 is an early marker of specified neural crest cells of the pigment lineage, and is not seen in nonectomesenchymal crest derivatives (i.e., craniofacial skeleton). The committed melanoblast temporally expresses sox10 then mitfa, followed by markers of differentiation such as dopachrome tautomerase (dct) and tyrosinase (tyr).
Melanocyte Development and Melanoma
Given the highly migratory nature of human melanoma, it is perhaps not surprising that many of the genes that are important for melanocyte development and emergence from the neural crest have pathogenic roles in cancer. An understanding of the developmental origins of the melanocyte lineage is likely to yield useful information regarding the cellular functions and corresponding genetic pathways that may be usurped in melanoma. The repertoire of genes involved in the development of the melanocyte lineage has been fully reviewed elsewhere, so this section will focus on several factors that have been implicated as having a pathogenic role in melanoma. These pathways may offer rational therapeutic targets in this otherwise chemoresistant disease.
Melanocytes are derived from the neural crest
Vertebrates exhibit as many as six pigmented cell types, referred to collectively as chromatophores, although most species only contain a subset of these. In the zebrafish, three primary pigment cells are commonly found: the black-brown melanocyte, the yellow xanthophore, and the reflective iridophore. All of these cell types are derived from the highly migratory embryonic neural crest. The neural crest is induced during gastrulation, at the border zone between the neural and nonneural ectoderm.9 The early events in neural crest induction depend upon intact BMP signaling, as the Bmp2 knockout mouse is essentially devoid of neural crest derivatives.10 Fate decisions of early progenitors toward the neural crest lineage are dependent upon intact Notch signaling, likely via DeltaA-dependent lateral inhibition.11 Before migration, neural crest cells undergo an epithelial–mesenchymal transition (EMT), in which cells undergo loss of adhesion to neighboring cells and acquire migratory and invasive properties. This is, in part, dependent upon the Snail/Slug family of transcription factors, due to their ability to transcriptionally repress E-cadherin.12 Onset of Snail transcription temporally coincides with neural crest migration.
In the zebrafish, fate mapping studies indicate that most neural crest cells fated to become chromatophores become lineage-restricted before migration.13 The potency of each premigratory neural crest cell ranges from a single lineage (i.e., melanocytes) to trilineage (i.e., melanocytes, iridophores, and xanthophores), although the latter is somewhat rare. Although the different types of chromatophores are closely related by lineage, a range of zebrafish mutants preferentially affects only one cell type. For example, the mitfa (a.k.a. nacre) mutant is primarily deficient in melanocytes,14 and the fms (a.k.a. panther) mutant is more severely affected in the xanthophore than the melanocyte lineage.15 These data indicate different genetic requirements for each chromotophore type. There are also mutants in which multiple lineages are affected (e.g., colorless zebrafish, which are mutant in sox10, show loss of all three pigment cell types16), supporting the notion that other genes are either required for specification of a multi-potent cell type or necessary for a postspecification function common to these cell types. There is only a single chromatophore lineage in mammals, the melanocyte, and below we focus on the genes required for development of the melanocyte lineage, with particular emphasis on those genes implicated in melanoma.
Melanocyte-specific genes involved in melanoma
Mitf
The central role of Mitf in melanocyte development has been demonstrated by loss-of-function studies in model organisms and by the pigmentary deficiencies evident in MITF mutant Waardenburg syndrome type IIa (WS2a) patients.17,18 Mouse mutants with pigment abnormalities caused by an Mitf mutation were originally described in 1942.19 Mouse Mitf mutants commonly lack pigmentation, are deaf, and have a reduction in eye size known as microphthalmia. In the zebrafish, the mitfa mutant, which lacks all neural crest–derived embryonic and adult melanocytes, provides clear evidence that this gene plays a critical role in specification of the lineage. Mitf is a myc-family basic helix-look-helix/leucine zipper protein,20 which is well conserved among vertebrate species, and closely related genes are present in invertebrates such as Caenorhabditis elegans as well.21 Abnormalities observed in WS2a patients and many Mitf mutant mouse strains are genetically dominant and are often produced by point mutations that are thought to disrupt MITF DNA or protein interactions. Whereas WS2a patients are often deaf and mouse Mitf mutants suffer from other defects such as microphthalmia and deafness, no such defects have been noted in the fish, possibly due to redundancies provided by the paralogous mitfb gene22 or to species-specific requirements of Mitf-dependent melanocytes in eye and ear development.
Molecularly, Mitf sits at a critical juncture between neural crest specification and promotion of melanogenesis.23 It is transcriptionally regulated by several other genes that are known to promote melanocyte development from the neural crest. Tcf/LEF binding sites, which are the mediators of canonical Wnt signaling, are found within the MITF promoters in humans and zebrafish.24 Sox10 is an sry-box transcription factor necessary for multiple neural crest derivatives that can bind to the Mitf promoter and activate its transcription.25–29 In mammals, recent evidence indicates that PAX3 can upregulate transcription of MITF.30 Finally, MITF can be posttranslationally modified.31,32 Activation of c-KIT by its ligand, SCF, promotes phosphorylation of MITF at serine-73 (via ERK2) and serine-409 (via p90RSK), which upregulates transactivation by MITF through recruitment of p300. In addition, MITF regulates the melanocyte lineage by transcriptionally activating several genes involved in melanin synthesis, including TYROSINASE,33 TYRP1,34 and MC1R.35 Mitf promotes survival of the melanocyte lineage by upregulation of Bcl2,36 which likely explains why some murine Mitf mutants specify melanocytes that ultimately die prematurely via apoptosis. More recently, an oncogenic role of MITF has been discovered.37 MITF is amplified and overexpressed in a subset of human melanomas, and overexpression may facilitate inappropriate cell cycle progression and survival of tumor cells. One area that awaits further clarification regards the importance of MITF in the melanocyte stem cell. Given increasing evidence that such stem cells exist, at least in murine hair follicles,38 it will be important to understand whether MITF is required for properties such as self-renewal and multipotency, particularly in the setting of tumor development. Allelic variants of Mitfa in the zebrafish, in combination with overexpression under inducible promoters, will help to define the precise role of MITF in adult melanocyte homeostasis and during tumor progression.
Kit
Kit is a type III receptor tyrosine kinase that is required for melanocyte development in mammals and fish. Humans with heterozygous mutations of the c-KIT gene have the pigment disorder piebaldism, and mouse mutants for either c-Kit or its ligand, Scf/Steel, exhibit varying degrees of pigmentary, hematopoietic, and germ cell abnormalities.39–41 In contrast to mouse mutants, the zebrafish kit (a.k.a sparse) mutant exhibits defects only in melanocytes.42 In kit mutant embryos melanocyte specification occurs properly, but these cells fail to migrate and subsequently undergo apoptosis. The kit mutant also exhibits defects in adult melanocytes, in that they fail to form dermal stripe melanocytes that arise early in the larval-to-adult metamorphic period and all of the epidermal, scale-associated melanocytes.
The role of c-KIT in human melanoma has come under renewed interest in the past few years. c-KIT is expressed in early melanocytic lesions, but downregulation of expression is typically seen with advancing stages of melanoma.43,44 Three initial trials of the c-KIT inhibitor imatinib mesylate in melanoma were disappointing, with essentially negative results.45–47 However, it has recently been demonstrated that a significant proportion of atypical melanomas, arising in areas such as mucosal surfaces, palms, and soles, harbor activating mutations of the c-KIT gene.48,49 Preliminary data from an ongoing phase II trial have reported a dramatic response to imatinib in one such patient,50 suggesting that inhibition of c-KIT might be useful in well-selected populations of patients. Some caution, however, is warranted in the use of this inhibitor in unselected patients (i.e., those without c-KIT mutations), because loss of c-KIT expression may be associated with invasion and/or metastatic progression. Because of the clinical importance of the invasion/metastasis question, it will be critical to define the role of c-KIT in adult melanocyte migration and self-renewal, and how this is altered in the oncogenic setting. The amenability to chemical approaches in the zebrafish melanoma model, along with the availability of numerous c-KIT inhibitors, may provide a unique opportunity to define the role of c-KIT inhibition in early and late stages of disease.
Endothelins
The endothelins are a family of vasoactive peptides (ET1, ET2, and ET3) that bind to the G-protein–coupled receptors EDNRA or EDNRB. The specific role of the endothelins in pigment cell biology was recognized over a decade ago, when knockout mice for either Ednrb or Et3 were found to have melanocyte defects.51,52 Defects in this pathway are also found in human patients with Waardenburg-Shah syndrome, which combines features of Hirschsprung's disease (aganglionic megacolon due to loss of neural crest–derived enteric neurons) and Waardenburg syndrome (white hair forelock, heterochromia irides, and vitiligo). Using the inducible tet-system in mice, Ednrb has been shown to be required during a narrow window of neural crest development, and its loss during this period leads to almost complete loss of the lineage.53 This indicates that the endothelins are necessary for the dispersal and survival of already-specified melanoblasts that are present in the neural crest during this window. The zebrafish ednrb1 (a.k.a. rose) mutant too has a melanocyte deficiency, although it is not as severe as in other species. Whereas mouse mutants have defects in embryonic melanocytes, ednrb1 mutants have normal embryonic and early stripe melanocytes. The pigment defect in ednrb1 mutants does not become apparent until mid-to-late metamorphosis, when severe defects in the late stripe melanocytes are observed.54,55
EDNRB is highly expressed in most human melanoma cell lines and some primary human melanomas as well.56,57 Treatment of melanoma cultures with the specific EDNRB antagonist BQ788 causes the cells to cease proliferation, and in some cases leads to apoptosis.58 Some evidence suggests that endothelins signal through the MAP kinase pathway.59 Endothelin signaling can also occur independently of MAP kinase, and it is expected that signaling by endothelins may synergize with BRAF activation, an important event in many melanomas. In support of this parallel pathway hypothesis, knockdown of oncogenic BRAF in melanoma cells led to a decrease in proliferation and colony formation, an effect that could be somewhat rescued by exposure to ET1.60 The endothelins may also promote melanoma cell invasiveness by transcriptionally upregulating the expression of Snail, which leads to a loss of E-cadherin and a recapitulation of the embryonic EMT program.61 These data led to a small phase II trial of the nonselective EDNRA/EDNRB antagonist bosentan in human melanoma. Disease stabilization was noted in 6/32 patients, which is typical for other small molecule inhibitors used on unselected patients.62 Whether endothelin blockade will synergize with BRAF inhibition in human melanoma remains an unanswered question. Little data currently exist as to the role of endothelin-receptor subtypes (i.e., EDNRA vs. EDNRB) in stages of melanoma development, and some data would suggest that EDNRB expression is lost with melanocytic transformation.63 Given the availability of EDNRA/EDNRB mutant zebrafish, and chemicals that specifically block each receptor type, zebrafish melanoma models will provide a powerful tool for dissecting the role of these genes during tumor evolution.
Snail/Slug
The Snail family of DNA binding proteins is encoded by several highly conserved genes in species from Drosophila to humans. In zebrafish and Xenopus, the Snail family gene slug is expressed in specified neural crest cells, and represents one of the earliest markers of this lineage.64 There is a reciprocal relationship within a group of neural crest specifiers that includes Slug, Sox10, and Foxd3. Morpholino knockdown of Sox10 leads to a loss of Slug expression, and overexpression of Sox10 leads to an expansion of the Slug expression domain.65 The converse is also true: Slug knockdown with a dominant negative form represses Sox10, and ectopic Slug activates expression of Sox10. The mechanism of this reciprocity is unclear; the Slug protein may require secondary intermediates rather than acting as a direct transcriptional regulator of Sox10. Slug is required for the onset of neural crest migration, likely via its ability to transcriptionally repress E-cadherin and activate EMT.12 Upon cessation of migration, when neural crest cells have reached their final destination in the embryo, expression of Slug is lost.66
Using a xenotransplantation approach, Weinberg and colleagues uncovered a central role for slug in the invasive and metastatic nature of human melanoma as well.67 In this study, primary human melanocytes were transformed with a defined set of factors (SV40ER, hTERT, and RAS) and gave rise to metastatic melanoma nodules in a mouse xenografts. In contrast, similarly transformed fibroblasts formed primary, but no metastatic tumors, suggesting that programs inherent to the melanocyte lineage mediated the migratory nature of these transformed cells. Slug may enable these programs to be used, as knockdown of SLUG in melanoma cells led to a small decrease in primary tumor growth in xenotransplants, but a marked reduction of metastatic potential of these cells. These data indicate that lineage-specific genes may play a broader role in the clinical behavior of melanoma, because early metastatic dissemination is one of the hallmarks of this disease. Advances in imaging techniques using transparent zebrafish adults in the setting of transgenic fluorescent lines, along with the possibility of in vivo gene targeting using morpholinos,68 will allow for a more complete understanding of which step in the invasion cascade (e.g., extravasation, extracellular matrix disruption, and endothelial adhesion) Slug transcription is needed.
Melanoma Genetics
Gene dysfunction contributes to the development of many cancers, including melanoma. A number of genes have been implicated in melanoma formation through the identification of somatic genetic defects in benign and malignant melanocytic lesions and germline defects in melanoma-prone families.6,69 Some of the common genetic lesions are discussed below.
Receptor tyrosine kinases including c-KIT
Receptor tyrosine kinase (RTK) overactivation has been linked to melanoma pathogenesis. Seminal work on melanomas that form in Xiphophorus platyfish interspecific hybrids70–72 led to the discovery that increased activity of Xmrk, an RTK related to mammalian epidermal growth factor receptors, promotes tumorigenesis.73 Various RTKs in mammals have been implicated in melanoma, most notably c-KIT. Point mutations that are thought to constitutively activate c-KIT signaling are found in nearly 30% of acral and mucosal melanomas.49 Tumors in these locations are quite rare, but the availability of the c-KIT inhibitor imatinib mesylate is likely to provide substantial benefit to this small patient cohort.50 There is an unresolved paradox regarding the role of c-KIT in melanoma: while the aforementioned rare melanomas are dependent on c-KIT overactivation, a majority of advanced melanomas lose c-KIT expression.44 The conservation of c-KIT function and availability of mutants offer an avenue to address this paradox in zebrafish.
Ras family GTPases
Ras family GTPases are among the most commonly mutated genes in cancers.74 In general, mutations increase the ratio of active GTP-bound to inactive GDP-bound Ras, causing constitutive downstream signaling. Although mutant HRAS is important in the formation of benign Spitz nevi75 and can promote melanoma formation in mice,76 NRAS is most relevant to human melanoma. NRAS activating mutations are present in roughly 20–30% of primary and metastatic melanomas.77,78 Ras family members signal through multiple downstream effectors, and a determination of the pathways important for NRAS oncogenic activity has been an area of considerable interest. Zebrafish and other models of melanoma, in which additional mutations can be overlaid on a defined genetic background, provide an excellent means of pathway analysis.
BRAF
A landmark cancer resequencing effort identified BRAF mutations in melanoma.79 Although the percentages vary from one study to another, roughly 50–60% of melanomas from intermittently sun-exposed skin, the most common site of disease, have mutations that activate BRAF. These BRAF mutations are somatically acquired as wild-type BRAF is found in normal tissue from melanoma patients. A V600E substitution is by far the most prevalent mutation, and it results in a 700-fold overactivation of intrinsic BRAF kinase activity.80 BRAF is an Ras effector that signals through ERKs to affect cell division, survival, and other processes. BRAF mutations are found not only in melanomas but also in nevi,81 indicating that mutant BRAF cooperates with other genetic lesions in tumor formation. Because mutant BRAF can induce senescence in cultured melanocytes and nevi,82 it is likely that some of these genetic lesions are needed to bypass a senescent arrest. BRAF and NRAS mutations are mutually exclusive in melanomas,79,83 suggesting that mutation in just one of these genes is enough to overactivate downstream ERK signaling. However, NRAS and BRAF mutations are not equivalent. Whereas BRAF signals primarily through ERKs, Ras activity is transduced through PI3 kinase and other pathways. The involvement of multiple Ras effector pathways is suggested by the finding that PTEN allelic loss, which is predicted to upregulate PI3 kinase activity, occurs in about 40% of BRAF mutant melanomas.78
The utility of zebrafish in melanoma research was strikingly demonstrated by studies in which human oncogenic BRAFV600E was expressed under the control of the melanocyte mitfa promoter.84 Wild-type zebrafish expressing BRAFV600E developed nevi that, like their human counterparts, were highly pigmented melanocyte clusters confined to the epidermis (Fig. 2A). BRAFV600E expression in p53 mutant fish led to the formation of melanomas (Fig. 2B) that histologically resembled human melanomas. Melanocytes expressing BRAFV600E and mutant for p53 did not always form melanomas, suggesting that BRAFV600E mutations may be necessary but are not sufficient for melanoma formation. This study was instrumental in validating that BRAF mutations play a causative role in melanoma formation and are not simply bystander events in human tumors.
FIG. 2.
Melanocytic lesions in zebrafish expressing BRAFV600E. (A) Spotted leopard zebrafish expressing human BRAFV600E under the control of the melanocyte mitfa promoter develop clusters of melanocytes akin to human nevi (arrow). (B) p53 Mutant zebrafish expressing the same mitfa:BRAFV600E transgene develop melanomas.
Mitf
MITF represents a lineage-specific oncogene in human melanoma. Integrative analysis of copy number and expression arrays demonstrated that MITF is amplified in a subset of human melanomas, and cooperates with oncogenic BRAFV600E to transform normal melanocytes.37 The oncogenic potential of mitf was strictly dependent upon activated MAP kinase signaling, suggesting that a combination of driver mutations (i.e., BRAF) plus lineage specification genes (MITF) is needed for the complete cancer phenotype. The mechanisms by which MITF acts as an oncogene are not completely resolved, but may involve dysregulation of the cell cycle85 and apoptotic response.36
CDKN2A
Studies of genomic alterations in melanoma-prone families led to the identification of mutations that inactivate the CDKN2A locus.86,87 CDKN2A is a complex locus that encodes the INK4A and ARF tumor suppressor genes.88 Each of these genes begins with its own coding sequence, but downstream exons are shared. Rather uniquely, the shared RNA coding sequence is translated in alternate reading frames (hence ARF for alternate reading frame), resulting in proteins with distinct, unrelated amino acid sequences. The INK4A protein inhibits cell cycle progression by negatively regulating CyclinD/CDK4,6 activity. Inhibition of CyclinD/CDK4,6 prevents its hyperphosphorylation and inactivation of the Retinoblastoma (RB) tumor suppressor protein. The ARF protein inhibits MDM2-mediated ubiquitylation and degradation of p53. Because of its unusual genetic structure, a single lesion that affects the CDKN2A locus can extinguish both RB and p53 pathways, impacting melanocyte cell cycle regulation, apoptosis, senescence, and other processes.
p53
p53 Mutations are present in about half of all solid tumors but are relatively rare (10–25%) in melanomas.89 However, a variety of data suggest that loss of p53 pathway activity is a critical contributor to melanoma pathogenesis. As noted above, ARF deficiency or silencing is common in melanomas, and dysfunction of ARF negatively impacts p53 activity. Further, general assessments of the p53 activity, for example by microarray monitoring of the DNA damage response, support a loss of pathway function in many melanomas.90 These data suggest that animal models of melanoma utilizing p53 loss-of-function mutations can accurately reflect human tumors.
Clinical Challenges
Although some progress has been made in understanding the basis of melanoma, significant challenges in the clinical management of this disease remain. These challenges are reflected in the fact that the overall prognosis for patients with advanced melanoma is very poor, largely because there are no effective therapies.91 Translational research that uses fundamental knowledge of a disease to develop therapeutic strategies holds promise for improving melanoma treatment. We believe that a zebrafish melanoma model has unique attributes that may lead to important insights at the interface of melanocyte biology and clinical medicine. Below we outline some of the areas in which zebrafish melanoma research may be most helpful.
Defining relevant target genes in human melanoma
Identifying the right proteins and pathways to target in melanoma is critical for developing successful therapies. We have seen substantial progress in other malignancies after drugs were developed to target critical genes.92 How can a zebrafish model of melanoma identify novel genes that might be good drug targets?
Genetic screens have proven invaluable in linking novel genes to a phenotype of interest.93 When this phenotype involves tissues, for example, melanocytes, and pathologies, for example, cancer, that are vertebrate-specific, the zebrafish can be a powerful platform. Screens in the zebrafish have typically focused on embryonic phenotypes, primarily because of practical advantages. However, it is possible to screen for an adult cancer phenotype, especially one that is externally visible. Although there are technical considerations that make this approach challenging, there are screen designs that could identify enhancers and suppressors of melanoma.
Validating candidate target genes in zebrafish melanomas
From a therapeutic perspective we are interested in drug targets that cancers rely on for their growth and metastasis.94 Human genetics and bioinformatics have created a rich body of data that can be mined for such targets. RNAi screening is a powerful approach for performing loss-of-function genetic analysis in cells and model organisms.95 We believe that it will be possible to develop a zebrafish melanoma model system in which RNAi can be used to target potential oncogenes. This approach could be used to determine which genes are necessary for continued tumor growth and do so in a setting, much like that of human tumors, with vasculature and cellular heterogeneity.
Which human precancerous lesions have the highest potential for malignancy?
Some melanomas arise from premalignant nevi, but most nevi do not give rise to melanomas. A similar situation exists in BRAFV600E transgenic zebrafish. Nevi in these animals are accessible by biopsy and may be subjected to gene expression and other analyses. By studying the same lesions as it progresses from a nevus to and early melanoma then, later, a more aggressive tumor, it will be possible to characterize the ontogeny of tumor development over time. This analysis could identify markers that are useful in the clinic in predicting whether certain nevi have high or low malignant potential. In addition, identifying markers that refine staging criteria of early melanomas to reflect propensity toward invasion and metastasis could be clinically useful.96
Transparent adult fish as tools for understanding metastasis
Recent work from our laboratory has described a relatively transparent zebrafish strain we refer to as casper.97 This fish, mutant in both the roy and mitfa genes, is devoid of all embryonic and adult melanocytes and iridophores, allowing for detailed visualization of internal structures. With this platform, migration and invasion of transplanted tumor cells (using either melanin or GFP as a marker) can be easily visualized and quantified at the single-cell level. This system may serve as an attractive tool for chemical or genetic screens to identify novel modifiers of the metastatic melanoma phenotype.
Testing compounds for their ability to prevent melanoma onset
Chemoprevention refers to the treatment of healthy subjects in an effort to prevent disease. For example, in a large human cohort study men who took aspirin regularly had lower rates of colorectal cancer than subjects who did not.98 It would be useful to have a model system in which compounds could be screened for their abilities to prevent tumor formation. We believe that it would be possible to use a zebrafish model of melanoma to screen compounds for efficacy in preventing melanoma. Mechanistically, the compounds might target tumor vasculature, metabolism, pathways that regulate growth and differentiation, or apoptosis.
Identifying new compounds to target melanoma
There is a great need for chemotherapies that are effective against melanoma. Large-scale chemical screening is possible with zebrafish99–101 and, when coupled with a melanoma phenotype, may enable the identification of such therapies. Chemical screening with limiting amounts of drug would be best performed on small zebrafish embryos or larvae, and tumor shrinkage and disappearance could serve as a readout. Ideally, an early onset, highly penetrant melanoma model can be developed for screening. Alternatively, it may be possible to screen for inappropriate melanocyte proliferation or survival in a zebrafish embryo. An advantage of performing chemical screens on a cancer phenotype in the zebrafish will be distinguishing between drugs that kill melanoma versus tumor stromal cells and testing how various genetic backgrounds affect drug efficacy. These experimental approaches make the zebrafish an attractive preclinical model for drug discovery and development.
Conclusions
Advanced melanoma is a devastating and lethal cancer. Significant progress in understanding the basis for this disease has been made. Further research, particularly the kind that translates knowledge of the disease into treatment options, will be required to improve the prognosis for melanoma patients. With great ease of studying melanocytes and a proven melanoma model, the zebrafish is poised to make great contributions toward this goal.
Acknowledgments
We are grateful to members of the Zon laboratory, especially Michael Dovey, for helpful discussions. C.J.C. is supported by a Charles A. King Trust Postdoctoral Fellowship, Bank of America, Co-Trustee; Y.H. by a National Institutes of Health K08 award (K08-DK075432); and R.M.W. by an American Society for Clinical Oncology Young Investigator Award and an Aid for Cancer Research Fellowship. Melanoma research by L.I.Z. is funded by NIH Grant R01-DK53298-08.
Disclosure Statement
No competing financial interests exist.
References
- 1.Jemal A. Siegel R. Ward E. Murray T. Xu J. Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 2.Lucas R. McMichael T. Smith W. Armstrong B World Health Organization. Solar ultraviolet radiation: global burden of disease from solar ultraviolet radiation. World Health Organization Environmental Burden of Disease Series, Vol. 13. 2006. http://www.who.int/uv/publications/solaradgbd/en/index.html http://www.who.int/uv/publications/solaradgbd/en/index.html
- 3.Ries LA. Wingo PA. Miller DS. Howe HL. Weir HK. Rosenberg HM, et al. The annual report to the nation on the status of cancer, 1973–1997, with a special section on colorectal cancer. Cancer. 2000;88:2398–2424. doi: 10.1002/(sici)1097-0142(20000515)88:10<2398::aid-cncr26>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 4.Purdue MP. Freeman LE. Anderson WF. Tucker MA. Recent trends in incidence of cutaneous melanoma among US Caucasian young adults. J Invest Dermatol. 2008;128:2905–2908. doi: 10.1038/jid.2008.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thompson JF. Scolyer RA. Kefford RF. Cutaneous melanoma. Lancet. 2005;365:687–701. doi: 10.1016/S0140-6736(05)17951-3. [DOI] [PubMed] [Google Scholar]
- 6.Chin L. Garraway LA. Fisher DE. Malignant melanoma: genetics and therapeutics in the genomic era. Genes Dev. 2006;20:2149–2182. doi: 10.1101/gad.1437206. [DOI] [PubMed] [Google Scholar]
- 7.Quigley IK. Parichy DM. Pigment pattern formation in zebrafish: a model for developmental genetics and the evolution of form. Microsc Res Tech. 2002;58:442–455. doi: 10.1002/jemt.10162. [DOI] [PubMed] [Google Scholar]
- 8.Rawls JF. Mellgren EM. Johnson SL. How the zebrafish gets its stripes. Dev Biol. 2001;240:301–314. doi: 10.1006/dbio.2001.0418. [DOI] [PubMed] [Google Scholar]
- 9.Erickson CA. Reedy MV. Neural crest development: the interplay between morphogenesis and cell differentiation. Curr Top Dev Biol. 1998;40:177–209. doi: 10.1016/s0070-2153(08)60367-1. [DOI] [PubMed] [Google Scholar]
- 10.Kanzler B. Foreman RK. Labosky PA. Mallo M. BMP signaling is essential for development of skeletogenic and neurogenic cranial neural crest. Development. 2000;127:1095–1104. doi: 10.1242/dev.127.5.1095. [DOI] [PubMed] [Google Scholar]
- 11.Cornell RA. Eisen JS. Delta signaling mediates segregation of neural crest and spinal sensory neurons from zebrafish lateral neural plate. Development. 2000;127:2873–2882. doi: 10.1242/dev.127.13.2873. [DOI] [PubMed] [Google Scholar]
- 12.Cano A. Perez-Moreno MA. Rodrigo I. Locascio A. Blanco MJ. del Barrio MG, et al. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 13.Raible DW. Eisen JS. Restriction of neural crest cell fate in the trunk of the embryonic zebrafish. Development. 1994;120:495–503. doi: 10.1242/dev.120.3.495. [DOI] [PubMed] [Google Scholar]
- 14.Lister JA. Robertson CP. Lepage T. Johnson SL. Raible DW. nacre Encodes a zebrafish microphthalmia-related protein that regulates neural-crest-derived pigment cell fate. Development. 1999;126:3757–3767. doi: 10.1242/dev.126.17.3757. [DOI] [PubMed] [Google Scholar]
- 15.Parichy DM. Ransom DG. Paw B. Zon LI. Johnson SL. An orthologue of the kit-related gene fms is required for development of neural crest-derived xanthophores and a subpopulation of adult melanocytes in the zebrafish, Danio rerio. Development. 2000;127:3031–3044. doi: 10.1242/dev.127.14.3031. [DOI] [PubMed] [Google Scholar]
- 16.Dutton KA. Pauliny A. Lopes SS. Elworthy S. Carney TJ. Rauch J, et al. Zebrafish colourless encodes sox10 and specifies non-ectomesenchymal neural crest fates. Development. 2001;128:4113–4125. doi: 10.1242/dev.128.21.4113. [DOI] [PubMed] [Google Scholar]
- 17.Steingrimsson E. Moore KJ. Lamoreux ML. Ferre-D'Amare AR. Burley SK. Zimring DC, et al. Molecular basis of mouse microphthalmia (mi) mutations helps explain their developmental and phenotypic consequences. Nat Genet. 1994;8:256–263. doi: 10.1038/ng1194-256. [DOI] [PubMed] [Google Scholar]
- 18.Tassabehji M. Newton VE. Read AP. Waardenburg syndrome type 2 caused by mutations in the human microphthalmia (MITF) gene. Nat Genet. 1994;8:251–255. doi: 10.1038/ng1194-251. [DOI] [PubMed] [Google Scholar]
- 19.Hertwig P. Neue Mutationen und Koppelungsgruppen bei der Hausmaus. Z Indukt Abstamm Vererbungsl. 1942;80:220–246. [Google Scholar]
- 20.Hodgkinson CA. Moore KJ. Nakayama A. Steingrimsson E. Copeland NG. Jenkins NA, et al. Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell. 1993;74:395–404. doi: 10.1016/0092-8674(93)90429-t. [DOI] [PubMed] [Google Scholar]
- 21.Rehli M. Den Elzen N. Cassady AI. Ostrowski MC. Hume DA. Cloning and characterization of the murine genes for bHLH-ZIP transcription factors TFEC and TFEB reveal a common gene organization for all MiT subfamily members. Genomics. 1999;56:111–120. doi: 10.1006/geno.1998.5588. [DOI] [PubMed] [Google Scholar]
- 22.Lister JA. Close J. Raible DW. Duplicate mitf genes in zebrafish: complementary expression and conservation of melanogenic potential. Dev Biol. 2001;237:333–344. doi: 10.1006/dbio.2001.0379. [DOI] [PubMed] [Google Scholar]
- 23.Widlund HR. Fisher DE. Microphthalamia-associated transcription factor: a critical regulator of pigment cell development and survival. Oncogene. 2003;22:3035–3041. doi: 10.1038/sj.onc.1206443. [DOI] [PubMed] [Google Scholar]
- 24.Dorsky RI. Raible DW. Moon RT. Direct regulation of nacre, a zebrafish MITF homolog required for pigment cell formation, by the Wnt pathway. Genes Dev. 2000;14:158–162. [PMC free article] [PubMed] [Google Scholar]
- 25.Bondurand N. Pingault V. Goerich DE. Lemort N. Sock E. Le Caignec C, et al. Interaction among SOX10, PAX3 and MITF, three genes altered in Waardenburg syndrome. Hum Mol Genet. 2000;9:1907–1917. doi: 10.1093/hmg/9.13.1907. [DOI] [PubMed] [Google Scholar]
- 26.Lee M. Goodall J. Verastegui C. Ballotti R. Goding CR. Direct regulation of the Microphthalmia promoter by Sox10 links Waardenburg-Shah syndrome (WS4)-associated hypopigmentation and deafness to WS2. J Biol Chem. 2000;275:37978–37983. doi: 10.1074/jbc.M003816200. [DOI] [PubMed] [Google Scholar]
- 27.Potterf SB. Furumura M. Dunn KJ. Arnheiter H. Pavan WJ. Transcription factor hierarchy in Waardenburg syndrome: regulation of MITF expression by SOX10 and PAX3. Hum Genet. 2000;107:1–6. doi: 10.1007/s004390000328. [DOI] [PubMed] [Google Scholar]
- 28.Verastegui C. Bille K. Ortonne JP. Ballotti R. Regulation of the microphthalmia-associated transcription factor gene by the Waardenburg syndrome type 4 gene, SOX10. J Biol Chem. 2000;275:30757–30760. doi: 10.1074/jbc.C000445200. [DOI] [PubMed] [Google Scholar]
- 29.Elworthy S. Lister JA. Carney TJ. Raible DW. Kelsh RN. Transcriptional regulation of mitfa accounts for the sox10 requirement in zebrafish melanophore development. Development. 2003;130:2809–2818. doi: 10.1242/dev.00461. [DOI] [PubMed] [Google Scholar]
- 30.Watanabe A. Takeda K. Ploplis B. Tachibana M. Epistatic relationship between Waardenburg syndrome genes MITF and PAX3. Nat Genet. 1998;18:283–286. doi: 10.1038/ng0398-283. [DOI] [PubMed] [Google Scholar]
- 31.Hemesath TJ. Price ER. Takemoto C. Badalian T. Fisher DE. MAP kinase links the transcription factor Microphthalmia to c-Kit signalling in melanocytes. Nature. 1998;391:298–301. doi: 10.1038/34681. [DOI] [PubMed] [Google Scholar]
- 32.Wu M. Hemesath TJ. Takemoto CM. Horstmann MA. Wells AG. Price ER, et al. c-Kit triggers dual phosphorylations, which couple activation and degradation of the essential melanocyte factor Mi. Genes Dev. 2000;14:301–312. [PMC free article] [PubMed] [Google Scholar]
- 33.Yasumoto K. Yokoyama K. Shibata K. Tomita Y. Shibahara S. Microphthalmia-associated transcription factor as a regulator for melanocyte-specific transcription of the human tyrosinase gene. Mol Cell Biol. 1994;14:8058–8070. doi: 10.1128/mcb.14.12.8058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yasumoto K. Yokoyama K. Takahashi K. Tomita Y. Shibahara S. Functional analysis of microphthalmia-associated transcription factor in pigment cell-specific transcription of the human tyrosinase family genes. J Biol Chem. 1997;272:503–509. doi: 10.1074/jbc.272.1.503. [DOI] [PubMed] [Google Scholar]
- 35.Aoki H. Moro O. Involvement of microphthalmia-associated transcription factor (MITF) in expression of human melanocortin-1 receptor (MC1R) Life Sci. 2002;71:2171–2179. doi: 10.1016/s0024-3205(02)01996-3. [DOI] [PubMed] [Google Scholar]
- 36.McGill GG. Horstmann M. Widlund HR. Du J. Motyckova G. Nishimura EK, et al. Bcl2 regulation by the melanocyte master regulator Mitf modulates lineage survival and melanoma cell viability. Cell. 2002;109:707–718. doi: 10.1016/s0092-8674(02)00762-6. [DOI] [PubMed] [Google Scholar]
- 37.Garraway LA. Widlund HR. Rubin MA. Getz G. Berger AJ. Ramaswamy S, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 38.Nishimura EK. Jordan SA. Oshima H. Yoshida H. Osawa M. Moriyama M, et al. Dominant role of the niche in melanocyte stem-cell fate determination. Nature. 2002;416:854–860. doi: 10.1038/416854a. [DOI] [PubMed] [Google Scholar]
- 39.Geissler EN. Ryan MA. Housman DE. The dominant-white spotting (W) locus of the mouse encodes the c-kit protooncogene. Cell. 1988;55:185–192. doi: 10.1016/0092-8674(88)90020-7. [DOI] [PubMed] [Google Scholar]
- 40.Giebel LB. Spritz RA. Mutation of the KIT (mast/stem cell growth factor receptor) protooncogene in human piebaldism. Proc Natl Acad Sci USA. 1991;88:8696–8699. doi: 10.1073/pnas.88.19.8696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Williams DE. Eisenman J. Baird A. Rauch C. Van Ness K. March CJ, et al. Identification of a ligand for the c-kit protooncogene. Cell. 1990;63:167–174. doi: 10.1016/0092-8674(90)90297-r. [DOI] [PubMed] [Google Scholar]
- 42.Parichy DM. Rawls JF. Pratt SJ. Whitfield TT. Johnson SL. Zebrafish sparse corresponds to an orthologue of c-kit and is required for the morphogenesis of a subpopulation of melanocytes, but is not essential for hematopoiesis or primordial germ cell development. Development. 1999;126:3425–3436. doi: 10.1242/dev.126.15.3425. [DOI] [PubMed] [Google Scholar]
- 43.Natali PG. Nicotra MR. Winkler AB. Cavaliere R. Bigotti A. Ullrich A. Progression of human cutaneous melanoma is associated with loss of expression of c-kit proto-oncogene receptor. Int J Cancer. 1992;52:197–201. doi: 10.1002/ijc.2910520207. [DOI] [PubMed] [Google Scholar]
- 44.Ryu B. Kim DS. Deluca AM. Alani RM. Comprehensive expression profiling of tumor cell lines identifies molecular signatures of melanoma progression. PLoS ONE. 2007;2:e594. doi: 10.1371/journal.pone.0000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eton O. Billings L. Kim K. Prieto V. Davis D. Frazier L, et al. Phase II trial of imatinib mesylate (STI-571) in metastatic melanoma (MM) Proc Am Soc Clin Oncol. 2004;22:7528. [Google Scholar]
- 46.Ugurel S. Hildenbrand R. Zimpfer A. La Rosee P. Paschka P. Sucker A, et al. Lack of clinical efficacy of imatinib in metastatic melanoma. Br J Cancer. 2005;92:1398–1405. doi: 10.1038/sj.bjc.6602529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wyman K. Atkins MB. Prieto V. Eton O. McDermott DF. Hubbard F, et al. Multicenter phase II trial of high-dose imatinib mesylate in metastatic melanoma: significant toxicity with no clinical efficacy. Cancer. 2006;106:2005–2011. doi: 10.1002/cncr.21834. [DOI] [PubMed] [Google Scholar]
- 48.Antonescu CR. Busam KJ. Francone TD. Wong GC. Guo T. Agaram NP, et al. L576P KIT mutation in anal melanomas correlates with KIT protein expression and is sensitive to specific kinase inhibition. Int J Cancer. 2007;121:257–264. doi: 10.1002/ijc.22681. [DOI] [PubMed] [Google Scholar]
- 49.Curtin JA. Busam K. Pinkel D. Bastian BC. Somatic activation of KIT in distinct subtypes of melanoma. J Clin Oncol. 2006;24:4340–4346. doi: 10.1200/JCO.2006.06.2984. [DOI] [PubMed] [Google Scholar]
- 50.Hodi FS. Friedlander P. Corless CL. Heinrich MC. Mac Rae S. Kruse A, et al. Major response to imatinib mesylate in KIT-mutated melanoma. J Clin Oncol. 2008;26:2046–2051. doi: 10.1200/JCO.2007.14.0707. [DOI] [PubMed] [Google Scholar]
- 51.Hosoda K. Hammer RE. Richardson JA. Baynash AG. Cheung JC. Giaid A, et al. Targeted and natural (piebald-lethal) mutations of endothelin-B receptor gene produce megacolon associated with spotted coat color in mice. Cell. 1994;79:1267–1276. doi: 10.1016/0092-8674(94)90017-5. [DOI] [PubMed] [Google Scholar]
- 52.Baynash AG. Hosoda K. Giaid A. Richardson JA. Emoto N. Hammer RE, et al. Interaction of endothelin-3 with endothelin-B receptor is essential for development of epidermal melanocytes and enteric neurons. Cell. 1994;79:1277–1285. doi: 10.1016/0092-8674(94)90018-3. [DOI] [PubMed] [Google Scholar]
- 53.Lee HO. Levorse JM. Shin MK. The endothelin receptor-B is required for the migration of neural crest-derived melanocyte and enteric neuron precursors. Dev Biol. 2003;259:162–175. doi: 10.1016/s0012-1606(03)00160-x. [DOI] [PubMed] [Google Scholar]
- 54.Johnson SL. Africa D. Walker C. Weston JA. Genetic control of adult pigment stripe development in zebrafish. Dev Biol. 1995;167:27–33. doi: 10.1006/dbio.1995.1004. [DOI] [PubMed] [Google Scholar]
- 55.Parichy DM. Mellgren EM. Rawls JF. Lopes SS. Kelsh RN. Johnson SL. Mutational analysis of endothelin receptor b1 (rose) during neural crest and pigment pattern development in the zebrafish Danio rerio. Dev Biol. 2000;227:294–306. doi: 10.1006/dbio.2000.9899. [DOI] [PubMed] [Google Scholar]
- 56.Bittner M. Meltzer P. Chen Y. Jiang Y. Seftor E. Hendrix M, et al. Molecular classification of cutaneous malignant melanoma by gene expression profiling. Nature. 2000;406:536–540. doi: 10.1038/35020115. [DOI] [PubMed] [Google Scholar]
- 57.Ross DT. Scherf U. Eisen MB. Perou CM. Rees C. Spellman P, et al. Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet. 2000;24:227–235. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- 58.Lahav R. Heffner G. Patterson PH. An endothelin receptor B antagonist inhibits growth and induces cell death in human melanoma cells in vitro and in vivo. Proc Natl Acad Sci USA. 1999;96:11496–11500. doi: 10.1073/pnas.96.20.11496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Simonson MS. Wang Y. Dunn MJ. Cellular signaling by endothelin peptides: pathways to the nucleus. J Am Soc Nephrol. 1992;2:S116–S125. doi: 10.1681/ASN.V210s116. [DOI] [PubMed] [Google Scholar]
- 60.Christensen C. Guldberg P. Growth factors rescue cutaneous melanoma cells from apoptosis induced by knockdown of mutated (V 600 E) B-RAF. Oncogene. 2005;24:6292–6302. doi: 10.1038/sj.onc.1208758. [DOI] [PubMed] [Google Scholar]
- 61.Bagnato A. Rosano L. Spinella F. Di Castro V. Tecce R. Natali PG. Endothelin B receptor blockade inhibits dynamics of cell interactions and communications in melanoma cell progression. Cancer Res. 2004;64:1436–1443. doi: 10.1158/0008-5472.can-03-2344. [DOI] [PubMed] [Google Scholar]
- 62.Kefford R. Beith JM. Van Hazel GA. Millward M. Trotter JM. Wyld DK, et al. A phase II study of bosentan, a dual endothelin receptor antagonist, as monotherapy in patients with stage IV metastatic melanoma. Invest New Drugs. 2007;25:247–252. doi: 10.1007/s10637-006-9014-7. [DOI] [PubMed] [Google Scholar]
- 63.Eberle J. Weitmann S. Thieck O. Pech H. Paul M. Orfanos CE. Downregulation of endothelin B receptor in human melanoma cell lines parallel to differentiation genes. J Invest Dermatol. 1999;112:925–932. doi: 10.1046/j.1523-1747.1999.00598.x. [DOI] [PubMed] [Google Scholar]
- 64.Aybar MJ. Nieto MA. Mayor R. Snail precedes slug in the genetic cascade required for the specification and migration of the Xenopus neural crest. Development. 2003;130:483–494. doi: 10.1242/dev.00238. [DOI] [PubMed] [Google Scholar]
- 65.Honore SM. Aybar MJ. Mayor R. Sox10 is required for the early development of the prospective neural crest in Xenopus embryos. Dev Biol. 2003;260:79–96. doi: 10.1016/s0012-1606(03)00247-1. [DOI] [PubMed] [Google Scholar]
- 66.Meulemans D. Bronner-Fraser M. Gene-regulatory interactions in neural crest evolution and development. Dev Cell. 2004;7:291–299. doi: 10.1016/j.devcel.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 67.Gupta PB. Kuperwasser C. Brunet JP. Ramaswamy S. Kuo WL. Gray JW, et al. The melanocyte differentiation program predisposes to metastasis after neoplastic transformation. Nat Genet. 2005;37:1047–1054. doi: 10.1038/ng1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li YF. Morcos PA. Design and synthesis of dendritic molecular transporter that achieves efficient in vivo delivery of morpholino antisense oligo. Bioconjug Chem. 2008;19:1464–1470. doi: 10.1021/bc8001437. [DOI] [PubMed] [Google Scholar]
- 69.Miller AJ. Mihm MC., Jr Melanoma. N Engl J Med. 2006;355:51–65. doi: 10.1056/NEJMra052166. [DOI] [PubMed] [Google Scholar]
- 70.Gordon M. The genetics of viviparous top-minnow Platypoecilus: the inheritance of two kinds of melanophores. Genetics. 1927;12:253–283. doi: 10.1093/genetics/12.3.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Häussler G. Über Melanombildungen bei Bastarden von Xiphophorus maculatus var. rubra. Klin Wochenschr. 1928;7:1561–1562. [Google Scholar]
- 72.Kosswig C. Über Kreuzungen zwischen den Teleostiern Xipphophorus helleri und Platypoecilus maculatus. Z Indukt Abstamm Vererbungsl. 1928;47:150–158. [Google Scholar]
- 73.Wittbrodt J. Lammers R. Malitschek B. Ullrich A. Schartl M. The Xmrk receptor tyrosine kinase is activated in Xiphophorus malignant melanoma. EMBO J. 1992;11:4239–4246. doi: 10.1002/j.1460-2075.1992.tb05518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bos JL. ras Oncogenes in human cancer: a review. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- 75.Bastian BC. Wesselmann U. Pinkel D. Leboit PE. Molecular cytogenetic analysis of Spitz nevi shows clear differences to melanoma. J Invest Dermatol. 1999;113:1065–1069. doi: 10.1046/j.1523-1747.1999.00787.x. [DOI] [PubMed] [Google Scholar]
- 76.Chin L. Pomerantz J. Polsky D. Jacobson M. Cohen C. Cordon-Cardo C, et al. Cooperative effects of INK4a and ras in melanoma susceptibility in vivo. Genes Dev. 1997;11:2822–2834. doi: 10.1101/gad.11.21.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Demunter A. Stas M. Degreef H. De Wolf-Peeters C. van den Oord JJ. Analysis of N- and K-ras mutations in the distinctive tumor progression phases of melanoma. J Invest Dermatol. 2001;117:1483–1489. doi: 10.1046/j.0022-202x.2001.01601.x. [DOI] [PubMed] [Google Scholar]
- 78.Lin WM. Baker AC. Beroukhim R. Winckler W. Feng W. Marmion JM, et al. Modeling genomic diversity and tumor dependency in malignant melanoma. Cancer Res. 2008;68:664–673. doi: 10.1158/0008-5472.CAN-07-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Davies H. Bignell GR. Cox C. Stephens P. Edkins S. Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 80.Wan PT. Garnett MJ. Roe SM. Lee S. Niculescu-Duvaz D. Good VM, et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116:855–867. doi: 10.1016/s0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 81.Pollock PM. Harper UL. Hansen KS. Yudt LM. Stark M. Robbins CM, et al. High frequency of BRAF mutations in nevi. Nat Genet. 2003;33:19–20. doi: 10.1038/ng1054. [DOI] [PubMed] [Google Scholar]
- 82.Michaloglou C. Vredeveld LC. Soengas MS. Denoyelle C. Kuilman T. van der Horst CM, et al. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436:720–724. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- 83.Goel VK. Lazar AJ. Warneke CL. Redston MS. Haluska FG. Examination of mutations in BRAF, NRAS, and PTEN in primary cutaneous melanoma. J Invest Dermatol. 2006;126:154–160. doi: 10.1038/sj.jid.5700026. [DOI] [PubMed] [Google Scholar]
- 84.Patton EE. Widlund HR. Kutok JL. Kopani KR. Amatruda JF. Murphey RD, et al. BRAF mutations are sufficient to promote nevi formation and cooperate with p53 in the genesis of melanoma. Curr Biol. 2005;15:249–254. doi: 10.1016/j.cub.2005.01.031. [DOI] [PubMed] [Google Scholar]
- 85.Carreira S. Goodall J. Aksan I. La Rocca SA. Galibert MD. Denat L, et al. Mitf cooperates with Rb1 and activates p21Cip1 expression to regulate cell cycle progression. Nature. 2005;433:764–769. doi: 10.1038/nature03269. [DOI] [PubMed] [Google Scholar]
- 86.Hussussian CJ. Struewing JP. Goldstein AM. Higgins PA. Ally DS. Sheahan MD, et al. Germline p16 mutations in familial melanoma. Nat Genet. 1994;8:15–21. doi: 10.1038/ng0994-15. [DOI] [PubMed] [Google Scholar]
- 87.Kamb A. Gruis NA. Weaver-Feldhaus J. Liu Q. Harshman K. Tavtigian SV, et al. A cell cycle regulator potentially involved in genesis of many tumor types. Science. 1994;264:436–440. doi: 10.1126/science.8153634. [DOI] [PubMed] [Google Scholar]
- 88.Lowe SW. Sherr CJ. Tumor suppression by Ink4a-Arf: progress and puzzles. Curr Opin Genet Dev. 2003;13:77–83. doi: 10.1016/s0959-437x(02)00013-8. [DOI] [PubMed] [Google Scholar]
- 89.Daniotti M. Oggionni M. Ranzani T. Vallacchi V. Campi V. Di Stasi D, et al. BRAF alterations are associated with complex mutational profiles in malignant melanoma. Oncogene. 2004;23:5968–5977. doi: 10.1038/sj.onc.1207780. [DOI] [PubMed] [Google Scholar]
- 90.Kaufmann WK. Nevis KR. Qu P. Ibrahim JG. Zhou T. Zhou Y, et al. Defective cell cycle checkpoint functions in melanoma are associated with altered patterns of gene expression. J Invest Dermatol. 2008;128:175–187. doi: 10.1038/sj.jid.5700935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lorigan P. Eisen T. Hauschild A. Systemic therapy for metastatic malignant melanoma—from deeply disappointing to bright future? Exp Dermatol. 2008;17:383–394. doi: 10.1111/j.1600-0625.2007.00673.x. [DOI] [PubMed] [Google Scholar]
- 92.Schiffer CA. BCR-ABL tyrosine kinase inhibitors for chronic myelogenous leukemia. N Engl J Med. 2007;357:258–265. doi: 10.1056/NEJMct071828. [DOI] [PubMed] [Google Scholar]
- 93.Patton EE. Zon LI. The art and design of genetic screens: zebrafish. Nat Rev Genet. 2001;2:956–966. doi: 10.1038/35103567. [DOI] [PubMed] [Google Scholar]
- 94.Strebhardt K. Ullrich A. Paul Ehrlich's magic bullet concept: 100 years of progress. Nat Rev Cancer. 2008;8:473–480. doi: 10.1038/nrc2394. [DOI] [PubMed] [Google Scholar]
- 95.Boutros M. Ahringer J. The art and design of genetic screens: RNA interference. Nat Rev Genet. 2008;9:554–566. doi: 10.1038/nrg2364. [DOI] [PubMed] [Google Scholar]
- 96.Ohsie SJ. Sarantopoulos GP. Cochran AJ. Binder SW. Immunohistochemical characteristics of melanoma. J Cutan Pathol. 2008;35:433–444. doi: 10.1111/j.1600-0560.2007.00891.x. [DOI] [PubMed] [Google Scholar]
- 97.White RM. Sessa A. Burke C. Bowman T. LeBlanc J. Ceol C, et al. Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell. 2008;2:183–189. doi: 10.1016/j.stem.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chan AT. Giovannucci EL. Meyerhardt JA. Schernhammer ES. Wu K. Fuchs CS. Aspirin dose and duration of use and risk of colorectal cancer in men. Gastroenterology. 2008;134:21–28. doi: 10.1053/j.gastro.2007.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Stern HM. Murphey RD. Shepard JL. Amatruda JF. Straub CT. Pfaff KL, et al. Small molecules that delay S phase suppress a zebrafish bmyb mutant. Nat Chem Biol. 2005;1:366–370. doi: 10.1038/nchembio749. [DOI] [PubMed] [Google Scholar]
- 100.Peterson RT. Link BA. Dowling JE. Schreiber SL. Small molecule developmental screens reveal the logic and timing of vertebrate development. Proc Natl Acad Sci USA. 2000;97:12965–12969. doi: 10.1073/pnas.97.24.12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.North TE. Goessling W. Walkley CR. Lengerke C. Kopani KR. Lord AM, et al. Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature. 2007;447:1007–1011. doi: 10.1038/nature05883. [DOI] [PMC free article] [PubMed] [Google Scholar]